Abstract
Osteosarcomas are sarcomas of the bone, derived from osteoblasts or their precursors, with a high propensity to metastasize. Osteosarcoma is associated with massive genomic instability, making it problematic to identify driver genes using human tumors or prototypical mouse models, many of which involve loss of Trp53 function. To identify the genes driving osteosarcoma development and metastasis, we performed a Sleeping Beauty (SB) transposon-based forward genetic screen in mice with and without somatic loss of Trp53. Common insertion site (CIS) analysis of 119 primary tumors and 134 metastatic nodules identified 232 sites associated with osteosarcoma development and 43 sites associated with metastasis, respectively. Analysis of CIS-associated genes identified numerous known and new osteosarcoma-associated genes enriched in the ErbB, PI3K-AKT-mTOR and MAPK signaling pathways. Lastly, we identified several oncogenes involved in axon guidance, including Sema4d and Sema6d, which we functionally validated as oncogenes in human osteosarcoma.
Osteosarcoma is the most common primary bone cancer and the third most common cancer in children and adolescents1. The current treatment options have not changed over the last three decades and rely on tumor resection and nonspecific combination chemotherapy, resulting in a 5-year survival rate of 0–29% if clinically apparent metastases are present2–6. Although these tumors are rare in humans, with ~900 cases annually, they are highly prevalent in canines, with over 50,000 cases annually, and have seemingly similar genetics7. Osteosarcomas are among the most disordered cancers in terms of whole-chromosome and gene copy number changes8. Furthermore, changes in gene copy number typically affect such large regions of the genome that it is usually impossible using these data alone to identify specific drivers whose low-level amplification or loss is selected for during tumor development. Although many genes have been proposed as drivers of osteosarcoma, only TP53, RB1, CDKN2A and MYC have been implicated with certainty9. Studies that have attempted to identify driver genes using copy number variation (CNV), mRNA expression and methylation data from human tumors have identified a limited number of candidate genes, with little to no overlap among studies, likely owing to the heterogeneity across tumors10–12. These findings suggest that there may be numerous driver genes altered at low frequency in osteosarcoma.
Collectively, these data highlight the need for functional genomics approaches to identify the genes and pathways driving osteosarcoma development and metastasis. Defining the processes underlying metastasis or the maintenance of metastatic nodules could have a major impact on osteosarcoma outcome. To this end, we performed a forward genetic screen using the conditional SB transposon mutagenesis system13. As TP53 is functionally inactivated in a large proportion of osteosarcomas, we performed the screen with both wild-type mice and mice harboring the conditional Trp53R270H dominant-negative allele1,8,14,15. Analysis of the recurrent transposon insertion sites identified 232 CIS-associated genes, including known and new osteosarcoma-related genes, from 119 primary tumors and 43 CIS-associated genes from 19 mice with metastatic disease. Comparative genomics and bioinformatic analyses of CIS-associated genes identified numerous cooperating genes and pathways associated with osteosarcoma development. Moreover, using unique transposon insertion sites as a molecular fingerprint, we were able to perform lineage tracing of metastases and primary tumors. These data indicate that metastasis is a complex phenomenon that does not strictly adhere to a single model of metastatic development.
RESULTS
Mutagenesis can accelerate or induce osteosarcoma
To identify genes involved in osteosarcoma development and metastasis, we generated mice undergoing SB mutagenesis on a wild-type background or a background predisposed to tumor formation (Supplementary Fig. 1a). Predisposed mice undergoing mutagenesis (Trp53-SBmut) contained the conditional SB system (loxP-STOP-loxP (LSL)–SB11 transposase and a T2/Onc concatemer), the conditional Trp53R270H allele (LSL-Trp53R270H) and Sp7-cre (also known as Osx-cre; expression restricted to committed osteoblast progenitors) (Supplementary Fig. 1b)13,16,17. Mice lacking the complete SB system served as controls (Trp53-C). We also performed mutagenesis in mice that were otherwise wild type (SBmut).
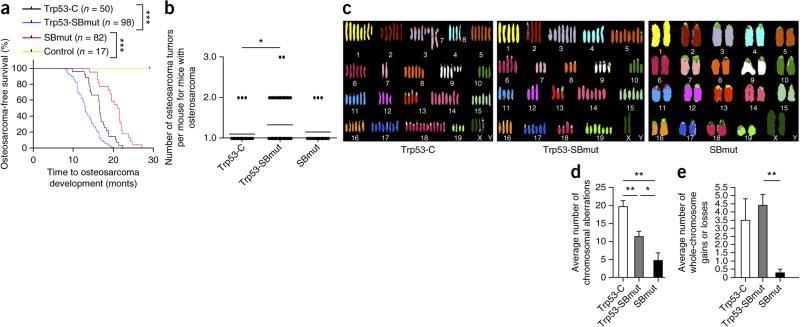
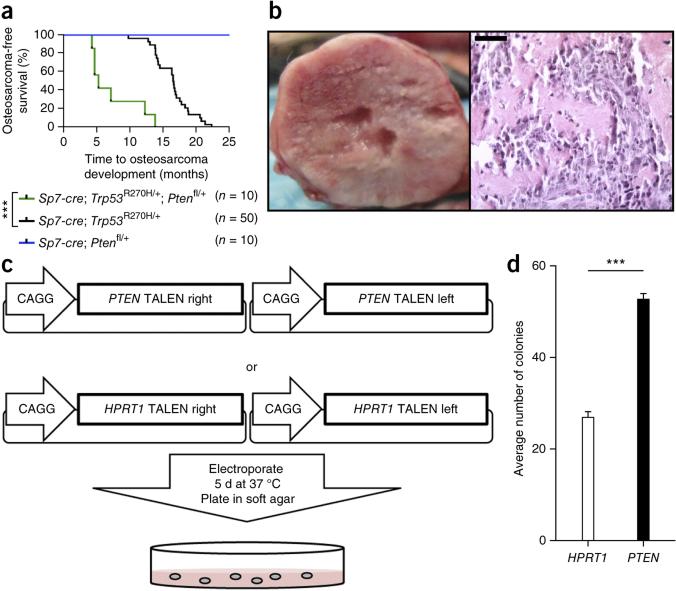
SB mutagenesis accelerated osteosarcomagenesis in Trp53-SBmut mice and increased tumor burden and penetrance (75% versus 59%) in comparison to Trp53-C mice (P < 0.0001 and 0.0159, respectively) (Fig. 1a,b). SBmut mice developed osteosarcomas with a longer latency and lower penetrance (24.4%). The majority of osteosarcomas were of the osteoblastic subtype (the most common subtype in humans), as determined on the basis of gross anatomy and histological appearance (Supplementary Fig. 1c–k)1. The site distribution of osteosarcoma development was similar to that in most models of osteosarcoma, with tumors arising in the skull, long bones and vertebrae (Supplementary Fig. 1l)15. We confirmed active SB mutagenesis in tumors by multiple methods (Supplementary Fig. 2a–c). These results demonstrate that SB mutagenesis can induce or accelerate development of the most prevalent subtype of osteosarcoma in humans.
Figure 1.
SB mutagenesis can accelerate or induce osteosarcoma development in cells with Sp7-cre expression. (a) Osteosarcoma-free survival curve depicting time to osteosarcoma development and survival endpoints in all cohorts. Control mice contained Sp7-cre with either SB11 or T2/Onc. ***P < 0.0001, log-rank test. (b) Histogram displaying the number of osteosarcomas per mouse. *P = 0.0159, Student's t test. (c) Representative SKY results from analysis of osteosarcoma tumor cells that developed in Trp53-C, Trp53-SBmut and SBmut mice. (d,e) Histograms demonstrating the number of chromosomal aberrations (d) and whole-chromosome gains and/or losses (e) identified by aCGH performed on Trp53-C (n = 4), Trp53-SBmut (n = 5) and SBmut (n = 4) osteosarcoma tumor DNA with matched normal tail DNA. *P < 0.05, **P < 0.001, Student's t test. Error bars, s.d.
Mutagenesis substitutes for genomic instability
We hypothesized that SB mutagenesis might substitute for alterations normally caused by changes in gene copy number due to Trp53 deficiency. Thus, we analyzed a subset of SB-mutagenized and control tumors by array comparative genomic hybridization (aCGH), spectral karyotyping (SKY) and G banding. All tumors had evidence of genomic instability, although some SBmut tumor cells had nearly perfect karyotypes and few whole-chromosome gains or losses; in contrast, Trp53-C tumors had the most genomic aberrations (Fig. 1c–e). These data demonstrate that osteosarcoma development in the context of Trp53 deficiency leads to selection of genomically unstable tumor clones, and this instability likely provides secondary molecular alterations required for tumor progression. In the presence of mutagenesis, the requirement for selection of these events is reduced, presumably because transposon-mediated mutations provide the changes required for tumor progression.
Identification of osteosarcoma driver genes
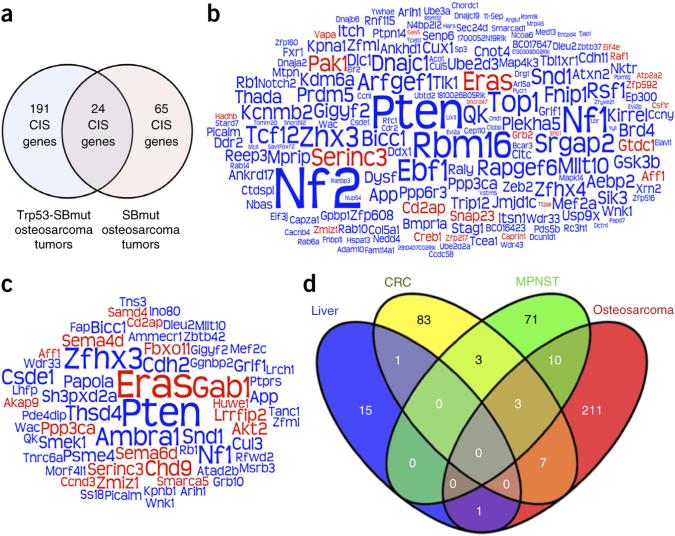
We analyzed the transposon insertions from 96 Trp53-SBmut and 23 SBmut osteosarcomas to identify CISs. As a disproportionate number of CISs were identified on the chromosome harboring the transposon concatemer (Supplementary Fig. 2d), we did not include these CISs in the analyses, although many likely harbor true driver genes, such as Myc and Cdkn2a (Supplementary Fig. 2e,f)13. In total, 36 candidate proto-oncogenes and 196 putative tumors-suppressor genes (TSGs) were identified (Fig. 2a–c and Supplementary Tables 1–3). The most commonly mutated genes in Trp53-SBmut tumors were Nf2 (26.0%; P = 2.0 × 10−38), Pten (24.0%; P = 1.82 × 10−28) and Nf1 (18.8%; P = 1.15 × 10−11) (Fig. 2b and Supplementary Table 1). Interestingly, Nf2 was not identified as a commonly mutated gene in SBmut tumors, although other top genes from Trp53-SBmut tumors, including Nf1 (26.1%; P = 2.84 × 10−5), Pten (39.1%; P = 1.3 × 10−10) and Eras (39.1%; P = 3.31 × 10−15), were identified in both cohorts (Fig. 2c and Supplementary Table 2). Notably, 20 genes previously implicated in osteosarcoma and the Rb1, Caprin1, Myc, Cdkn2a and Akt2 genes with established roles were identified, validating the screen18–39. Lastly, we compared our CIS-associated genes with those identified in three published SB screens to assess the relevance of our genes specifically to osteosarcoma (Fig. 2d). Interestingly, there was sparse overlap among tumor types, indicating that SB forward genetic screens identify genes that are cancer type specific and not just generally involved in cancer.
Figure 2.
CIS analysis identifies osteosarcoma driver genes. (a) Venn diagram depicting the number of CIS-associated genes from Trp53-SBmut and SBmut tumors and the number of genes identified in both groups. (b,c) Word cloud diagrams depicting Trp53-SBmut (b) and SBmut (c) CIS-associated genes. The size of each gene name is representative of the percentage of tumors with insertions in the CIS-associated gene identified by TAPDANCE or gene-centric analysis; if CISs in the gene were identified by both analyses, the larger percentage was used. Predicted proto-oncogenes are depicted in red, and candidate TSGs are depicted in blue. (d) Venn diagram depicting the overlap of the identified osteosarcoma-associated CIS genes with the CIS genes identified in previous SB screens for colorectal (CRC), liver and malignant peripheral nerve sheath (MPNST) tumors.
Pathways and upstream regulators of CIS genes
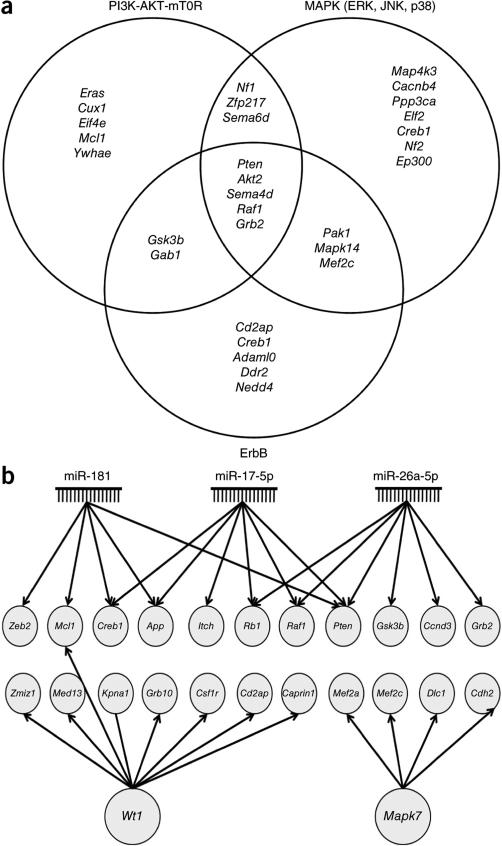
As all but two SBmut CIS-associated genes were also mutated in Trp53-SBmut tumors but not significantly, the gene lists were combined for further investigation (Supplementary Fig. 3). We used Ingenuity Pathway Analysis (IPA) and the Database for Annotation, Visualization and Integrated Discovery (DAVID) to identify signaling pathways and cellular processes enriched for candidate genes. The top cellular process was the cell cycle (IPA: n = 46 genes, P = 1.21 × 10−7 to 4.26 × 10−3; DAVID: n = 6 genes, P = 0.015) (Supplementary Tables 4 and 5). IPA also identified enrichment in cancer-associated pathways (n = 164 genes, P = 6.73 × 10−8 to 4.26 × 10−3), including phosphoinositide 3-kinase (PI3K)-AKT-mTOR (P = 1.35 × 10−5), ErbB (P = 3.09 × 10−3) and extracellular signal–regulated kinase–mitogen-activated protein kinase (ERK-MAPK; P = 6.45 × 10−3) signaling (Supplementary Table 4). Notably, a large number of CIS-associated genes were common to all three pathways (Fig. 3a).
Figure 3.
Analysis of CIS-associated genes identifies cooperating mutations, genetic pathways and upstream regulators in osteosarcoma development. (a) Venn diagram depicting the overlapping CIS genes found in three well-known cancer-associated signaling pathways. CIS genes were categorized into specific pathways on the basis of IPA, DAVID, GeneCards and a literature review. (b) Gene module depicting upstream regulators found to be regulating a significant number of CIS-associated genes by IPA (Supplementary Table 4).
IPA also identified enrichment in upstream regulators of CIS-associated genes. Interestingly, the top three most significantly enriched upstream regulators were microRNAs (miRNAs) implicated in human osteosarcoma, namely the genes encoding miR-181 (P = 2.18 × 10−5), miR-17-5p (P = 3.42 × 10−5) and miR-26a-5p (P = 3.06 × 10−7) (Fig. 3b and Supplementary Table 4)40–42. The remaining top regulators were MAPK7 (P = 7.64 × 10−5) and WT1 (P = 8.50 × 10−5), both implicated in osteosarcoma as prognostic markers of poor outcome43,44. These data demonstrate that CIS-associated genes are enriched in cancer signaling pathways and cellular processes, and a significant number are regulated by miRNAs and genes implicated in human osteosarcoma.
CIS genes are altered in cancer, including osteosarcoma
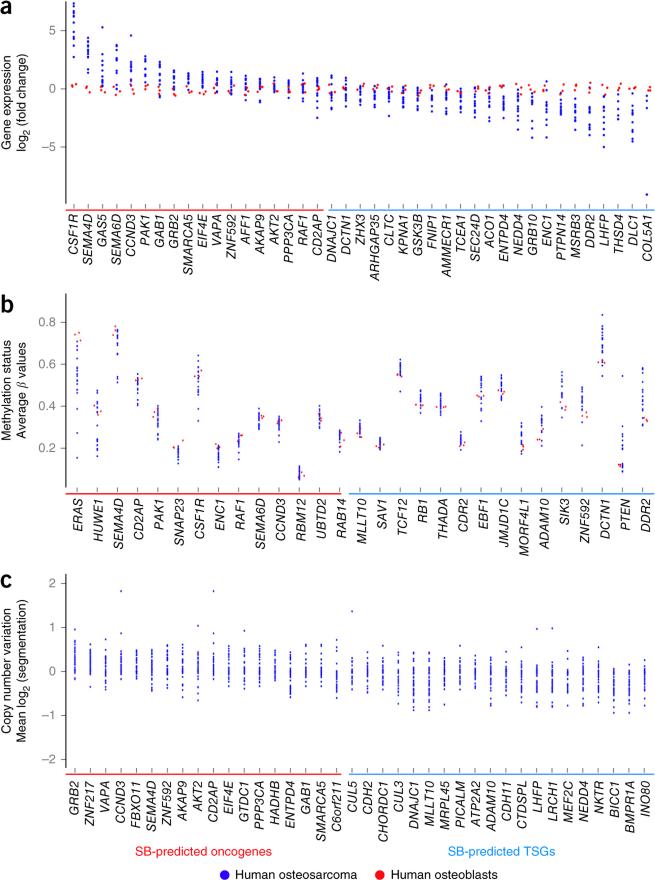
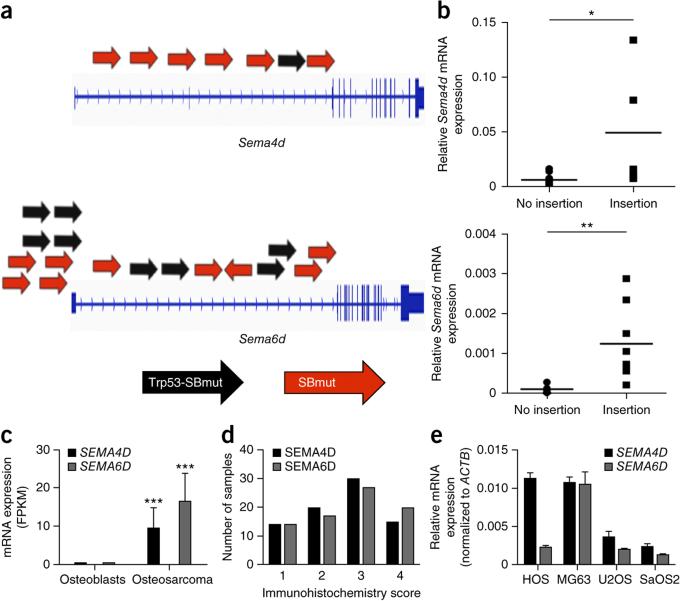
We queried the Catalogue of Somatic Mutations in Cancer (COSMIC) to identify CIS-associated genes frequently mutated in human cancer. Over half (57.1%) of the CIS-associated genes were mutated in >250 samples, and 31 genes were mutated in >1,000 samples (P = 2.14 × 10−20 and 9.24 × 10−12, respectively, hypergeometric test) (Supplementary Table 6). Moreover, 28 CIS-associated genes are in the Cancer Gene Census (CGC), which curates known cancer-related genes (P = 1.03 × 10−11, hypergeometric test) (Supplementary Table 6). To assess the relevance of CIS-associated genes in osteosarcoma gene expression, we queried CNV, structural variation and methylation data from human osteosarcoma samples. We considered only genes having significant changes consistent with their prediction by SB mutagenesis as candidate oncogenes or TSGs. Expression analysis demonstrated that 41 CIS-associated genes were significantly (P < 0.05) up- or downregulated in osteosarcomas in comparison to normal human osteoblasts (Fig. 4a and Supplementary Table 7). As promoter hypermethylation is associated with gene silencing and hypomethylation is associated with gene expression, we investigated methylation status. This analysis identified 29 CIS-associated genes with significant (P < 0.05) differential methylation patterns (Fig. 4b and Supplementary Table 8). aCGH analysis identified CIS-associated genes with a tendency for CNV gains (n = 16) and losses (n = 32), whereas others were mutated by structural variation (n = 10; NF1, KDM6A and BRD4) (Fig. 4c and Supplementary Tables 9 and 10). Overall, ~78% of candidate oncogenes and ~34% of candidate TSGs were altered in human osteosarcoma in a manner consistent with SB prediction as an oncogene or TSG in one or more data sets analyzed. These genes included known (RB1 and PTEN) and new (SEMA4D, SEMA6D, GRB10, ZNF217, ZNF592, GAB1 and PICALM) drivers of osteosarcoma. Although numerous CIS-associated genes are altered in human osteosarcoma, the conservation of specific gene alterations across species would increase the chance that these changes drive osteosarcoma.
Figure 4.
Comparative genomics analysis of CIS-associated genes in osteosarcoma. (a–c) RNA sequencing, whole-methylome and CNV data from human osteosarcoma samples were queried for alterations in CIS genes (Supplementary Tables 7–9). Only gene alterations that substantiate SB prediction of candidate oncogenes or TSGs were considered and appear in the presented scatterplots. (a) Scatterplot depicting CIS genes with significant (P < 0.05) differences in gene expression in human osteosarcomas (n = 12) in comparison to normal human osteoblasts. Genes are ordered on the basis of decreasing fold change in expression for oncogenes followed by TSGs. (b) Scatterplot depicting CIS genes with significant (P < 0.05) differences in methylation, including hypomethylation (negative values) or hypermethylation (positive values), in human osteosarcomas (n = 21) in comparison to normal human osteoblasts. Genes are ordered by increasing differential methylation. (c) Scatterplot depicting CIS genes in regions with tendencies for copy number gains or losses observed in human osteosarcoma (n = 56). Genes are ordered by decreasing number of amplifications from oncogenes to TSGs.
To complete our cross-species analysis, we queried a well-annotated data set of canine osteosarcoma gene expression, which was previously found to be highly conserved in human osteosarcoma, allowing us to examine the relationship of altered expression of CIS-associated genes with disease outcome45. Unsupervised hierarchical clustering of CIS-associated genes having appreciable variance in expression among canine samples generated two groups with different survival times (P = 0.05, log-rank test) (Supplementary Fig. 4a,b). This finding suggests that some CIS-associated genes are coordinately regulated or work cooperatively to establish cell-autonomous traits that mediate tumor behavior and, hence, patient outcomes. The observation that 22 CIS-associated genes independently predict survival in the same cohort supports this interpretation (Supplementary Fig. 4c). Moreover, 7 of these 22 genes are misregulated in a similar fashion in human and SB-mutagenized mouse osteosarcomas (APP, COL5A1, HADHB, LHFP, MEF2A, PAK1 and SEMA6D). These data demonstrate that CIS-associated genes are frequently altered in human osteosarcoma and predict outcome in canine osteosarcoma, supporting the conclusion that they are likely true drivers of osteosarcoma and are highly relevant to the human disease.
Cooperative loss of PTEN and TP53 in osteosarcoma
The most frequently mutated gene in our screen was Pten. The fact that the majority of SB-mutagenized tumors had loss of Trp53 function and the loss of TP53 in many human osteosarcomas strongly suggest that loss of both these genes is a cooperative event driving osteosarcomagenesis14. Additionally, copy number loss of PTEN has been observed in >60% of human osteosarcomas, and oncogenic miRNAs implicated in osteosarcoma target PTEN40–42,46. To validate this cooperation in osteosarcoma and generate a new model genetically relevant to human disease, we combined conditional Pten (loxP-flanked exons 4 and 5) and Trp53R270H alleles with Sp7-cre mice (Trp53-Pten mice)40,46,47. Pten loss accelerated osteosarcoma development and slightly increased penetrance in comparison to controls (P < 0.0001, log-rank test) (Fig. 5a). Tumors in these mice had a gross and histological appearance consistent with osteosarcoma (Fig. 5b).
Figure 5.
Pten loss accelerates osteosarcoma development in mice and PTEN loss enhances the anchorage-independent growth of immortalized human osteoblast cells. (a) Osteosarcoma-free survival curve depicting time to osteosarcoma development and survival endpoints for Trp53-C, Trp53-Pten and Pten mice. ***P < 0.0001, log-rank test. (b) Representative gross necropsy image of a primary osteosarcoma demonstrating typical gross morphology (left) and a section stained with hematoxylin and eosin (right) demonstrating a histological appearance consistent with osteosarcoma. Scale bar, 200 μm. (c) Diagram of the experimental procedure used to knock out PTEN with TALENs in immortalized human osteoblast cells. (d) Average number of colonies formed in soft agar by immortalized osteoblast cells treated with PTEN or HPRT1 TALENs. Data are the means ± s.e.m. of five independent experiments; ***P < 0.0001, Student's t test. Error bars, s.d.
To validate this cooperation in human osteosarcoma, we implemented transcription activator-like effector nucleases (TALENs) to knock out PTEN in an immortalized osteoblast cell line expressing the large T antigen, a known inhibitor of TP53 function. Cells were targeted with either PTEN or control HPRT1 TALENs and assayed for their ability to form colonies in soft agar (Fig. 5c). A Surveyor nuclease assay demonstrated robust mutation of PTEN (~60%) only when cells were treated with PTEN-targeting TALENs (data not shown), which significantly increased colony formation in soft agar (P < 0.0001; Fig. 5d). These results demonstrate that losses of Trp53 and Pten cooperate to accelerate osteosarcomagenesis in genetically engineered mice and enhance the transformed phenotype of immortalize osteoblast cells.
Neurofibromin 1 and 2 in osteosarcoma
Nf1 and Nf2 (encoding neurofibromin 1 and 2) were among the most frequently mutated genes in our screen. Interestingly, Nf2 was the top CIS-associated gene in Trp53-SBmut tumors, but no insertions in this gene were found in SBmut tumors (Supplementary Fig. 3). Both of these genes have been implicated but never validated as frank drivers of human osteosarcoma. In an attempt to validate NF1 and NF2 in human osteosarcoma, we performed TALEN-mediated knockout in immortalize osteoblast cells and found that NF1 but not NF2 knockout increased colony formation in soft agar (Supplementary Fig. 5; NF2 data not shown). Further, reexamination of published data sets demonstrated NF1 copy number losses, mutation by structural variation, hypermethylation and reduced expression in a few human osteosarcoma cell lines and tumors14,48. These data suggest that loss of NF1 is likely functionally important in human osteosarcoma, whereas the role of NF2 remains unclear.
Axon guidance genes in osteosarcoma
Pathway analysis of CIS-associated genes also found an enrichment for genes involved in axon guidance (IPA: n = 12 genes, P = 4.78 × 10−3; DAVID: n = 6 genes, P = 1.74 × 10−2) (Supplementary Tables 4 and 5). Of these genes, Sema4d and Sema6d are strong candidate proto-oncogenes (Fig. 6a). Fusion mRNA transcripts between the transposon murine stem cell virus (MSCV) promoter and splice donor site were detected in tumors containing insertions, which led to overexpression of Sema4d and Sema6d relative to tumors without insertions (Fig. 6b). Human osteosarcoma tumors demonstrated upregulation of SEMA4D and SEMA6D transcripts in comparison to normal human osteoblasts (Fig. 6c). Other CIS-associated genes involved in axon guidance were also significantly differentially expressed (Supplementary Fig. 6). Immunohistochemical analysis of SEMA4D and SEMA6D using tissue microarrays confirmed that these proteins are expressed in nearly all human osteosarcomas: these proteins were expressed at high levels (immunohistochemistry score of 3+ on a scale of 0–4) in over half of the tumors tested (Fig. 6d and Supplementary Table 11). SEMA4D and SEMA6D were also expressed at variable levels in human osteosarcoma cell lines (Fig. 6e). To functionally validate SEMA4D and SEMA6D as proto-oncogenes in human osteosarcoma, we overexpressed the corresponding cDNAs in human osteosarcoma cell lines.
Figure 6.
Axon guidance–related genes are implicated in osteosarcoma. (a) Diagram depicting T2/Onc insertion sites driving overexpression of Sema4d and Sema6d. Black and red arrows represent T2/Onc insertions identified in tumors from Trp53-SBmut and SBmut mice, respectively. (b) Relative mRNA levels of Sema4d and Sema6d in tumors with or without T2/Onc insertions in the respective genes. *P = 0.05, **P = 0.001, Student's t test. (c) Relative mRNA levels of SEMA4D and SEMA6D in normal human osteoblasts and human osteosarcoma tumors analyzed by RNA sequencing (n = 12 human osteosarcoma and 3 normal osteoblast samples). ***P < 0.0001, Student's t test. Error bars, s.d. (d) Immunohistochemistry scores for SEMA4D and SEMA6D in 80 osteosarcoma tumor samples. (e) Relative mRNA expression of SEMA4D and SEMA6D in the HOS, MG63, U2OS and SaOS2 human osteosarcoma cell lines normalized to ACTB expression. Data are means ± s.e.m.; n ≥ 3. Error bars, s.d.
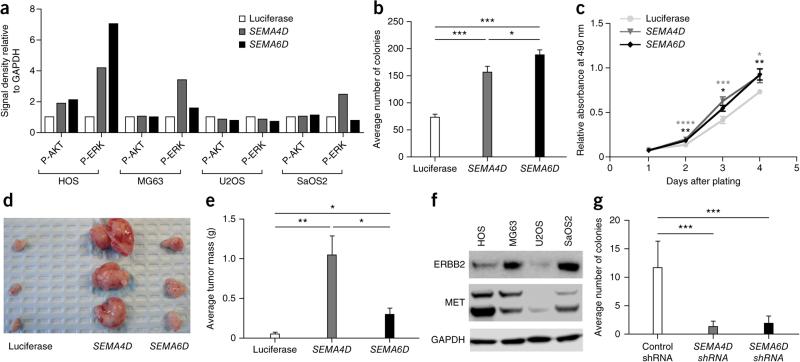
SEMA4D and SEMA6D signaling activates MET or ERBB2 and VEGFR2, respectively, which can lead to activation of PI3K-AKT-mTOR and MAPK signaling. SEMA4D overexpression increased the levels of phosphorylated AKT and/or ERK in HOS, MG63 and SaOS2 cells, whereas SEMA6D overexpression increased the levels of phosphorylated AKT and/or ERK in HOS and MG63 cells (Fig. 7a and Supplementary Fig. 7). To assess the phenotypic changes associated with these observations, we performed soft agar, proliferation and xenograft assays. Overexpression of SEMA4D or SEMA6D significantly increased colony formation in soft agar, proliferation and xenograft formation only in HOS cells (Fig. 7b–e). Interestingly, HOS was the only cell line with upregulation of both phosphorylated AKT and ERK. For SEMA4D, it is possible that the lack of phenotypic change is due to the expression levels of MET and ERBB2, as the expression of MET relative to ERBB2 can result in a negative or positive effect on tumorigenic properties that is cell type dependent49. Confirming this hypothesis, immunoblot analysis demonstrated that HOS cells expressed high MET and low ERBB2 levels in comparison to SaOS2 and MG63 cells, whereas U2OS cells had low levels of both receptors (Fig. 7f). To assess the effect of loss of function for these genes, we performed short hairpin RNA (shRNA)-mediated knockdown of SEMA4D and SEMA6D; again, only knockdown in HOS cells had the expected effect on soft agar colony formation (Fig. 7g and Supplementary Fig. 6). These data demonstrate that SEMA4D and SEMA6D are highly expressed in a large fraction of human osteosarcoma tumors and cell lines and, in some contexts, can increase the tumorigenic properties of human osteosarcoma cell lines when overexpressed. In contrast, reduced expression of these genes can inhibit some tumorigenic properties.
Figure 7.
SEMA4D and SEMA6D overexpression increases the levels of phosphorylated AKT and/or ERK. (a) Densitometry of immunoblot analysis (Supplementary Fig. 7) for cell lines overexpressing the luciferase gene, SEMA4D or SEMA6D. P-AKT, phosphorylated AKT; P-ERK, phosphorylated ERK. (b) Average number of colonies formed in soft agar by HOS cells overexpressing luciferase control, SEMA4D or SEMA6D cDNA. Data are the means ± s.e.m. of three independent experiments; *P < 0.05, ***P < 0.0001, Student's t test. (c) MTS proliferation assay of HOS cells overexpressing luciferase control, SEMA4D or SEMA6D cDNA. Data are the means ± s.e.m. of three independent experiments; *P < 0.05, **P < 0.001, ***P < 0.0001, ****P < 0.00001, Student's t test. (d) Representative image of the xenografts developing from subcutaneous injection of HOS cells overexpressing luciferase control, SEMA4D or SEMA6D cDNA into nude mice 3 weeks after injection. (e) Average tumor mass of the xenograft tumors in c. Data are means ± s.e.m.; n ≥ 6; *P < 0.05, **P < 0.001, Student's t test. (f) Immunoblot analysis for the indicated proteins in lysates from HOS cells overexpressing luciferase control, SEMA4D or SEMA6D cDNA. (g) Average number of colonies formed in soft agar by HOS cells treated with non-silencing, control, SEMA4D or SEMA6D shRNA pools. Data are the means ± s.e.m. of three independent experiments; ***P < 0.0001, t test.
Osteosarcoma metastases are clonal and identify CIS genes
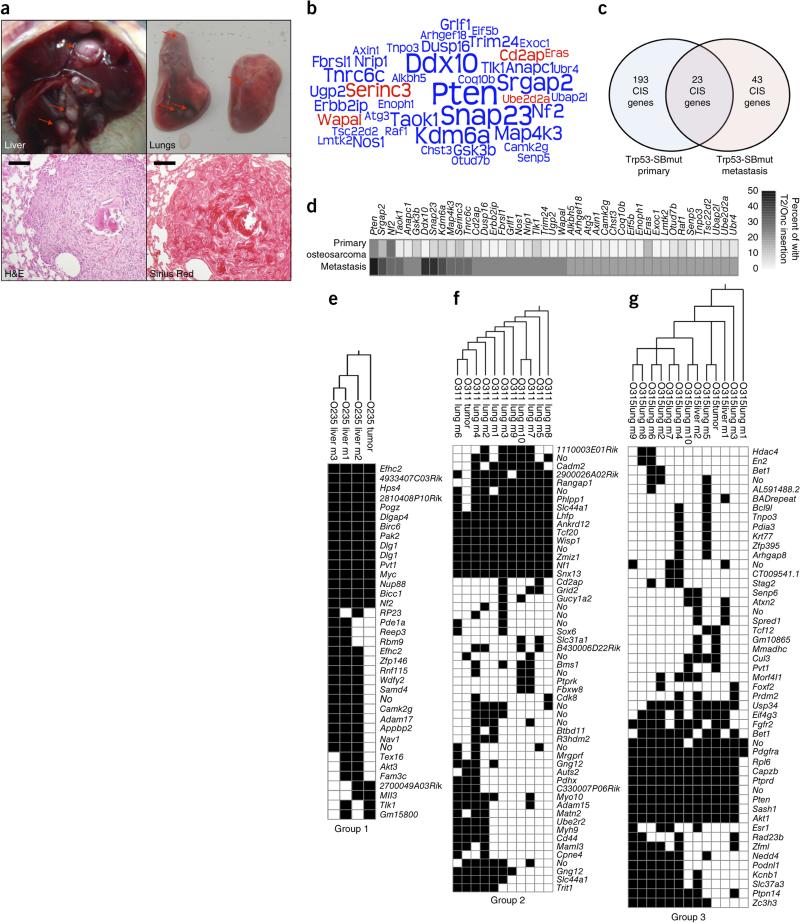
In addition to osteosarcomagenesis, 7 of 28 Trp53-C mice (25%) and 19 of 72 Trp53-SBmut mice (26.4%) had metastases in the liver and/or lungs (1–17 metastases per mouse, average of ~6). Metastases had a gross and histological appearance consistent with osteosarcoma metastases and were positive for collagen by Sirius Red staining (Fig. 8a). We identified transposon insertions for 134 metastases from 19 mutagenized mice, which identified 5 candidate proto-oncogenes and 38 candidate TSGs (Fig. 8b and Supplementary Table 12). Some metastasis CIS-associated genes were recurrently mutated in primary tumors; others were unique to metastases (Fig. 8c,d). The observation that 9 candidate metastasis genes (Pten, Gsk3b, Snap23, Map4k3, Arhgap35 (Grlf1), Arhgef18, Axin1, Raf1 and Ubap2l) have been implicated in metastasis of other cancers supports our conclusion that we have identified genes truly central to osteosarcoma metastasis50–58.
Figure 8.
Osteosarcoma metastases are clonal in nature and identify CIS-associated metastasis genes. (a) Top, representative gross necropsy images of osteosarcoma metastases in liver (left) and lung (right) demonstrating typical gross morphology. Red arrows indicate independent macroscopic metastases. Bottom left, section stained with hematoxylin and eosin (H&E) showing representative metastasis morphology with surrounding normal lung tissue. Scale bar, 100 μm. Bottom right, section stained with Sirius Red validating the presence of collagen deposits in osteosarcoma metastases. Scale bar, 100 μm. (b) Word cloud diagram depicting Trp53-SBmut metastasis CIS-associated genes. The size of each gene name is representative of the percentage of metastasis sets with insertions in the CIS-associated gene identified by TAPDANCE or gene-centric analysis; if a gene was identified by both analyses, the larger percentage was used. Predicted proto-oncogenes are depicted in red, and candidate TSGs are depicted in blue. (c) Venn diagram depicting the overlap in CIS-associated genes identified in primary osteosarcomas and metastases. (d) Heat map comparing the percentages of primary tumors and sets of metastases harboring T2/Onc insertions in metastasis CIS-associated genes. (e–g) Representative phylogeny analysis generated from parsimony and hierarchical clustering of the T2/Onc insertion sites identified in primary osteosarcoma tumors (tumor) and matched metastases (m). Group 1 demonstrates low overlap of T2/Onc insertions for primary tumors and metastases (e), group 2 shows many shared insertions (f) and group 3 appears to have an intermediate number of shared insertions (g). Phylogenetic trees and hierarchical clustering for all mice that developed metastases and had more than two macroscopic metastases can be found in Supplementary Figures 8 and 9.
Using transposon integration sites as a molecular fingerprint for individual metastases and their matched primary tumors, we performed a general parsimony analysis for 16 mice having more than 2 macroscopic metastases (Fig. 8e–g and Supplementary Fig. 8)59,60. The parsimony analysis produced three distinct groups of relatedness. The metastases for the first group (2/16 mice) shared few insertions with the corresponding primary tumors, suggesting an early metastatic event and supporting the early dissemination or parallel evolution model (Fig. 8e)61. The tumors for the second group (8/16 mice) shared many insertions with the metastases, indicating a model of clonal evolution of tumor and metastasis during tumorigenesis (Fig. 8f)61. The tumors for the third group (4/16 mice) were not the most common ancestor and did not share as many insertions with the metastases as seen in group 2, suggesting an intermediate model (Fig. 8g). The remaining mice had two tumors clustering with separate sets of metastases, which fell into different parsimony groups, indicating the presence of two independent metastatic osteosarcomas in a single mouse. In fact, five mice with metastatic disease harbored two tumors each. In some cases, all the metastases clustered with one tumor, and in other cases the metastases and both primary tumors clustered together; the latter situation is likely to be the result of a bone metastasis from the original osteosarcoma (Supplementary Figs. 8 and 9, O103 and O399). It is conceivable that these observations are partially due to insertions missed by linker-mediated PCR (LM-PCR); however, previous SB studies analyzing matched tumors and metastases performed extensive PCR analysis of specific insertions and found LM-PCR results to be highly coordinate60. These data demonstrate that SB-mutagenized osteosarcoma metastases are highly clonal and do not strictly adhere to a single model of metastasis development. Furthermore, candidate metastasis driver genes were identified.
DISCUSSION
We report a SB forward genetic screen identifying hundreds of significant CIS-associated genes for osteosarcoma. Notably, we identified dozens of genes previously implicated in osteosarcoma and numerous new genes. Candidate genes are frequently altered in human osteosarcoma and are enriched in cancer-associated signaling pathways, including PI3K-AKT-mTOR, ErbB, MAPK and axon guidance signaling. These pathways have previously been implicated in osteosarcoma, but the genes identified in these pathways are likely driving these pathways in human osteosarcoma and represent potential therapeutic targets62–64.
Interestingly, we identified a disproportionate number of TSGs relative to oncogenes. This phenomenon has been observed in previous solid tumor SB screens59,60,65,66. One possibility is that the MSCV promoter is not highly active in cells expressing Sp7-cre. One study performed whole-body somatic mutagenesis in mice using a strong CAG promoter in place of the MSCV promoter, with the mice developing more solid tumors than in MSCV studies67. The fact that T2/Onc has the ability to inactivate genes when inserted in either orientation also likely contributes to this observation. Regardless, we identified previously validated oncogenic drivers (MYC, CAPRIN1 and AKT2)1,20,37, new candidate oncogenes overexpressed and amplified in osteosarcoma (SEMA4D, ZNF217 and ZNF592) and TSGs under-expressed with copy number loss (LHFP, ENC1 and ENTPD4).
Nf1 and Nf2 were among the most frequently targeted CIS-associated genes in our screen. There have been reports of patients with neurofibromatosis type 1 (NF1) developing primary bone tumors. In addition, a retrospective study found that patients with NF1 had an eightfold higher risk of bone tumors, and whole-genome sequencing of human osteosarcomas found multiple deleterious structural variations in NF1 (refs. 14,68–70). These findings, in combination with our functional data, support a role for NF1 loss in human osteosarcoma. It has been reported that Nf2 loss induces osteosarcoma development in mice and is enhanced by Trp53 deficiency71. However, follow-up studies using human osteosarcoma tumors found no alteration in NF2 (ref. 72). Notably, the osteosarcomas seen in Nf2-null mice were not extensively tested for spontaneous Trp53 loss. The strong cooperation demonstrated with loss of both genes and the chromosomal proximity of these genes in mice suggest that there might be an obligate loss of both or of one allele of each for osteosarcoma formation in mice. The fact that these genes are on separate chromosomes in humans may partially explain why NF2 loss is not seen in human osteosarcoma.
Interestingly, Nf1 and Pten were also identified as top CIS-associated genes in our previously published SB screen for peripheral nerve sheath tumors65. This observation raises the possibility that these and/or other CIS-associated genes are hotspots for transposon insertion. To investigate this possibility, we compared genes identified in our previous screens with our osteosarcoma CIS-associated genes and found sparse overlap among the four different cancers, indicating that SB mutagenesis identifies cancer type–specific genes and not genes involved in cancer in general. Conversely, there are genes altered in specific human cancers that are not identified by forward genetic screens, demonstrating that there are limitations to screens in mice. For instance, whole-genome sequencing of pediatric osteosarcoma found that DLG2 and ATRX were altered in a significant portion of the samples analyzed14. Dlg2 was not significantly mutated in our screen, although many tumors harbored an inactivating insertion (n = 8 Trp53-SBmut and 3 SBmut tumors); the lack of significance is likely due to our stringent CIS calling. Virtually no insertions were identified in Atrx, which could be attributed to a species-specific difference, as ATRX is involved in telomere maintenance and telomere length is seemingly less critical in mouse models of cancer73. These data demonstrate that not all human cancer-related genes are readily identifiable through forward genetic screens in mice.
One of the new pathways identified by our screen was axon guidance, with the SEMA4D and SEMA6D components investigated. SEMA4D has been implicated in other cancers, including soft tissue sarcoma (STS), pancreatic cancer and glioma74. Less is known about SEMA6D, although it reportedly signals through PLXNA1 and its co-receptor, KDR (VEGFR2), and has been implicated in mesothelioma74,75. In normal bone homeostasis, osteoclasts express high levels of Sema4d, whereas osteoblasts do not express Sema4d but instead express its cognate receptor and co-receptor, Plxnb1 and Erbb2, respectively76. Thus, it is possible that osteoblasts misexpressing SEMA4D and MET might give rise to a subset of osteosarcomas. Similarly, the tumorigenic properties induced by overexpression of SEMA4D in human osteosarcoma cells were dependent on MET and ERBB2 levels, which has been reported49. Previous studies in osteosarcoma showed that high levels of ERBB2 are associated with a good outcome and that overexpression of MET can directly transform osteoblasts into osteosarcomas77,78. Perhaps analogous mechanisms regulating SEMA6D signaling exist or unrecognized crosstalk between SEMA4D and SEMA6D signaling occurs. Finally, the fact that SEMA4D and SEMA6D are cell surface receptors makes them attractive candidates for novel therapies. In fact, antibody therapy targeting SEMA4D was shown to reduce tumor growth in a xenograft model of STS when combined with antibody therapy for VEGF79. Currently, there is a fully humanized antibody targeting SEMA4D in phase I clinical trials for other solid tumors, perhaps allowing for an expedited trial in human patients with osteosarcoma (VX15, Vaccinex). Thus, it is possible that osteosarcomas expressing high levels of MET and SEMA4D, with low ERBB2 levels, may be susceptible to SEMA4D antibody therapy. Preclinical testing of XV15 antibody using human osteosarcoma tumor or cell line xenografts with this profile will need to be performed to assess the potential of this novel targeted therapy in osteosarcoma.
A subset of mutagenized mice also developed macroscopic metastases. Analysis of the CIS-associated genes from metastases showed that some genes are unique to metastases, whereas others are mutated in both primary tumors and metastases. This observation is consistent with the human disease, where metastasis-specific genes have been termed metastasis virulence genes, that is, genes only expressed after seeding of distal organs to initiate metastasis progression80. The genes shared by metastases and primary tumors have been proposed to be mediators of metastatic activities, such as angiogenesis, invasiveness and cell mobility80. These genes support tumor development via one effect and exhibit another effect at distant metastases. This hypothesis suggests that our metastasis CIS-associated genes likely consist of both gene types, whereas the genes identified in primary tumors are potentially mediators of metastatic activities. It is possible that if more mice with metastatic disease were generated that the mediators of metastatic activities identified in our primary tumors would also be identified in metastases. As osteosarcoma metastasis is the most prominent feature of poor outcome, validating metastasis-associated genes and developing new therapeutics to target them may be the most effective method to develop successful treatments to increase patient survival.
Overall, functional genomics using SB mutagenesis identified candidate osteosarcoma-related genes with functional relevance in human osteosarcoma that predict outcome in canine osteosarcoma and are capable of accelerating osteosarcomagenesis in mice. In addition, we generated a new mouse model of osteosarcoma and validated the involvement of axon guidance–related genes in human osteosarcoma. Further testing of candidate driver genes in development and metastasis may lead to the identification of new pathways or critical targets for novel osteosarcoma treatments.
URLs
Integrative Genomics Viewer, http://www.broadinstitute.org/igv/; Ingenuity Pathway Analysis (IPA), http://www.ingenuity.com/; Catalogue of Somatic Mutations in Cancer (COSMIC), http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/; DAVID, http://david. abcc.ncifcrf.gov/; Gene-centric CIS analysis, http://ias.eng.uiowa.edu/uploader/.
ONLINE METHODS
Mice
All mouse work was carried out under University of Minnesota Institutional Animal Care and Use Committee guidelines. All mouse strains used in this study have been previously described13,16,17,47. Because of the use of many transgenes, experimental and control mice were on a mixed genetic background. The two concatemer lines used had concatemers located on chromosomes 4 and 15 harboring ~300 and ~25 copies of T2/Onc, respectively; equal numbers of mice were generated for the chromosome 4 and 15 lines. The Trp53R270H allele was chosen over the Trp53R172H allele, as its analogous substitution is more prevalent in human osteosarcoma (5.1% versus 3.4%; International Agency for Research on Cancer (IARC) TP53 Database), and the dominant-negative Trp53 allele was chosen over the traditional loxP-flanked allele to reduce the number of transgenes needed to achieve impaired Trp53 function81. Mice were aged until tumors developed or until moribund. Upon necropsy, tumors were divided into three portions, with one portion each snap frozen for DNA extraction, fixed in 10% formalin for histological analysis and snap frozen in RNAlater (Sigma) for RNA extraction. Pathological analysis and tumor classification were performed at the US National Institutes of Health/National Cancer Institute. Osteosarcoma-free survival graphs were generated using the Prism statistical software package.
CIS analysis
CIS analysis of T2/Onc insertion sites was performed using TAPDANCE and gene-centric software, as previously described65,82,83. Because of the ‘local hopping’ phenomenon in SB transposition and in accordance with all previously published SB models, CIS-associated genes identified on the chromosome with the transposon concatemer were removed from all core analyses, although they are listed in Supplementary Table 3. CIS-associated genes were considered proto-oncogenes if all insertions were upstream or in early introns of the gene with the transposon's MSCV promoter and splice donor site in the same orientation as the ORF, driving overexpression. Conversely, candidate TSGs had an insertion profile lacking bias for the orientation of the MSCV promoter or the location of the insertion within the gene and disrupted gene transcription by virtue of the transposon's splice acceptor and polyadenylation signals (Supplementary Tables 1–3). Because of the high clonality of the metastases, the insertion sites of all metastases in a single mouse were collapsed to represent a distinct sample for TAPDANCE and gCIS analyses.
Vector construction
PB/SB-CAG-GOI-GFP-PGK-PURO–based overexpression vectors were generated with the LR Clonase Gateway cloning system (Invitrogen) using cDNAs in Entry vectors with the previously described PB/SB-CAG-DEST-GFP-PGK-PURO vector, following the manufacturer's instructions65. SEMA4D and SEMA6D cDNAs were purchased from Open Biosystems and cloned into the Gateway system by standard PCR and BP Clonase reaction (Invitrogen), following the manufacturer's instructions.
Cell culture and electroporation
HOS, U2OS, MG63 and SaOS2 cells were acquired from the American Type Culture Collection (ATCC). HOS and MG63 cells were maintained in EMEM medium supplemented with 10% FBS and 1% penicillin-streptomycin. U2OS and SaOS2 cells were maintained in McCoy's 5A medium supplemented with 10% FBS and 1% penicillin-streptomycin. Immortalized human osteoblast cells were maintained in OGM BulletKit medium (Lonza) with 500 ng/μl puromycin and 100 ng/μl neomycin. These cells were generated by transducing human osteoblasts (human mesenchymal stem cells were purchased from Lonza and differentiated into osteoblasts) with cDNA for human TERT and SV40 large T antigen (data not shown). Electroporation was carried out using the NEON transfection system (Invitrogen), following the manufacturer's protocol. Stable lines were generated by electroporation with 2 μg of PB7 transposase and 2 μg of transposon plasmid. Cell lines were tested for mycoplasma using the Lonza kit. All lines were negative.
Canine osteosarcoma analysis
CIS-associated genes were evaluated against survival in a set of 27 canine osteosarcoma cell lines established from canine patients with a known outcome (overall survival time), with their expression evaluated on the Affymetrix Canine Genome 2.0 Array. The 186 CIS genes identified in primary tumors that were present on the canine platform were filtered for the most differentially expressed genes in the canine samples by setting the coefficient of variance to 0.5. Unsupervised hierarchical clustering of the canine samples on the basis of 48 differentially expressed genes was performed as previously described45. Genedata Analyst was used for Kaplan-Meier survival analysis.
To establish whether loss or gain of expression for individual CIS genes correlated with a poor outcome in canine patients, we first determined the mean gene expression values for each of the CIS genes. The canine samples were then grouped according to their gene expression relative to the mean expression for each CIS gene (samples with gene expression above the mean were designated as having ‘high’ expression, whereas samples with gene expression below the mean were designated as having ‘low’ expression). Genedata Analyst was used for Kaplan-Meier survival analysis of the canine patient groups with high and low expression. Log-rank Mantel-Cox P values of <0.05 were considered significant.
Genotyping and quantitative RT-PCR analysis
Upon weaning of the mice, a small portion of the tail (~0.5 cm) was collected, and DNA was extracted using standard phenol:chloroform:isoamyl alcohol extraction after overnight digestion in SDS extraction buffer containing proteinase K as described84. The sequences for the genotyping primers can be found in Supplementary Table 13. PCR was performed using 1.1× ReddyMix (Thermo Scientific) with an initial denaturation step at 95 °C for 2 min; 35 cycles of denaturing at 95 °C for 25 s, annealing at 55 °C for 35 s and extension at 72 °C for 1.5 min; and a final extension step at 72 °C for 5 min. PCR amplicons were resolved on 1% agarose gels and analyzed for the presence of the transgenes. RNA was extracted from cell culture pellets using the RNA Mini kit (Invitrogen) and reverse transcribed into cDNA using the SuperScript III reverse-transcriptase kit (Invitrogen), following the manufacturer's instructions. Quantitative RT-PCR was performed using FastStart Universal SYBR Green Master (Rox) mix (Roche) on an Eppendorf Realplex 2 MasterCycler EP Gradient S. Primer sequences are listed in Supplementary Table 13.
aCGH, SKY and G banding
aCGH, SKY and G-banding analyses were performed by the University of Minnesota's Cytogenomics Laboratory. For aCGH analysis, mouse tumor DNA was digested by restriction enzyme and labeled with the Cy5 fluorochrome using random primers and exo Klenow fragment DNA polymerase. Control tail DNA from the same mouse was concurrently labeled with Cy3. The sample and control DNA were combined, and aCGH was performed with a microarray constructed by Agilent Technologies that contains approximately 170,000 distinct biological oligonucleotides spaced at an average interval of 10.9 kb. The ratio of sample to control DNA for each oligonucleotide was calculated using Feature Extraction software 10.5 (Agilent Technologies). The abnormal threshold was applied using Genomics Workbench 7.0 (Agilent Technologies). A combination of several statistical algorithms was applied. A minimum of three oligonucleotides with a minimum absolute ratio of 0.1 (based on the log2 ratio) was required for a copy number loss or gain to be reported.
SB excision assays
SB excision assays were performed as previously described85.
Linker-mediated PCR
LM-PCR was performed as previously described65. Briefly, genomic DNA was extracted from tumor samples by standard phenol: chloroform:isoamyl alcohol extraction. DNA (100 ng) was digested with BfaI and Nlalll separately to clone regions adjacent to the left and right sides of the transposon, respectively. After overnight digestion at 37 °C, reactions were heat inactivated at 80 °C for 20 min. Linkers for the left and right sides were then prepared by annealing sense and antisense primers (sequences are listed in Supplementary Table 13). The prepared linkers were then ligated overnight at 16 °C to genomic DNA digested with the corresponding restriction enzyme. Ligated DNA was purified using Qiagen MinElute 96 UF Plates, following the manufacturer's instructions. The purified DNA was digested with BamHI overnight at 37 °C to remove any transposons still in the T2/Onc concatemer array. The BamHI-digested DNA was purified using Qiagen MinElute 96 UF Plates, following the manufacturer's instructions. Primary PCR was performed using 2× ReddyMix with an annealing temperature of 55 °C for 30 cycles (primer sequences are listed in Supplementary Table 13). The primary PCR reaction was then diluted 1:75 in water, and 1 μl of this dilution was used for secondary PCR with Long-Range Taq (Roche), applying an annealing temperature of 53 °C for 35 cycles (primer sequences are listed in Supplementary Table 13). The primers used for secondary PCR were barcoded and designed for Illumina sequencing; these have been published previously65.
Immunohistochemistry and immunoblot analysis
Immunohistochemistry was performed using the mouse-on-mouse (M.O.M.) kit (Vector Laboratories) and developed using the ABC Vectastain peroxidase system (Vector Laboratories) as previously described84. Antibody to SB11 (mouse monoclonal; clone 324622, R&D Systems) was used at 2.5 μg/ml. Unstained tumor microarray sections (4 μm thick) were deparaffinized and rehydrated using standard methods. For antigen retrieval, slides were incubated in pH 6.0 buffer (Reveal Decloaking reagent, Biocare Medical) in a steamer for 30 min at 95–98 °C, followed by a 20-min cooling period. Subsequent steps were automated using an immunohistochemical staining platform (Nemesis, Biocare). Endogenous peroxidase activity was quenched by immersion of slides in 3% hydrogen peroxide solution (Peroxidazed, Biocare) for 10 min followed by a TBST rinse. A serum-free blocking solution (Background Sniper, Biocare) Medical) was placed on sections for 10 min. Blocking solution was removed, and slides were incubated in primary antibody diluted in a solution comprising 10% blocking solution and 90% TBST. Mouse monoclonal antibody to CD100 (610671, BD Transduction; 1:10,000 dilution) and rabbit polyclonal antibody to SEMA6D (HPA043109, Sigma Prestige; 1:200 dilution) were incubated with slides for 60 min at room temperature followed by rinsing in TBST and detection with the Novocastra Novolink Polymer kit (Leica Microsystems) using the manufacturer's specifications. All slides were then rinsed with TBST, and signal was detected with diaminobenzidine (DAB; Covance). Slides were incubated for 5 min, rinsed in TBS and counterstained with CAT Hematoxylin (Biocare) for 5 min. Slides were then dehydrated and coverslipped.
Immunoblot analysis was performed on whole-cell lysates collected using RIPA buffer (Sigma) supplemented with phosphatase inhibitors (Sigma) and protease inhibitors (Roche). Lysates were separated by SDS-PAGE, transferred to a PVDF membrane (Invitrogen), incubated in 5% BSA in TBST to block nonspecific binding and probed with appropriate antibodies. Proteins were detected on a LI-COR Odyssey Fc instrument using a chemiluminescent horseradish peroxidase (HRP) substrate (Advansta). Antibodies against ERK (4696), phosphorylated ERK(4370), AKT (4685), phosphorylated AKT (4060), MET (8198), ERBB2 (4290) and GAPDH (2118) were purchased from Cell Signaling Technology.
Sirius Red staining
Slides were dehydrated in a standard ethanol gradient, washed once in 1× PBS and stained with hematoxylin (Fisher Scientific). Following counterstaining, slides were incubated in Sirius Red solution (0.5 g Direct Red 80 in 500 ml of saturated 1.2% picric acid) for 1 h at room temperature while gently shaking. Slides were washed twice for 5 min each wash in acidified water (5 ml of glacial acetic acid in 1 l of water). Slides were then dehydrated and coverslipped.
MTS proliferation assays
Cells were trypsinized using 0.25% trypsin in 2.21 mM EDTA (CellGro) and quantified using the Countess cell counter (Invitrogen); 1,200 cells were plated per well of 96-well plates in sextuplicate. Twenty-four hours after plating, 20 μl of a 1:20 mixture of phenazine methosulfate:3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (PMS:MTS) was added to each well, and cells were allowed to incubate for 2.5 h. Absorbance at 490 nm and 650 nm was read using the BioTek SynergyMx fluorescence plate reader. Data were analyzed using Microsoft Excel and graphed using the Prism software package.
Soft agar assays
Cells (1 × 104) were seeded into soft agar in single wells of six-well plates and allowed to incubate for 2–3 weeks. The resultant colonies were stained with 0.5% crystal violet in buffered formalin for 1 h. Each well was divided into four quadrants and photographed. Colonies were quantified using ImageJ software using a standard colony quantification macro.
RNA sequencing and analysis
Fastq data from 12 juvenile primary osteosarcoma RNA sequencing samples were analyzed14. RNA was extracted from three human osteoblast samples (PromoCell) using an RNA Midi extraction kit (Qiagen), and quality was tested using a Bioanalyzer 2100 (Agilent Technologies). The RNA was converted into sequence libraries using the TruSeq RNA Sample Preparation kit (Illumina). Lastly, each library was size selected, and the selection process was validated and the libraries quantified by capillary electrophoresis and quantitative PCR, respectively. Sequencing was accomplished on a HiSeq 2000 (Illumina), with the goal of generating a minimum of 70 million paired-end 50-bp reads. The fastq files for both the osteosarcomas and the osteoblast controls were loaded into Galaxy. FastQC: Read QC (version 0.51) was used to evaluate the quality of the reads. TopHat 2.0.9 was used to map the paired reads to the GRCh37/hg19 assembly of the human genome. Gene expression analysis of the RNA sequencing data was conducted using Cuffdiff 2.1.1 by performing paired comparisons of the OB2 sample and all other samples. Cuffdiff mapped the reads to the GRCh37/hg19 human genome assembly and presented the data in terms of fragments per kilobase of transcript per million mapped reads (FPKM). Expression of the CIS-associated genes was verified by visualization of the mapped reads using the Integrative Genomics Viewer. The FPKM values for the CIS-associated genes are available in Supplementary Table 7.
Parsimony analysis of metastases
To generate a formal and mathematically rigorous phylogenetic tree describing the relationship between the primary osteosarcoma tumors and the metastases, all non-redundant insertions found in two or more samples in each mouse were used as input for the phylogeny calculation program Pars, a part of PHYLIP (PHYLIP-3.69). Pars is a general parsimony program that carries out the Wagner parsimony method with multiple states. Wagner parsimony allows changes among all states, and the criterion is to find the tree that requires the minimum number of changes.
Supplementary Material
ACKNOWLEDGMENTS
B.S.M. was funded by US National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases Musculoskeletal Training Grant AR050938. This research was funded by the Sobiech Osteosarcoma Fund Award, the Children's Cancer Research Fund, an American Cancer Center Research Professor Grant (123939) and National Cancer Institute grant R01 CA113636 (to D.A.L.). We extend our thanks to the University of Minnesota resources involved in our project. The University of Minnesota Genomics Center provided services for RNA sequencing, oligonucleotide preparation and Sanger sequencing. The Minnesota Supercomputing Institute maintains the Galaxy software platform, as well as provides data management services and training. The cytogenetic analyses were performed in the Cytogenomics Shared Resource at the University of Minnesota with support from the comprehensive Masonic Cancer Center (US National Institutes of Health grant P30 CA077598).
Footnotes
AUTHOR CONTRIBUTIONS
B.S.M., G.M.O., E.P.R., S.K.R., N.K.W., M.T.W., L.A.M. and K.J.H. performed laboratory experiments and/or analyzed the data. N.A.T. and K.C. performed bioinformatic data analysis of RNA sequencing, methylome and copy number analysis data. M.A.D. provided RNA sequencing and methylation data for human osteosarcoma samples. C.L.F. performed immunohistochemistry staining on osteosarcoma tumor microarrays. M.C.S. and J.F.M. provided data on canine osteosarcoma gene expression and outcome. A.L.S. analyzed the deep sequencing data for CIS analysis. R.K. and S.D.M. acquired and analyzed data from COSMIC and CGC. R.S.L. performed comparative analysis of CIS genes among SB screens. S.M.H. and C.K. assessed the histology of mouse tumors. R.G. and Y.Y. generated the immortalized osteoblast cells. D.A.L. supervised laboratory experiments and assisted in writing the manuscript. B.S.M. wrote the manuscript.
Accession codes. Raw data files for the osteoblast samples are available at the Gene Expression Omnibus (GSE57925). Methylation data for primary tumors and human osteoblasts have been deposited in the Gene Expression Omnibus (GSE58770). Gene expression data for canine osteosarcoma evaluated on the Affymetrix Canine Genome 2.0 Array are available in the Gene Expression Omnibus (GSE27217).
Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- 2.Allison DC, et al. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma. 2012;2012:704872. doi: 10.1155/2012/704872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaffe N, Puri A, Gelderblom H. Osteosarcoma: evolution of treatment paradigms. Sarcoma. 2013;2013:203531. doi: 10.1155/2013/203531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsuchiya H, et al. Effect of timing of pulmonary metastases identification on prognosis of patients with osteosarcoma: the Japanese Musculoskeletal Oncology Group study. J. Clin. Oncol. 2002;20:3470–3477. doi: 10.1200/JCO.2002.11.028. [DOI] [PubMed] [Google Scholar]
- 5.Kager L, et al. Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols. J. Clin. Oncol. 2003;21:2011–2018. doi: 10.1200/JCO.2003.08.132. [DOI] [PubMed] [Google Scholar]
- 6.Meyers PA, et al. Osteogenic sarcoma with clinically detectable metastasis at initial presentation. J. Clin. Oncol. 1993;11:449–453. doi: 10.1200/JCO.1993.11.3.449. [DOI] [PubMed] [Google Scholar]
- 7.Dobson JM. Breed-predispositions to cancer in pedigree dogs. ISRN Vet. Sci. 20132013:941275. doi: 10.1155/2013/941275. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overholtzer M, et al. The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc. Natl. Acad. Sci. USA. 2003;100:11547–11552. doi: 10.1073/pnas.1934852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat. Rev. Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 10.Kuijjer ML, et al. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosom. Cancer. 2012;51:696–706. doi: 10.1002/gcc.21956. [DOI] [PubMed] [Google Scholar]
- 11.Sadikovic B, et al. Identification of interactive networks of gene expression associated with osteosarcoma oncogenesis by integrated molecular profiling. Hum. Mol. Genet. 2009;18:1962–1975. doi: 10.1093/hmg/ddp117. [DOI] [PubMed] [Google Scholar]
- 12.Kresse SH, et al. Integrative analysis reveals relationships of genetic and epigenetic alterations in osteosarcoma. PLoS ONE. 2012;7:e48262. doi: 10.1371/journal.pone.0048262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat. Rev. Cancer. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7:104–112. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones KB. Osteosarcomagenesis: modeling cancer initiation in the mouse. Sarcoma. 2011;2011:694136. doi: 10.1155/2011/694136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 17.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Zhang G, Li M, Zhu X, Bai Y, Yang C. Knockdown of Akt sensitizes osteosarcoma cells to apoptosis induced by cisplatin treatment. Int. J. Mol. Sci. 2011;12:2994–3005. doi: 10.3390/ijms12052994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark JC, Dass CR, Choong PF. A review of clinical and molecular prognostic factors in osteosarcoma. J. Cancer Res. Clin. Oncol. 2008;134:281–297. doi: 10.1007/s00432-007-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabile AA, et al. Caprin-1, a novel Cyr61-interacting protein, promotes osteosarcoma tumor growth and lung metastasis in mice. Biochim. Biophys. Acta. 2013;1832:1173–1182. doi: 10.1016/j.bbadis.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Guo W, et al. Expression of bone morphogenetic proteins and receptors in sarcomas. Clin. Orthop. Relat. Res. 1999;365:175–183. doi: 10.1097/00003086-199908000-00023. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima G, et al. CDH11 expression is associated with survival in patients with osteosarcoma. Cancer Genomics Proteomics. 2008;5:37–42. [PubMed] [Google Scholar]
- 23.Chueh FS, et al. Bufalin-inhibited migration and invasion in human osteosarcoma U-2 OS cells is carried out by suppression of the matrix metalloproteinase-2, ERK, and JNK signaling pathways. Environ. Toxicol. 2014;29:21–29. doi: 10.1002/tox.20769. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, et al. miR-16 inhibits cell proliferation by targeting IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS Lett. 2013;587:1366–1372. doi: 10.1016/j.febslet.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Osborne TS, et al. Evaluation of eIF4E expression in an osteosarcoma-specific tissue microarray. J. Pediatr. Hematol. Oncol. 2011;33:524–528. doi: 10.1097/MPH.0b013e318223d0c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H, Mei Q, Xiong C, Zhao J. Tumor-suppressing effects of miR-141 in human osteosarcoma. Cell Biochem. Biophys. 2014;69:319–325. doi: 10.1007/s12013-013-9801-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhao R, Ni D, Tian Y, Ni B, Wang A. Aberrant ADAM10 expression correlates with osteosarcoma progression. Eur. J. Med. Res. 2014;19:9. doi: 10.1186/2047-783X-19-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toledo SRC, et al. Bone deposition, bone resorption, and osteosarcoma. J. Orthop. Res. 2010;28:1142–1148. doi: 10.1002/jor.21120. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, et al. Rapamycin increases pCREB, Bcl-2, and VEGF-A through ERK under normoxia. Acta Biochim. Biophys. Sin. (Shanghai) 2013;45:259–267. doi: 10.1093/abbs/gmt002. [DOI] [PubMed] [Google Scholar]
- 30.Zhang P, Yang Y, Zweidler-McKay PA, Hughes DP. Critical role of Notch signaling in osteosarcoma invasion and metastasis. Clin. Cancer Res. 2008;14:2962–2969. doi: 10.1158/1078-0432.CCR-07-1992. retracted 15 September 2013. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Patiño-García A, Zalacaín M, Marrodán L, San-Julián M, Sierrasesúmaga L. Methotrexate in pediatric osteosarcoma: response and toxicity in relation to genetic polymorphisms and dihydrofolate reductase and reduced folate carrier 1 expression. J. Pediatr. 2009;154:688–693. doi: 10.1016/j.jpeds.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 32.Kim KO, et al. Proteomic identification of 14-3-3ε as a linker protein between pERK1/2 inhibition and BIM upregulation in human osteosarcoma cells. J. Orthop. Res. 2014;32:848–854. doi: 10.1002/jor.22598. [DOI] [PubMed] [Google Scholar]
- 33.de Nigris F, et al. YY1 overexpression is associated with poor prognosis and metastasis-free survival in patients suffering osteosarcoma. BMC Cancer. 2011;11:472. doi: 10.1186/1471-2407-11-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu XY, et al. Cell cycle regulator gene CDC5L, a potential target for 6p12-p21 amplicon in osteosarcoma. Mol. Cancer Res. 2008;6:937–946. doi: 10.1158/1541-7786.MCR-07-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan D, et al. Overexpression of fibroblast activation protein and its clinical implications in patients with osteosarcoma. J. Surg. Oncol. 2013;108:157–162. doi: 10.1002/jso.23368. [DOI] [PubMed] [Google Scholar]
- 36.Sato H, et al. Repression of p53-dependent sequence-specific transactivation by MEF2c. Biochem. Biophys. Res. Commun. 1995;214:468–474. doi: 10.1006/bbrc.1995.2310. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Y, Zhou J, Ji Y, Yu B. Elevated expression of AKT2 correlates with disease severity and poor prognosis in human osteosarcoma. Mol. Med. Rep. 2014;10:737–742. doi: 10.3892/mmr.2014.2314. [DOI] [PubMed] [Google Scholar]
- 38.Han K, et al. microRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. Int. J. Oncol. 2014;45:1437–1449. doi: 10.3892/ijo.2014.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K, Zhang X. Inhibition of SENP5 suppresses cell growth and promotes apoptosis in osteosarcoma cells. Exp. Ther. Med. 2014;7:1691–1695. doi: 10.3892/etm.2014.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones KB, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y, Luo L, Li S, Yang C. miR-17 inhibitor suppressed osteosarcoma tumor growth and metastasis via increasing PTEN expression. Biochem. Biophys. Res. Commun. 2014;444:230–234. doi: 10.1016/j.bbrc.2014.01.061. [DOI] [PubMed] [Google Scholar]
- 42.Song QC, et al. Downregulation of microRNA-26a is associated with metastatic potential and the poor prognosis of osteosarcoma patients. Oncol. Rep. 2014;31:1263–1270. doi: 10.3892/or.2014.2989. [DOI] [PubMed] [Google Scholar]
- 43.Tesser-Gamba F, et al. MAPK7 and MAP2K4 as prognostic markers in osteosarcoma. Hum. Pathol. 2012;43:994–1002. doi: 10.1016/j.humpath.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava A, et al. High WT1 expression is associated with very poor survival of patients with osteogenic sarcoma metastasis. Clin. Cancer Res. 2006;12:4237–4243. doi: 10.1158/1078-0432.CCR-05-2307. [DOI] [PubMed] [Google Scholar]
- 45.Scott MC, et al. Molecular subtypes of osteosarcoma identified by reducing tumor heterogeneity through an interspecies comparative approach. Bone. 2011;49:356–367. doi: 10.1016/j.bone.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freeman SS, et al. Copy number gains in EGFR and copy number losses in PTENare common events in osteosarcoma tumors. Cancer. 2008;113:1453–1461. doi: 10.1002/cncr.23782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao A, et al. Somatic induction of Pten loss in a preclinical astrocytoma model reveals major roles in disease progression and avenues for target discovery and validation. Cancer Res. 2005;65:5172. doi: 10.1158/0008-5472.CAN-04-3902. [DOI] [PubMed] [Google Scholar]
- 48.Neale G, et al. Molecular characterization of the pediatric preclinical testing panel. Clin. Cancer Res. 2008;14:4572–4583. doi: 10.1158/1078-0432.CCR-07-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swiercz JM, Worzfeld T, Offermanns S. ErbB-2 and Met reciprocally regulate cellular signaling via plexin-B1. J. Biol. Chem. 2008;283:1893–1901. doi: 10.1074/jbc.M706822200. [DOI] [PubMed] [Google Scholar]
- 50.Jiang L, et al. CLDN3 inhibits cancer aggressiveness via Wnt-EMT signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget. 2014;5:7663–7676. doi: 10.18632/oncotarget.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 52.Williams KC, Coppolino MG. SNARE-dependent interaction of Src, EGFR and β1 integrin regulates invadopodia formation and tumor cell invasion. J. Cell Sci. 2014;127:1712–1725. doi: 10.1242/jcs.134734. [DOI] [PubMed] [Google Scholar]
- 53.Zhao B, et al. MicroRNA let-7c inhibits migration and invasion of human non–small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 54.Kusama T, et al. Inactivation of Rho GTPases by p190 RhoGAP reduces human pancreatic cancer cell invasion and metastasis. Cancer Sci. 2006;97:848–853. doi: 10.1111/j.1349-7006.2006.00242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song C, et al. Expression of p114RhoGEF predicts lymph node metastasis and poor survival of squamous-cell lung carcinoma patients. Tumour Biol. 2013;34:1925–1933. doi: 10.1007/s13277-013-0737-8. [DOI] [PubMed] [Google Scholar]
- 56.Zhou CX, Gao Y. Frequent genetic alterations and reduced expression of the Axin1 gene in oral squamous cell carcinoma: involvement in tumor progression and metastasis. Oncol. Rep. 2007;17:73–79. [PubMed] [Google Scholar]
- 57.Li X, et al. PHLPP is a negative regulator of RAF1, which reduces colorectal cancer cell motility and prevents tumor progression in mice. Gastroenterology. 2014;146:1301–1312. doi: 10.1053/j.gastro.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li D, Huang Y. Knockdown of ubiquitin associated protein 2–like inhibits the growth and migration of prostate cancer cells. Oncol. Rep. 2014;32:1578–1584. doi: 10.3892/or.2014.3360. [DOI] [PubMed] [Google Scholar]
- 59.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat. Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu X, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klein CA. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 62.Loh AH, et al. Dissecting the PI3K signaling axis in pediatric solid tumors: novel targets for clinical integration. Front. Oncol. 2013;3:93. doi: 10.3389/fonc.2013.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yue B, Ren Q, Su T, Wang L, Zhang L. ERK5 silencing inhibits invasion of human osteosarcoma cell via modulating the Slug/MMP-9 pathway. J. Clin. Exp. Pathol. 2014;4:2640–2647. [PubMed] [Google Scholar]
- 64.Geryk-Hall M, Hughes DPM. Critical signaling pathways in bone sarcoma: candidates for therapeutic interventions. Curr. Oncol. Rep. 2009;11:446–453. doi: 10.1007/s11912-009-0061-z. [DOI] [PubMed] [Google Scholar]
- 65.Rahrmann EP. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies novel genes and genetic pathways driving tumorigenesis. Nat. Genet. 2013;45:756–766. doi: 10.1038/ng.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dupuy AJ, et al. A modified Sleeping Beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cinamon U, Avinoach I, Harell M. Neurofibromatosis type 1, hyperparathyroidism, and osteosarcoma: interplay? Eur. Arch. Otorhinolaryngol. 2002;259:540–542. doi: 10.1007/s00405-002-0497-3. [DOI] [PubMed] [Google Scholar]
- 69.Ferrari A, Bisogno G, Carli M. Management of childhood malignant peripheral nerve sheath tumor. Paediatr. Drugs. 2007;9:239–248. doi: 10.2165/00148581-200709040-00005. [DOI] [PubMed] [Google Scholar]
- 70.Chowdhry M, et al. Bone sarcomas arising in patients with neurofibromatosis type 1. J. Bone Joint Surg. Br. 2009;91:1223. doi: 10.1302/0301-620X.91B9.22299. [DOI] [PubMed] [Google Scholar]
- 71.McClatchey AI, et al. Mice heterozygous for a mutation at the Nf2 tumor suppressor locus develop a range of highly metastatic tumors. Genes Dev. 1998;12:1121–1133. doi: 10.1101/gad.12.8.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stemmer-Rachamimov AO, et al. The NF2 gene and merlin protein in human osteosarcomas. Neurogenetics. 1998;2:73–74. doi: 10.1007/s100480050054. [DOI] [PubMed] [Google Scholar]
- 73.Rudolph KL, et al. Longevity, stress response, and cancer in aging telomerasedeficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 74.Law JWS, Lee AYW. The role of semaphorins and their receptors in gliomas. J. Signal Transduct. 2012;2012:902854. doi: 10.1155/2012/902854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Catalano A, Lazzarini R, Di Nuzzo S, Orciari S, Procopio A. The plexin-A1 receptor activates vascular endothelial growth factor–receptor 2 and nuclear factor– κB to mediate survival and anchorage-independent growth of malignant mesothelioma cells. Cancer Res. 2009;69:1485–1493. doi: 10.1158/0008-5472.CAN-08-3659. [DOI] [PubMed] [Google Scholar]
- 76.Negishi-Koga T, et al. Suppression of bone formation by osteoclastic expression of semaphorin 4D. Nat. Med. 2011;17:1473–1480. doi: 10.1038/nm.2489. [DOI] [PubMed] [Google Scholar]
- 77.Patanè S, et al. MET overexpression turns human primary osteoblasts into osteosarcomas. Cancer Res. 2006;66:4750–4757. doi: 10.1158/0008-5472.CAN-05-4422. [DOI] [PubMed] [Google Scholar]
- 78.Akatsuka T, et al. ErbB2 expression is correlated with increased survival of patients with osteosarcoma. Cancer. 2002;94:1397–1404. doi: 10.1002/cncr.10360. [DOI] [PubMed] [Google Scholar]
- 79.Zhou H, Binmadi NO, Yang Y, Proia P, Basile JR. Semaphorin 4D cooperates with VEGF to promote angiogenesis and tumor progression. Angiogenesis. 2012;15:391–407. doi: 10.1007/s10456-012-9268-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 81.Ognjanovic S, Olivier M, Bergemann TL, Hainaut P. Sarcomas in TP53germline mutation carriers. Cancer. 2012;118:1387–1396. doi: 10.1002/cncr.26390. [DOI] [PubMed] [Google Scholar]
- 82.Sarver AL, Erdman J, Starr T, Largaespada DA, Silverstein KA. TAPDANCE: an automated tool to identify and annotate transposon insertion CISs and associations between CISs from next generation sequence data. BMC Bioinformatics. 2012;13:154. doi: 10.1186/1471-2105-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brett BT, et al. Novel molecular and computational methods improve the accuracy of insertion site analysis in Sleeping Beauty–induced tumors. PLoS ONE. 2011;6:e24668. doi: 10.1371/journal.pone.0024668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moriarity B, Largaespada DA. A comprehensive guide to Sleeping Beauty–based somatic transposon mutagenesis in the mouse. Curr. Protoc. Mouse Biol. 2011:347–368. doi: 10.1002/9780470942390.mo110087. [DOI] [PubMed] [Google Scholar]
- 85.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.