Abstract
Dexras1 is a small GTPase and plays a central role in neuronal iron trafficking. We have shown that stimulation of glutamate receptors activates neuronal nitric oxide synthase, leading to S-nitrosylation of Dexras1 and a physiological increase in iron uptake. Here we report that Dexras1 is phosphorylated by PKA on serine 253, leading to a suppression of iron influx. These effects were directly associated with the levels of S-nitrosylated Dexras1, whereby PKA activation reduced Dexras1 S-nitrosylation in a dose dependent manner. Moreover, we found that adiponectin modulates Dexras1 via PKA. Hence these findings suggest the involvement of the PKA pathway in modulating glutamate-mediated ROS in neurons, and hint to a functional crosstalk between S-nitrosylation and phosphorylation.
Introduction
Living cells are constantly exposed to numerous stimuli from their environment and hence need to adapt to rapidly changing surroundings. Among various modes to responds to ever-changing environment, post-translational modification (PTM) is one of the most dynamic and rapid processes compared to transcriptional or translational responses. S-nitrosylation is one of such modifications which occurs on the cysteine of protein by attachment of a nitric oxide (NO) [1,2]. S-nitrosylation is a result of a non-enzymatically catalyzed chemical reaction [3]. Nevertheless, there appears to be some specificity in this reaction. More than a hundred proteins have been identified as targets of S-nitrosylation which is associated with modification of their functions or structures [1–5]. But there are many other modifications and hence PTM becomes more complex than ever [6,7]. It is important to understand how each modification communicates and modulates enzyme function.
Dexras1 (RASD1) belongs to the Ras family of small G proteins that is selectively induced by dexamethasone [8,9]. Dexras1 has a high homology with the Ras subfamily of proteins and has all of the conserved domains of regular GTPases. Unlike conventional GTPases, Dexras1 contains a 7kDA C-terminal tail. Dexras1 is also known as an activator of G-protein signaling (AGS1) or RASD1 [10]. Glutamate via NMDA receptors triggers cellular calcium entry with calcium-calmodulin activating nNOS [11–13], whose binding to C-terminal associated protein of nNOS (CAPON) provides a means for NO delivery to Dexras 1, leading to S-nitrosylation of Dexras1 on cysteine-11 [8,14]. We have shown that an activated Dexras1 further interacts with an iron import channel, divalent metal transporter 1 (DMT1) via a scaffolding protein called Acyl-CoA Binding Protein 3 (ACBD3) modulating neuronal iron trafficking. This pathway further plays a critical role in glutamate excitotoxicity as a deletion of Dexras1 dramatically reduced NMDA toxicity in vitro and in vivo [15,16]. Moreover, Dexras1 is shown to regulate circadian rhythm, adenylyl cyclase and G-protein-linked neurotransmitter [17–20].
ACBD3, a scaffolding protein between Dexras1 and DMT1, is reported to interact with protein kinase A (PKA) regulatory subunit [21–23], hence we wondered whether PKA pathway contributes to Dexras1-mediated neuronal iron trafficking. Here we report that Dexras1 is indeed phosphorylated by PKA at Serine 253 residue. Surprisingly, we found that PKA-mediated phosphorylation of Dexras1 reduces iron influx unlike its homolog, Dexras2 (also known as Rhes or RASD2) in which PKA-mediated phosphorylation upregulates iron influx [24]. We have further examined the crosstalk between PKA-mediated phosphorylation and S-nitrosylation as S-nitrosylation of Dexras1 is required to activate its GTPase activity and increase iron influx into the cells. We found that phosphorylation of Dexras1 blocks NO-mediated modulation of iron trafficking. Hence these findings suggest that there is a functional crosstalk between two different PTMs and may explain a potential mechanism by which PKA pathway reduces iron influx and ROS generation leading to a neuronal protection against NMDA excitotoxicity.
Experimental Procedures
Material and Methods
2.1. Cells and generation of mutant constructs
HEK 293T cells were maintained in DMEM with 10% FBS, 2 mM L-glutamine and 100 U/ml penicillin-streptomycin at 37°C with 5% CO2 atmosphere in a humidified incubator. Wild type Rhes was cloned into pCMV-Myc (Clonotech, Mountain View, CA) and subsequently S293A and S293E mutants were created with QuickChange (Stratagene, Ipswich, MA) method according to manufacturer’s instruction.
2.2. Iron uptake assay
Non-transferrin-bound iron (NTBI) uptake assays were performed as previously described [15,25–27]. In brief, HEK293T cells were transfected with Dexras1-Myc or mutants using Polyfect reagent (Qiagen, Valencia, CA). After 48 hr, the cells were washed with phosphate-buffered saline (PBS) then resuspended into iron uptake buffer (25 mM Tris, 25 mM MES, 140 mM NaCl, 5.4 mM KCl, 5 mM glucose, 1.8 mM CaCl2 [pH 5.5]) and transferred to glass test tubes. Ascorbic acid was added to 1 mM FeSO4 at a 44:1 ratio. 55FeCl3 (PerkinElmer Life Science, Waltham, MA) was added to the iron/ascorbic acid mixture, which was then added to the cells in iron uptake buffer to a final concentration of 20 μM. Cells were incubated at 37°C with shaking for 15 min. The cells were washed twice with cold PBS plus 0.5 mM EDTA and harvested. An aliquot of resuspended cells was taken for protein assay using the Bio-Rad Protein Assay Reagent; the protein concentrations of individual samples were used to quantitate 55Fe incorporation (cpm/μg protein). Samples were normalized to control. Statistical comparisons of iron uptake were performed by student’s t-test. All NBTI uptake experiments were repeated at least three times, each sample in triplicate.
2.3 GST Pull-down assay
GST or GST-tagged ACBD3 constructs were cotransfected with Dexras1-Myc constructs into HEK293T cells using PolyFect (Qiagen, Valencia, CA), with a transfection efficiency of greater than 90%. Cells were lysed 48 hr after transfection in buffer A (100 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton X-100, 15% glycerol, 1 mM PMSF, 25 mg/ml antipain, 50 mg/ml leupeptin, 50 mg/ml aprotinin, 25 mg/ml chymostatin, and 25 mg/ml pepstatin). Lysates were precleared with pansorbin cells (Calbiochem, Billerica, MA), then 1 mg of total protein was incubated with Glutathione-Sepharose beads overnight at 4 C. Beads were washed with wash buffer (50 mM Tris [pH 7.4], 500 mM NaCl, 10 mM b-glycerophosphate) twice, then once with buffer A. Beads were quenched in sample buffer (100 mM Tris [pH 6.8], 10% glycerol, 250 mM b-mercaptoethanol, 2% sodium dodecyl sulfate, and bromophenol blue). Total protein (50 mg) was loaded as input. Rhes-Myc binding was examined using an anti-myc antibody (EMD Millipore, Billerica, MA) followed by incubation with anti-mouse secondary conjugated to horseradish peroxidase (HRP) (Jackson Immunoresearch Laboratories, West Grove, PA); blots were then stripped and probed with an anti-GST antibody conjugated to HRP to detect ACBD3. Pierce Chemiluminescence (Life Technologies, Grand Island, NY) was used to detect bands on the Western blot.
2.4. in vitro phosphorylation
Immunoprecipitation and in vitro kinase assay were performed as previously described [24]. Cells transfected with Dexras1-Myc or Myc were lysed in buffer A, and then centrifuged at 12,000 ×g for 10 min at 4 °C. After preclearing with 125 μl of Protein A beads prepared as a 20% (v/v) suspension for 1 h at 4 °C, supernatants were incubated with anti-Myc antibody and the immunocomplexes were precipitated by addition of Protein A bead suspensions. The immunoprecipitates were collected by centrifugation and washed twice with buffer A and twice with PBS. The kinase assay was performed by incubating cell lysates in phosphatase buffer containing 20 mM MgCl2 with or without λ phosphatase (λ PPase) for 3 hours at 30°C. After thoroughly washing with PBS three times, protein bound beads were incubated with kinase buffer containing 1 mM MgATP, λ PPase inhibitors, and trace mount of [γ-32P] ATP for 30 min at 30°C. When indicated, protein kinases, such as PKA, protein kinase C (PKC), and casein kinase 2 (CK2) were added to the reaction mixture. The kinase reaction was stopped by adding 4 × SDS loading buffer and the samples were subjected to SDS-PAGE. Protein transferred blot was exposed to X-ray film for autoradiography. The blot was incubated with blocking buffer of 10% skim milk then proved with anti-Myc antibody for detection of input signal. To block PKA activities in cells, 1 μM H89 was added to the kinase buffer.
S-nitrosylation biotin switch assay
The assay will be performed as previously described [28]. In brief, cells will be lysed and reduced cysteines will be blocked with 4 mM methyl methanethionsulfonate (MMTS). Subsequently, S-nitrosylated cysteines will be reduced with 1 mM ascorbate and biotinylated with 1 mM Biotin-HPDP (Life Technologies, Grand Island, NY). The biotinylated proteins will be pulled down with streptavidin agarose and analyzed by Western blotting.
Data analysis
All quantitative data are presented as mean±S.E.M., if they were derived from at least three experiments. For comparison of multiple groups, the data were analysed by one-way ANOVA followed by a Tukey post hoc test. Student's t test was employed for directly testing the difference between two sets of independent samples. If the P-value less than 0.05, the difference was defined as significant. All statistical analyses were performed using GraphPad Prism (GraphPad Software).
Results
PKA phosphorylates Dexras1
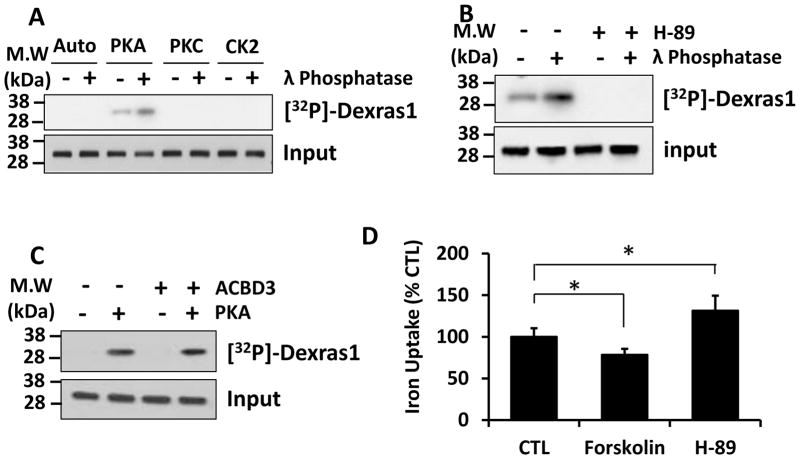
We previously reported that Dexras1 modulates neuronal iron trafficking via an iron import channel DMT1 [15]. Importantly, a scaffolding protein, ACBD3 between Dexras1 and DMT1 is required for Dexras1 to regulate iron uptake [15]. It has been shown that ACBD3 also interacts with various other proteins [21,22,29], one of which is protein kinase A (PKA) regulatory subunit and yet PKA does not directly phosphorylate ACBD3 (data not shown). Hence we wondered whether Dexras1 can be phosphorylated by especially PKA. We performed in vitro phosphorylation assay utilizing recombinant Dexras1 isolated from HEK293T overexpressed with Dexras1. Purified Dexras1 was incubated with various kinases and was only phosphorylated by PKA. Also we observed that pretreatment of Dexras1 with γ-phosphatase further increased phosphorylation of Dexras1 by PKA suggesting that Dexras1 was basally phosphorylated by PKA (Fig 1A). Moreover, treatment of cells with H-89, a well-known PKA inhibitor, completely abrogated PKA-mediated phosphorylation of Dexras1 indicating that PKA mediates the phosphorylation of Dexras1 (Fig 1B). Since ACBD3 binds to a regulatory subunit of PKA, we investigated whether ACBD3 had any effect on PKA-mediated phosphorylation of Dexras1 by performing in vitro phosphorylation assay with or without purified recombinant ACBD3. We found ACBD3 does not affect PKA-mediated phosphorylation of Dexras1 indicating that ACBD3 is not involved in the phosphorylation of Dexras1 by PKA (Fig 1C). Previously we reported that Dexras1 regulates iron influx [15,16] and hence we wondered whether PKA-mediated phosphorylation modulates Dexras1-mediated iron trafficking. HEK293T cells were overexpressed with wild type (WT) Dexras1 and treated with either forskolin (PKA activator, 10 uM) or H-89 (PKA inhibitor, 10 uM) for 1 hour. We found that PKA activator reduced iron influx while an inhibitor of PKA increased it suggesting that PKA regulates iron influx via Dexras1 (Fig 1D).
Figure 1. PKA phosphorylates and inactivates Dexras1.
Dexras1-myc was transfected into HEK293T and was immunoprecipitated by anti-myc antibody. (A). In vitro phosphorylation assay was performed in the presence of various kinases with or without pretreatment of λ phosphatase. (B) PKA-mediated phosphorylation assay was performed in the presence or absence of PKA inhibitor, H-89 (10 uM). (C) In vitro phosphorylation assay was performed with or without purified recombinant ACBD3. (D) HEK293T cells transfected with Dexras1-myc was treated with either forskolin (10 uM) or H-89 (10 uM) for 1 hour. Then iron uptake assay was performed (* P<0.05). Phosphorylation assays were performed three times. Iron uptake experiments were repeated five times, each sample in at least triplicate.
PKA phosphorylates only serine-253 amino acid on Dexras1
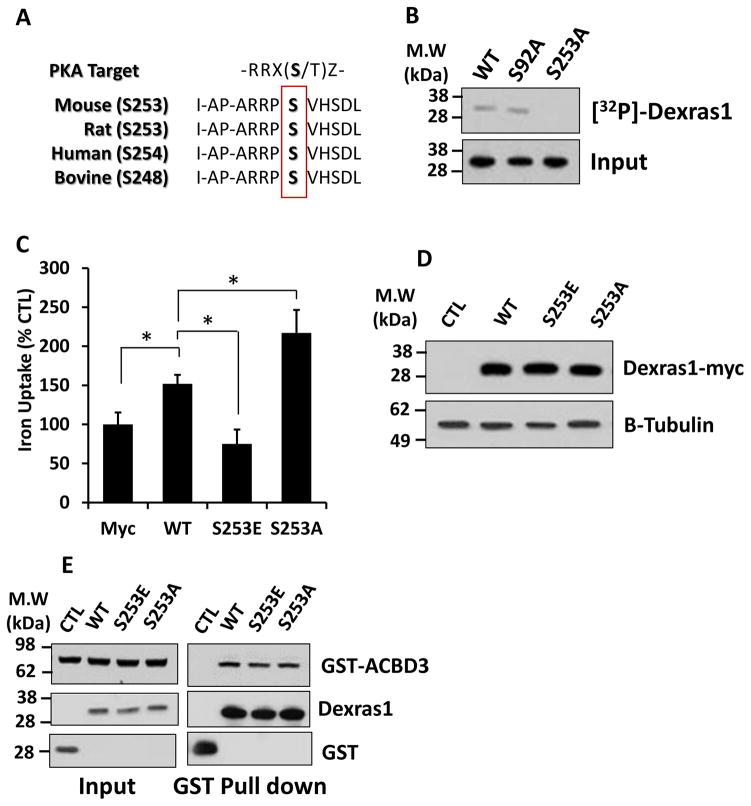
We have performed a primary sequence analysis by ScanSite (http://scansite.mit.edu) and revealed that Dexras1 indeed contains putative PKA phosphorylation sites at serine 92 and serine 253. The motif is highly conserved in Dexras1 protein from all the species examined, suggesting these two may be potential targets for PKA-mediated phosphorylation of Dexras1 (Fig 2A). To directly examine which amino acid is phosphorylated by PKA, we mutated each serine (S92, and S253) into alanine that cannot be phosphorylated by a kinase. From the in vitro phosphorylation assay utilizing HEK293T cells overexpressed with phosphodead mutants of either S92A or S253A we generated, we found that PKA-mediated phosphorylation was completely eliminated from S253A and not from S92A, demonstrating that serine-253 is the single amino acid phosphorylated by PKA (Fig. 2B). To further understand whether PKA-mediated phosphorylation of Dexras1 has any functional consequence, we have generated phosphomimetic Dexras1 mutant by changing Serine 253 to glutamic acid (S253E) and examined its effect on iron uptake. HEK293T cells were overexpressed with WT, phosphomimetic (S253E) or phosphodead (S253A) and iron uptake was measured. We found that a phosphodead mutant, S253A significantly increased iron uptake while a phosphomimetic mutant, S253E decreased it (Fig 2C). We have confirmed that expression levels of each mutant and WT Dexras1 were comparable (Fig 2D). We further investigated whether any of mutants show a different affinity to ACBD3 as an interaction between ACBD3 and Dexras1 is necessary to up-regulate iron uptake. However, we did not find any difference on protein-protein interaction between ACBD3 and Dexras1 mutants (Fig 2E) suggesting that phosphorylation of Dexras1 may affect Dexras1 GTPase activity and modulate iron influx.
Figure 2. Serine 253 in Dexras1 is phosphorylated by PKA.
(A) Amino acid sequence alignment of Dexras1. (B) Either WT Dexras1-myc or phosphodead mutants (S92A or S253A) were transfected into HEK293T cells and were immunoprecipitated by anti-myc antibody. In vitro phosphorylation assay was performed and the results were visualized by autoradiography (top). Western blot image shows the loading of Myc-tagged Dexras1 protein in the reactions (bottom). (C) HEK293T cells were transfected with WT, S253E or S253A mutant Dexras1 and were subjected to iron uptake assays. (D) HEK293T cells transfected with WT, S253E or S253A mutant Dexras1 were subjected to western blotting. (E) HEK293T cells were transfected with GST-tagged Dexras1 (WT, 253E or S253A) and ACBD3. GST-pull down assay was performed. Immunoassays were performed three times. Iron uptake experiments were repeated five times, each sample in at least triplicate.
PKA regulates S-nitrosylation status of Dexras1
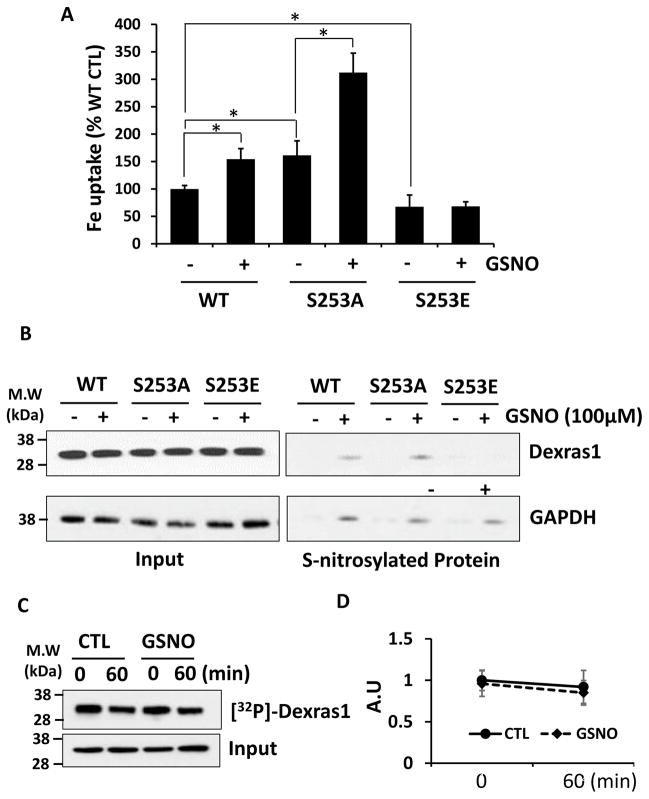
It has been shown that Dexras1 GTPase is activated by NO-mediated S-nitrosylation on cysteine 11 and hence NO basically functions as guanidine exchange factor for Dexras1 [8,15]. Also, we demonstrated that this modification indeed potentiates iron trafficking in the neurons [15]. Hence we wondered whether phosphorylation status of Dexras1 affects NO-mediated Dexras1 function. First, HEK293T cells were overexpressed with WT, S253A and S253E Dexras1 and treated with 100 uM GSNO for 1 hour, followed by iron uptake assay. We found that cells overexpressed with S253A showed an increase of iron uptake which was equivalent to the result from cells overexpressed with WT Dexras1 plus NO treatment (Fig 3A). Moreover, NO treatment further augmented iron uptake in cells overexpressed with S253A. On the other hand, cells overexpressed with S253E were resistant to NO treatment and did not display any change in iron uptake after NO treatment (Fig 3A).
Figure 3. Phosphorylation in Dexras1 blocks S-nitrosylation of Dexras1.
HEK293T cells transfected with WT, S253E or S253A mutant Dexras1 and were treated with GSNO (100 uM) for 1 hour. (A) Then iron uptake assay was performed. (B) Biotin switch assay was performed. (C) HEK293T cells were transfected with Dexras1-myc and labeled with inorganic phosphate. Then cells were treated with GSNO (100 uM) for 1 hour. Dexras1-myc was immunoprecipitated by anti-myc antibody and visualized by autoradipgraphy. Input was detected by immunoblotting. (D) Quantification of phosphorylated Dexras1 after GSNO treatment. Iron uptake experiments were repeated five times, each sample in at least triplicate. Phosphorylation and S-nitrosylation assays were performed three times.
Since S-nitrosylation of Dexras1 is necessary to activate GTPase activity of Dexras1 leading to an increase in iron influx, and yet S253E phosphomimetic mutant is resistant to NO treatment, we have interrogated whether phosphorylation status affects S-nitrosylation of Dexras1. HEK293T cells were transfected with WT, S253A and S253E Dexras1 and treated with 100 uM GSNO for 1 hour, which was identical condition as described above experiment. Then biotin-switch assay was performed to examine the levels of S-nitrosylated Dexras1. We observed that S-nitrosylation levels of a phosphodead mutant S253A was twice higher than that of WT while S253E mutant was not S-nitrosylated by NO treatment even though an internal control protein, GAPDH was all S-nitrosylated throughout the conditions (Fig 3B). These data suggest that Serine 253 residue needs to be a free amino acid without phosphorylation for S-nitrosylation to occur in Dexras1.
We have shown above that Dexras1 is basally phosphorylated and hence we wondered whether NO-treatment would promote dephosphorylation of Dexras1 to activate its GTPase activity. To test this, HEK293T cells were overexpressed with WT Dexras1 and first pre-labeled with 32P-orthophosphate for 30 min. Then these cells were washed with cold PBS and were treated with 100 uM GSNO for additional 1 hour, which is sufficient to S-nitrosylate and activate Dexras1., First, we noticed that levels of phosphorylation was slightly decreased even in the absence of NO treatment but this was not statistically significant. More importantly, we found that NO-treatment did not facilitate dephosphorylation of Dexras1 suggesting that S-nitrosylation-mediated activation of Dexras1 does not promote its dephosphorylation and may preferentially occur on unphosphorylated Dexras1 (Fig 3C and D).
Phosphorylation prevents S-nitrosylation of Dexras1
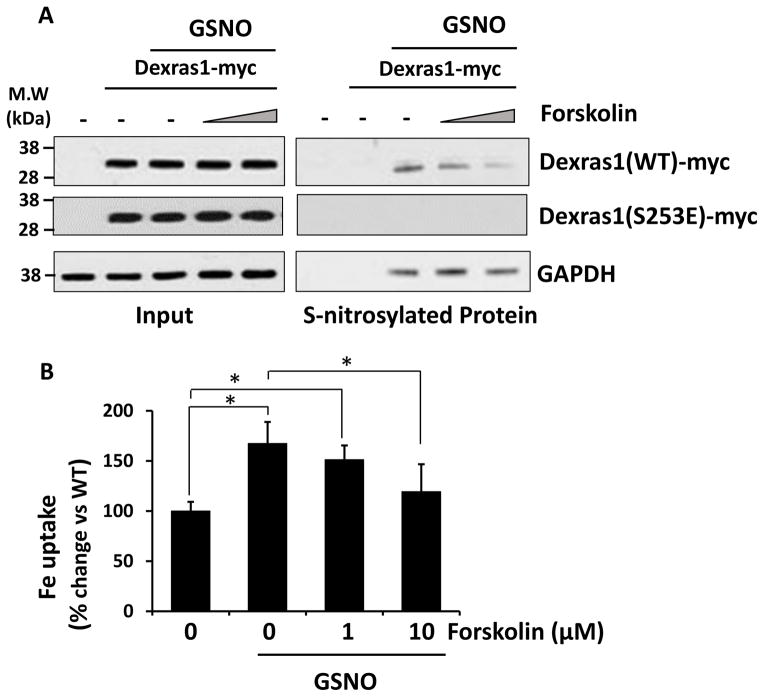
To further understand a crosstalk between phosphorylation and S-nitrosylation of Dexras1, we investigated whether PKA activation affects status of S-nitrosylated Dexras1 and iron uptake. HEK293T cells were transfected with either WT or S253E Dexras1 and pretreated with different doses of forskolin for 1 hour and followed by additional GSNO treatment (100 uM, 1 h). We found that PKA activator, forskolin facilitates de-nitrosylation of WT-Dexras1 in a dose-dependent manner while S253E mutant was neither S-nitrosylated or modulated by PKA treatment any more (Fig 4A). Under the same condition, we examined iron uptake into the cells and found that forskolin treatment reduces iron uptake as similar to a decrease in S-nitrosylated protein (Fig 4B) suggesting that PKA pathway actively prevent NO-mediated iron uptake pathway via Dexras1.
Figure 4. Dexras1 phosphorylation attenuates NO-mediated iron uptake via Dexras1.
HEK293T cells were transfected with Dexras1-myc and pre-treated with forskolin (1 and 10 uM) for 1 hour and then further treated with GSNO (100 uM) for 1 hour. (A) Cells were subjected to biotin switch assay or (B) iron uptake assay. Iron uptake experiments were repeated five times, each sample in at least triplicate. S-nitrosylation assay was performed three times.
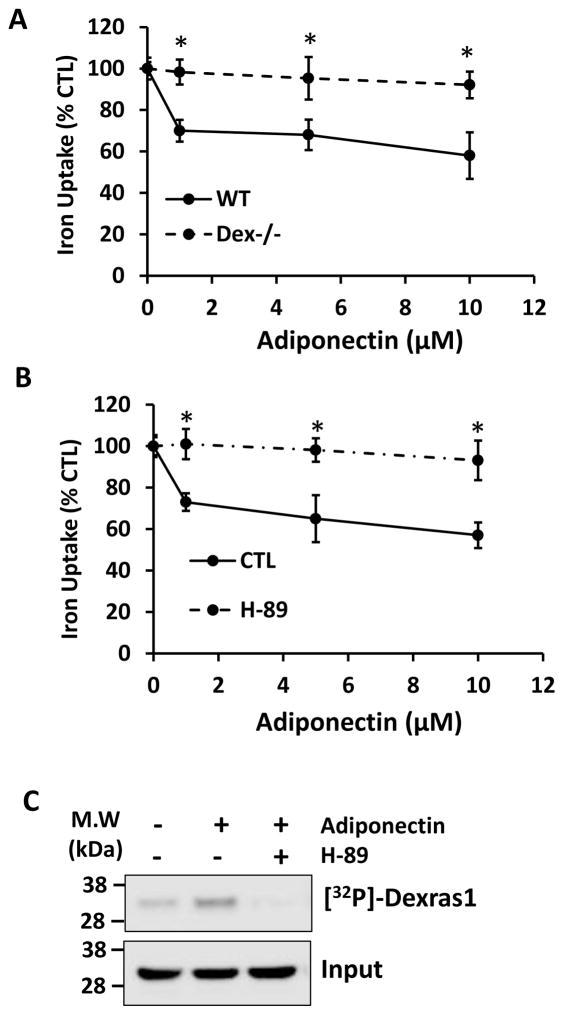
Adiponectin modulates neuronal iron influx via PKA/Dexras1 pathway
We have shown that Dexras1 plays a crucial role in oxidative stress-mediated neuronal cell death. Hence an inhibition of Dexras1 via PKA may contribute to the cell survival. Adiponectin is synthesized and secreted by adipocytes and circulates as various molecular forms [30,31]. Obesity decreases adiponectin levels and blunts adiponectin sensitivity, leading to insulin resistance, diabetes, steatosis and inflammation [31,32]. Many studies have shown that adiponectin is present in cerebrospinal fluid, and exerts potent actions on neurons in the hypothalamus and brainstem [33–37]. Adiponectin also has neuroprotective effects in stroke, seizure and other pathologies in mouse models [38–42]. Because adiponectin activates PKA pathway in peripheral tissues [43,44], we wondered whether adiponectin could modulate Dexras1-mediated iron trafficking in neurons via PKA. We treated primary hippocampal neuronal cultures with various doses of adiponectin and measured iron uptake [15,27]. We confirmed that adiponectin treatment attenuated iron influx and yet this effect was abolished in the cultures prepared from Dexras knockout mice, suggesting that adiponectin may modulate iron trafficking in neurons via Dexras1 (Fig. 5A). Dexras1 is a small GTPase requiring GTP binding for its activity and it has been shown that nitric oxide-mediated post-translational modification (S-nitrosylation) is the only way to activate Dexras1 GTPase activity [14]. Hence, we examined whether adiponectin affects the basal levels of NO production from nNOS and found that adiponectin treatment did not influence it (data not shown), suggesting that Dexras1 GTPase activity is suppressed by other mechanisms. Moreover, we found that adiponectin’s ability to inhibit iron uptake was blocked by H-89 treatment which inhibits PKA activity (Fig. 5B). We next performed inorganic phosphate metabolic labeling in primary hippocampal neurons and found that adiponectin treatment indeed increased the phosphorylation of Dexras1 while PKA inhibitor H-89 pretreatment blocked this modification suggesting that adiponectin modulates iron trafficking via PKA/Dexras1 pathway. (Fig. 5C). Our data show that adiponectin reduces iron influx via PKA/Dexra1 pathway in neurons.
Figure 5. Adiponectin regulates iron trafficking in neurons via PKA/Dexras1 dependent manner.
(A) Primary hippocampal neurons were treated with adiponectin for 8 hours and iron uptake assay was performed w/out H-89, PKA inhibitor. Data represent triplicate samples. (B) Primary hippocampal cultures were prepared from either WT or Dexras1 knockout mice and iron uptake assay was performed after treating cells with adiponectin for 4 hours. (C) Primary hippocampal neurons are treated with adiponectin 10 uM for 8 hours and metabolic labeling was performed. Dexras1 protein was immunoprecipitated and detected by autoradiography. Input levels were detected by immunoblotting. Phosphorylation assays were performed three times. Iron uptake experiment was repeated five times, each sample in at least triplicate.
Discussion
In the present study, we have identified that adiponectin, which is produced by adipose tissue phosphorylates Dexras1 via PKA and this modification reduces Dexras1-mediated iron uptake process, which is dependent on its GTPase activity. Importantly, phosphorylation of Dexras1 inhibits its S-nitrosylation, which is essential for Dexras1 activity and our study showed that a functional crosstalk between two different modifications further modifies an enzyme activity.
Dexras1 belongs to Ras superfamily and Rhes (Dexras2) is the only RAS homology that closely resembles Dexras1 with −62% amino acid homology. Both of them contain an extra 7 kDa tail in C-terminal region and its roles are not fully understood. Whereas Dexars1 is stimulated by glucocorticoid, Rhes is selectively induced by thyroid hormone. Most strikingly, Rhes is selectively expressed in striatum and involved in pathogenesis of Huntingdon’s disease. Moreover, it mediates mTOR signaling and L-DOPA-induced dyskinesia [45] while Dexras1 does not regulate mTOR pathway (unpublished results, Chen, Y. and Kim, SF.). Previously we have reported that Rhes can indeed bind to ACBD3 and modulate iron trafficking in the neuron and iron uptake is up-regulated by PKA [24]. Interestingly, our new study examining the relationship between PKA and Dexras1 demonstrated that indeed Dexras1 is basally phosphorylated and yet its phosphorylation by PKA led to a decrease of iron influx unlike Rhes. It has been shown before that S-nitrosylation, NO-mediated PTM, acts as a guanidine exchange factor (GEF) for Dexras1 and activates its GTPase activity, leading to an increase of iron influx in neurons [8]. However, the phosphorylation status of Dexras1 seems to limit the degree of Dexras1 activation via S-nitrosylation. In contrast, we have demonstrated that Rhes is not S-nitrosylated via NO treatment even though putative S-nitrosylation target cysteine is conserved in both Dexras1 and Rhes. Moreover, NO-treatment did not affect iron uptake in HEK293T cells overexpressed with Rhes [24]. Hence even though both Rhes and Dexras1 participates in iron uptake and are regulated by PKA, their precise mechanisms are not likely identical. Therefore, the converse regulation of enzyme activity by phosphorylation and S-nitrosylation is very unique for Dexras1. However, it is not clear how a negatively charged S253 inhibits S-nitrosylation at C11 in Dexras1. Nevertheless, it is interesting to point out that the phosphorylation occurs in the unique tail region. It is less likely that phosphorylation of S253 is directly blocking the access of NO to C11. Instead, we speculate that phosphorylation at S253 may slightly change the structure of Dexras1 and possibly bury C11 site. In fact, C11 site is also conserved in Dexras2 (also known as Rhes) with high homology and yet we found Dexras2 is not S-nitrosylated by NO treatment.
Our previous studies demonstrated that Dexras1 plays a central role in either glutamate/NMDA or NO-mediated cell death pathway as a genetic deletion of Dexras1 drastically reduced cell death induced by either NMDA excitotoxicity or excess NO treatment suggesting that Dexras1 may be a viable target to treat glutamate toxicity [15,16]. Our current study demonstrated that Dexras1 activity, or Dexras1-mediated iron influx can be reduced by a PKA dependent manner. In fact many studies demonstrated that PKA pathway is involved in neuroprotection mechanism via various pathways. The transcription factor myocyte-enhancer factor 2 (MEF2) is a known to be required for neuronal survival and it has been shown that PKA-mediated phosphorylation of MEF2 is a key step in modulating its DNA binding activity and ability to promote neuronal survival [46]. Moreover, Dore’s group identified that neuroprotective effect of prostaglandin system also requires an activation of PKA pathway [47]. It appears that PKA-mediated survival pathway is not limited to neurons. Cell death in rat rod photoreceptor can be blocked by interleukin-4 in a PKA dependent manner [48]. Hence it is tempting to speculate that a stimulus or reagent which can induce PKA pathway may serve as a neuroprotective reagents by reducing iron-catalyzed oxidative stress via a suppression of Dexras1. In fact, it has been shown that PKA activation can reduce iron uptake pathway [49]. However it is important to mention that PKA pathway is not always associated with a cell survival pathway [50,51]. An excess lipid can lead to a neuronal cell death and an initiation of apoptotic pathway was triggered by PKA-dependent manner [52]. Hence it appears important to examine what triggered a cell death pathway and this may determine whether PKA pathway is involved with cytoprotective or cytotoxic pathways. Our study identified that activation of PKA can reduce iron uptake and consequently ROS generation induced by NMDA/NO pathway.
In this study, we have identified that adiponectin, a cytokine produced by adipose tissue, indeed down-regulates Dexras1 iron flux pathway via PKA. Adiponectin is one of the most abundant hormone in the plasma and its level is inversely related to body fat content. It is known to regulate various metabolic processes such as glucose homeostasis or fatty acid oxidation. However, recent studies showed that adiponectin may have some non-metabolic functions in particular in the brain. Adiponectin has neuroprotective effects in stroke, seizure and other pathologies in mouse models, which are strongly associated with oxidative stress-mediated damages [38–42]. Therefore, our findings further strengthen a notion that PKA/Dexras1 pathway may serve as a target to protect neurons against neurotoxicity. Also, other studies have reported that adiponectin promotes neurogenesis, decreases depression, and mediates the effects of exercise on hippocampal neurons [37]. It has been shown that nNOS/CAPON-ERK pathway is upregulated in anxiogenic condition and Dexras1 is mediating this pathway [53]. Our studies showing adiponectin as a negative regulator of Dexras1 pathway may provide a molecular link between energy balance and anxiety.
NO is a gaseous molecule which is produced by a group of enzymes called nitric oxide synthase and can diffuse through a membrane to some degree. Hence it can function as a signaling molecule that acts in many tissues to regulate a diverse range of physiological processes, which include vasodilation, neuronal function, inflammation and immune function [11,13,54]. One of the key cellular roles of NO is reversible protein modification, such as S-nitrosylation, a covalent addition of an NO group to a cysteine thiol/sulfhydryl (RSH or, more properly, thiolate anion, RS-), resulting in formation of an S-nitrosothiol derivative (RSNO) [1,55]. In analogy to phosphorylation by kinases, S-nitrosylation by NOSs influences protein activity, protein–protein interactions, and protein location, thus serving as the prototypical redox-based signal [55]. S-Nitrosylation is readily reversible with high spatial and temporal specificity. In the current study, we have revealed that two analogous PTMs influence each other and provide an extra layer of complexity on how neuronal iron status or iron-catalyzed redox status is modulated.
In conclusion, we have identified that Dexras1-mediated iron trafficking is not only regulated by NO but also PKA pathway which is associated with various neurotransmitters or cytokines [44,56]. In particular we have shown that Dexras1 is negatively modulated by adiponectin, which plays crucial roles on energy balance, neuroprotection, and neurogenesis. Hence our studies may provide potential mechanism by which PKA pathway may protect neurons from NMDA excitotoxicity. Moreover, there are numerous studies describing that iron status in the brain influence neuro-development while there is very few studies describing the role of neuronal iron or its trafficking on any specific behavior. Our studies will provide a promising groundwork which can be further extended to investigate how Dexras1/iron pathway modulate neural function and consequently afford a novel therapeutic strategies to treat patients with neuropsychiatric diseases.
Highlights.
PKA phosphorylates Dexras1 on serine 253.
PKA phosphorylation of Dexras1 prevents NO-mediated activation of Dexras1
Adiponectin phosphorylates Dexras1 via PKA.
Acknowledgments
We are grateful for Catherine Steenstra for laboratory support.
Funding
This work was supported by HD026979, MH079614 and DK084336 (SFK).
Abbreviation
- NO
Nitric oxide
- PKA
Protein Kinase A
- PTM
Post Translational Modification
- ACBD3
Acyl CoA Binding Domain 3
- GST
Glutathione-S-transferase
- NTBI
Non-transferrin bound iron
Footnotes
Contribution
Yong Chen and Sangwon Kim were involved in the concept, design and interpretation of Data. Lauren Mathias performed iron uptake assays and Juliana M. Falero Perez did biotin switch assays. The paper was written by Yong Chen and Sangwon Kim with contributions from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hess DT, Stamler JS. Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem. 2012;287:4411–8. doi: 10.1074/jbc.R111.285742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seth D, Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol. 2011;15:129–36. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura T, Lipton SA. Emerging role of protein-protein transnitrosylation in cell signaling pathways. Antioxid Redox Signal. 2013;18:239–49. doi: 10.1089/ars.2012.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura T, Tu S, Akhtar MW, Sunico CR, Okamoto S, Lipton SA. Aberrant protein s-nitrosylation in neurodegenerative diseases. Neuron. 2013;78:596–614. doi: 10.1016/j.neuron.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodsmith J, Stelzl U. Studying post-translational modifications with protein interaction networks. Curr Opin Struct Biol. 2014;24:34–44. doi: 10.1016/j.sbi.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Venne AS, Kollipara L, Zahedi RP. The next level of complexity: crosstalk of posttranslational modifications. Proteomics. 2014;14:513–24. doi: 10.1002/pmic.201300344. [DOI] [PubMed] [Google Scholar]
- 8.Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 9.Kemppainen RJ, Behrend EN. Dexamethasone rapidly induces a novel ras superfamily member-related gene in AtT-20 cells. J Biol Chem. 1998;273:3129–3131. doi: 10.1074/jbc.273.6.3129. [DOI] [PubMed] [Google Scholar]
- 10.Blumer JB, Cismowski MJ, Sato M, Lanier SM. AGS proteins: receptor-independent activators of G-protein signaling. Trends Pharmacol Sci. 2005;26:470–6. doi: 10.1016/j.tips.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- 12.Bredt DS, Snyder SH. Nitric oxide mediates glutamate-linked enhancement of cGMP levels in the cerebellum. Proc Natl Acad Sci U S A. 1989;86:9030–3. doi: 10.1073/pnas.86.22.9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bredt DS, Snyder SH. Nitric oxide: a physiologic messenger molecule. Annu Rev Biochem. 1994;63:175–95. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- 14.Jaffrey SR, Snowman AM, Eliasson MJ, Cohen NA, Snyder SH. CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron. 1998;20:115–124. doi: 10.1016/s0896-6273(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 15.Cheah JH, Kim SF, Hester LD, Clancy KW, Patterson SE, III, Papadopoulos V, Snyder SH. NMDA receptor-nitric oxide transmission mediates neuronal iron homeostasis via the GTPase Dexras1. Neuron. 2006;51:431–440. doi: 10.1016/j.neuron.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, et al. Dexras1, a Small GTPase, Is Required for Glutamate-NMDA Neurotoxicity. J Neurosci. 2013;33:3582–3587. doi: 10.1523/JNEUROSCI.1497-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng HY, et al. Dexras1 potentiates photic and suppresses nonphotic responses of the circadian clock. Neuron. 2004;43:715–728. doi: 10.1016/j.neuron.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 18.Graham TE, Prossnitz ER, Dorin RI. Dexras1/AGS-1 inhibits signal transduction from the Gi-coupled formyl peptide receptor to Erk-1/2 MAP kinases. J Biol Chem. 2002;277:10876–10882. doi: 10.1074/jbc.M110397200. [DOI] [PubMed] [Google Scholar]
- 19.Harrison LM, He Y. Rhes and AGS1/Dexras1 affect signaling by dopamine D1 receptors through adenylyl cyclase. J Neurosci Res. 2011;89:874–882. doi: 10.1002/jnr.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sang A, Cheng Y, Lu H, Chen D, Gao R, Shen A. Light-induced retinal ganglion cell damage in vivo involves Dexras1. Mol Vis. 2011;17:134–143. [PMC free article] [PubMed] [Google Scholar]
- 21.Fan J, Liu J, Culty M, Papadopoulos V. Acyl-coenzyme A binding domain containing 3 (ACBD3; PAP7; GCP60): an emerging signaling molecule. Prog Lipid Res. 2010;49:218–234. doi: 10.1016/j.plipres.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Degenhardt B, Tobin D, Yao ZX, Tasken K, Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor- and PKA (RIalpha)-associated protein. Mol Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Li H, Papadopoulos V. PAP7, a PBR/PKA-RIalpha-associated protein: a new element in the relay of the hormonal induction of steroidogenesis. J Steroid Biochem Mol Biol. 2003;85:275–283. doi: 10.1016/s0960-0760(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 24.Choi BR, Bang S, Chen Y, Cheah JH, Kim SF. PKA modulates iron trafficking in the striatum via small GTPase, Rhes. Neuroscience. 2013;253:214–20. doi: 10.1016/j.neuroscience.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S, Ponka P. Control of transferrin receptor expression via nitric oxide-mediated modulation of iron-regulatory protein 2. J Biol Chem. 1999;274:33035–42. doi: 10.1074/jbc.274.46.33035. [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Ponka P. Effects of interferon-gamma and lipopolysaccharide on macrophage iron metabolism are mediated by nitric oxide-induced degradation of iron regulatory protein 2. J Biol Chem. 2000;275:6220–6. doi: 10.1074/jbc.275.9.6220. [DOI] [PubMed] [Google Scholar]
- 27.Kim S, Ponka P. Nitrogen monoxide-mediated control of ferritin synthesis: implications for macrophage iron homeostasis. Proc Natl Acad Sci USA. 2002;99:12214–12219. doi: 10.1073/pnas.192316099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 29.Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res. 1999;59:831–842. [PubMed] [Google Scholar]
- 30.Kusminski CM, McTernan PG, Schraw T, Kos K, O'Hare JP, Ahima R, Kumar S, Scherer PE. Adiponectin complexes in human cerebrospinal fluid: distinct complex distribution from serum. Diabetologia. 2007;50:634–42. doi: 10.1007/s00125-006-0577-9. [DOI] [PubMed] [Google Scholar]
- 31.Turer AT, Scherer PE. Adiponectin: mechanistic insights and clinical implications. Diabetologia. 2012;55:2319–26. doi: 10.1007/s00125-012-2598-x. [DOI] [PubMed] [Google Scholar]
- 32.Yamauchi T, et al. Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat Med. 2007;13:332–9. doi: 10.1038/nm1557. [DOI] [PubMed] [Google Scholar]
- 33.Fry M, Smith PM, Hoyda TD, Duncan M, Ahima RS, Sharkey KA, Ferguson AV. Area postrema neurons are modulated by the adipocyte hormone adiponectin. J Neurosci. 2006;26:9695–702. doi: 10.1523/JNEUROSCI.2014-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoyda TD, Fry M, Ahima RS, Ferguson AV. Adiponectin selectively inhibits oxytocin neurons of the paraventricular nucleus of the hypothalamus. J Physiol. 2007;585:805–16. doi: 10.1113/jphysiol.2007.144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubota N, et al. Adiponectin stimulates AMP-activated protein kinase in the hypothalamus and increases food intake. Cell Metab. 2007;6:55–68. doi: 10.1016/j.cmet.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Qi Y, Takahashi N, Hileman SM, Patel HR, Berg AH, Pajvani UB, Scherer PE, Ahima RS. Adiponectin acts in the brain to decrease body weight. Nat Med. 2004;10:524–9. doi: 10.1038/nm1029. [DOI] [PubMed] [Google Scholar]
- 37.Yau SY, Li A, Hoo RL, Ching YP, Christie BR, Lee TM, Xu A, So KF. Physical exercise-induced hippocampal neurogenesis and antidepressant effects are mediated by the adipocyte hormone adiponectin. Proc Natl Acad Sci U S A. 2014;111:15810–5. doi: 10.1073/pnas.1415219111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Geng J, Liu J. Adiponectin offers protection against L166P mutant DJ-1-induced neuronal cytotoxicity mediated by APPL1-dependent AMPK activation. Int J Neurosci. 2014;124:350–61. doi: 10.3109/00207454.2013.846340. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura M, et al. Adiponectin prevents cerebral ischemic injury through endothelial nitric oxide synthase dependent mechanisms. Circulation. 2008;117:216–23. doi: 10.1161/CIRCULATIONAHA.107.725044. [DOI] [PubMed] [Google Scholar]
- 40.Sekiyama K, et al. Disease-Modifying Effect of Adiponectin in Model of alpha-Synucleinopathies. Ann Clin Transl Neurol. 2014;1:479–489. doi: 10.1002/acn3.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song W, et al. Globular adiponectin elicits neuroprotection by inhibiting NADPH oxidase-mediated oxidative damage in ischemic stroke. Neuroscience. 2013;248:136–44. doi: 10.1016/j.neuroscience.2013.05.063. [DOI] [PubMed] [Google Scholar]
- 42.Thundyil J, et al. Evidence that adiponectin receptor 1 activation exacerbates ischemic neuronal death. Exp Transl Stroke Med. 2010;2:15. doi: 10.1186/2040-7378-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medina EA, Oberheu K, Polusani SR, Ortega V, Velagaleti GV, Oyajobi BO. PKA/AMPK signaling in relation to adiponectin's antiproliferative effect on multiple myeloma cells. Leukemia. 2014;28:2080–9. doi: 10.1038/leu.2014.112. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Adiponectin inhibits oxidative/nitrative stress during myocardial ischemia and reperfusion via PKA signaling. Am J Physiol Endocrinol Metab. 2013;305:E1436–43. doi: 10.1152/ajpendo.00445.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramaniam S, et al. Rhes, a striatal-enriched small G protein, mediates mTOR signaling and L-DOPA-induced dyskinesia. Nat Neurosci. 2012;15:191–3. doi: 10.1038/nn.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Tang X, Li M, Marshall J, Mao Z. Regulation of neuroprotective activity of myocyte-enhancer factor 2 by cAMP-protein kinase A signaling pathway in neuronal survival. J Biol Chem. 2005;280:16705–13. doi: 10.1074/jbc.M501819200. [DOI] [PubMed] [Google Scholar]
- 47.Echeverria V, Clerman A, Dore S. Stimulation of PGE receptors EP2 and EP4 protects cultured neurons against oxidative stress and cell death following beta-amyloid exposure. Eur J Neurosci. 2005;22:2199–206. doi: 10.1111/j.1460-9568.2005.04427.x. [DOI] [PubMed] [Google Scholar]
- 48.Adao-Novaes J, de Guterres CC, da Silva AG, Campello-Costa P, Linden R, Sholl-Franco A. Interleukin-4 blocks thapsigargin-induced cell death in rat rod photoreceptors: involvement of cAMP/PKA pathway. J Neurosci Res. 2009;87:2167–74. doi: 10.1002/jnr.22026. [DOI] [PubMed] [Google Scholar]
- 49.Du F, Qian ZM, Gong Q, Zhu ZJ, Lu L, Ke Y. The iron regulatory hormone hepcidin inhibits expression of iron release as well as iron uptake proteins in J774 cells. J Nutr Biochem. 2012;23:1694–700. doi: 10.1016/j.jnutbio.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Edwards HV, Scott JD, Baillie GS. PKA phosphorylation of the small heat-shock protein Hsp20 enhances its cardioprotective effects. Biochem Soc Trans. 2012;40:210–4. doi: 10.1042/BST20110673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Insel PA, Zhang L, Murray F, Yokouchi H, Zambon AC. Cyclic AMP is both a pro-apoptotic and anti-apoptotic second messenger. Acta Physiol (Oxf) 2012;204:277–87. doi: 10.1111/j.1748-1716.2011.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou MH, Yang G, Jiao S, Hu CL, Mei YA. Cholesterol enhances neuron susceptibility to apoptotic stimuli via cAMP/PKA/CREB-dependent up-regulation of Kv2.1. J Neurochem. 2012;120:502–14. doi: 10.1111/j.1471-4159.2011.07593.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhu LJ, et al. CAPON-nNOS coupling can serve as a target for developing new anxiolytics. Nat Med. 2014;20:1050–4. doi: 10.1038/nm.3644. [DOI] [PubMed] [Google Scholar]
- 54.Nathan C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992;6:3051–3064. [PubMed] [Google Scholar]
- 55.Stamler JS, Lamas S, Fang FC. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–83. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 56.Svenningsson P, Nishi A, Fisone G, Girault JA, Nairn AC, Greengard P. DARPP-32: an integrator of neurotransmission. Annu Rev Pharmacol Toxicol. 2004;44:269–96. doi: 10.1146/annurev.pharmtox.44.101802.121415. [DOI] [PubMed] [Google Scholar]