Abstract
We introduce efficient Markov chain Monte Carlo methods for inference and model determination in multivariate and matrix-variate Gaussian graphical models. Our framework is based on the G-Wishart prior for the precision matrix associated with graphs that can be decomposable or non-decomposable. We extend our sampling algorithms to a novel class of conditionally autoregressive models for sparse estimation in multivariate lattice data, with a special emphasis on the analysis of spatial data. These models embed a great deal of flexibility in estimating both the correlation structure across outcomes and the spatial correlation structure, thereby allowing for adaptive smoothing and spatial autocorrelation parameters. Our methods are illustrated using a simulated example and a real-world application which concerns cancer mortality surveillance. Supplementary materials with computer code and the datasets needed to replicate our numerical results together with additional tables of results are available online.
Keywords: CAR model, G-Wishart distribution, Markov chain Monte Carlo (MCMC) simulation, Spatial statistics
1. INTRODUCTION
Graphical models (Lauritzen 1996), which encode the conditional independence among variables using a graph, have become a popular tool for sparse estimation in both the statistics and machine learning literatures (Dobra et al. 2004; Meinshausen and Bühlmann 2006; Yuan and Lin 2007; Banerjee, El Ghaoui, and D’Aspremont 2008; Drton and Perlman 2008; Friedman, Hastie, and Tibshirani 2008; Ravikumar, Wainwright, and Lafferty 2010). Implementing model selection approaches in the context of graphical models typically allows a dramatic reduction in the number of parameters under consideration, preventing overfitting and improving predictive capability. In particular, Bayesian approaches to inference in graphical models generate regularized estimators that incorporate model structure uncertainty.
The focus of this article is Bayesian inference in Gaussian graphical models (Dempster 1972) using the G-Wishart prior (Roverato 2002; Atay-Kayis and Massam 2005; Letac and Massam 2007). This class of distributions is extremely attractive since it represents the conjugate family for the precision matrix whose elements associated with edges not in the underlying graph are constrained to be equal to zero. Many recent articles have described various stochastic search methods for Gaussian graphical models (GGMs) with the G-Wishart prior based on marginal likelihoods which, in this case, are given by the ratio of the normalizing constants of the posterior and prior G-Wishart distributions—see the works of Atay-Kayis and Massam (2005), Jones et al. (2005), Carvalho and Scott (2009), Armstrong et al. (2009), Lenkoski and Dobra (2011) and the references therein. Wang and West (2009) proposed a MCMC algorithm for model determination and estimation in matrix-variate GGMs that also involves marginal likelihoods. Although the computation of marginal likelihoods for decomposable graphs is straightforward, similar computations for non-decomposable graphs or matrix-variate GGMs raise significant numerical challenges. This leads to the idea of devising Bayesian model determination methods that avoid the computation of marginal likelihoods.
The contributions of this article are threefold. First, we develop a new Metropolis–Hastings method for sampling from the G-Wishart distribution associated with an arbitrary graph. We discuss our algorithm in the context of the related sampling methods of Wang and Carvalho (2010) and Mitsakakis, Massam, and Escobar (2011), and empirically show that it scales better to graphs with many vertices. Second, we propose novel reversible jump MCMC samplers (Green 1995) for model determination and estimation in multivariate and matrix-variate GGMs. We contrast our approaches with the algorithms of Wong, Carter, and Kohn (2003) and Wang and West (2009) which focus on arbitrary GGMs and matrix-variate GGMs, respectively. Third, we devise a new flexible class of conditionally autoregressive models (CAR) (Besag 1974) for lattice data that rely on our novel sampling algorithms. The link between conditionally autoregressive models and GGMs was originally pointed out by Besag and Kooperberg (1995). However, since typical neighborhood graphs are non-decomposable, fully exploiting this connection within a Bayesian framework requires that we are able to estimate Gaussian graphical models based on general graphs. Our main focus is on applications to multivariate lattice data, where our approach based on matrix-variate GGMs provides a natural approach to create sparse multivariate CAR models.
The organization of the article is as follows. Section 2 formally introduces GGMs and the G-Wishart distribution, along with a description of our novel sampling algorithm for the G-Wishart distribution. Section 3 describes our reversible jump MCMC algorithm for GGMs. This algorithm represents a generalization of the work of Giudici and Green (1999) and is applicable beyond decomposable GGMs. Section 4 discusses inference and model determination in matrix-variate GGMs. Unlike the related framework developed by Wang and West (2009) which involves exclusively decomposable graphs, our sampler operates on the joint distribution of the row and column precision matrices, the row and column conditional independence graphs, and the auxiliary variable that needs to be introduced to solve the underlying non-identifiability problem associated with matrix-variate normal distributions. Section 5 reviews conditional autoregressive priors for lattice data and their connection to GGMs. This section discusses both univariate and multivariate models for continuous and discrete data based on generalized linear models. Section 6 presents two illustrations of our methodology: a simulation study and a cancer mortality mapping model. Finally, Section 7 concludes the article by discussing future directions for our work.
2. GAUSSIAN GRAPHICAL MODELS AND THE G–WISHART DISTRIBUTION
We let X = XVp, Vp = {1, 2, …, p}, be a random vector with a p-dimensional multivariate normal distribution Np(0, K−1). We consider a graph G = (Vp, E), where each vertex i ∈ V corresponds with a random variable Xi and E ⊂ Vp × Vp are undirected edges. Here “undirected” means that (i, j) ∈ E if and only if (j, i) ∈ E. We denote by 𝒢p the set of all 2p(p−1)/2 undirected graphs with p vertices. A Gaussian graphical model with conditional independence graph G is constructed by constraining to zero the off-diagonal elements of K that do not correspond with edges in G (Dempster 1972). If (i, j) ∉ E, Xi and Xj are conditionally independent given the remaining variables. The precision matrix K = (Kij)1≤i,j≤p is constrained to the cone PG of symmetric positive definite matrices with off-diagonal entries Kij = 0 for all (i, j) ∉ E.
We consider the G-Wishart distribution WisG(δ, D) with density
| (1) |
with respect to the Lebesgue measure on PG (Roverato 2002; Atay-Kayis and Massam 2005; Letac and Massam 2007). Here 〈A, B〉 = tr(ATB) denotes the trace inner product. The normalizing constant IG(δ, D) is finite if δ > 2 and D positive definite (Diaconnis and Ylvisaker 1979). If G is the full graph (E = Vp × Vp), WisG(δ, D) is the Wishart distribution Wisp(δ, D) (Muirhead 2005).
Since our sampling methods rely on perturbing the Cholesky decompositions of the matrix K, we review some key results. We write K ∈ PG as K = QT (ΨTΨ)Q where Q = (Qij)1≤i≤j≤p and Ψ = (Ψij)1≤i≤j≤p are upper triangular, while D−1 = QTQ is the Cholesky decomposition of D−1. We see that K = ΦTΦ where Φ = ΨQ is the Cholesky decomposition of K. The zero constraints on the off-diagonal elements of K associated with G induce well-defined sets of free elements Φν(G) = {Φij : (i, j) ∈ ν(G)} and Ψν(G) = {Ψij : (i, j) ∈ ν(G)} of the matrices Φ and Ψ—see proposition 2, page 320, and lemma 2, page 326, of the article by Atay-Kayis and Massam (2005). Here ν(G) = ν=(G) ∪ ν<(G), ν=(G) = {(i, i) : i ∈ Vp} and ν<(G) = {(i, j) : i < j and (i, j) ∈ E}.
We denote by Mν(G) the set of incomplete triangular matrices whose elements are indexed by ν(G) and whose diagonal elements are strictly positive. Both Φν(G) and Ψν(G) must belong to Mν(G). The non-free elements of Ψ are determined through the completion operation (Atay-Kayis and Massam 2005, lemma 2) as a function of the free elements Ψν(G). Each element Ψij with i < j and (i, j) ∉ E is a function of the other elements Ψi′j′ that precede it in lexicographical order. Roverato (2002) proved that the Jacobian of the transformation that maps K ∈ PG to Φν(G) ∈ Mν(G) is , where . Here |A| denotes the number of elements of the set A. Atay-Kayis and Massam (2005) showed that the Jacobian of the transformation that maps Φν(G) to Ψν(G) is given by , where . We have det and Φii = ΨiiQii. It follows that the density of Ψν(G) with respect to the Lebesgue measure on Mν(G) is
| (2) |
We note that represents the number of neighbors of vertex i in the graph G.
2.1 Existing Methods for Sampling From the G-Wishart Distribution
The problem of sampling from the G-Wishart distribution has received considerable attention in the recent literature. Piccioni (2000) followed by Asci and Piccioni (2007) exploited the theory of regular exponential families with cuts and proposed the block Gibbs sampler algorithm. Their iterative method generates one sample from WisG(δ, D) by performing sequential adjustments with respect to submatrices associated with each clique of G. The adjustment with respect to a clique C involves the inversion of a (p − |C|) × (p − |C|) matrix. If G has several cliques involving a small number of vertices but has a large number of vertices p, repeated inversions of high-dimensional matrices must be performed at each iteration. This makes the block Gibbs sampler algorithm impractical for being used in the context of large graphs.
Carvalho, Massam, and West (2007) gave a direct sampling method for WisG(δ, D) that works well for decomposable graphs. This algorithm was extended by Wang and Carvalho (2010) to non-decomposable graphs by introducing an accept– reject step as follows. Based on theorem 1, page 328, of the article of Atay-Kayis and Massam (2005), Wang and Carvalho (2010) and Mitsakakis, Massam, and Escobar (2011) wrote the density of Ψν(G) given in (2) as
| (3) |
where
is a function of the non-free elements of Ψ which, in turn, are uniquely determined from the free elements in Ψν(G). Furthermore, h(Ψν(G)) is the product of mutually independent chi-squared and standard normal distributions, that is, for i ∈ Vp and Ψij ~ N(0, 1) for (i, j) ∈ ν<(G). The expectation Eh[f (Ψν(G))] is calculated with respect to h(·). Since f(Ψν(G)) ≤ 1 for any Ψν(G) ∈ Mν(G), we have
| (4) |
where . From algorithm A.4 of the book by Robert and Casella (2004), an accept–reject method for sampling from WisG(δ, D) proceeds by sampling Ψν(G) ~ h(·) and U ~ Uni(0, 1) until U ≤ f(Ψν(G)). The probability of acceptance of a random sample from h(·) as a random sample from WisG(δ, D) is
| (5) |
Wang and Carvalho (2010) further decomposed G in its maximal prime subgraphs (Tarjan 1985), applied the accept–reject method for marginal or conditional distributions of WisG(δ, D) corresponding with each maximal prime subgraph, and generated a sample from WisG(δ, D) by putting together the resulting lower-dimensional sampled matrices.
Mitsakakis, Massam, and Escobar (2011) proposed a Metropolis–Hastings method for sampling from the G-Wishart WisG(δ, D) distribution. We denote by K[s] = QT(Ψ[s])TΨ[s]Q the current state of their Markov chain, where (Ψ[s])ν(G) ∈ Mν(G). Mitsakakis, Massam, and Escobar (2011) generated a candidate K′ = QT (Ψ′)TΨ′Q for the next state by sampling (Ψ′)ν(G) ~ h(·) and determining the non-free elements of Ψ′ from the free sampled elements (Ψ′)ν(G). The chain moves to K′ with probability
| (6) |
The inequality (4) implies that this independent chain is uniformly ergodic (Mengersen and Tweedie 1996) and its expected acceptance probability is greater than or equal to Eh[f(Ψν(G))]—see lemma 7.9 in the book by Robert and Casella (2004). Hence, the Markov chain of Mitsakakis, Massam, and Escobar (2011) is more efficient than the method of Wang and Carvalho (2010) when the latter algorithm is employed without graph decompositions or when the graph G has only one maximal prime subgraph. On the other hand, the method of Mitsakakis, Massam, and Escobar (2011) involves changing the values of all free elements Ψν(G) in a single step. As a result, if their Markov chain is currently in a region of high probability, it could potentially have to generate many candidates from h(·) before moving to a new state with comparable or larger probability.
2.2 Our Algorithm for Sampling From the G-Wishart Distribution
We introduce a new Metropolis–Hastings algorithm for sampling from the G-Wishart WisG(δ, D) distribution. In contrast to the work of Mitsakakis, Massam, and Escobar (2011), our approach makes use of a proposal distribution that depends on the current state of the chain and leaves all but one of the free elements in Ψν(G) unchanged. The distance between the current and the proposed state is controlled through a Gaussian kernel with a precision parameter σm.
We denote by K[s] = QT (Ψ[s])TΨ[s]Q the current state of the chain with (Ψ[s])ν(G) ∈ Mν(G). The next state K[s+1] = QT(Ψ[s+1])TΨ[s+1]Q is obtained by sequentially perturbing the free elements (Ψ[s])ν(G). A diagonal element is updated by sampling a value γ from a distribution truncated below at zero. We define the upper triangular matrix Ψ′ such that for (i, j) ∈ ν(G) \ {(i0, i0)} and . The non-free elements of Ψ′ are obtained through the completion operation (Atay-Kayis and Massam 2005, lemma 2) from (Ψ′)ν(G). The Markov chain moves to K′ = QT(Ψ′)TΨ′Q with probability min{Rm, 1}, where
| (7) |
Here φ(·) represents the CDF of the standard normal distribution and
| (8) |
A free off-diagonal element is updated by sampling a value . We define the upper triangular matrix Ψ′ such that for (i, j) ∈ ν(G) \ {(i0, j0)} and . The remaining elements of Ψ′ are determined by the completion operation from lemma 2 in the article of Atay-Kayis and Massam (2005) from (Ψ′)ν(G). The proposal distribution is symmetric , thus we accept the transition of the chain from K[s] to K′ = QT(Ψ′)TΨ′Q with probability , where is given in Equation (7). Since (Ψ[s])ν(G) ∈ Mν(G), we have (Ψ′)ν(G) ∈ Mν(G) which implies K′ ∈ PG. We denote by K[s+1] the precision matrix obtained after completing all the updates associated with the free elements indexed by ν(G).
A key computational aspect is related to the dependence of Cholesky decompositions on a particular ordering of the variables involved. Empirically we noticed that the mixing times of Markov chains that make use of our sampling approach can be improved by changing the ordering of the variables in Vp at each iteration. More specifically, a permutation υ is uniformly drawn from the set of all possible permutations ϒp of Vp. The row and columns of D are reordered according to υ and a new Cholesky decomposition of D−1 is determined. The set ν(G) and { : i ∈ V} are recalculated given the ordering of the vertices induced by υ. Although the random shuffling of the ordering of the indices worked well for the particular applications we have considered in this article, we do not have any theoretical justification that explains why it improves the computational efficiency of our sampling approaches.
Our later developments from Section 4 involve sampling K ~ WisG(δ, D) subject to the constraint K11 = 1. We have . We subsequently obtain the next state K[s+1] of the Markov chain by perturbing the free elements (Ψ[s])ν(G)\{(1,1)}. When defining the triangular matrix Ψ′ we set which implies that the corresponding candidate matrix K′ has . Thus K[s+1] also obeys the constraint . The random orderings of the variables need to be drawn from the set of permutations υ ∈ ϒp such that υ(1) = 1. This way the (1, 1) element of K always occupies the same position.
2.3 The Scalability of Sampling Methods From the G-Wishart Distribution
Practical applications of our novel framework for analyzing multivariate lattice data from Section 5 involve sampling from G-Wishart distributions associated with arbitrary graphs with tens or possibly hundreds of vertices—see Section 6.2 as well as relevant examples from the works of Elliott et al. (2001), Banerjee, Carlin, and Gelfand (2004), Rue and Held (2005), Lawson (2009), Gelfand et al. (2010). We perform a simulation study to empirically compare the scalability of our approach (DLR) for simulating from G-Wishart distributions with respect to the algorithms of Wang and Carvalho (2010) (WC) and Mitsakakis, Massam, and Escobar (2011) (MME).We consider the cycle graph Cp ∈ 𝒢p with edges {(i, i + 1) : 1 ≤ i ≤ p − 1} ∪ {(p, 1)} and the matrix Ap ∈ PCp such that (Ap)ii = 1 (1 ≤ i ≤ p), (Ap)i,i−1 = (Ap)i−1,i = 0.5 (2 ≤ i ≤ p), and (Ap)1,p = (Ap)p,1 = 0.4. We chose Cp because it is the sparsest graph with p vertices and only one maximal prime subgraph, hence no graph decompositions can be performed in the context of the WC algorithm.
We employ the three sampling methods to sample from the G-Wishart WisCp (103, Dp) distribution, where and Ip is the p-dimensional identity matrix. This is representative of a G-Wishart posterior distribution corresponding with 100 samples from a and a G-Wishart prior WisCp (3, Ip). The acceptance probability for generating one sample with the DLR algorithm is defined as the average acceptance probabilities of the updates corresponding with diagonal and off-diagonal free elements in Ψν(G). We calculate Monte Carlo estimates and their standard errors of the acceptance probabilities for our method by running 100 independent chains of length 2500 for each combination (σm, p) ∈ {0.1, 0.5, 1, 2} × {4, 6, …, 20}. The same number of chains of the same length have been run with the MME algorithm for p = 4, 6, …, 20. We calculate Monte Carlo estimates of the acceptance probabilities (5) associated with the WC algorithm, that is,
The corresponding standard errors were determined by calculating 100 such estimates for each p = 4, 6, …, 20. The results are summarized in Table 1. As we would expect, the acceptance rates for the DLR algorithm decrease as σm increases since the proposed jumps in PCp become larger. More importantly, for each value of σm, the acceptance rates for the DLR algorithm decrease very slowly as the number of vertices p grows. This shows that our Metropolis–Hastings method is likely to retain its efficiency for graphs that involve a large number of vertices. On the other hand, the acceptance rates for the WC algorithm are extremely small even for p = 6 vertices. This implies that a large number of samples from the instrumental distribution h(·) would need to be generated before one sample from WisCp (103, Dp) is obtained. The MME algorithm gives slightly better acceptance rates, but they are still small and are indicative of a simulation method that constantly attempts to make large jumps in areas of low probability of WisCp (103,Dp). Therefore the WC and MME methods might not scale well despite being perfectly valid in theory, hence they cannot be used in the context of the multivariate lattice data models from Section 5. The acceptance rates of the DLR algorithm are a function of the precision parameter σm which can be easily adjusted to yield jumps that are not too short but also not too long in the target cone of matrices. This flexibility is key for successfully employing our algorithm for graphs with many vertices.
Table 1.
Monte Carlo estimates and their standard errors (in parentheses) of the acceptance probabilities for our proposed algorithm (DLR) for sampling from the G-Wishart distribution, the Wang and Carvalho (2010) algorithm (WC), and the Mitsakakis, Massam, and Escobar (2011) algorithm (MME)
| p | DLR
|
WC | MME | |||
|---|---|---|---|---|---|---|
| σm = 0.1 | σm = 0.5 | σm = 1 | σm =2 | |||
| 4 | 0.953 (2.0e–3) | 0.776 (3.0e–3) | 0.600 (3.2e–3) | 0.389 (3.8e–3) | 0.340 (6.4e–3) | 0.473 (1.0e–2) |
| 6 | 0.947 (2.0e–3) | 0.751 (2.4e–3) | 0.565 (2.9e–3) | 0.356 (2.3e–3) | 5.08e–2 (2.2e–3) | 0.185 (1.0e–2) |
| 8 | 0.944 (1.8e–3) | 0.740 (2.1e–3) | 0.551 (2.3e–3) | 0.343 (2.6e–3) | 7.94e–3 (7.1e–4) | 0.078 (0.9e–2) |
| 10 | 0.943 (1.7e–3) | 0.734 (2.0e–3) | 0.543 (2.1e–3) | 0.336 (2.1e–3) | 1.18e–3 (1.7e–4) | 0.035 (8.0e–3) |
| 12 | 0.942 (1.4e–3) | 0.729 (1.8e–3) | 0.537 (2.1e–3) | 0.331 (2.2e–3) | 1.56e–4 (5.1e–5) | 0.013 (6.0e–3) |
| 14 | 0.940 (1.5e–3) | 0.725 (1.6e–3) | 0.532 (1.8e–3) | 0.327 (1.9e–3) | 2.27e–5 (1.4e–5) | 0.005 (4.0e–3) |
| 16 | 0.940 (1.3e–3) | 0.721 (1.6e–3) | 0.527 (1.7e–3) | 0.324 (1.7e–3) | 2.19e–6 (1.8e–6) | 0.002 (3.0e–3) |
| 18 | 0.939 (1.2e–3) | 0.717 (1.4e–3) | 0.523 (1.5e–3) | 0.321 (1.5e–3) | 3.87e–7 (8.0e–7) | 0.001 (2.0e–3) |
| 20 | 0.938 (1.0e–3) | 0.714 (1.4e–3) | 0.520 (1.6e–3) | 0.318 (1.5e–3) | 2.08e–8 (3.5e–8) | 5.4e–4 (9.0e–4) |
It may initially appear that failure to handle graphs with a maximal prime subgraph with 20 vertices is not a major shortcoming of an algorithm that relies on graph decompositions. However, the example we consider in Section 6.2 involves a fixed graphical model whose underlying graph has a maximal prime subgraph with 36 vertices (see the Supplementary Materials). Since this graph is one that is constructed from the neighborhood structure of the United States, we can see the importance of the ability to scale when applying the GGM framework to spatial statistical problems.
3. REVERSIBLE JUMP MCMC SAMPLERS FOR GGMS
The previous section was concerned with sampling from the G-Wishart distribution when the underlying conditional independence graph is fixed. In contrast, this section is concerned with performing Bayesian inference in GGMs, which involves determination of the graph G. Giudici and Green (1999) proposed a reversible jump Markov chain algorithm that is restricted to decomposable graphs and employs a hyper inverse Wishart prior (Dawid and Lauritzen 1993) for the covariance matrix Σ = K−1 = (Σij)1≤i,j≤p. The efficiency of their approach comes from representing decomposable graphs through their junction trees and from a specification of Σ as an incomplete matrix Γ = (Γij)1≤i,j≤p such that Σij = Γij if i = j or if (i, j) is an edge in the current graph (i.e., Kij is not constrained to 0); the remaining elements of Γ are left unspecified but can be uniquely determined using the iterative proportional scaling algorithm (Dempster 1972; Speed and Kiiveri 1986). Brooks, Giudici, and Roberts (2003) presented techniques for calibrating the precision parameter of the normal kernel used by Giudici and Green (1999) to sequentially update the elements of Γ and the edges of the underlying decomposable graph which lead to improved mixing times of the resulting Markov chain. Related work (Scott and Carvalho 2008; Carvalho and Scott 2009; Armstrong et al. 2009) has also focused on decomposable graphs. Graphs of this type have received attention mainly because their special structure is convenient from a computational standpoint. However, decomposability is a serious constraint as it drastically reduces 𝒢p to a much smaller subset of graphs: the ratio between the number of decomposable graphs and the total number of graphs decreases from 0.95 for p = 4 to 0.12 for p = 8 (Armstrong 2005).
Roverato (2002), Atay-Kayis and Massam (2005), Jones et al. (2005), Dellaportas, Giudici, and Roberts (2003), Moghaddam et al. (2009), Lenkoski and Dobra (2011) explored various methods for numerically calculating marginal likelihoods for non-decomposable graphs. Besides being computationally expensive, stochastic search algorithms that traverse 𝒢p based on marginal likelihoods output a set of graphs that have the highest posterior probabilities from all the graphs that have been visited, but do not produce estimates of the precision matrix K unless a direct sampling algorithm from the posterior distribution of K given a graph is available. As we have seen in Section 2.3, current direct sampling algorithms (e.g., Wang and Carvalho 2010) might not scale well to graphs with many vertices. Since our key motivation comes from modeling multivariate lattice data (see Section 5), we want to develop a Bayesian method that samples from the joint posterior distribution of precision matrices K ∈ PG and graphs G ∈ 𝒢p, thereby performing inference for both K and G. Wong, Carter, and Kohn (2003) developed such a reversible jump Metropolis–Hastings algorithm by decomposing K = TΔT, where and Δ = (Δij)1≤i,j≤p is a correlation matrix with Δii = 1 and Δij = Kij/(KiiKjj)1/2, for i < j. Their prior specification for K involves independent priors for Tii and a joint prior for the off-diagonal elements of Δ whose normalizing constant is associated with all the graphs with the same number of edges.
We propose a new reversible jump Markov chain algorithm that is based on a G-Wishart prior for K. While we do not empirically explore the efficiency of our approach with respect to the Wong, Carter, and Kohn (2003) algorithm, we state that the key advantage of our framework lies in its generalization to matrix-variate data—see Section 4.We are unaware of any similar extension of the Wong, Carter, and Kohn (2003) approach. Another benefit of our method with respect to that of Wong, Carter, and Kohn (2003) is related to its flexibility with respect to prior specifications on 𝒢p—see the discussion below.
We let 𝒟= {x(1), …, x(n)} be the observed data of n independent samples from Np(0, K−1). Given a graph G ∈ 𝒢p, we assume a G-Wishart prior WisG(δ0, D0) for the precision matrix K ∈ PG. We take δ0 = 3 > 2 and D0 = Ip. With this choice the prior for K is equivalent with one observed sample, while the observed variables are assumed to be a priori independent of each other. Since the G-Wishart prior for K is conjugate to the likelihood p(𝒟| K), the posterior of K given G is WisG(n + δ0, U + D0) where . We also assume a prior Pr(G) on 𝒢p. We develop a MCMC algorithm for sampling from the joint posterior distribution
that is well-defined if and only if K ∈ PG. We sequentially update the precision matrix given the current graph and the edges of the graph given the current precision matrix—see Appendix A. The addition or deletion of an edge involves a change in the dimensionality of the parameter space since the corresponding element of K becomes free or constrained to zero, hence we make use of the reversible jump MCMC methodology of Green (1995). The graph update step also requires the calculation of the normalizing constants of the G-Wishart priors corresponding with the current and the candidate graph. To this end, we make use of the Monte Carlo method of Atay-Kayis and Massam (2005) which converges very fast when computing IG(δ, D) [see Equation (1)] even for large graphs when δ is small and D is set to the identity matrix (Lenkoski and Dobra 2011).
Our framework accommodates any prior probabilities Pr(G) on the set of graphs 𝒢p which is a significant advantage with respect to the covariance selection prior from Wong, Carter, and Kohn (2003). Indeed, the prior in the work of Wong, Carter, and Kohn (2003) induces fixed probabilities for each graph and does not allow the possibility of further modifying these probabilities according to prior beliefs. A usual choice is the uniform prior Pr(G) = 2−m with , but this prior is biased toward middle-size graphs and gives small probabilities to sparse graphs and to graphs that are almost complete. Here the size of a graph G is defined as the number of edges in G and is denoted by size(G) ∈ {0, 1, …, m}. Dobra et al. (2004) and Jones et al. (2005) assumed that the probability of inclusion of any edge in G is constant and equal to ψ ∈ (0, 1), which leads to the prior
| (9) |
Sparser graphs can be favored with prior (9) by choosing a small value for ψ. Since ψ could be difficult to elicit in some applications, Carvalho and Scott (2009) integrated out ψ from (9) by assuming a conjugate beta distribution Beta(a, b), which leads to the prior
| (10) |
where B(·, ·) is the beta function. From the work of Scott and Berger (2006) it follows that prior (10) has an automatic multiplicity correction for testing the inclusion of spurious edges. Armstrong et al. (2009) suggested a hierarchical prior on 𝒢p (the size-based prior) that gives equal probability to the size of a graph and equal probability to graphs of each size, that is,
| (11) |
which is also obtained by setting a = b = 1 in (10). We note that the expected size of a graph under size-based prior is m/2, which is also the expected size of a graph under the uniform prior on 𝒢p.
4. REVERSIBLE JUMP MCMC SAMPLER FOR MATRIX–VARIATE GGMS
We extend our framework to the case when the observed data 𝒟 = {x(1), …, x(n)} are associated with a pR × pC random matrix X = (Xij) that follows a matrix-variate normal distribution
with probability distribution function (Gupta and Nagar 2000):
| (12) |
Here KR is a pR ×pR row precision matrix and KC is a pC × pC column precision matrix. Furthermore, we assume that KR ∈ PGR and KC ∈ PGC where GR = (VpR, ER) and GC = (VpC, EC) are two graphs with pR and pC vertices, respectively. We consider the rows X1*, …, XpR* and the columns X*1, …, X*pC of the random matrix X. From theorem 2.3.12 of the book by Gupta and Nagar (2000) we have and . The graphs GR and GC define graphical models for the rows and columns of X (Wang and West 2009):
| (13) |
Any prior specification for KR and KC must take into account the fact that the two precision matrices are not uniquely identified from their Kronecker product which means that, for any z > 0, (z−1KR) ⊗ (zKC) = KR ⊗ KC represents the same precision matrix for vec(X)—see Equation (12). We follow the idea laid out in the article of Wang and West (2009) and impose the constraint (KC)11 = 1.We define a prior for KC through parameter expansion by assuming a G-Wishart prior WisGC (δC, DC) for the matrix zKC with z > 0, δC > 2, and DC ∈ PGC. It is immediate to see that the Jacobian of the transformation from zKC to (z, KC) is J((zKC)→(z, KC)) = z|ν(GC)|−1. It follows that our joint prior for (z, KC) is given by
The elements of KR ∈ PGR are not subject to any additional constraints, hence we assume a G-Wishart prior WisGR (δR, DR) for KR. We take δC = δR = 3, DC = IpC, and DR = IpR. We complete our prior specification by choosing two priors Pr(GC) and Pr(GR) for the row and column graphs, where GR ∈ 𝒢pR and GC ∈ 𝒢pC—see our discussion from Section 3.
We perform Bayesian inference for matrix-variate GGMs by developing a MCMC algorithm for sampling from the joint posterior distribution of the row and column precision matrices, the row and column graphs, and the auxiliary variable z:
| (14) |
that is defined for KR ∈ PGR, KC ∈ PGC with (KC)11 = 1 and z > 0. Equation (14) is written as
| (15) |
The details of our sampling scheme are presented in Appendix B. Updating the row and column precision matrices involves sampling from their corresponding G-Wishart conditional distributions using the Metropolis–Hastings approach from Section 2.2, while updating z involves sampling from its gamma full conditional. The updates of the row and column graphs involve changes in the dimension of the parameter space and require a reversible jump step (Green 1995).
It is relevant to discuss how our MCMC sampler from Appendix B is different from the methodology proposed by Wang and West (2009). First of all, Wang and West (2009) allowed only decomposable row and column graphs which makes their framework inappropriate for modeling multivariate lattice data where the graph representing the neighborhood structure of the regions involved is typically not decomposable. Second of all, they proposed a Markov chain sampler for the marginal distribution associated with (GR, GC) of the joint posterior distribution (15):
| (16) |
which involves the marginal likelihood
| (17) |
Wang and West (2009) employed the candidate’s formula (Besag 1989; Chib 1990) to approximate the marginal likelihood (17). They sampled from (16) by sequentially updating the row and column graphs. The update of the row graph proceeds by sampling a candidate from an instrumental distribution and accepting it with Metropolis–Hastings probability
| (18) |
The update of the column graph is done in a similar manner. Therefore resampling GR or GC requires the computation of a new marginal likelihood (17) which entails a considerable computational effort even for graphs with a small number of vertices. We avoid the computation of marginal likelihoods by sampling from the joint distribution (15) in which the row and column precision matrices have not been integrated out. Sampling from the joint marginal distribution (16) seems appealing because it involves the reduced space of row and column graphs. However, the numerical difficulties associated with repeated calculations of marginal likelihoods (17) outweigh the benefits of working in a smaller space. We empirically compare the efficiency of our inference approach for matrix-variate GGMs with the work of Wang and West (2009) in Section 6.1.
5. BAYESIAN HIERARCHICAL MODELS FOR MULTIVARIATE LATTICE DATA
Conditional autoregressive (CAR) models (Besag 1974; Mardia 1988) are routinely used in spatial statistics to model lattice data. In the case of a single observed outcome in each region, the data are associated with a vector X = (X1, …, XpR)T where Xi corresponds to region i. The zero-centered CAR model is implicitly defined by the set of full conditional distributions
| (19) |
Therefore, CAR models are just two-dimensional Gaussian Markov random fields. According to Brook’s (1964) theorem, this set of full-conditional distributions implies that the joint distribution for X satisfies
where and B is a pR × pR matrix such that B = (bij) and bii = 0. In order for Λ−1(I−B) to be a symmetric matrix we require that for i ≠ j; therefore the matrix B and vector λ must be carefully chosen. A popular approach is to begin by constructing a symmetric proximity matrix W= (wij), and then set bij = wij/wi+ and where wi+ = Σj wij and τ2 > 0. In that case, Λ−1(I − B) = τ−2(EW −W), where EW = diag{w1+, …, wpR+}. The proximity matrix W is often constructed by first specifying a neighborhood structure for the geographical areas under study; for example, when modeling state or county level data it is often assumed that two geographical units are neighbors if they share a common border. This neighborhood structure can be summarized in a graph GR ∈ 𝒢pR whose vertices correspond to geographical areas, while its edges are associated with areas that are considered neighbors of each other. The proximity matrix W is subsequently specified as
| (20) |
where ∂GR(i) denotes the set of vertices that are linked by an edge in the graph GR with the vertex i.
Specifying the joint precision matrix for X using the proximity matrix derived from the neighborhood structure is very natural; essentially, it implies that observations collected on regions that are not neighbors are conditionally independent from each other given the rest. However, note that the specification in (20) implies that (EW − W)1pR = 0 and therefore the joint distribution on X is improper. Here 1l is the column vector of length l with all elements equal to 1. Proper CAR models (Cressie 1973; Sun et al. 2000; Gelfand and Vounatsou 2003) can be obtained by including a spatial autocorrelation parameter ρ, so that
| (21) |
The joint distribution on X is then multivariate normal where VW(τ−2, ρ) = τ−2(EW − ρW) ∈ PGR. This distribution is proper as long as ρ is between the reciprocals of the minimum and maximum eigenvalues for W. In particular, note that taking ρ = 0 leads to independent random effects.
In the spirit of Besag and Kooperberg (1995), an alternative but related approach to the construction of models for lattice data is to let X ~ NpR (0, K−1) and assign K a G-Wishart prior WisGR(δR, (δR − 2)DR), where . The mode of WisGR (δR, (δR − 2)DR) is the unique matrix K = (Kij) ∈ PGR that satisfies the relations
| (22) |
The matrix verifies the system (22), hence it is the mode of WisGR(δR, (δR − 2)DR). As such, the mode of the prior for KR induces the same prior specification for X as (21). It is easy to see that, conditional on KR ∈ PGR, we have
| (23) |
Hence, by modeling X using a Gaussian graphical model and restricting the precision matrix K to belong to the cone PGR, we are inducing a mixture of CAR priors on X where the priors on
and [see Equation (19)] are induced by the G-Wishart prior WisGR (δR, (δR − 2)DR).
The specification of CAR models through G-Wishart priors solves the impropriety problem of intrinsic CAR models and preserves the computational advantages derived from standard CAR specifications while providing greater flexibility. Indeed, the prior is trivially proper because the matrix K ∈ PGR is invertible by construction. The computational advantages are preserved because the full conditional distributions for each Xi can be easily computed for any matrix K without the need to perform matrix inversion, and they depend only on a small subset of neighbors {Xj:j ∈ ∂GR(i)}. Additional flexibility is provided because the weights bij for j ∈ ∂GR (i) and smoothing parameters are being estimated from the data rather than being assumed fixed, allowing for adaptive spatial smoothing. Our approach provides what can be considered as a nonparametric alternative to the parametric estimates of the proximity matrix of Cressie and Chan (1989).
A similar approach can be used to construct proper multivariate conditional autoregressive (MCAR) models (Mardia 1988; Gelfand and Vounatsou 2003). In this case, we are interested in modeling a pR × pC matrix X = (Xij) where Xij denotes the value of the jth outcome in region i. We let X follow a matrix-variate normal distribution with row precision matrix KR capturing the spatial structure in the data (which, as in univariate CAR models, is restricted to the cone PGR defined by the neighborhood graph GR), and column precision matrix KC, which controls the multivariate dependencies across outcomes. It can be easily shown that the row vector Xi* of X depends only on the row vectors associated with those regions that are neighbors with i—see also Equation (13):
Given the matrix-variate GGMs framework from Section 4, we can model the conditional independence relationships across outcomes through a column graph GC ∈ 𝒢pC and require KC ∈ PGC As opposed to the neighborhood graph GR which is known and considered fixed, the graph GC is typically unknown and needs to be inferred from the data.
The matrix-variate GGM formulation for spatial models can also be used as part of more complex hierarchical models. Indeed, CAR and MCAR models are most often used as a prior for the random effects of a generalized linear model (GLM) to account for residual spatial structure not accounted for by covariates. When no covariates are available, the model can be interpreted as a spatial smoother where the spatial covariance matrix controls the level of spatial smoothing in the underlying (latent) surface. Similarly, MCAR models can be used to construct multivariate spatial GLMs. More specifically, consider the pR × pC matrix Y = (Yij) of discrete or continuous outcomes, and let Yij ~ hj(·|ηij) where hj is a probability mass or probability density function that belongs to the exponential family with location parameter ηij. The spatial GLM is then defined through the linear predictor
| (24) |
where g(·) is the link function, μj is an outcome-specific intercept, Xij is a zero-centered spatial random effect associated with location i, Zij is a matrix of observed covariates for outcome j at location i, and βj is the vector of fixed effects associated with outcome j. As an example, by choosing yij ~ Poi(ηij), g−1(·) = log(·) we obtain a multivariate spatial log-linear model for count data, which is often used for disease mapping in epidemiology (see Section 6.2). We further assign a matrix-variate normal distribution for X with independent G-Wishart priors for KR and zKC, where z > 0 is an auxiliary variable needed to impose the identifiability constraint (KC)11 = 1:
| (25) |
Prior elicitation for this class of spatial models is relatively straightforward. Indeed, elicitation of the matrix DR = (EW − ρW)−1 requires only the elicitation of the neighborhood matrix W which also defines the neighborhood graph GR, along with reasonable values for the spatial autocorrelation parameter ρ. In particular, in the application we discuss in Section 6.2 we assume that, a priori, there is a strong degree of positive spatial association, and choose a prior for ρ that gives higher probabilities to values close to 1 (Gelfand and Vounatsou 2003):
| (26) |
In the case of MCAR models, it is common to assume that the prior value for the conditional covariance between variables is zero, which leads to choosing DC from the G-Wishart prior for zKC to be a diagonal matrix. We note that the scale parameter τ2 is no longer identifiable in the context of the joint prior (25), hence it is then sensible to pick DC = IpC .
At this point, a word of caution about the interpretation of the models seems appropriate. The graphs GR = (VpR, ER) and GC = (VpC, EC) induce conditional independence relationships associated with the rows and columns of the random-effects matrix X—see Equation (13). Similar conditional independence relationships hold for the rows and columns of the matrix of location parameters η = (ηij), but these extend to the observed outcomes Y only if its entries are continuous (for a more thorough discussion, see the Supplementary Materials). Hence, the reader must be careful when interpreting the results of these models when binary or count data are involved; in these cases, any statement about conditional independence needs to be circumscribed to the location parameters η.
6. ILLUSTRATIONS
6.1 Simulation Study
We empirically compare the reversible jump MCMC sampler for matrix-variate GGMs proposed in Section 4 (RJ-DLR) with the methodology of Wang and West (2009) (WW). We consider the row graph ḠR = (VpR, ĒR) with pR = 5 vertices and with edges ĒR = {(1, i) : 1 < i ≤ 5} ∪ {(2, 3), (3, 4), (4, 5), (2, 5)}. We take the column graph ḠC to be the cycle graph with pC = 10 vertices (see Section 2.3). We note that both the row and column graphs are non-decomposable, with ḠR being relatively dense and with ḠC being relatively sparse. We define the row and column precision matrices K̄R ∈ PḠR and K̄C ∈ PḠC to have diagonal elements equal to 1 and nonzero off-diagonal elements equal with 0.4. We generate 100 datasets each comprising n = 100 observations sampled from the 5 × 10 matrix-variate normal distribution p(·|K̄R, K̄C)—see Equation (12).
For each dataset we attempted to recover the edges of ḠR and ḠC with the RJ-DLR and the WW algorithms. The two MCMC samplers were run for 10,000 iterations with a burn-in of 1000 iterations. We implemented the RJ-DLR algorithm in R and C++ (code available as Supplemental Material). We used the code developed by Wang and West (2009) for their method. In the RJ-DLR algorithm we set σm,R = σm,C = σg,R = σg,C = 0.5, which yields average rejection rates on Metropolis–Hastings updates of about 0.3 for both K̄C and K̄R. We assumed independent uniform priors for the row and column graphs.
Tables 2 and 3 show the average posterior edge inclusion probabilities for ḠR and ḠC. Our RJ-DLR method recovers the structure of both graphs very well: the edges that belong to each graph receive posterior inclusion probabilities of 1, while the edges that are absent from each graph receive posterior inclusion probabilities below 0.1. The ability of the RJ-DLR algorithm to recover the structures of a dense row graph and of a sparse column graph is quite encouraging since the uniform priors for the row and column graphs favored middle-size graphs.
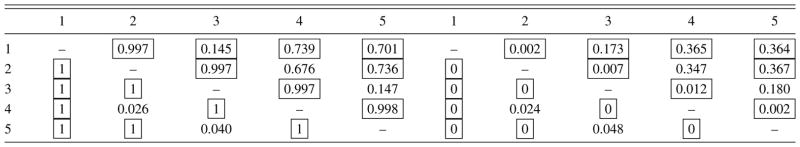
Table 2.
Average estimates (left panel) and their standard errors (right panel) of posterior edge inclusion probabilities for the row graph ḠR obtained using the RJ-DLR algorithm (below diagonal) and the WW algorithm (above diagonal). The boxes identify the edges that are in ḠR
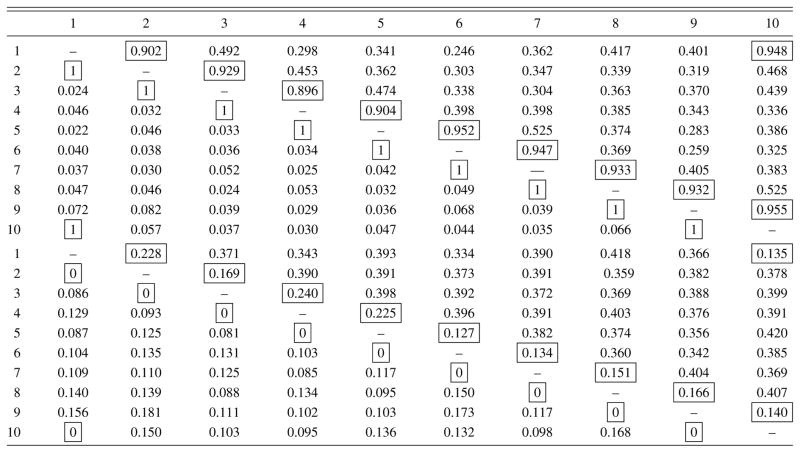
Table 3.
Average estimates (upper panel) and their standard errors (lower panel) of posterior edge inclusion probabilities for the column graph ḠC obtained using the RJ-DLR algorithm (below diagonal) and the WW algorithm (above diagonal). The boxes identify the edges that are in ḠC
The WW method shows diminished performance in recovering graphical structures. Edges (1, 4), (1, 5), and (2, 5), which are actually in ḠR, are given average inclusion probabilities of only 0.7. Edge (1, 3), which is in ḠR, receives a very low inclusion probability, while edge (2, 4), which is not in ḠR, is given a high inclusion probability. The WW algorithm includes edges that are in the column graph with relatively high probabilities (typically above 0.9). However, edges that do not belong to ḠC also appear to be included quite often, with an average inclusion probability between 0.3 and 0.4. These tables also show a large standard deviation of edge inclusion probabilities across the 100 datasets using the WW method. The decreased performance of the WW algorithm versus our own RJ-DLR algorithm is likely attributable to the fact that Wang and West (2009) accounted only for decomposable graphs in their framework, which may be poor at approximating the non-decomposable graphs considered in this example.
6.2 Mapping Cancer Mortality in the United States
Accurate and timely counts of cancer mortality are very useful in the cancer surveillance community for purposes of efficient resource allocation and planning. Estimation of current and future cancer mortality broken down by geographic area (state) and tumor has been discussed in a number of recent articles, including those by Tiwari et al. (2004), Ghosh and Tiwari (2007), Ghosh et al. (2007), and Ghosh, Ghosh, and Tiwari (2008). This section considers a multivariate spatial model on state-level cancer mortality rates in the United States for 2000. These mortality data are based on death certificates that are filed by certifying physicians. They are collected and maintained by the National Center for Health Statistics (http://www.cdc.gov/nchs) as part of the National Vital Statistics System. The data are available from the Surveillance, Epidemiology, and End Results (SEER) program of the National Cancer Institute (http://seer.cancer.gov/seerstat).
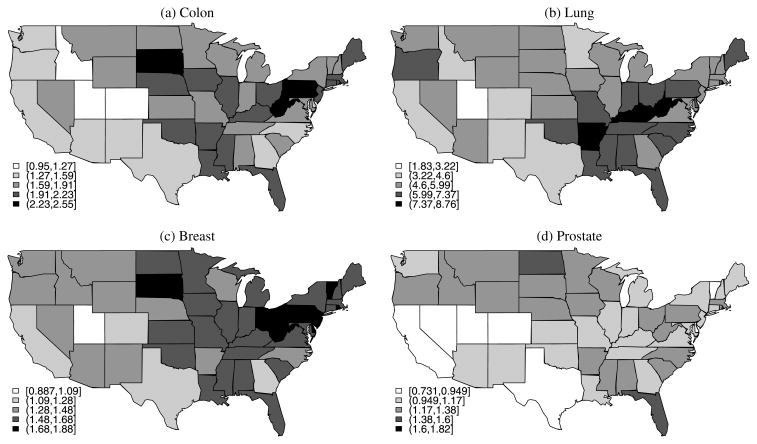
The data we analyze consist of mortality counts for 11 types of tumors recorded in the 48 continental states plus the District of Columbia. Since mortality counts below 25 are often deemed unreliable in the cancer surveillance community, we treated them as missing. Along with mortality counts, we obtained population counts in order to model death risk. Figure 1 shows raw mortality rates for four of the most common types of tumors (colon, lung, breast, and prostate). Although the pattern is slightly different for each of these cancers, a number of striking similarities are present; for example, Colorado appears to be a state with low mortality for all of these common cancers, while West Virginia, Pennsylvania, and Florida present relatively high mortality rates.
Figure 1.
Mortality rates (per 10,000 habitants) in the 48 continental states and the D.C. area corresponding to four common cancers during 2000.
We consider modeling these data using Poisson multivariate log-linear models with spatial random effects. More concretely, we let Yij be the number of deaths in state i = 1, …, pR = 49 for tumor type j = 1, …, pC = 11. We set
| (27) |
Here mi is the population of state i, μj is the intercept for tumor type j, and Xij is a zero-mean spatial random effect associated with location i and tumor j. We denote X̃ij = μj + Xij. We further model X̃ = (X̃ij) with a matrix-variate normal prior:
| (28) |
where μ = (μ1, …, μpC)T.
We consider four models in total. The first two models we consider (GGM-U and GGM-S) are based on the sparse models for multivariate aerial data described in Section 5, and differ only in terms of the prior assigned on the space of graphs [GGM-U uses a uniform prior, while GGM-S uses the size-based prior from Equation (11)]. Hence, the joint prior for the row and column precision matrices is given in (25) for both models. The column graph GC is unknown and allowed to vary in 𝒢11. The row graph GR is fixed and derived from the incidence matrix W corresponding with the neighborhood structure of the U.S. states. More explicitly, each state is associated with a vertex in GR. Two states are linked by an edge in GR if they share a common border. The Supplementary Materials describe the neighborhood structure of GR and provide its decomposition into maximal prime subgraphs. What is interesting about the graph GR, and relevant in light of the results of Section 2.3, is that its decomposition yields 13 maximal prime subgraphs. Most of these prime components are complete and only contain two or three vertices; however, one maximal prime subgraph has 36 vertices (states), making GR non-decomposable with a sizable maximal prime subgraph. The degrees of freedom for the G-Wishart priors are set as δR = δC = 3, while the centering matrices are chosen as DC = IpC and DR = (EW − ρW)−1.
The third model, which we call model FULL, is obtained by keeping the column graph GC fixed to the full graph. This is equivalent to replacing the G-Wishart prior for (zKC) from Equation (25) with a Wishart prior WispC (δC, DC). Finally, model MCAR is obtained from model FULL by substituting the G-Wishart prior for KR from Equation (25) with KR = EW − ρW. The resulting prior distribution for the spatial random effects X̃ in model MCAR is precisely the MCAR(ρ, Σ) prior of Gelfand and Vounatsou (2003). In all cases, we complete the model specification by choosing the prior from Equation (26) for the spatial autocorrelation parameter ρ and a multivariate normal prior for the mean rates vector μ ~ NpC (μ0, Ω−1) where μ0 = μ01pC and Ω = ω−2IpC. We set μ0 to be the median log incidence rate across all cancers and regions, and ω to be twice the interquartile range in raw log incidence rates.
Posterior inferences for models GGM-U and GGM-S are obtained by extending the sampling algorithm from Appendix B; details are presented in the Supplemental Materials. MCMC samplers for models FULL and MCAR are similarly derived in a straightforward manner. Missing counts were sampled from their corresponding predictive distributions. We monitored the chains to ensure convergence for each model. We set σm,R = σg,R = 0.2 and σm,C = σg,C = 0.5, which achieved rejection rates of roughly 0.3 when updating elements of both KR or KC. Furthermore, the average acceptance rate of a reversible jump move for the column graph GC was around 0.22 for both GGM-U and GGM-S.
To assess the out-of-sample predictive performance of the four models, we performed a 10-fold cross-validation exercise. The exercise was run by randomly dividing the non-missing counts (those above 25) in ten bins. For each bin j, we used the samples from the other nine bins as data and treated the samples from bin j as missing. In this comparison, the MCMC sampler for each of the four models was run ten times for 160,000 iterations and the first half of the run was discarded as burn-in. In the sequel, we denote the predicted counts for model ℳ ∈ {GGM-U, GGM-S, FULL, MCAR} by . We employ the goodness-of-fit (mean squared error of the posterior predictive mean or MSE) and the variability/penalty (mean predictive error or VAR) terms of Gelfand and Ghosh (1998), that is,
to compare the predictive mean and variance of the four models. We also calculated the ranked probability score (RPS) (discussed by Czado, Gneiting, and Held 2009 in the context of count data), which measures the accuracy of the entire predictive distribution. The results, which are summarized in Table 4, reveal an interesting progression in the out-of-sample predictive performance of the four methods. Under either the MSE, VAR, or the RPS criteria, model MCAR performs considerably worse than model FULL, which in turn is outperformed by both models GGM-U and GGM-S. Also, the effect of the choice of prior on the column graph space appears negligible as GGM-U and GGM-S have roughly the same predictive performance. It is noteworthy to mention that the improvement in the RPS in the sequence of models MCAR, FULL, GGM-U, and GGM-S comes both because of a better prediction of the means as well as because the predictive distributions become sharper—see the article by Gneiting and Raftery (2007) for a discussion of the trade-off between sharpness and calibration in the formation of predictive distributions. The dramatic improvement in predictive performance that results from moving from model MCAR to model FULL is the result of using the G-Wishart distribution to allow greater flexibility in the spatial interactions over the CAR specification suggested by Gelfand and Vounatsou (2003). On the other hand, the additional gain yielded by the use of models GGM-U or GGM-S versus model FULL would seem to come from the increased parsimony of the GGM models.
Table 4.
Ten-fold cross-validation predictive scores in the U.S. cancer mortality example. Model GGM-S is best with respect to fit (MSE) and ranked probability score (RPS), while model GGM-U is best with respect to variability (VAR) of the out-of-sample predicted counts
| Model | MSE | VAR | RPS |
|---|---|---|---|
| GGM-U | 17,379.9 | 23,685.2 | 62.1 |
| GGM-S | 16,979.8 | 24,361.1 | 61.8 |
| FULL | 18,959.6 | 24,530.4 | 63.2 |
| MCAR | 19,211.1 | 47,568.7 | 76.7 |
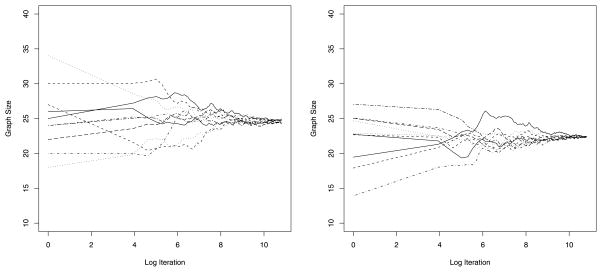
We also conducted an in-sample comparison of the four models. For each model, we ran ten Markov chains for 160,000 iterations and discarded the first half as burn-in based; Figure 2 is indicative of the performance of the MCMC sampler we employed. These runs used the entire dataset (with the exception of counts below 25, which are still treated as missing), rather than the cross-validation datasets. The Supplemental Materials give detailed tables of fitted values and 95% credible intervals for all counts in the dataset obtained using each of the four models. Table 5 summarizes these results by presenting the empirical coverage rates and the average lengths of these in-sample 95% credible intervals for each of the four models. We see that the methods based on the G-Wishart priors for the spatial component (GGM-U, GGM-S, FULL) have very similar coverage probabilities that are close to the nominal 95%. These three models also tend to have relatively short credible intervals, with GGM-U and GGM-S presenting the smallest (and almost identical) values. In contrast, model MCAR returns credible intervals that are too wide, containing 99% of all observed values. As with the out-of-sample exercise, these results suggest that both the additional flexibility in modeling the spatial dependence and the sharper estimates of the dependence across cancer types provided by the matrix-variate GGMs are key to prevent overfitting.
Figure 2.
Convergence plot of the average size of the column graph GC by log iteration for model GGM-U (left panel) and model GGM-S (right panel).
Table 5.
Nominal coverage rates and mean length of the in-sample 95% credible intervals for the U.S. cancer mortality example obtained using the four Poisson multivariate log-linear models
| Model | Coverage rate | Mean length |
|---|---|---|
| GGM-U | 0.960 | 65.464 |
| GGM-S | 0.964 | 65.474 |
| FULL | 0.964 | 65.885 |
| MCAR | 0.990 | 69.239 |
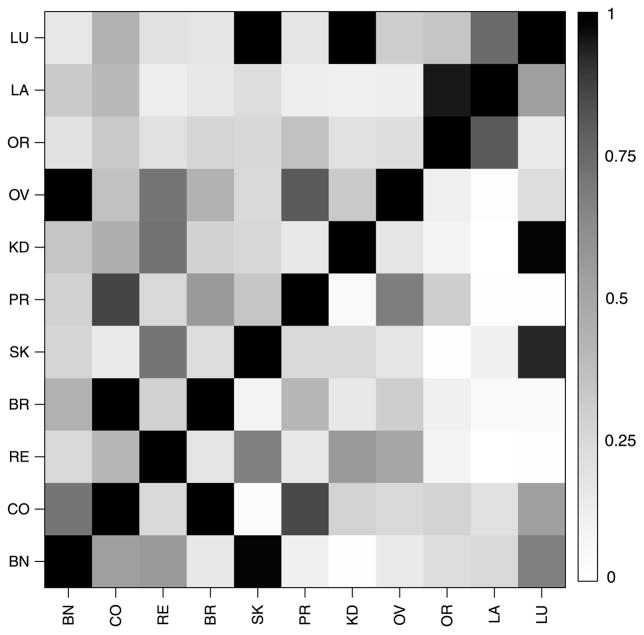
Finally, we compare the estimates of the column graph GC under GGM-U and GGM-S to assess the sensitivity of the model to the prior on graph space. Figure 3 shows a heatmap of the posterior edge inclusion probabilities in the column graph GC, with the lower triangle corresponding to GGM-U and the upper triangle to GGM-S. Overall there is substantial coherence in terms of the effects of the uniform and size-based priors on 𝒢11 on the frequency on which edges are added or deleted from the column graph. From the left panel of Figure 2 we see that the average size of the column graph GC in GGM-U is 24.8 edges while the right panel shows that the average graph size in model GGM-S is 22.5 edges. Both of these numbers are quite close to 27.5—the average graph size under the uniform prior and the size-based prior on 𝒢11. This suggests that there is little sensitivity to the prior in terms of graph selection.
Figure 3.
Edge inclusion probabilities for model GGM-U (lower triangle) and model GGM-S (upper triangle) in the U.S. cancer mortality example. The acronyms used are explained in the Supplementary Materials.
7. DISCUSSION
In this article we have developed and illustrated a surprisingly powerful computational approach for multivariate and matrix-variate GGMs that scales well to problems with a moderate number of variables. Convergence seems to be achieved quickly (usually within the first 5000 iterations) and acceptance rates are within reasonable ranges and can be easily controlled. In the context of the sparse multivariate models for aerial data discussed in Section 5 the running times, although longer than for conventional MCAR models, are still short enough to make routine implementation feasible (e.g., in the cancer surveillance example from Section 6.2, about 22 hours on a dual-core 2.8 GHz computer running Linux). However, computations can still be quite challenging when the number of units in the lattice is very high. We plan to explore more efficient implementations that exploit the sparse structure of the Cholesky decompositions of precision matrices induced by GGMs.
We believe that our application to the construction of spatial models for lattice data makes for particularly appealing illustrations. On one hand, the U.S. cancer mortality example we have considered suggests that the additional flexibility in the spatial correlation structure provided by our approach is necessary to accurately model some spatial datasets. Indeed, our approach allows for “nonstationary” CAR models, where the spatial autocorrelation and spatial smoothing parameters vary spatially. On the other hand, to the best of our knowledge, we are unaware of any approach in the literature to construct and estimate potentially sparse MCAR, particularly under a Bayesian approach. In addition to providing insights into the mechanisms underlying the data generation process, the model offers drastically improved predictive performance and sharper estimation of model parameters.
SUPPLEMENTARY MATERIALS
Code
Computer code to run all routines discussed in this article, as well as the U.S. cancer mortality data. (DLR-archive.tar.gz.zip, GNU zipped tar file)
Results
Theoretical results related to conditional independence relationships in sparse latent Gaussian processes; the description of the MCMC algorithm used to fit the sparse multivariate spatial Poisson count model from Section 6.2; tables describing the U.S. cancer mortality data, the graphical model constructed from the neighborhood structure of the United States, as well as fitted values and 95% credible intervals for the models considered in the article in Section 6.2. (DLR-supplement.pdf, PDF file)
Acknowledgments
The first author was supported in part by the National Science Foundation (DMS 1120255). The second author gratefully acknowledges support by the joint research project “Spatio/Temporal Graphical Models and Applications in Image Analysis” funded by the German Science Foundation (DFG), grant GRK 1653, as well as the MAThematics Centre Heidelberg (MATCH). The work of the third author was supported in part by the National Science Foundation (DMS 0915272) and the National Institutes of Health (R01GM090201-01). The authors thank the editor, the associate editor, and three reviewers for helpful comments.
APPENDIX A: DETAILS OF THE REVERSIBLE JUMP MCMC SAMPLER FOR GGMS
The MCMC algorithm from Section 3 sequentially updates the elements of the precision matrix and the edges of the underlying graph as follows. We denote the current state of the chain by (K[s], G[s]), K[s] ∈ PG[s]. Its next state (K[s+1], G[s+1]), K[s+1] ∈ PG[s+1], is generated by sequentially performing the following two steps. We make use of two strictly positive precision parameters σm and σg that remain fixed at some suitable small values. We assume that the ordering of the variables has been changed according to a permutation υ selected at random from the uniform distribution on ϒp. We denote by (U + D0)−1 = (Q*)TQ* the Cholesky decomposition of (U + D0)−1, where the rows and columns of this matrix have been permuted according to υ.
We denote by the graphs that can be obtained by adding an edge to a graph G ∈ 𝒢p and by the graphs that are obtained by deleting an edge from G. We call the one-edge-way set of graphs the neighborhood of G in 𝒢p. These neighborhoods connect any two graphs in 𝒢p through a sequence of graphs such that two consecutive graphs in this sequence are each others’ neighbors.
Step 1: Resample the graph
We sample a candidate graph G′ ∈ nbdp(G[s]) from the proposal
| (A.1) |
where Uni(A) represents the uniform distribution on the discrete set A. The distribution (A.1) gives an equal probability of proposing to delete an edge from the current graph and of proposing to add an edge to the current graph. We favor (A.1) over the more usual proposal distribution Uni(nbdp(G[s])) that is employed, for example, by Madigan and York (1995). If G[s] contains a very large or a very small number of edges, the probability of proposing a move that adds or, respectively, deletes an edge from G[s] is extremely small when sampling from Uni(nbdp(G[s])), which could lead to poor mixing in the resulting Markov chain.
We assume that the candidate G′ sampled from (A.1) is obtained by adding the edge (i0, j0), i0 < j0, to G[s]. Since we have . We consider the decomposition of the current precision matrix K[s] = (Q*)T ((Ψ[s])T Ψ[s])Q* with (Ψ[s])ν(G[s]) ∈ Mν(G[s]). Since the vertex i0 has one additional neighbor in G′, we have , and ν(G′) = ν(G[s]) ∪ {(i0, j0)}. We define an upper triangular matrix Ψ′ such that for (i, j) ∈ ν(G[s]). We sample and set . The rest of the elements of Ψ′ are determined from (Ψ′)ν(G′) through the completion operation. The value of the free element was set by perturbing the non-free element . The other free elements of Ψ′ and Ψ[s] coincide.
We take K′ = (Q*)T ((Ψ′)T Ψ′)Q*. Since (Ψ′)ν(G′) ∈ Mν(G′), we have K′ ∈ PG′. The dimensionality of the parameter space increases by 1 as we move from (K[s], G[s]) to (K′, G′). Since (Ψ′)ν(G[s]) = (Ψ[s])ν(G[s]), the Jacobian of the transformation from ((Ψ[s])ν(G[s]), γ) to (Ψ′)ν(G′) is equal to 1. Moreover, Ψ[s] and Ψ′ have the same diagonal elements, hence . The Markov chain moves to (K′, G′) with probability where is given by Green (1995)
| (A.2) |
Otherwise the chain stays at (K[s], G[s]).
Next we assume that the candidate G′ is obtained by deleting the edge (i0, j0) from G[s]. We have , and ν(G′) = ν(G[s])\{(i0, j0)}. We define an upper triangular matrix Ψ′ such that for (i, j) ∈ ν(G′). The rest of the elements of Ψ′ are determined through completion. The free element becomes non-free in Ψ′, hence the parameter space decreases by 1 as we move from (Ψ[s])ν(G[s]) to (Ψ′)ν(G′) ∈ Mν(G′). As before, we take K′= (Q*)T ((Ψ′)T Ψ′)Q*. The acceptance probability of the transition from (K[s], G[s]) to (K′, G′) is where
| (A.3) |
We denote by (K[s+1/2], G[s+1]), K[s+1/2] ∈ G[s+1], the state of the chain at the end of this step.
Step 2: Resample the precision matrix
Given the updated graph G[s+1], we update the precision matrix K[s+1/2] = (Q*)T × (Ψs+1/2)T Ψs+1/2Q* by sequentially perturbing the free elements (Ψ[s+1/2])ν(G[s+1]). For each such element, we perform one iteration of the Metropolis–Hastings algorithm from Section 2.2 with δ = n + δ0, D = U + D0, and Q = Q*. The standard deviation of the normal proposals is σm. We denote by K[s+1] ∈ PG[s+1] the precision matrix obtained after all the updates have been performed.
APPENDIX B: DETAILS OF THE REVERSIBLE JUMP MCMC SAMPLER FOR MATRIX-VARIATE GGMS
Our sampling scheme from the joint posterior distribution (15) is composed of the following five steps that explain the transition of the Markov chain from its current state ( , z[s]) to its next state ( , z[s+1]). We use four strictly positive precision parameters σm,R, σm,C, σg,R, and σg,C.
Step 1: Resample the row graph
We denote and . We generate a random permutation υR ∈ ϒpR of the row indices VpR and reorder the row and columns of the matrix according to υR. We determine the Cholesky decomposition . We proceed as described in Step 1 of Appendix A. Given the notations from Appendix A, we take p = pR, , δ0 = δR, D0 = DR, and σg = σg,R. We denote the updated row precision matrix and graph by .
Step 2: Resample the row precision matrix
We denote and . We determine the Cholesky decomposition after permuting the row and columns of according to a random ordering in ϒpR.. The conditional distribution of is G-Wishart . We make the transition from to using Metropolis–Hastings updates described in Section 2.2. Given the notations we used in that section, we take p = pR, , and σm = σm,R.
Step 3: Resample the column graph
We denote and . We sample a candidate column graph from the proposal
| (B.1) |
where 1A is equal to 1 if A is true and is 0 otherwise. The proposal (B.1) gives an equal probability that the candidate graph is obtained by adding or deleting an edge from the current graph.
We assume that is obtained by adding the edge (i0, j0) to . We generate a random permutation of the row indices VpC and reorder the row and columns of the matrix according to υC. The permutation υC is such that υC(1) = 1, hence the (1, 1) element of remains in the same position. We determine the Cholesky decomposition of . We consider the decomposition of the column precision matrix with . We define an upper triangular matrix such that for . We sample and set . The rest of the elements of are determined from through the completion operation (Atay-Kayis and Massam 2005, lemma 2). We consider the candidate column precision matrix
| (B.2) |
We know that must satisfy . The last equality implies , hence . Therefore we have and .
We make the transition from ( ) to ( ) with probability where
| (B.3) |
Next we assume that is obtained by deleting the edge (i0, j0) from . We define an upper triangular matrix such that for . The candidate is obtained from as in Equation (B.2). We make the transition from ( ) to ( ) with probability where
| (B.4) |
We denote the updated column precision matrix and graph by ( ).
Step 4: Resample the column precision matrix
We denote and . We determine the Cholesky decomposition after permuting the row and columns of according to a random ordering in . The conditional distribution of with (KC)11 = 1 is G-Wishart . We make the transition from to using the Metropolis–Hastings updates from Section 2.2. Given the notations we used in that section, we take p = pC, , and σm = σm,C. The constraint (KC)11 = 1 is accommodated as described at the end of Section 2.2.
Step 5: Resample the auxiliary variable
The conditional distribution of z > 0 is
| (B.5) |
Here Gamma(α, β) has density f(x|α, β) ∝ βαxα−1 exp(−βx). We sample z[s+1] from (B.5).
Contributor Information
Adrian Dobra, Email: adobra@uw.edu, Assistant Professor, Departments of Statistics, Biobehavioral Nursing, and Health Systems and the Center for Statistics and the Social Sciences, Box 354322, University of Washington, Seattle, WA 98195.
Alex Lenkoski, Email: alex.lenkoski@uni-heidelberg.de, Postdoctoral Research Fellow, Institut für Angewandte Mathematik, Universität Heidelberg, 69115 Heidelberg, Germany.
Abel Rodriguez, Email: abel@soe.ucsc.edu, Assistant Professor, Department of Applied Mathematics and Statistics, University of California, Santa Cruz, CA 95064.
References
- Armstrong H. PhD thesis. The University of New South Wales; 2005. Bayesian Estimation of Decomposable Gaussian Graphical Models. [Google Scholar]
- Armstrong H, Carter CK, Wong KF, Kohn R. Bayesian Covariance Matrix Estimation Using a Mixture of Decomposable Graphical Models. Statistics and Computing. 2009;19:303–316. [Google Scholar]
- Asci C, Piccioni M. Functionally Compatible Local Characteristics for the Local Specification of Priors in Graphical Models. Scandinavian Journal of Statistics. 2007;34:829–840. [Google Scholar]
- Atay-Kayis A, Massam H. A Monte Carlo Method for Computing the Marginal Likelihood in Nondecomposable Gaussian Graphical Models. Biometrika. 2005;92:317–335. [Google Scholar]
- Banerjee O, El Ghaoui L, D’Aspremont A. Model Selection Through Sparse Maximum Likelihood Estimation for Multivariate Gaussian or Binary Data. Journal of Machine Learning Research. 2008;9:485–516. [Google Scholar]
- Banerjee S, Carlin BP, Gelfand AE. Hierarchical Modeling and Analysis for Spatial Data. Boca Raton, FL: Chapman & Hall/CRC; 2004. [Google Scholar]
- Besag J. Spatial Interaction and the Statistical Analysis of Lattice Systems” (with discussion) Journal of the Royal Statistical Society, Ser B. 1974;36:192–236. [Google Scholar]
- Besag J. A Candidate’s Formula: A Curious Result in Bayesian Prediction. Biometrika. 1989;76:183. [Google Scholar]
- Besag J, Kooperberg C. On Conditional and Intrinsic Autoregressions. Biometrika. 1995;82:733–746. [Google Scholar]
- Brook D. On the Distinction Between the Conditional Probability and the Joint Probability Approaches in the Specification of the Nearest Neighbour Systems. Biometrika. 1964;51:481–489. [Google Scholar]
- Brooks SP, Giudici P, Roberts GO. Efficient Construction of Reversible Jump Markov Chain Monte Carlo Proposals Distributions. Journal of the Royal Statistical Society, Ser B. 2003;65:3–55. [Google Scholar]
- Carvalho CM, Scott JG. Objective Bayesian Model Selection in Gaussian Graphical Models. Biometrika. 2009;96:1–16. [Google Scholar]
- Carvalho CM, Massam H, West M. Simulation of Hyper-Inverse Wishart Distributions in Graphical Models. Biometrika. 2007;94:647–659. [Google Scholar]
- Chib S. Marginal Likelihood From the Gibbs Output. Journal of the American Statistical Association. 1990;90:1313–1321. [Google Scholar]
- Cressie NAC. Statistics for Spatial Data. New York: Wiley; 1973. [Google Scholar]
- Cressie NAC, Chan NH. Spatial Modeling of Regional Variables. Journal of American Statistical Association. 1989;84:393–401. [Google Scholar]
- Czado C, Gneiting T, Held L. Predictive Model Assessment for Count Data. Biometrics. 2009;65:1254–1261. doi: 10.1111/j.1541-0420.2009.01191.x. [DOI] [PubMed] [Google Scholar]
- Dawid AP, Lauritzen SL. Hyper Markov Laws in the Statistical Analysis of Decomposable Graphical Models. The Annals of Statistics. 1993;21:1272–1317. [Google Scholar]
- Dellaportas P, Giudici P, Roberts G. Bayesian Inference for Nondecomposable Graphical Gaussian Models. Sankhyā: The Indian Journal of Statistics. 2003;65:43–55. [Google Scholar]
- Dempster AP. Covariance Selection. Biometrics. 1972;28:157–175. [Google Scholar]
- Diaconnis P, Ylvisaker D. Conjugate Priors for Exponential Families. The Annals of Statistics. 1979;7:269–281. [Google Scholar]
- Dobra A, Hans C, Jones B, Nevins JR, Yao G, West M. Sparse Graphical Models for Exploring Gene Expression Data. Journal of Multivariate Analysis. 2004;90:196–212. [Google Scholar]
- Drton M, Perlman MD. A SINful Approach to Gaussian Graphical Model Selection. Journal of Statistical Planning and Inference. 2008;138:1179–1200. [Google Scholar]
- Elliott P, Wakefield J, Best N, Briggs D. Spatial Epidemiology: Methods and Applications. New York: Oxford University Press; 2001. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Sparse Inverse Covariance Estimation With the Graphical Lasso. Biostatistics. 2008;9:432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand AE, Ghosh SK. Model Choice: A Minimum Posterior Predictive Loss Approach. Biometrika. 1998;85:1–11. [Google Scholar]
- Gelfand AE, Vounatsou P. Proper Multivariate Conditional Autoregressive Models for Spatial Data Analysis. Biostatistics. 2003;4:11–25. doi: 10.1093/biostatistics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Gelfand AE, Diggle PJ, Fuentes M, Guttorp P. Handbook of Spatial Statistics. Boca Raton, FL: Chapman & Hall/CRC; 2010. [Google Scholar]
- Ghosh K, Tiwari RC. Prediction of U.S. Cancer Mortality Counts Using Semiparametric Bayesian Techniques. Journal of the American Statistical Association. 2007;102:7–15. [Google Scholar]
- Ghosh K, Ghosh P, Tiwari RC. Comment on “The Nested Dirichlet Process,” by A. Rodriguez, D. B. Dunson, and A. E. Gelfand. Journal of the American Statistical Association. 2008;103(483):1147–1149. [Google Scholar]
- Ghosh K, Tiwari RC, Feuer EJ, Cronin K, Jemal A. Predicting U.S. Cancer Mortality Counts Using State Space Models. In: Khattree R, Naik DN, editors. Computational Methods in Biomedical Research. Boca Raton, FL: Chapman & Hall/CRC; 2007. pp. 131–151. [Google Scholar]
- Giudici P, Green PJ. Decomposable Graphical Gaussian Model Determination. Biometrika. 1999;86:785–801. [Google Scholar]
- Gneiting T, Raftery AE. Strictly Proper Scoring Rules, Prediction and Estimation. Journal of the American Statistical Association. 2007;102:359–378. [Google Scholar]
- Green PJ. Reversible Jump Markov Chain Monte Carlo Computation and Bayesian Model Determination. Biometrika. 1995;82:711–732. [Google Scholar]
- Gupta AK, Nagar DK. Matrix Variate Distributions. Monographs and Surveys in Pure and Applied Mathematics. Vol. 104. London: Chapman & Hall/CRC; 2000. [Google Scholar]
- Jones B, Carvalho C, Dobra A, Hans C, Carter C, West M. Experiments in Stochastic Computation for High-Dimensional Graphical Models. Statistical Science. 2005;20:388–400. [Google Scholar]
- Lauritzen SL. Graphical Models. New York: Oxford University Press; 1996. [Google Scholar]
- Lawson AB. Bayesian Disease Mapping: Hierarchical Modeling in Spatial Epidemiology. Boca Raton, FL: Chapman & Hall/CRC; 2009. [Google Scholar]
- Lenkoski A, Dobra A. Computational Aspects Related to Inference in Gaussian Graphical Models With the G-Wishart Prior. Journal of Computational and Graphical Statistics. 2011;20:140–157. [Google Scholar]
- Letac G, Massam H. Wishart Distributions for Decomposable Graphs. The Annals of Statistics. 2007;35:1278–1323. [Google Scholar]
- Madigan D, York J. Bayesian Graphical Models for Discrete Data. International Statistical Review. 1995;63:215–232. [Google Scholar]
- Mardia KV. Multi-Dimensional Multivariate Gaussian Markov Random Fields With Application to Image Processing. Journal of Multivariate Analysis. 1988;24:265–284. [Google Scholar]
- Meinshausen N, Bühlmann P. High Dimensional Graphs and Variable Selection With the Lasso. The Annals of Statistics. 2006;34:1436–1462. [Google Scholar]
- Mengersen KL, Tweedie RL. Rates of Convergence of the Hastings and Metropolis Algorithms. The Annals of Statistics. 1996;24:101–121. [Google Scholar]
- Mitsakakis N, Massam H, Escobar MD. A Metropolis–Hastings Based Method for Sampling From the G-Wishart Distribution in Gaussian Graphical Models. Electronic Journal of Statistics. 2011;5:18–30. [Google Scholar]
- Moghaddam B, Marlin B, Khan E, Murphy K. Accelerating Bayesian Structural Inference for Non-Decomposable Gaussian Graphical Models. In: Bengio Y, Schuurmans D, Lafferty J, Williams CKI, Culotta A, editors. Advances in Neural Information Processing Systems. Vol. 22. San Mateo, CA: Morgan Kaufmann; 2009. pp. 1285–1293. [Google Scholar]
- Muirhead RJ. Aspects of Multivariate Statistical Theory. New York: Wiley; 2005. [Google Scholar]
- Piccioni M. Independence Structure of Natural Conjugate Densities to Exponential Families and the Gibbs Sampler. Scandinavian Journal of Statistics. 2000;27:111–127. [Google Scholar]
- Ravikumar P, Wainwright MJ, Lafferty JD. High-Dimensional Ising Model Selection Using l1-Regularized Logistic Regression. The Annals of Statistics. 2010;38:1287–1319. [Google Scholar]
- Robert C, Casella G. Monte Carlo Statistical Methods. 2. New York: Springer-Verlag; 2004. [Google Scholar]
- Roverato A. Hyper Inverse Wishart Distribution for Non-Decomposable Graphs and Its Application to Bayesian Inference for Gaussian Graphical Models. Scandinavian Journal of Statistics. 2002;29:391–411. [Google Scholar]
- Rue H, Held L. Gaussian Markov Random Fields: Theory and Applications. Boca Raton, FL: Chapman & Hall/CRC; 2005. [Google Scholar]
- Scott JG, Berger JO. An Exploration of Aspects of Bayesian Multiple Testing. Journal of Statistical Planning and Inference. 2006;136:2144–2162. [Google Scholar]
- Scott JG, Carvalho CM. Feature-Inclusion Stochastic Search for Gaussian Graphical Models. Journal of Computational and Graphical Statistics. 2008;17:790–808. [Google Scholar]
- Speed TP, Kiiveri HT. Gaussian Markov Distributions Over Finite Graphs. The Annals of Statistics. 1986;14:138–150. [Google Scholar]
- Sun D, Tsutakawa RK, Kim H, He Z. Bayesian Analysis of Mortality Rates With Disease Maps. Statistics and Medicine. 2000;19:2015–2035. doi: 10.1002/1097-0258(20000815)19:15<2015::aid-sim422>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Tarjan RE. Decomposition by Clique Separators. Discrete Mathematics. 1985;55:221–232. [Google Scholar]
- Tiwari RC, Ghosh K, Jemal A, Hachey M, Ward E, Thun MJ, Feuer EJ. A New Method for Predicting U.S., and State-level Cancer Mortality Counts for the Current Calendar Year. CA: A Cancer Journal for Clinicians. 2004;54(1):30–40. doi: 10.3322/canjclin.54.1.30. [DOI] [PubMed] [Google Scholar]
- Wang H, Carvalho CM. Simulation of Hyper-Inverse Wishart Distributions for Non-Decomposable Graphs. Electronic Journal of Statistics. 2010;4:1470–1475. [Google Scholar]
- Wang H, West M. Bayesian Analysis of Matrix Normal Graphical Models. Biometrika. 2009;96:821–834. doi: 10.1093/biomet/asp049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F, Carter CK, Kohn R. Efficient Estimation of Covariance Selection Models. Biometrika. 2003;90:809–830. [Google Scholar]
- Yuan M, Lin Y. Model Selection and Estimation in the Gaussian Graphical Model. Biometrika. 2007;94:19–35. [Google Scholar]