Abstract
Background
Identification of genetic changes in CNS tumors is important for the appropriate clinical management of patients. Our objective was to develop a next-generation sequencing (NGS) assay for simultaneously detecting the various types of genetic alterations characteristic for adult and pediatric CNS tumors that can be applied to small brain biopsies.
Methods
We report an amplification-based targeted NGS assay (GlioSeq) that analyzes 30 genes for single nucleotide variants (SNVs) and indels, 24 genes for copy number variations (CNVs), and 14 types of structural alterations in BRAF, EGFR, and FGFR3 genes in a single workflow. GlioSeq performance was evaluated in 54 adult and pediatric CNS tumors, and the results were compared with fluorescence in-situ hybridization, Sanger sequencing, and reverse transcription PCR.
Results
GlioSeq correctly identified 71/71 (100%) genetic alterations known to be present by conventional techniques, including 56 SNVs/indels, 9 CNVs, 3 EGFRvIII, and 3 KIAA1549-BRAF fusions. Only 20 ng of DNA and 10 ng of RNA were required for successful sequencing of 100% frozen and 96% formalin-fixed, paraffin-embedded tissue specimens. The assay sensitivity was 3%–5% of mutant alleles for SNVs and 1%–5% for gene fusions. The most commonly detected alterations were IDH1, TP53, TERT, ATRX. CDKN2A, and PTEN in high-grade gliomas, followed by BRAF fusions in low-grade gliomas and H3F3A mutations in pediatric gliomas.
Conclusions
GlioSeq NGS assay offers accurate and sensitive detection of a wide range of genetic alterations in a single workflow. It allows rapid and cost-effective profiling of brain tumor specimens and thus provides valuable information for patient management.
Keywords: CNS tumors, gene fusions, mutations, next generation sequencing, paraffin
The American Cancer Society estimates that 22 850 new primary adult and pediatric CNS tumors will be diagnosed in 2015 in the United States alone. Survival rates vary widely depending on the tumor type, and it is projected that as many as 15 320 people will die of brain tumor in 2015. In children, these tumors are the second most common cancers after leukemia and account for about 1 out of 5 childhood cancers.
Among primary adult and pediatric CNS tumors, diffuse gliomas are the largest and most diverse group. They usually arise in the cerebral hemispheres and are defined by their widely infiltrative properties and their tendency for biological progression. According to the World Health Organization (WHO) criteria, gliomas are classified as astrocytomas, oligodendrogliomas, or oligoastrocytomas grades II to III and as grade IV glioblastoma multiforme (GBM), which is the most aggressive astrocytic tumor and has a dismal prognosis.1 Less infiltrative gliomas of children and young adults include WHO grade I pilocytic astrocytomas and gangliogliomas. Other major classes of CNS tumors include cerebellar medulloblastomas and extra-axial meningiomas.
Historically, the diagnosis of CNS tumor has been based primarily on histopathologic features. However, patients with morphologically identical tumors may experience different clinical outcomes and responses to treatment because the underlying genetic characteristics of the tumors differ. Over the last decade, our knowledge of the molecular composition of CNS tumors has increased dramatically. A number of genetic alterations have been discovered including IDH, TP53, and ATRX, which are most characteristic of grades II and III infiltrating astrocytomas of adults and secondary GBMs.2–4 In contrast, primary GBMs typically lack IDH mutations but demonstrate EGFR amplification as well as EGFRvIII and TERT promoter mutations. Like astrocytomas, oligodendrogliomas have IDH mutations but with 1p/19q co-deletion and mutations in CIC, FUBP1, and TERT.5,6 Pediatric gliomas are unique and feature mutations in H3F3A, ATRX, and BRAF, but IDH mutations are rare unless the patient is an adolescent.7,8 Circumscribed lower-grade gliomas such as pilocytic astrocytoma, pilomyxoid astrocytoma, ganglioglioma, and pleomorphic xanthoastrocytoma often harbor mutations or gene fusions in BRAF.9 Medulloblastomas have been recently divided into 4 groups (WNT [wingless], SHH [sonic hedgehog], group 3, and group 4) based on molecular profiling and clinical outcome.10–13 Finally, recurrent mutations in KLF4, AKT1, and SMO genes are often present in NF2-negative sporadic meningiomas.14 These and other genetic alterations can serve as diagnostic, prognostic, and predictive biomarkers for tumor classification, patient risk stratification, and targeted therapies. Therefore, broad molecular profiling is important in CNS tumors and improves their clinical management.15,16
Next-generation sequencing (NGS) technology fulfills this need as it enables analysis of large areas of the human genome in a massively paralleled manner. NGS analysis can be developed for sequencing at various scales (whole genome, whole exome, or targeted sequencing). Sequencing of whole genome/exome is expensive, time-consuming, and difficult to perform on small brain biopsy specimens because of the amount of DNA needed. Targeted panel-based NGS is designed to sequence selected cancer genes or genetic regions and can be used for detecting a number of genetic alterations including point mutations, insertions and deletions, copy number changes, and gene fusions.17 This methodology allows for a fast turnaround time and is cost-effective. Several targeted NGS panels are available commercially, although none of them is designed to specifically target the important alterations in CNS tumors.
Overall, an NGS assay for the routine workup of CNS tumors must address several specific goals. (i) The assay should include a broad set of genes and mutational hotspots occurring in adult and pediatric brain tumors that can be tested for single nucleotide variations (SNVs), insertions and deletions, and more complex genetic alterations such as gene fusions and copy number variations (CNVs). (ii) The analysis should be reliable in formalin-fixed, paraffin-embedded (FFPE) tissues; otherwise, its utility in routine patient care would be greatly diminished. (iii) The assay should require a small input of nucleic acids to successfully analyze small stereotactic brain biopsies because more extensive tumor resection is not possible in many patients.
In this study, we describe the design and validation of a custom amplification-based targeted NGS assay (GlioSeq) for simultaneous detection of various types of genetic alterations in adult and pediatric CNS tumors in a variety of clinical samples.
Material and Methods
Brain Tissue Samples
Snap-frozen tissue and FFPE tissue from surgically removed CNS tumors were collected at the Department of Pathology, University of Pittsburgh Medical Center, and the CNS Tumor Bank, University of Kentucky, following approval by their institutional review boards. All tumors were classified according to WHO diagnostic criteria. A total of 54 CNS tumors collected in 2010–2015 were analyzed including 4 pilocytic astrocytomas, 2 pilomyxoid astrocytomas), 6 grade II astrocytomas, 4 anaplastic astrocytomas, 17 glioblastomas (15 adults and 2 pediatric), 5 grade II oligodendrogliomas, 4 anaplastic oligodendrogliomas, 3 medulloblastomas, 4 meningiomas, 1 grade II ependymoma, 1 gliosarcoma, 1 malignant neurocytoma, 1 low-grade glioma not otherwise classifiable, and 1 high-grade neoplasm not otherwise classifiable. Twenty-six analyzed specimens were snap-frozen tissues, and 28 specimens were FFPE tissues.
Nucleic Acids Isolation
For FFPE tissues, tumor-rich areas (>50% of neoplastic cells) were microdissected. Initially, board-certified pathologists selected the best area for microdissection using hematoxylin & eosin-stained (H&E) slides, and the tissue was then microdissected from 3–6 4 µm unstained histologic sections under stereomicroscopic visualization with an Olympus SZ61 microscope (Olympus). For all frozen tumors, matching FFPE tissue was used to verify tumor diagnosis and cellularity. Only specimens with at least 50% cellularity were included (Supplementary material, Table S3). Total nucleic acids were isolated from each target with the DNeasy Blood and Tissue Kit on the automated QIAcube (Qiagen) instrument according to the manufacturer's instructions, albeit with some modifications (ie, RNAse treatment reagent was omitted from the protocol). From frozen tissue specimens, total nucleic acid extraction was performed using the MagNA Lyser instrument (Roche) according to the manufacturer's instructions and the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche) on Compact MagNaPure (Roche). Extracted DNA and RNA were quantitated on the Qubit 2.0 Fluorometer using the dsDNA HS Assay Kit and the RNA HS Assay Kit (Invitrogen).
Targeted Next-generation Sequencing
For GlioSeq-targeted NGS analysis, the custom primer pools were designed using Life Technologies’ design tool. For each sample, libraries were generated to test separately for (i) point mutations, indels, and copy number changes (GlioSeq-DNA) and (ii) structural alterations and gene expression (GlioSeq-RNA).
The GlioSeq-DNA custom library was designed to include 396 total primer pairs in 2 primer pools for amplification and sequencing of genomic regions of interest. In more detail, 10–20 ng of DNA were amplified by PCR using the premixed primer pools and Ion AmpliSeq HiFi Master Mix (Ion AmpliSeq Library Kit 2.0). Amplicons were treated with FuPa reagent to partially digest primer sequences and phosphorylate the amplicons. The amplicons were then ligated to adapters with the addition of barcodes from the Ion Xpress Barcode Adapters 1–96 Kit according to manufacturer's instructions (Life Technologies). After ligation, the amplicons underwent nick-translation and additional library amplification by PCR to complete the linkage between adapters and amplicons. Library concentration and amplicon size were determined using the Tapestation 2200 (Agilent). Equal molar volumes of libraries from the 2 primer pools were combined, and multiplexed barcoded libraries were enriched by clonal amplification using emulsion PCR on Ion Sphere particles (Ion PGMTemplate OT2 200 kit or Ion PI OT2 200 kit v3) and loaded on an Ion 318 Chip or Ion P1 Chip (Life Technologies). Massively parallel sequencing was carried out on a Personal Genome Machine Sequencer (Ion Torrent) or Ion Proton according to the manufacturer's instructions (Life Technologies).
The GlioSeq-RNA custom library was designed to generate an 18-primer pair pool to detect BRAF and FGFR3 gene fusions, EGFRvIII structural alteration, and expression of housekeeping genes (GUSB, PGK, HPRT1). Briefly, 5–10 ng of RNA derived from either FFPE or snap-frozen samples were reverse-transcribed into cDNA and amplified using the custom-designed primer pool. Amplicons were partially digested, phosphorylated, ligated to Ion adapters and Ion Xpress barcodes, and bead-purified. Region analysis was done using the High Sensitivity D1000 tapes and reagents and the TapeStation 2200 (Agilent). Barcoded libraries were diluted and mixed for template preparation using the Ion PGM Template OT2 200 kit or Ion PI OT2 200 kit v3 (Life Technologies). Then, barcoded samples were sequenced on the Ion Torrent PGM or Ion Proton (Life Technologies).
The validation of the GlioSeq was performed as recommended by the College of American Pathologists18 in order to establish analytical accuracy of the assay. The analytical sensitivity or limits of detection for the GlioSeq NGS panel was determined using serial dilutions of (i) brain tumor DNA carrying IDH1 (p.R132H), TP53 (p.A138V), PIK3CA (p.I391M), and MET (p.A374T) mutations diluted in DNA from normal tissue and (ii) brain tumor RNA expressing an EGFRvIII structural alteration diluted in RNA from normal tissue.
Next-generation Sequencing Data Analysis
For GlioSeq-DNA sequencing, the raw signal data were analyzed using Torrent Suite (version 4.0.1) to generate (.bam) files after signal processing, base-calling adapter trimming, and alignment to the reference human genome (hg19). Variants were called with Torrent Suite Variant Caller and were further analyzed using an internally created software suite based on ANNOVAR,19 which annotates each variant with SIFT20 and Polyphen-221 scores predicting its effect on the protein function, and with existing databases including dbSNP22 and COSMIC.23 Analysis of copy number alterations and establishment of the copy number ratio (CNR) was performed as previously reported.24 For GlioSeq-RNA sequencing, a custom bioinformatics pipeline was used to detect and quantify the fusion transcripts and expression levels of specific gene regions from the sequencing data generated by Ion Torrent instruments. First, raw data in FASTQ format were aligned to a custom reference genome using TMAP (https://github.com/iontorrent/TMAP; accessed June 26, 2014) after adapter sequences were removed by CutAdapt.25 The custom reference genome contains sequences of the fusion transcripts, normal transcripts of the genes involved in the fusions, gene regions for differential expression analysis, and the entire hg19. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; accessed June 26, 2014) was used for quality control of the raw FASTQ data, and alignStats and SAMStat26 were used to examine the quality of alignment. Visual inspection of the aligned reads for the fusions was performed in Integrative Genomics Viewer (IGV, Broad Institute)27 using the custom reference sequences above.
Confirmation of mutations was performed by Sanger sequencing analysis. Sequencing of the TERT promoter region was performed as previously described28 using AmpliTaq Gold 360 Master Mix and GC Enhancer (Life Technologies). EGFR copy number changes and BRAF fusions were confirmed by fluorescence in-situ hybridization (FISH) and EGFRvIII by reverse transcription (RT-)PCR.29,30
Results
GlioSeq Next-generation Sequencing Panel Design and Validation
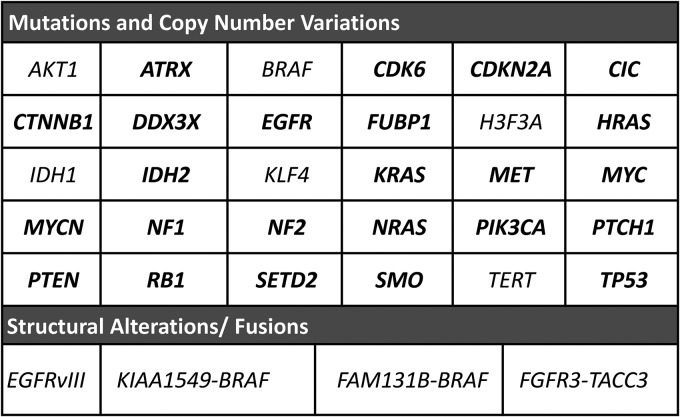
The aim of this study was to create a NGS assay that would allow simultaneous detection of the major mutations, gene fusions, and gene copy number changes known to occur in CNS tumors, including adult and pediatric gliomas, medulloblastomas, meningiomas, and others. To achieve this goal, we identified 30 genes with genetic alterations repeatedly found in CNS tumors (Fig. 1) and designed custom DNA primer pools to generate libraries and sequence more than 1360 CNS tumor-related hot spots (>13 000 all cancer hot spots listed in COSMIC database v.68) and for copy number alterations in 24 genes (Fig. 1). In addition, we designed an RNA custom primer pool to detect BRAF and FGFR3 fusions, EGFRvIII structural alteration (Fig. 1), and expression of 3 housekeeping genes (GUSB, HPRT1, PGK) for evaluation of nucleic acid integrity.
Fig. 1.
GlioSeq next-generation sequencing (NGS) panel design. It includes 30 genes (>1360 CNS tumor- related hot spots) analyzed for point mutations and small insertions and deletions, 24 genes (in bold) for copy number changes, 16 subtypes of BRAF and FGFR3 gene fusions, and EGFRvIII structural alterations. In addition, it tests for the expression of 3 housekeeping genes (GUSB, HPRT1, PGK) for evaluation of RNA integrity.
The initial performance of the GlioSeq NGS panel was determined using HT-29, SW620, and HT-1080 cell lines with known genetic alterations in the BRAF (p.V600E), KRAS (p.G12V), NRAS (p.Q61K), TP53 (p.R273H), IDH1 (p.R132C), and PIK3CA (p.P449T) genes, normal specimens (7 blood, 6 normal FFPE tissue, and a HapMap cell line GM12878). Very small amounts of DNA (20 ng) and RNA (10 ng) were sufficient for the successful preparation of DNA and RNA libraries in 100% of specimens. GlioSeq was able to correctly detect all pathogenic mutations in cell lines, whereas normal samples were negative for mutations. The analytical sensitivity or limits of detection for the GlioSeq NGS panel was 3%–5% allele frequency for SNVs and 1%–5% frequency for structural alterations. A minimum of 50 reads spanning the fusion break point or 0.1% of mapped reads was sufficient for fusion detection.
GlioSeq Next-generation Sequencing Analysis of CNS Tumors
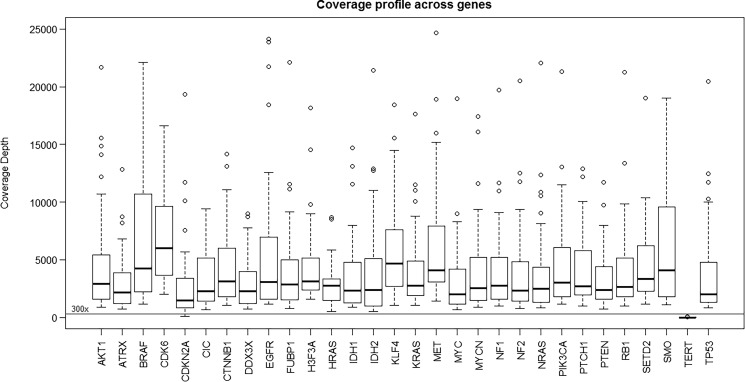
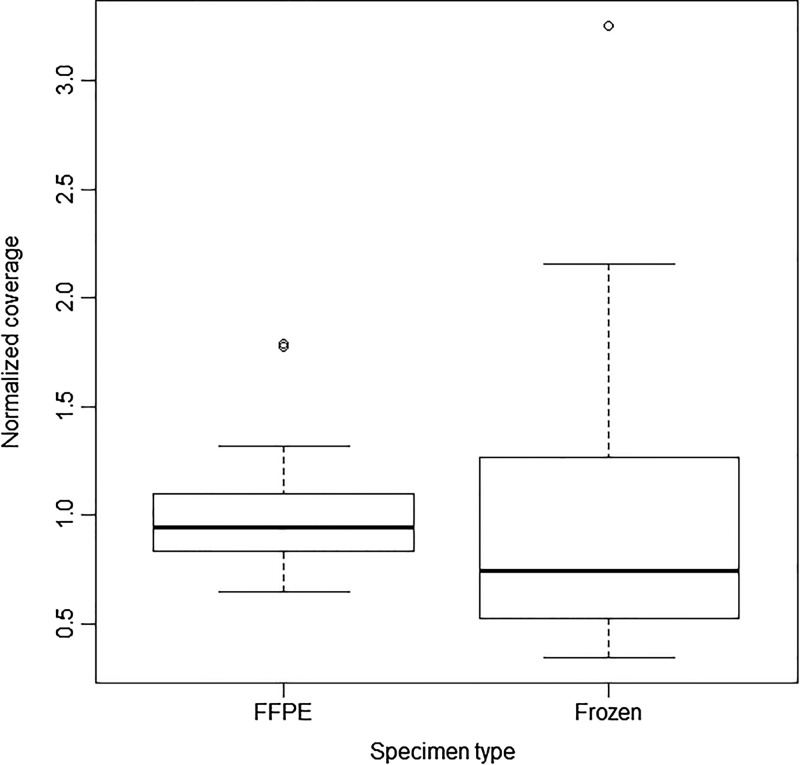
Next, the GlioSeq performance was evaluated in 54 CNS tumor specimens collected in 2012–2015, including 28 FFPE and 26 snap-frozen tissues. DNA library preparation and sequencing were successful in 54 of 54 (100%) specimens tested, yielding an average of 137 000 000 AQ20 bases, 115 bp mean read length, 1 303 502 mean mapped reads per specimen, and 2972 mean sequencing depth per amplicon. The coverage depth values for each sequenced gene are shown in Fig. 2. The established minimal sequencing depth of 300x was achieved for all genes with the exception for TERT promoter region, which had variable coverage and was therefore subsequently analyzed by Sanger sequencing. The assay worked equally well on DNA isolated from FFPE and frozen tissue specimens and demonstrated comparable depth of coverage for sequenced amplicons (Fig. 3). Fifty-three tumor specimens had sufficient amounts of RNA for GlioSeq analysis. RNA library preparation and sequencing were successful in 51 of 53 (96.2%) tested tumors. RNA library failed for one tumor, and another specimen demonstrated unacceptable expression of housekeeping genes (<15 000 mapped reads). Similar to DNA, the sequencing performance was similar for RNA isolated from FFPE and frozen tissues and demonstrated on average 64 403 (97%) and 109 401 (98%) reads mapped to housekeeping genes for FFPE and frozen tissue, respectively.
Fig. 2.
Boxplot demonstrating the range of coverage depth values for known (hot spot) variants detected in 30 genes in GlioSeq next-generation sequencing panel. Horizontal line indicates the threshold for minimum coverage (300x) for reporting sequence variants.
Fig. 3.
Boxplot comparing the normalized depth of coverage for sequenced amplicons between formalin-fixed paraffin embedded (FFPE) and frozen tissue specimens.
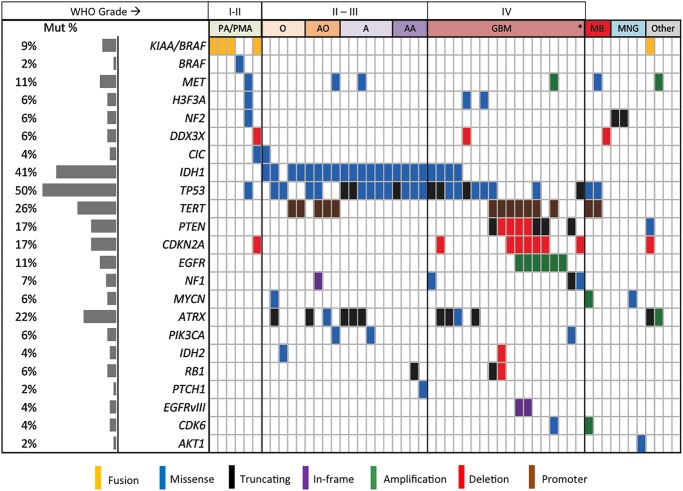
GlioSeq analysis identified a variety of genetic alterations in tested CNS tumors including SNVs, insertions, deletions, gene fusions, and CNVs (Fig. 4 and Supplementary material, Table S1). The most commonly mutated genes across all sample types were IDH1, TP53, TERT, and ATRX. Only 2 of 54 (3.7%) of all tumors were negative for all genetic alterations included in the panel, both demonstrating unusual morphologic features and diagnosed as low-grade glioma not otherwise classifiable and high-grade neoplasm not otherwise classifiable. GlioSeq correctly detected 71 of 71 (100%) genetic alterations, which had been previously identified by conventional methods including 56 SNVs by Sanger sequencing analysis, 9 CNVs by FISH (6 EGFR amplifications, 2 1p/19q co-deletions, 1 CDKN2A deletion), 3 EGFRvIII structural alterations by RT-PCR, and 3 KIAA1549-BRAF fusions detected by FISH.
Fig. 4.
Genomic landscape of 54 CNS tumors profiled using the GlioSeq panel. Left pane indicates the mutational rate for each target across all the samples. Each vertical bar represents an individual case. A, astrocytoma (WHO grade II); AA, anaplastic astrocytoma (WHO grade III); AO, anaplastic oligodendroglioma (WHO grade III); GBM, glioblastoma multiforme (WHO grade IV); MB, medulloblastoma (WHO grade IV); MNG, meningioma; O, oligodendroglioma (WHO grade II); PA, pilocytic astrocytoma (WHO grade I); PMA, pilomyxoid astrocytoma (WHO grade II; *, gliosarcoma.
Detected genomic alterations were appropriate for each tested tumor class and subtype. Five of the 6 grade I-II PAs and PMAs harbored BRAF alterations (one with a BRAF p.V600E point mutation and the other 4 with KIAA1549-BRAF fusions) (Fig. 4). The KIAA1549-BRAF fusions showed different fusion subtypes with the most common being KIAA1549 exon 16 and BRAF exon 9 transcripts. In addition to the BRAF fusion, one PMA showed presence of CIC mutation and CDKN2A copy number loss. In contrast, one tumor from a 44 year-old female (a septal lesion initially diagnosed as a grade I PA) contained mutations in the TP53, H3F3A, and MET genes. Within one year, this case recurred as a high-grade astrocytoma showing necrosis, microvascular proliferation, and mitoses.
All 10 grade II-III astrocytomas were positive for IDH1 (8 R132H, 1 R132G, 1 R132S) and TP53 mutations (Fig. 4). In addition, one grade II astrocytoma harbored a PIK3CA E545K mutation. No TERT mutations were detected in this group, and 3 tumors were positive for ATRX mutations. Out of 15 adult GBMs, 6 (40%) were positive for EGFR amplification, and 3 of 6 (50%) amplified cases showed presence of EGFRvIII structural alteration (Fig. 4). The amplification levels were different between GBMs with 2 to 37 CNR. Only highly amplified cases (CNR = 22 to 37) were positive for EGFRvIII. IDH1/2 mutations were detected in combination with mutations in ATRX and TP53 genes in 4 GBMs, suggesting secondary GBM origin, and 7 of 11 (64%) IDH wild-type GBMs had TERT mutations. Other detected alterations were 2 EGFR point mutations in EGFR-amplified tumors, 4 PTEN, 1 PIK3CA, 6 CDKN2A mutations, and 6 PTEN gene copy number losses. Two pediatric gliomas were positive for H3F3A K28M and G35R mutations in combination with TP53 mutations (Fig. 4 and Supplementary material, Table S1). In addition, we performed GlioSeq NGS analysis on 12 pediatric brain tumor specimens (1 Juvenile Pilocytic Astrocytoma, 3 gangliogliomas, 3 low-grade gliomas, 1 anaplastic astrocytoma, 3 GBMs, and 1 medulloblastoma). All grade I tumors were positive for BRAF V600E. One low-grade glioma had IDH1 and PIK3CA mutations, and 3 high-grade gliomas showed mutations in H3F3A in combination with ATRX and TP53. Interestingly, GBM from the patient with history of bilateral retinoblastoma showed a different genomic profile with deletion in the RB1 gene and TP53 and TERT mutations (Supplementary material, Table S1).
All 9 grade II-III oligodendrogliomas demonstrated the presence of IDH1/2 mutations including 8 IDH1 R132H and one IDH2 R172M mutation. Five (56%) had TERT mutations. In contrast, 3 of the 4 oligodendrogliomas lacking TERT mutations showed mutant TP53 with or without ATRX mutations.
Three tested medulloblastomas revealed copy number increases of the MYCN and CDK6 genes in one tumor and copy number loss for the DDX3X gene in another tumor (Fig. 4). In addition, 2 medulloblastomas were positive for TERT in combination with TP53 mutations. Meningiomas showed presence of AKT1 E17K in one case and NF2 mutations in 2 other cases. A single case of gliosarcoma was positive for TP53 and NF1 mutations and CDKN2A copy number loss, and a case of ependymoma showed copy number gain of MET and ATRX. A case of recurrent disseminated atypical neurocytoma was surprisingly positive for the KIAA1549-BRAF fusion (confirmed by FISH), PTEN and ATRX mutations, and CDKN2A copy number loss.
We compared the GlioSeq cost of reagents with the cost of reagents using conventional techniques (eg, Sanger sequencing, RT-PCR, and single nucleotide polymorphism array) that are needed to detect all types of genetic alterations and determined that conventional methods would cost 15 times more than GlioSeq analysis (Supplementary material, Table S4). It takes 7 days to perform GlioSeq NGS testing (allowing for fast turnaround time) (Supplementary material, Figure S1).
Discussion
In this study, we report the development and validation of the targeted amplification-based NGS panel (GlioSeq) for detecting clinically useful genetic alterations in the most common pediatric and adult brain tumors.
The diagnosis of CNS tumors has historically been performed using light microscopy, and the WHO has long established a series of criteria that improve pathologic classification and prognostic stratification of gliomas, primarily based on histologic features of tumors.1 However, morphologically identical tumors can have different molecular profiles that define their biological behavior and clinical outcome. Therefore, many molecular markers became deeply integrated into the diagnosis of CNS tumors and are now used to guide patient prognostication and treatment. The growth in the number of clinically relevant molecular alterations has greatly reduced the practicality and cost-effectiveness of single-gene assays, and there is a need for high-throughput technology that can rapidly assess a variety of genetic alterations from limited neuropathology specimens. Exome sequencing currently has limitations for routine use in neuro-oncology specimens; these limitations include the need for a large quantity and high-quality DNA (frequently not obtainable from FFPE brain biopsies), the inability to detect gene fusions, high cost, and slow turnaround time. We created a targeted amplification-based NGS panel (GlioSeq) that allows simultaneous analysis of a broad spectrum of genetic alterations including point mutations in 30 CNS tumor-related genes (>1360 hot spots), copy number alterations in 24 genes, and 14 fusion types involving BRAF, FGFR3, and EGFRvIII. A key advantage to this assay is that it requires only a minimal amount of nucleic acids, which can be readily harvested from the majority of FFPE brain tissue biopsies or resected tumor specimens. It also allows for sensitive detection of multiple genetic alterations in a single workflow. GlioSeq can be performed for approximately the same cost as performing Sanger sequencing and copy number analysis of a single gene by conventional methods, thereby eliminating the need for ordering multiple molecular tests.
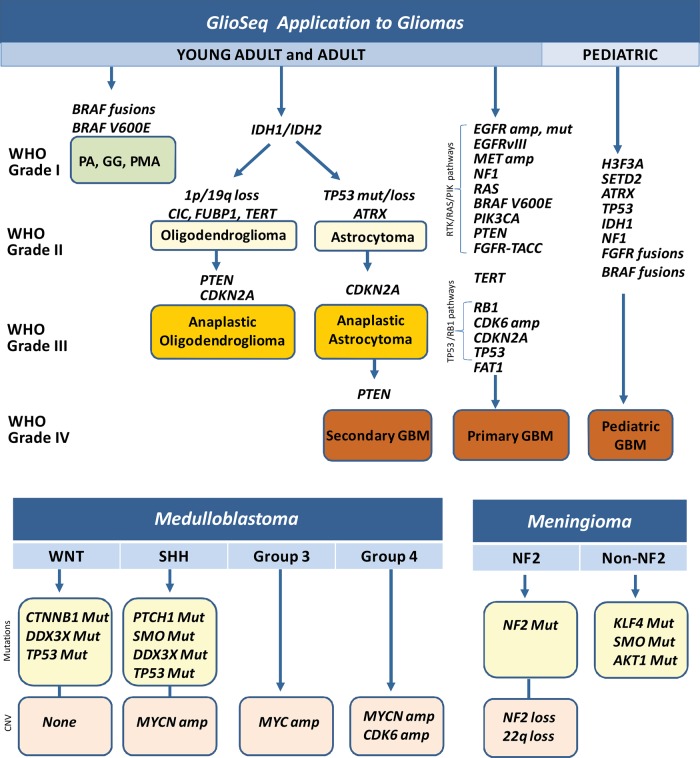
The GlioSeq panel was designed to include genetic alterations that are relevant to different glioma subtypes and grades in both adult and pediatric cohorts (Fig. 5). For example, fusions of BRAF and KIAA1549 or FAM131B, and the BRAF V600E mutation are characteristic for pilocytic astrocytomas.8,31 In children, however, a BRAF alteration with concomitant CDKN2A deletion raises the possibility of transformation into a more aggressive tumor.32,33 Point mutations in IDH1/2 are highly distinctive for diffusely infiltrative grades II and III astrocytomas and oligodendrogliomas, as well as secondary grade IV GBMs.34 It can often be very difficult to differentiate between a noninvasive grade I tumor and a more aggressive, incurable grade II-IV glioma, especially in a small biopsy using light microscopy alone. In fact, the presence of IDH1/2 mutation should preclude the diagnosis of a benign grade I tumor or non-neoplastic reactive astrocytosis.35,36 Somatic mutations in the H3.3-ATRX chromatin remodeling pathway have been identified in addition to TP53 mutations in pediatric gliomas. The GlioSeq assay allows detection of an H3F3A point mutation, which strongly favors an infiltrative glioma over a noninfiltrative tumor in children (Fig. 5).7,8,37,38 Of note, our pediatric tumor subset was small, and conclusions about the GlioSeq performance in pediatric tumors have to be made with some caution.
Fig. 5.
Application of GlioSeq panel for detection of genetic alterations relevant to different subtypes and grades of both adult and pediatric gliomas, medulloblastomas, and meningiomas. GBM, glioblastoma; GG; PA, pilocytic astrocytoma; PMA, pilomyxoid astrocytoma; SHH, sonic hedgehog; WNT, wingless.
Even when the diagnosis of a diffusely infiltrative glioma is unequivocal by light microscopy, it can still be challenging to accurately distinguish between an astrocytoma and an oligodendroglioma. This distinction is important because oligodendrogliomas tend to be less aggressive than their grade-matched astrocytic counterparts, and clinical trials usually make assignments based on tumor subtype. Both types of gliomas contain IDH1/2 mutations, but astrocytomasalso usually have mutations in ATRX, while oligodendrogliomas feature TERT promoter mutations and 1p/19q co-deletion (Fig. 5).6,39 Furthermore, mutations in CIC and FUBP1 are also characteristic of many oligodendrogliomas.40
Several decades of molecular studies have identified important genetic events in human GBMs that lead to dysregulation of 3 critical signaling pathways including the following: (i) receptor tyrosine kinase (RTK)/RAS/PI(3)K pathway via amplification and mutation in EGFR, PIK3CA, RAS, NF1, EGFR, and MET and (ii) TP53 and RB1 pathways via mutations/loss of TP53, CDKN2A, and RB1 genes (Fig. 5).41 In addition, key driver mutations are associated with certain expression subtypes in GBMs, including the IDH1/2 subset of proneural tumors, the classical pathway that is enriched for EGFR amplification and aberrant exon 1-8 junction characteristic of EGFRvIII, and the mesenchymal subtype with NF1 mutations.2 Therefore, analysis of gliomas for multiple molecular markers not only helps establish the correct morphological diagnosis but also highlights the biological differences between morphologically similar tumors and can help with the clinical management of patients. For example, patients with IDH1/CIC/FUBP1 mutations have a significantly longer median overall survival than patients with IDH1/ATRX mutations.5 A recent study by Eckel-Passov et al showed that 3 tumor markers (IDH, TERT, and 1p/19q loss) classify gliomas into 5 groups that have different outcomes.42 Although very few molecular biomarkers are truly predictive of response to targeted therapies in gliomas, some do show promise. For example, clinical trials are now open for EGFRvIII-mutated GBMs, vemurafenib is being evaluated in BRAF V600E mutant gliomas, and a clinical response has already been observed in FGFR3-TACC3-positive patients treated with an FGFR inhibitor.43
Like gliomas, meningiomas are stratified according to WHO criteria, with a higher grade denoting a more aggressive clinical course.1 Fewer genomic studies focus on meningiomas compared with gliomas, although they are comparable to gliomas in frequency and can also be lethal. Mutually exclusive mutations in NF2, AKT1, SMO, and KLF4 (Fig. 5B) were reported in a large cohort of meningiomas, each of which was associated with distinct locations in the meninges.44 Moreover, NF2-driven meningiomas are far more likely to exhibit atypical grade II features than the other subtypes. While it remains to be seen whether this molecular subclassification is comparable to or exceeds traditional WHO grading in terms of prognostic power, our GlioSeq NGS panel is capable of distinguishing these subtypes.
Unlike gliomas and meningiomas, medulloblastomas are automatically assigned a WHO grade IV based on their extremely aggressive growth and dismal outcome if untreated.1 However, large-scale genomic studies showed that, like gliomas, medulloblastomas can be divided into several subtypes based on specific driver mutations. Those with mutations in CTNNB1 or DDX3X are of particular interest because they identify Wnt pathway medulloblastomas, which tend to have a much better prognosis (Fig. 5). In contrast, MYC or MYCN/CDK6 amplifications are characteristic of group 3 and 4 medulloblastomas, respectively, and are far more likely to metastasize and have a poor prognosis, even with therapy (Fig. 5). Tumors with PTCH and SMO belong in the Shh class and have a relatively intermediate prognosis between Wnt and group 3 and 4 tumors.10–12 Considering that these subtypes and their driver mutations remain stable even at posttherapy recurrence,13 upfront characterization of medulloblastomas has lasting clinical value.
Several newly discovered targets were not included in the current version of GlioSeq; however, they may be of interest for future developments. Recent studies of embryonal tumors have revealed SMARCB1 (INI1/BAF47) as the principal recurrent genomic alteration in atypical teratoid rhabdoid tumor.45 Ependymal tumors have been demonstrated to harbor prototypic gene fusions involving the genes RELA and YAP1. For example, the C11orf95/RELA fusion transcript is particularly enriched in supratentorial ependymomas and is associated with a poor prognosis.46
In summary, we present a new GlioSeq NGS assay that offers accurate and sensitive detection of a wide spectrum of point mutations, small insertions and deletions, copy number variations, and gene fusions in the most common pediatric and adult brain tumors. This versatile assay allows rapid and cost-effective profiling of brain tumor specimens, including small FFPE tissue biopsies, and provides valuable information for diagnosis, prognostication, and treatment of patients.
Supplementary Material
Funding
The National Institutes of Health (K08CA155764 to C.H. and R01NS37704 to I.F.P.)
Supplementary Material
Acknowledgments
We thank Dr. Wayne Ernst, MGP laboratory for participation in validating GlioSeq-RNA and Drs. Clayton Wiley and Geoffrey Murdoch, Department of Pathology, UPMC at for their valuable comments. The National Institutes of Health (K08CA155764 to C.H. and R01NS37704 to I.F.P.) provided partial support for this study.
Conflict of interest statement. None declared.
References
- 1.Kliehues P, Birger PC, Aldape KD, et al. Glioblastoma. In: Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, eds. WHO Classification of Tumours of the Central Nervous System. 4th ed Lyon: IARC; 2007:33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110(15):6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Wu G, Miller CP, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45(6):602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. [DOI] [PubMed] [Google Scholar]
- 10.Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123(4):473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones DT, Jager N, Kool M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramaswamy V, Remke M, Bouffet E, et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 2013;14(12):1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45(3):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huse JT, Aldape KD. The evolving role of molecular markers in the diagnosis and management of diffuse glioma. Clin Cancer Res. 2014;20(22):5601–5611. [DOI] [PubMed] [Google Scholar]
- 16.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):829–848. [DOI] [PubMed] [Google Scholar]
- 17.Nikiforova MN, Wald AI, Roy S, Durso MB, Nikiforov YE. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013;98(11):E1852–E1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jennings L, Van Deerlin VM, Gulley ML. Recommended principles and practices for validating clinical molecular pathology tests. Arch Pathol Lab Med. 2009;133(5):743–755. [DOI] [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grasso C, Butler T, Rhodes K, et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17(1):53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10–12. [Google Scholar]
- 26.Lassmann T, Hayashizaki Y, Daub CO. SAMStat: monitoring biases in next generation sequencing data. Bioinformatics. 2011;27(1):130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14(2):178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Bishop J, Shan Y, et al. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat. Cancer. 2013;20(4):603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korshunov A, Meyer J, Capper D, et al. Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol. 2009;118(3):401–405. [DOI] [PubMed] [Google Scholar]
- 30.Faulkner C, Palmer A, Williams H, et al. EGFR and EGFRvIII analysis in glioblastoma as therapeutic biomarkers. Br J Neurosurg. 2014:1–7. [DOI] [PubMed] [Google Scholar]
- 31.Cin H, Meyer C, Herr R, et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol. 2011;121(6):763–774. [DOI] [PubMed] [Google Scholar]
- 32.Mistry M, Zhukova N, Merico D, et al. BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J Clin Oncol. 2015;33(9):1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horbinski C, Nikiforova MN, Hagenkord JM, Hamilton RL, Pollack IF. Interplay among BRAF, p16, p53, and MIB1 in pediatric low-grade gliomas. Neuro Oncol. 2012;14(6):777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horbinski C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol. 2013;125(5):621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horbinski C, Kofler J, Yeaney G, et al. Isocitrate dehydrogenase 1 analysis differentiates gangliogliomas from infiltrative gliomas. Brain Pathol. 2011;21(5):564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camelo-Piragua S, Jansen M, Ganguly A, et al. A sensitive and specific diagnostic panel to distinguish diffuse astrocytoma from astrocytosis: chromosome 7 gain with mutant isocitrate dehydrogenase 1 and p53. J Neuropathol Exp Neurol. 2011;70(2):110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 38.Wu G, Broniscer A, McEachron TA, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koelsche C, Sahm F, Capper D, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013;126(6):907–915. [DOI] [PubMed] [Google Scholar]
- 40.Yip S, Butterfield YS, Morozova O, et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol. 2012;226(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Stefano AL, Fucci A, Frattini V, et al. Detection, Characterization, and Inhibition of FGFR-TACC Fusions in IDH Wild-type Glioma. Clin Cancer Res. 2015;21(14):3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kieran MW, Roberts CW, Chi SN, et al. Absence of oncogenic canonical pathway mutations in aggressive pediatric rhabdoid tumors. Pediatr Blood Cancer. 2012;59(7):1155–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506(7489):451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.