Abstract
Melanoma prognosis is dictated by tumor-infiltrating lymphocytes, the migratory and functional behavior of which is guided by chemokine or cytokine gradients. Here, we retrospectively analyzed the expression patterns of 9 homing receptors (CCR/CXCR) in naive and memory CD4+ and CD8+ T lymphocytes in 57 patients with metastatic melanoma (MMel) with various sites of metastases to evaluate whether T cell CCR/CXCR expression correlates with intratumoral accumulation, metastatic progression, and/or overall survival (OS). Homing receptor expression on lymphocytes strongly correlated with MMel dissemination. Loss of CCR6 or CXCR3, but not cutaneous lymphocyte antigen (CLA), on circulating T cell subsets was associated with skin or lymph node metastases, loss of CXCR4, CXCR5, and CCR9 corresponded with lung involvement, and a rise in CCR10 or CD103 was associated with widespread dissemination. High frequencies of CD8+CCR9+ naive T cells correlated with prolonged OS, while neutralizing the CCR9/CCL25 axis in mice stimulated tumor progression. The expansion of CLA-expressing effector memory CD8+ T cells in response to a single administration of CTLA4 blockade predicted disease control at 3 months in 47 patients with MMel. Thus, specific CCR/CXCR expression patterns on circulating T lymphocytes may guide potential diagnostic and therapeutic approaches.
Introduction
Metastatic melanoma (MMel) therapy has been revolutionized by the recent availability of BRAF inhibitors and immune checkpoint blockers. However, there is still a need for prognostic or predictive factors aimed at guiding the clinical management of metastases.
Chemokines are critical regulators of leukocyte trafficking and immune functions (1). They act on G protein–coupled receptors, which are abundantly expressed in a variegated manner on all immune cells to regulate their migration and differentiation (2). The magnitude and duration of the signal depend on the intensity of the exposure to chemokines and on subsequent chemokine receptor desensitization, phosphorylation, and internalization (3). The human chemokine system consists of 23 receptors and 48 ligands (4). The chemokine receptor–ligand interaction is redundant, in that chemokine receptors can be activated by several chemokines, while a given chemokine can bind multiple receptors. Immune responses guided by chemokine receptor signaling represent a complex network of regulations at different levels. At the systemic level, there is a spatiotemporal and geographic, tissue-specific expression of chemokine receptors and their respective ligands. At the cellular level, coexpression of several chemokine receptors with other immune regulators determines the final functional outcome. At the molecular level, signaling through a given chemokine receptor may be fine tuned by different ligands and intracellular proteins (2). Adding more complexity to the system, chemokine receptor signaling can be affected by glycoaminoglycans, which may alter the affinities of their ligands, as well as by scavenger molecules (2). Chemokine receptors have been classified as XC, CC, CXC, and CX3C receptors on the basis of the chemokine subclass ligand to which they bind. Chemokine expression patterns affect the pathophysiology of major diseases, such as cancer (5), inflammation (6), allergy, transplantation, and infections, as they influence the migration of various T lymphocyte subpopulations to specific tissues and polarize T cell function. Chemokine receptors identify functional subsets within the human peripheral CD4+ and CD8+ T cell pool. In response to TCR stimulation or homeostatic cytokines, CXCR3+ or CCR4+CD4+ central memory T cells (TCMs) fully differentiate into CCR7– Th1 and Th2 cells, respectively (7). A population of human skin-homing memory CD4+ T cells expressing the chemokine receptors CCR10, CCR6, and CCR4 produce IL-22, but these cells do not produce IL-17 or IFN-γ (8). CCR4 is also highly expressed in a major fraction of blood α4β7– CD4+ TCMs, including almost all skin memory CD4+ cells expressing cutaneous lymphocyte antigen (CLA). Approximately 25% of human peripheral blood TCMs express the CXCR5 chemokine receptor, a hallmark of follicular Th cells. CXCR5+ TCMs express high levels of the chemokine CXCL13 and efficiently induce Ig secretion. These cells are characterized by a high responsiveness to ICOS ligand costimulation, leading to IL-10 release (9).
The prognosis of a variety of human malignancies, such as melanoma (10), colorectal cancer (11), and ovarian cancer (12), is dictated by the abundance, spatial trafficking, and quality of intratumoral T cell infiltrates (13). Gene expression profiling performed on a series of MMels and other malignancies revealed a major segregation of samples based on the presence of T cell–associated transcripts correlating with the expression of defined chemokine genes (5, 14). Immune-mediated regression of melanoma, whether spontaneous or therapy induced, supposedly relies on the elicitation of antitumor effector Th1 and Th17 or pathogenic Th17 (pTh17) cells and their subsequent homing to malignant lesions.
Salerno et al. (15) recently studied whether T cell expression of the chemokine receptors CCR4, CCR5, and CXCR3 and integrins could account for T cell retention in MMel lesions. With the exception of bowel metastases, in which T cells showed high expression levels of CCR9, we observed a limited tissue site–specific homing to human melanoma metastases. We revisited this notion and performed retrospective analyses of the phenotype and functions of circulating T lymphocytes and tumor-infiltrating lymphocytes (TILs) from tumoral lymph nodes (LNs) in 3 independent cohorts of MMel patients treated before the era of efficient therapies, with the aim of correlating homing receptor expression with the pattern of metastatic spreading (skin and LNs, lung, distant organs, with or without lung dissemination) and overall survival (OS). We found (a) an orchestrated deregulation of CCR/CXCR expression according to the various sites of disease dissemination; (b) substantial correlations with OS, mainly for 3 markers (CD8+CCR6+ effector memory T cells [TEMs], CD4+CCR10+ TCMs and TEMs, and CD8+CCR9+ naive T cells [TNs]); (c) an upregulation of CXCR3, CXCR4, CD103, and CLA expression in terminally differentiated (T effector memory cells expressing CD45RA [TEMRAs]) TILs compared with that detected in circulating T cells; (d) a functional role of the CCR9/CCL25 axis in mice and patients bearing tumors; and (e) pharmacodynamic biomarkers of response to ipilimumab such as circulating numbers of CLA+CD8+ TEMs.
Results
Chemokine receptor expression patterns.
We performed flow cytometric analyses of peripheral blood CD4+ and CD8+ T cell subsets based on their expression of CCR7 and CD45RA, which allowed us to distinguish TNs (CCR7+CD45RA+), TEMRAs (CCR7–CD45RA+), TEMs (CCR7–CD45RA–), and TCMs (CCR7+CD45RA–) and then to assess, for each subset, the expression (9 homing receptors) or coexpression (23 combinatorions in pooled CD4+ or CD8+ T cells) of a variety of CC and CXC receptors (Supplemental Table 1 and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI80071DS1) in 57 patients with stage IIIB-C or stage IV melanoma (Supplemental Table 2 and Supplemental Figure 2) compared with 26 healthy volunteers (HVs).
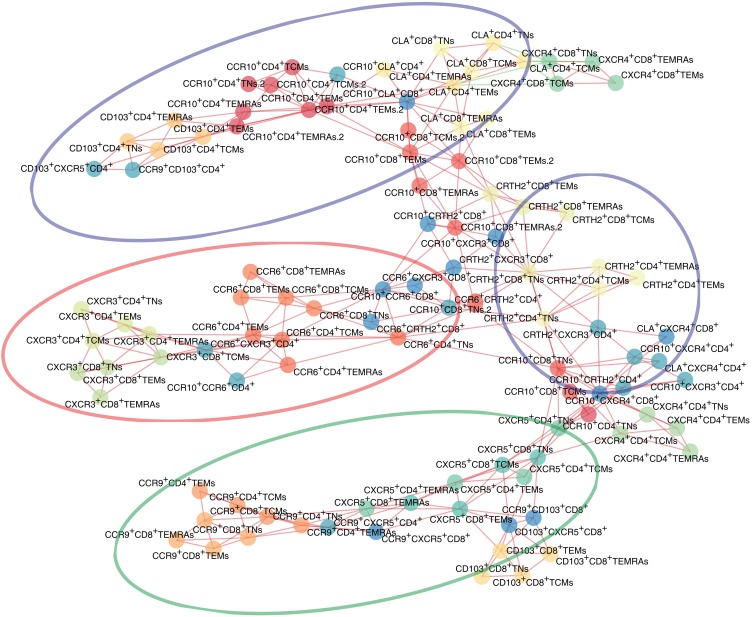
Correlation matrices indicated similar chemokine receptor expression on CD4+ and CD8+ T cells, except for CXCR4 and CD103, and concordant patterns in TNs, TEMs, and TCMs, and TEMRAs (Supplemental Figure 3). In accordance with previous reports, skin-homing receptors (CCR10 and CLA) and functional receptors (CXCR3 on Th1 and CCR6 on Th17) appeared to be associated with each other, as indicated on the planar representation of the correlation matrix of chemokine receptors obtained for all samples analyzed in this study. However, unexpectedly, we found no correlation between CD103 and CCR9 expression (Figure 1).
Figure 1. Planar maximally filtered graph representation of the matrix of correlation among the 104 parameters used in this study.
Interconnecting lines are in red or green if the 2 nodes were positively and negatively correlated, respectively. Ellipses represent 4 groups of chemokine receptors associated with the metastatic spreading pattern described in Figure 2.
Orchestration of chemokine receptor expression patterns according to melanoma metastatic spreading.
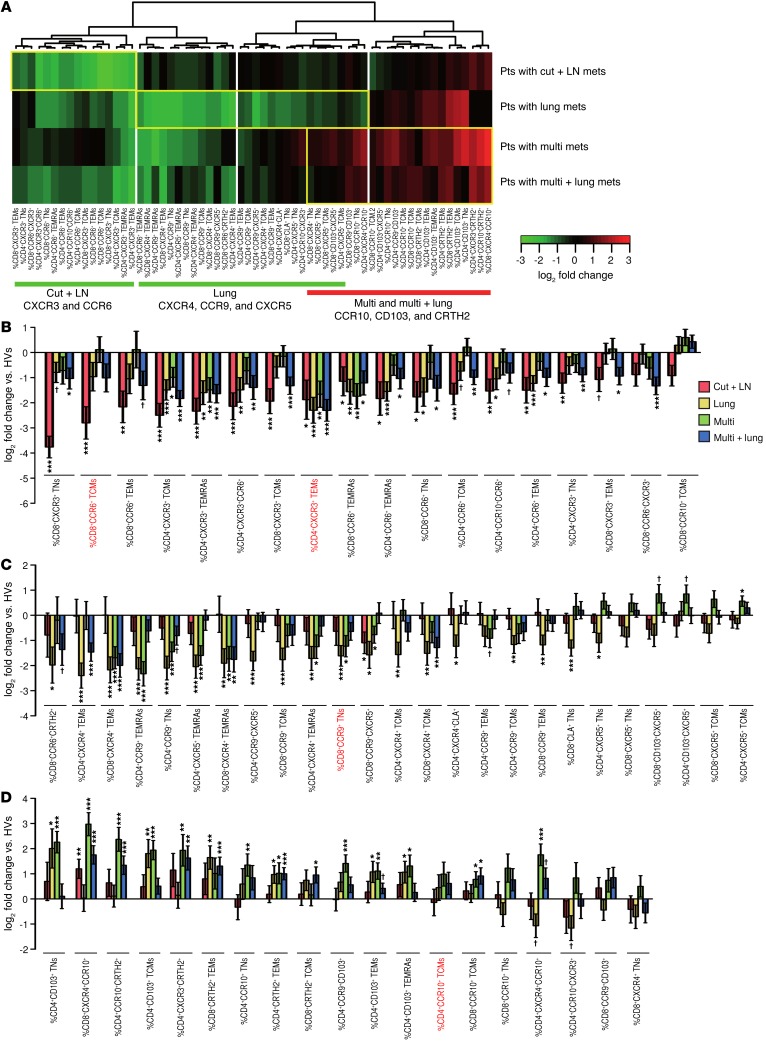
The organization pattern of T cell CCR/CXCR expression was not randomly distributed among patient groups but rather mirrored metastatic spreading. Of 104 parameters, 62 were found to differ significantly (FDR <0.1) among patients bearing metastases in distinct locations. Expression levels were normalized with respect to those of HVs, indicating that specific signatures could identify preferential metastasis to skin and LNs (Cut + LN), lung, or other distant organs (Multi mets), without (Multi), or with lung involvement (Multi + lung) (Figure 2A). Loss of CXCR3 or CCR6 expression on both CD4+ and CD8+ T cells was associated with s.c. or secondary lymphoid organ metastases (Figure 2, A and B).
Figure 2. Nonhierarchical clustering of the expression of CCR and CXCR studied in circulating T cells from patients with MMel after normalization to HV-related values.
(A) Heatmap representing 62 markers that were found significantly altered among patient groups at an FDR below 0.1. Each cell depicts the fold change between the metastatic group and a cohort of HVs (n = 26): red indicates an increase and green a decrease in marker levels compared with those in the reference population. The dendrogram classifies 57 patients (featured according to their metastatic dissemination patterns at diagnosis) for CCR and CXCR expression on all 4 subsets of CD4+ and CD8+ T lymphocytes (featured for each marker), highlighting that most chemokine receptor families segregated according to metastatic spreading (summarized at the bottom), with a decrease (green) or increase (red) in peripheral blood levels compared with those in HVs. From left to right: primary sites of invasion (skin and LNs), followed by additional locations of metastatic spreading (such as to lungs and other distant organs). As for liver metastases, CD103 upregulation may constitute a hallmark (Supplemental Figure 4). Pts, patients. (B–D) log2-based fold changes (and 95% CI) in parameter levels between the metastatic groups and the HV cohort for the 62 markers found altered among the patient groups. Markers that were significantly associated with patient prognosis (detailed in the subsequent figures) are highlighted in red. Asterisks indicate significance (as compared with HVs), after adjusting for multiple tests: †P < 0.1, *P < 0.05, **P < 0.01, and ***P < 0.001, by beta regression.
We observed a 4- to 8-fold decrease in the percentages of circulating (a) CXCR3+ TCMs, (b) CD8+CCR6+ memory T cells, and (c) CXCR3/CCR6–coexpressing CD4+ T cells in patients with MMel compared with that seen in HVs (Figure 2B). Loss of CXCR4 or CCR9 expression on both CD4+ and CD8+ T cells as well as a drop in CXCR5 expression levels on CD4+ and CD8+ TNs compared with HVs indicated lung dissemination (Figure 2C). Loss of CXCR4 expression on CD4+ and CD8+ TEMs was associated with patients who had pulmonary lesions. Similarly, lung tumor deposits were associated with decreased CCR9 expression on almost all CD4 and CD8 T cell subsets, including CXCR5/CCR9-coexpressing T cells. Moreover, a 2- to 4-fold decrease in CRTH2 and CCR6 expression on CD8+ T cells in MMel patients compared with HVs was observed when lung metastases were diagnosed (Figure 2C).
As a general pattern, oligometastatic spread (skin and LN or lung) was associated with decreased CCR/CXCR expression on effector and memory T lymphocytes, while polymetastatic dissemination favored the increase in CCR/CXCR expression on naive T lymphocytes. A rise in the number of CD4+CD103+ and CD4+CCR10+ TNs as well as CCR10 and CRTH2 CD4+ T cells and CCR10 and CXCR4 CD8+ T cells indicated wide spreading of melanoma (Figure 2D). A rise in expression of CRTH2 and CXCR3 in pooled CD4+ T cells was also observed (Figure 2D). Of note, an increase in CD4+CD103+ TNs was strongly associated with liver metastases (but not in bone, brain, or gut locations, Supplemental Figure 4, A and B). Moreover, CD103 expression with CCR9 or CXCR5 was also associated with the ulceration status of primitive tumors (Supplemental Figure 4, C and D). Polymetastatic locations were also associated with a loss of effector cells. Hence, a more than 4-fold decrease in CXCR3+ TCMs, TEMs, and TEMRAs (Figure 2B) as well as CD8+CXCR4+ TEMs, TEMRAs, and CD4+CCR9+ TEMRAs (Figure 2C) was associated with distant metastases. Interestingly, CXCR5 alone or in association with CD103 on both CD4+ and CD8+ T cells allowed the distinction of lung-free disseminated disease, in that there was a greater than 2-fold increase in the number of these cells in the blood of patients presenting with distant metastases without lung involvement (Figure 2C).
We conclude that CCR/CXCR chemokine expression patterns on peripheral T lymphocytes correlate with metastatic dissemination during melanoma progression.
LN metastases–associated chemokine receptors, lymphocyte functions, and prognosis.
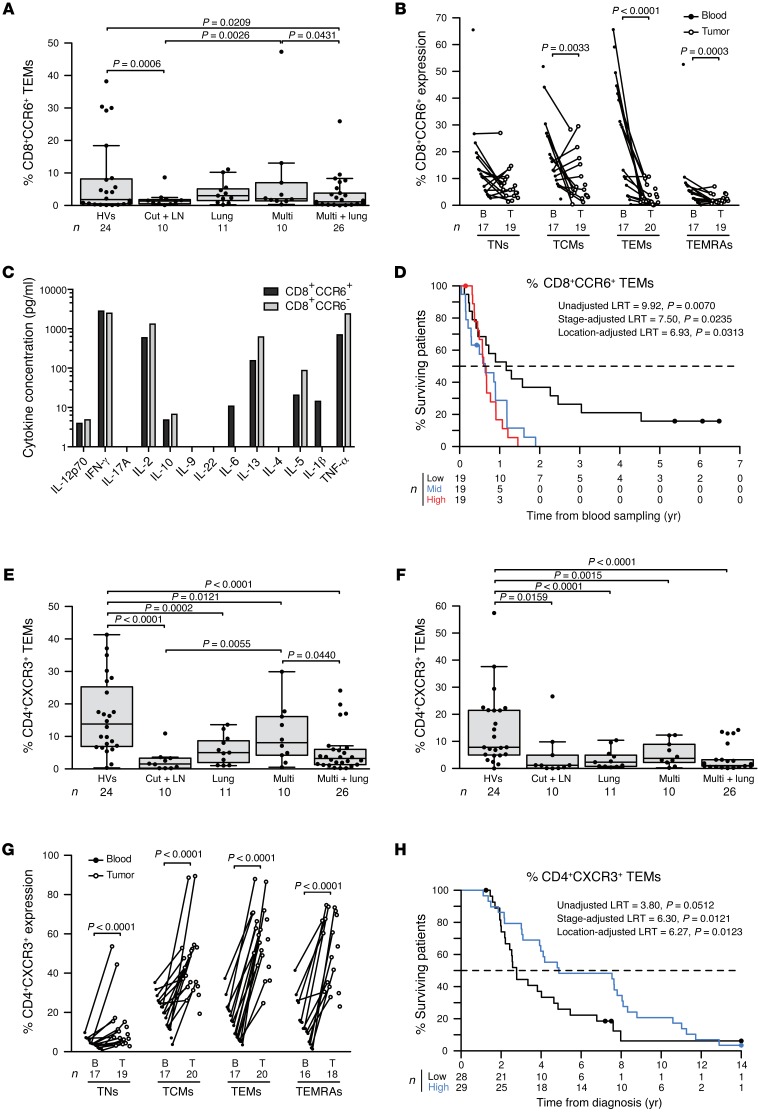
Loss of circulating CD8+CCR6+ TEMs is one hallmark of patients with skin and LN metastases (Figure 2B and Figure 3A). This loss could not be explained by preferential trafficking and accumulation of such cells to the affected LNs, which contained fewer CD8+CCR6+ TEMs than did the blood (Figure 3B). However, when the tumor continued to invade distant organs, not just the lungs, this cell population marker increased to reach the levels found in HVs (Figure 3A). Flow cytometry–based cell sorting of circulating CD8+CCR6+ T cells revealed that such cells expressed less IL-2, IL-5, IL-13, and TNF-α (but similar levels of IFN-γ) as compared with the levels in CD8+CCR6– control T cells (Figure 3C). Low frequencies of circulating CD8+CCR6+ TEMs were associated with prolonged survival (considered a continuous variable, adjusted according to localization group, P = 0.0317; with the median as a cut-off value, adjusted according to localization group, P = 0.0305; with tertiles as cut-off values, adjusted according to localization group, P = 0.0313; and adjusted according to stage, P = 0.0235, Figure 3D.
Figure 3. LN metastases–associated chemokine receptor CCR6 and CXCR3 expression, function, and survival.
(A) CCR6 expression on CD8+ TEMs in HVs and in patients presenting with only cutaneous and LN metastases (Cut + LN), additional lung involvement, disseminated disease (Multi), and in those with metastases in lungs and other distant organs (Multi + lung) at the time of inclusion in 1 of the 3 protocols described in the Methods. (B) Match-paired comparison of CCR6 expression (performed by flow cytometry on fresh tissues) in all CD8+ T cell subsets from blood (B) and tumors (T) at the time of surgery in the prospective cohort of 20 patients with MMel. (C) CCR6+CD8+ T cell cytokine profile. Flow cytometry–guided sorting based on CCR6 expression in blood CD8+ T cells from 1 representative patient (out of 2 patients, yielding similar results; data represent the mean from duplicate wells) to analyze cytokine release after a 40-hour CD3/CD28 mAb–driven stimulation. (D) Kaplan-Meier survival curves representing data from 57 MMel patients according to their proportions of circulating CCR6+CD8+ TEMs, segregated in tertiles. (E and F) Same as in A, showing expression of the CXCR3 subset on CD4 TCMs (E) and TEMs (F). (G) Same as in B, showing CXCR3 expression on CD4 T cell subsets. (H) Kaplan-Meier survival curves for 57 MMel patients according to the median of the proportions of circulating CXCR3+CD4+ TEMs. Each point represents 1 patient specimen, and the total number is indicated for all subpopulations studied. Statistical analyses were performed by beta regression (A, E, and F), linear mixed-effects (B and G), and Cox regression (D and H) modeling. Raw P values are indicated. LRT, likelihood ratio test.
A significant decrease in the frequency of circulating CD4+CXCR3+ and CD8+CXCR3+ TNs and TCMs as well as CXCR3+CCR6+ double-positive CD4+ T cells was the second fingerprint of cutaneous and LN (and other) metastases (Figure 2B and Figure 3, E and F). CD4+CXCR3+ T cells accumulated in metastatic LNs, perhaps explaining their decrease in the blood (Figure 3G). As already reported in the context of MMel, CXCR3+ T cells have a Th1 profile, home to inflammatory lesions, and are expanded by vaccine adjuvant–based immunotherapies (16–18). In the present study, high circulating levels of CD4+CXCR3+ TEMs indicated a favorable prognosis for MMel patients (considered with the median for the cut-off value, adjusted according to localization group, P = 0.0123, or according to stage, P = 0.0121, Figure 3H).
Unexpectedly, CLA expression on circulating T cells was not modulated by skin or LN metastatic dissemination (Supplemental Figure 5, A and B), although CLA+CD4+ TEMRAs accumulated in LN tumors (Supplemental Figure 5, C and D). In the polymetastatic configuration, the numbers of CD4+CLA+ TEMRAs or CD8+CLA+ TCMs eventually increased in the blood (Supplemental Figure 5, A and B).
Altogether, since CXCR3 and CCR6 expression on CD4+ and CD8+ T cells correlated with each other (Figure 1 and Supplemental Figure 3), we propose that a significant drop in CCR6+ and CXCR3+ TCM numbers (the dominant subset in terms of numbers; Figure 2) represents a hallmark of metastatic dissemination into LNs.
Lung metastases–associated chemokine receptors, lymphocyte functions, and survival.
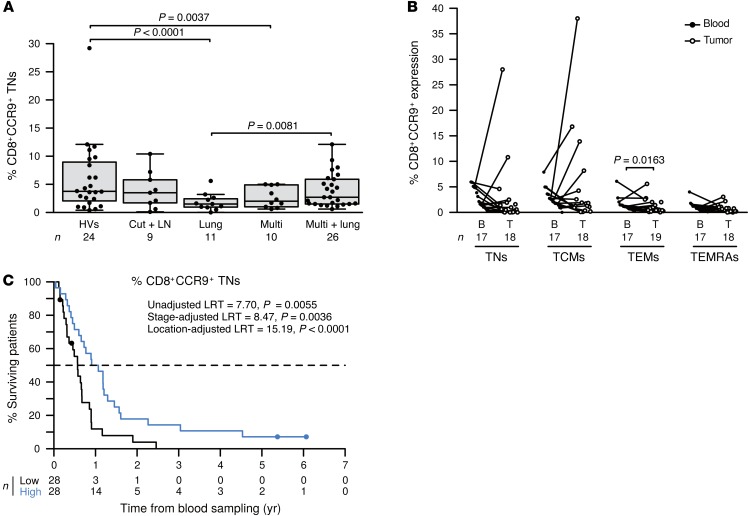
Eleven melanoma patients presented with metastases in the lung, skin, and LNs. Circulating CD4+ TEM, TEMRA, and TCM lymphocytes from these patients showed reduced CXCR4 expression levels (Supplemental Figure 6, A and B, and data not shown). CD4+CXCR4+ TEMRAs tended to accumulate in LNs infiltrated by melanoma cells (Supplemental Figure 6C). Even more specific to isolated lung metastases, CXCR5 expression was reduced in circulating CD4+ TEMRAs or CD4+CCR9+ T cells (Supplemental Figure 7, A and B, and data not shown). Cells with this phenotype did not accumulate in metastatic LNs (Supplemental Figure 7C). They exhibited a Th1 cytokine production profile upon TCR engagement (Supplemental Figure 7D). Importantly, CCR9 in circulating CD4+ (not shown) and CD8+ TNs was strongly decreased in patients with lung metastases (Figure 2C and Figure 4A) but rarely accumulated in metastatic LNs (Figure 4B). High levels of circulating CCR9+CD8+ TNs were associated with a favorable prognosis (Figure 4C, using localization group–adjusted continuous variables [P = 0.0084]; median values [P < 0.0001], or tertile values [P = 0.0009] as cut-offs or using stage-adjusted values [P = 0.0036]). Of note, the numbers of CRTH2/CCR6-coexpressing CD8+ T cells were also reduced in patients with lung metastases (Figure 2C).
Figure 4. CD8+CCR9+ TNs leave the blood during lung metastasis and dictate MMel prognosis.
(A) CCR9 expression on CD8+ TNs is depicted for HVs, patients presenting with only cutaneous/LN (Cut + LN) metastases, those with additional lung (Lung) involvement, those with disseminated disease (Multi), and those with distant metastases plus lung involvement (Multi + lung) at the time of inclusion. Box plots summarize data from 57 patients with MMel and 24 HVs. (B) Match-paired comparison of CCR9 expression levels (performed by flow cytometry on fresh tissues) in all CD8+ T cell subsets from blood and tumors at the time of surgery in the prospective cohort of 20 patients with MMel. (C) Kaplan-Meier survival curves of 57 patients with MMel according to their proportions of circulating CCR9+CD8+ TNs segregated with the median. Each point represents 1 patient specimen, and the total number is indicated for all subpopulations studied. Statistical analyses were performed by beta regression (A), linear mixed-effects (B), and Cox regression (C) modeling. Raw P values are indicated.
Altogether, loss of CXCR4, CXCR5, and CCR9 in TNs appears to be a hallmark of metastatic dissemination into lungs.
Distant metastases–associated chemokine receptors, lymphocyte functions, and survival.
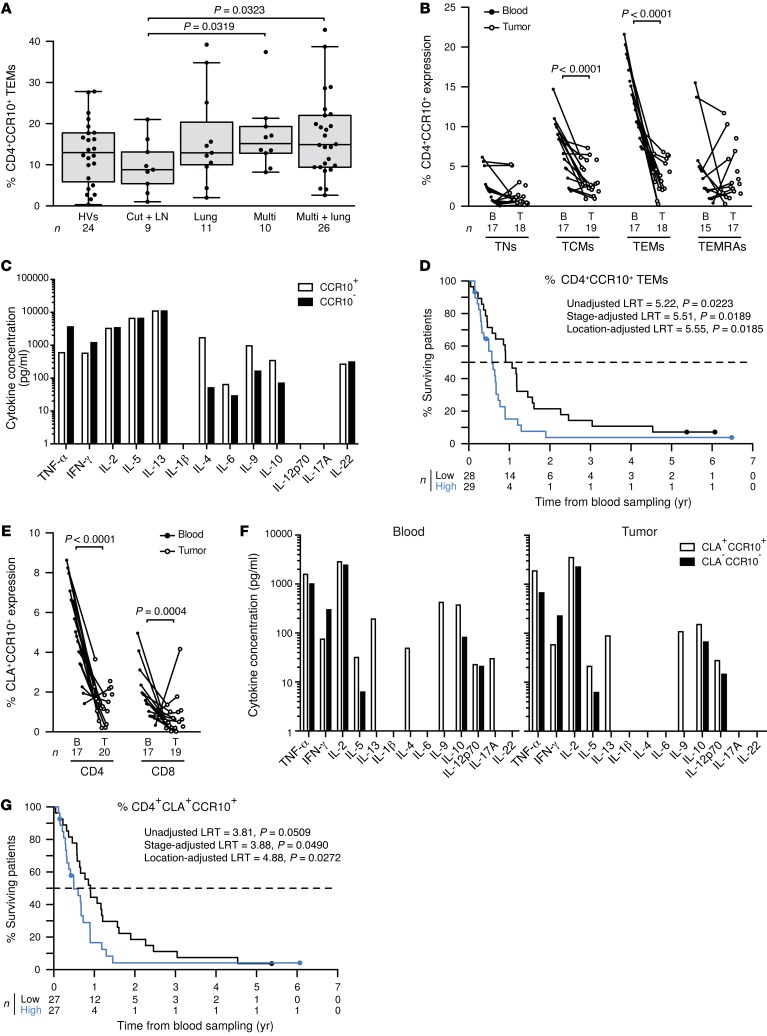
Melanoma dissemination is associated with a major loss of CXCR3 in CD4+ TCMs, TEMs, and TEMRAs (>4-fold, Figure 2B) as well as of CD4+CCR9+ TEMRAs and CD8+CXCR4+ TEMs and TEMRAs (Figure 2C). In parallel, a broad spectrum of metastases was accompanied with a significant rise in circulating CD4+CCR10+ TNs, TCMs, and TEMs (Figure 2D, Figure 5A, and data not shown) and CCR10+CRTH2+, CCR10+CXCR3+, or CRTH2+CXCR3+ CD4+ T cells (Supplemental Figure 8, A–C). CD4+CCR10+ TCMs and TEMs were severely reduced in melanoma LN metastases (Figure 5B), and circulating cells expressed higher levels of IL-9, IL-4, and IL-10, but low levels of Th1 cytokines in polymetastatic patients (Figure 5C). Low frequencies of CCR10+CD4+ TCMs (data not shown) and TEMs were associated with prolonged survival (Figure 5D, using localization group–adjusted median values [P = 0.0185] or tertile values [P = 0.0083] as cut-offs). Moreover, CLA+CCR10+ T cells were reduced in metastatic LNs (Figure 5E). Circulating CLA+CCR10+ T cells expressed higher levels of IL-5, IL-9, IL-10, and IL-13 (Figure 5F). Reduced numbers of circulating CCR10+CLA+ T cells predicted a prolonged OS (considered a continuous variable, adjusted according to localization group [P = 0.0359], with the median as the cut-off value, adjusted according to localization group [P = 0.0272], and tertiles as cut-off values, adjusted according to localization group [P = 0.0060]; Figure 5G). Interestingly, as with CXCR5, CD103 expression on CD4+ T cells correlated with distant metastases outside of the lung (Supplemental Figure 9A). CD4+CD103+ T cells exhibited a Lag3-like profile (19) characterized by IL-10, TNF-α, and IL-2 production (Supplemental Figure 9B), failed to accumulate in LN tumor lesions (Supplemental Figure 9C), and did not express FOXP3 (Supplemental Figure 9D). In addition, the frequencies of circulating CD4+CXCR4+CCR10+ T cells were selectively decreased in patients with lung metastases but were increased in patients with widespread dissemination (Figure 2C).
Figure 5. Levels of CD4+CCR10+ TEMs and CCR10+CLA+ T cells modulated by distant metastases dictate MMel prognosis.
(A) CCR10 expression on CD4+ TEMs is depicted for HVs and patients presenting with only cutaneous/LN (Cut + LN) metastases, additional lung (Lung) involvement, disseminated disease (Multi), and distant metastases plus lung involvement (Multi + lung) at the time of inclusion in 1 of the 3 protocols described in the Methods. (B) Match-paired comparisons of CCR10 expression (performed by flow cytometry) in all CD4+ T cell subsets from blood and tumors at the time of surgery in the prospective cohort of 20 patients with MMel. (C) CD4+CCR10+ T cell cytokine profile. Flow cytometry–guided sorting based on CCR10 expression in blood CD4+ T cells in 1 representative patient (out of 2 yielding similar results) to analyze cytokine release after a 40-hour CD3/CD28 bead–driven stimulation. (D) Kaplan-Meier survival curves of 57 patients with MMel according to the median of their proportions of circulating CD4+CCR10+ TEMs (of note, identical results with TCMs are not shown). (E and F) Same as as in B and C, showing the subset of double-positive CCR10+CLA+CD4+ T cells. (G) Same as in D, analyzing OS as a function of CCR10/CLA CD4+ T cell subsets. Each point represents 1 patient specimen, and the total number is indicated for all subpopulations studied. Statistical analyses were performed by beta regression (A), linear mixed-effects (B and E), and Cox regression (D and G) modeling. Raw P values are indicated.
In summary, a concomitant rise in CCR10 and CD103 in naive or memory CD4+ T cells with a decrease in CCR9, CXCR4, or CXCR3 TEMs or TEMRAs are hallmarks of distant metastases.
Significant changes during immune checkpoint blockade by ipilimumab.
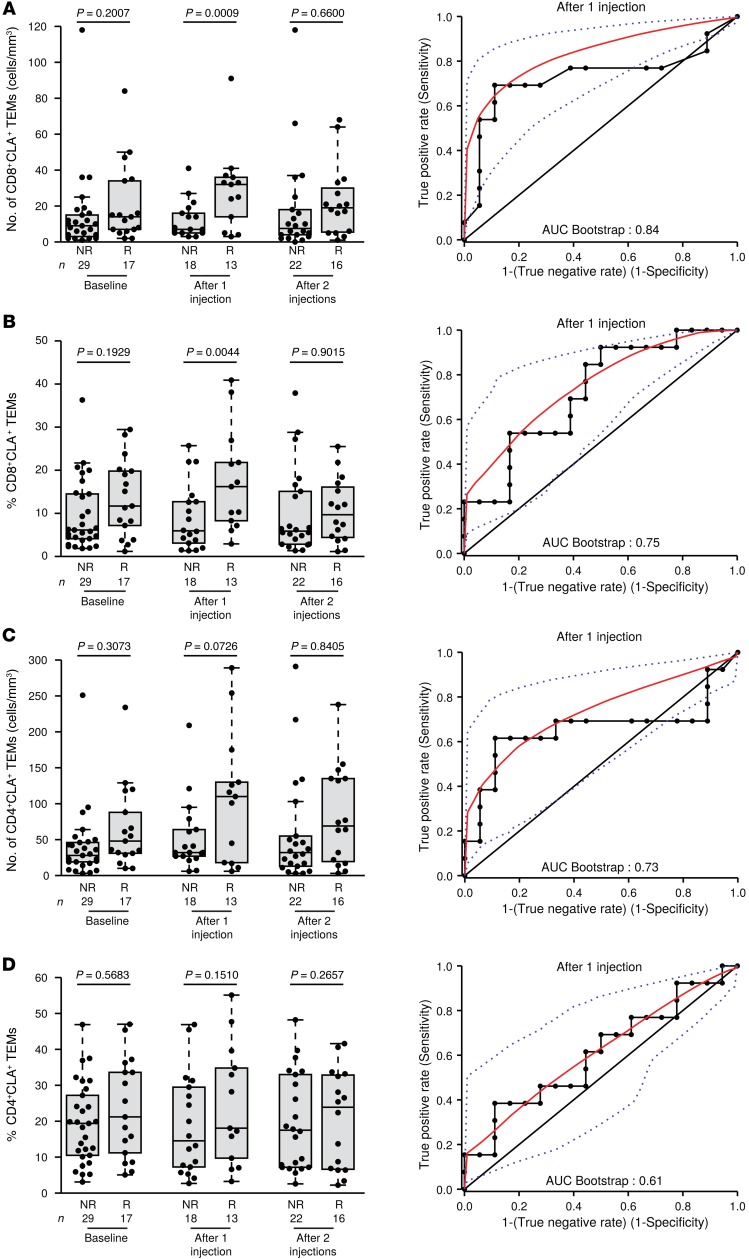
CTLA4 blockade by the FDA-approved and the European Medicines Agency–approved (EMA-approved) drug ipilimumab induces significant and prolonged (>7 years) antitumor effects in approximately 20% of patients with MMel (20, 21). We analyzed all the CC and CXC chemokine receptors described herein (Supplemental Table 1) in 47 patients diagnosed with stage IV MMel who were treated with ipilimumab (mainly 3 mg/kg [87%]) and enrolled at 4 clinical centers (Supplemental Table 3). Interestingly, although we analyzed most of the above-detailed markers were, we found that only CLA expression on CD8+ TEMs (cell numbers, shown in Figure 6A, as well as proportions, shown in Figure 6B), and not on the CD4 T cell population (Figure 6, C and D) monitored after 1 injection of ipilimumab, was significantly associated with clinical responses, as evaluated according to Response Evaluation Criteria In Solid Tumors (RECIST) criteria 3 months after treatment commencement and taking into account the inter-center variations (Figure 6A, left and right panels, AUC = 0.84; and Figure 6B, left and right panels, AUC = 0.75).
Figure 6. Ipilimumab-induced CLA+CD8+ TEMs are associated with favorable clinical outcome.
Absolute numbers (A, left) and proportions (B, left) of CLA-expressing CD8+ TEMs over time (left panel) are depicted for a cohort of 47 ipilimumab-treated patients with MMel and then segregated by nonresponders (NR) and responders (R) evaluated 3 months (4 injections) after commencement of therapy. (A and B, right panels) Receiver operating characteristics (ROC) curves depicting the predictive properties of each parameter determined after 1 ipilimumab injection and associated with the AUC. (C and D) Same as in A and B, except C shows the absolute number and D the proportions of CLA-expressing CD4+ TEMs. Each point represents 1 patient specimen, and the total number is indicated for all subpopulations studied. Statistical analyses were performed by logistic regression and adjusted according to the study center.
Altogether, we found that CTLA4 blockade modulated the numbers and/or proportions of CLA+ TEMs, and such changes might constitute pharmacodynamic markers or predictors of therapeutic response.
Relevance of CCR9 expression by T cells in a spontaneous melanoma model.
Transgene-enforced expression of the Ret protooncogene under control of the metallothionein 1 promoter drives spontaneous melanomagenesis. This model mimics human melanoma progression, in that the primary melanoma lesions develop along with LN metastases and invade the bone marrow and multiple distant organs. In the Ret-Tg melanoma model, TRP2-specific TEMs can be monitored at early stages (in primary tumors, metastatic LNs, and bone marrow) before chronic inflammatory processes promote the accumulation of myeloid-derived suppressor cells and Tregs and tumor progression in an uncontrolled fashion (22–26).
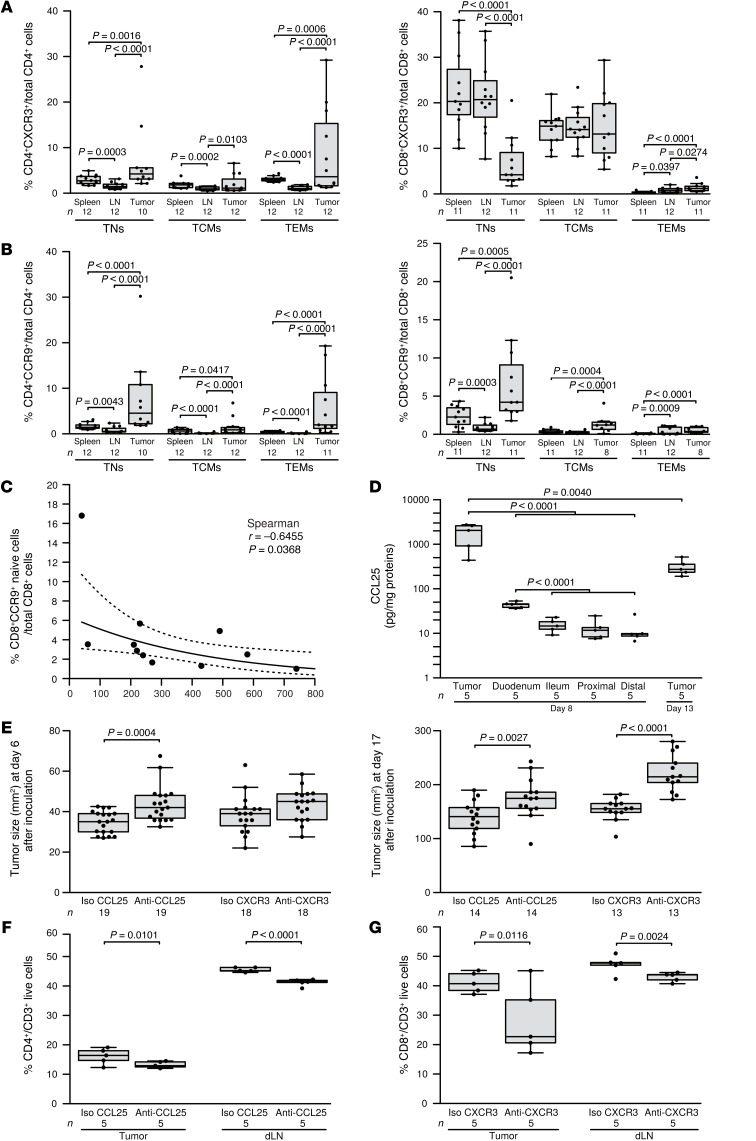
We took advantage of this well-characterized model to analyze CCR9 and CXCR3 expression on TILs from primary and LN metastatic lesions, bearing in mind that these 2 chemokine receptors are associated with prognosis in humans. The proportions of CXCR3+CD4+ TEMs and TCMs decreased in metastatic LNs (Figure 7A) compared with the proportions observed in the spleen, exactly as was observed in the MMel patients (Figure 3, E and F). CCR9+CD8+ TNs accumulated in primary lesions, recirculated to the spleen (Figure 7B), and were inversely correlated with tumor size (Figure 7C), paralleling the observation that the frequency of circulating CCR9+CD8+ TNs was correlated with the OS of MMel patients (Figure 4C). To further elucidate the clinical significance of the CCR9/CCL25 axis in cancer bearers, we neutralized the murine CCL25 chemokine using a specific Ab in mice bearing MCA205 sarcoma. This transplantable tumor model was selected on the basis of the high levels of CCL25 released into the MCA205 tumor microenvironment of the primary tumor in vivo (Figure 7D), levels that were much higher than those in the small intestine (Figure 7D). Systemic administration of the neutralizing anti-CCL25 Abs was rapidly (by day 6) capable of accelerating tumor outgrowth (compared with neutralizing anti-CXCR3 Ab), while blocking CXCR3 interfered at later stages (at day 17) of natural tumor immunosurveillance (Figure 7E). Moreover, neutralization of CCL25 or CXCR3 differentially affected the proportions of CD4+ and CD8+ T cells in tumor beds (mainly effector T cells; data not shown) and tumor-draining LNs, respectively (Figure 7, F and G). Hence, the CCL25/CCR9 and the CXCR3/CXCL9-CXCL10-CXCL11 axes are biologically relevant and act at different levels to control the tumor.
Figure 7. Neutralizing CCL25 or CXCR3 independently impacted tumor progression.
(A and B) Naive and memory CD4+ and CD8+ T cells from RET-induced mouse melanomas, metastatic LNs, and spleens analyzed by multicolor flow cytometry. TNs were characterized as CD4+CD45RB+ and CD8+CD44lo, central memory (CM), CD4+CD45RB–CD62L+, and CD8+CD44hiCD62L+ cells, while effector memory (EM) cells were defined as CD4+CD45RB–CD62L– and CD8+CD44hiCD62L– cells. Cumulative data showing expression levels of CCR9 and CXCR3 are depicted as percentages of the respective cell subset within live total CD4+ or CD8+ T cells from 3 independent experiments in the 3 locations. (C) Frequencies of CD8+CD44loCCR9+ T cells within total live CD8+ T cells in skin lesions in relation to the skin tumor weight (n = 11, 3 independent experiments). (D) Ex vivo explanted MCA205 sarcoma tumor as well as gut explants were dosed for CCL25 and normalized to total protein content. (E) MCA205 cells were inoculated in C57BL/6 mice that were concomitantly treated with i.p. administration of anti-CCL25 Ab, positive control anti-CXCR3 Ab, and their respective isotype controls (Iso) (5 injections, every 3 days). Tumor sizes were monitored until sacrifice and are depicted at day 6 (left panel) and day 17 (right panel) from 2 independent experiments. (F and G) Flow cytometric monitoring of CD4+ and CD8+ T cells in tumor beds (left panel) and tumor-dLNs (right panel) after neutralization of CCL25 or CXCR3 at the time of sacrifice (day 13). Each point represents 1 specimen, and the total number is indicated for all subpopulations studied. Statistical analyses were performed by beta regression (A and B), linear mixed-effects (D, F, and G), and linear (D and E) modeling, or by Spearman’s test. Raw P values are indicated.
Discussion
Despite the recirculation of tumor-specific CD8+ TEMs or TCMs in the peripheral blood, MMel progresses by evading immune-mediated destruction. Since the migratory phenotype of tumor antigen–specific T cells determines their antitumor efficacy, we hypothesized that the expression of CC or CXC chemokine receptors on circulating CD4+ and CD8+ T cells might define the phenotypic patterns associated with effective immunosurveillance against melanoma and produce tangible clinical benefit (27). Although such research endpoints were already addressed in MMel (28, 29), no previous investigation has been as comprehensive as the present study, in which we include 104 flow cytometric parameters, provide correlates between the blood and tumor beds, and make functional assessments of defined subsets. In summary, this study (a) unravels, for the first time to our knowledge, the orchestrated regulation of CCR/CXCR expression on peripheral lymphocytes as a function of the metastatic pattern of the disease; (b) highlights correlates between CCR/CXCR expression and OS; (c) unravels the biological significance of the CCR9/CCL25 axis in human and murine tumors; and (d) highlights new pharmacodynamic parameters (CLA-expressing TEMs) during immune checkpoint blockade with ipilimumab (Supplemental Table 4).
In a retrospective evaluation of CCR/CXCR expression in cytotoxic T lymphocytes (CTLs) from 52 patients with stage III or IV MMel, Mullins et al. (18) reported that high expression levels of CXCR3 by CD8+CD45RO+ cells were significantly associated with enhanced survival in stage III, but not stage IV, disease. Here, we confirm that CXCR3 expression is readily lost (on CD8+ TNs, TCMs, and CD4+ TCMs) when the skin and LNs are invaded, and even more so when distant metastases appear and the numbers of CD4+CXCR3+ TEMRAs, TEMs, and TCMs are strongly reduced. We found that these cells have the propensity to accumulate in tumor beds and that blockade of CXCR3 in mice severely compromises natural cancer immunosurveillance. CXCR3+ T cells have been reported to preferentially expand post–incomplete Freund’s adjuvant– (post-IFA–) or granulocyte macrophage–CSF–based (GM-CSF–based) vaccination and to traffic to vaccine-induced lesions, correlating with Th1 or T cytotoxic 1 (Tc1) immune responses (17, 30). In these vaccine studies and in the context of psoriasis, CLA and CXCR3 expression levels were often associated with CD8+ CTLs and identified a subset of cells expressing IFN-γ, T-bet, and IL-12Rβ1 (16). It has been suggested that an appropriate Th1-like chemokine pattern in melanoma may promote CXCR3+ T cell homing to lesions and protect the host against melanoma progression (5, 14). However, melanoma infiltration of LNs is insufficient to induce production of the CXCR3 chemokines CXCL9, CXCL10, and CXCL11. Some melanomas may be capable of secreting chemokines upon exogenous stimulation with IFN-α or IFN-γ (31). Despite harboring a prognostic value in many studies (32, 33), CXCR3 expression on T cells or CXCL10 release into the tumor microenvironment does not appear to be a pharmacodynamic marker or a predictive factor of response to anti-CTLA4 Abs.
CXCR4 and CCR9 are highly regulated during lung metastasis, and both receptors may facilitate the homing of lymphocytes to tumors. CXCR4 expression on CD8+ T cells is inversely correlated with CCR10 and CLA expression on CD4+ and CD8+ T cells. Indeed, in contrast to CCR10 (which increases with tumor dissemination), we observed a decrease in CXCR4 in the blood of patients with lung metastasis, but an accumulation of CXCR4 in metastatic LNs. The CXCR4/CXCL12 axis is involved not only in the retention of hematopoietic progenitors in the bone marrow, but also in the trafficking of Tregs to lesions (34). CXCL12 expression levels in bone and lung metastatic niches can be regulated by a variety of inflammatory cytokines such as IL-17A, IL-6, and M-CSF (35). Interestingly, chemokine receptors also bind factors other than chemokines. Noncanonical CXCR4 ligands include the evolutionarily conserved, proinflammatory macrophage migration inhibitory factor (MIF) (36) and the alarmin family member HMGB1, which interacts with CXCL12, which may mediate mononuclear cell recruitment (37). It is conceivable that these forms of nonconventional ligands are present in melanoma-invaded lungs. HMGB1 expression by tumor cells has been associated with DC and T cell infiltration of lung tumors in a cohort of 82 patients with advanced non–small-cell lung cancers (38). Hence, enforcing the CXCR4-mediated migration of TEMRA T cells to lesions (for instance, upon adoptive T cell transfer of engineered chimeric antigen receptor–Tg [CAR-Tg] or TCR-Tg T cells) could be a strategy for treating MMel.
Our findings indicate that (a) elevated numbers of blood CCR9+CD8+ TNs are associated with prolonged survival in humans and anticorrelated with tumor size in mice; (b) neutralizing CCR9 ligand accelerates the early phase of tumor exponential growth; (c) blockade of CCL25 reduces CD4+ TIL and draining LN (dLN) infiltrates; and (d) CCL25 and CXCR3 modulate immunosurveillance of tumors with different kinetics and mechanisms. CCL25 and CCR9 constitute a chemokine-receptor pair that is involved in both T cell development and gut-associated immune responses (39, 40). CCL25 is indispensable for the trafficking of CD8+ TEMs to the epithelium and lamina propria of the small intestine, but is dispensable for the priming in mesenteric LNs that imprints gut-homing properties (41). CCR9 is also expressed by Tregs, and CCR9 signaling inhibits the differentiation of FOXP3+ T cells (42). CCR9 is upregulated by retinoic acid, which also regulates CD103 expression on DCs and primes proinflammatory CD4+ and CD8+ T cells in the mesenteric LNs for their homing to intestinal mucosa. In the present study, circulating CCR9+CD8+ TNs or TCMs did not correlate with metastases in the gut or peritoneum. Instead, they tended to accumulate in primary tumors and, to a lesser extent, in metastatic LNs in mice and humans. Neutralization of CCL25 did not reduce tumor infiltration by CD4+CCR9+ TEMs (data not shown), but reduced CD4+ TIL content (Figure 7F) and significantly augmented the recirculation of CD4+CCR9+ TEMs and TCMs into the spleen (data not shown), perhaps facilitating their passive homing to the gut. Supporting this assumption, effector Th1 cells generated from a previous viral lung infection could cause intestinal damage following CCR9/CCL25-dependent trafficking of Th1 cells to the small intestine and their IL-15Rα–dependent transdifferentiation into pathogenic Th17 cells (43). Likewise, macrophages can express CCR9 and become proinflammatory after ligation with CCL25, producing TNF-α and activating hepatic stellate cells to eventually accelerate concanavalin A–induced (Con A–induced) acute hepatitis (44). In our model, interruption of crosstalk between intratumoral macrophages and CD4+ TILs following infusion of anti-CCL25 Abs may have impaired TNF-α production by macrophages and hence reset the tumor microenvironment toward an antiinflammatory phenotype, which would account for the tumor progression. In conclusion, disruption of the CCR9/CCL25 axis in tumor bearers may affect homeostasis of the common lymphoid system and compromise the delicate balance between gut and systemic immunity (45). Nonetheless, several observations point to a cell-autonomous role of CCR9/CCL25 in cancer cell migration, invasion, and metastatic dissemination as well as in PI3K-dependent resistance to chemotherapy (46–51). Enforced CCR9 expression on colorectal cancer cells dictated their niching in mouse intestines after xenografting, whereas CCL25 blockade enhanced liver metastases (52). Hence, therapeutic interventions aimed at disrupting the CCR9/CCL25 axis in the context of inflammation-induced carcinogenesis (such as in inflammatory bowel disease [IBD]) are worthy of more in-depth investigation (53).
CCR10-CTAK/CCL27 interactions may be involved in the selective recruitment of CTLs to the skin in a variety of pathophysiological disorders involving cutaneous tissues such as that seen in graft-versus-host disease (54). We observed increased recirculation of CCR10-expressing CD4+ TNs, TEMs, and TCMs, with or without CRTH2 expression, contrasting with many s.c. metastatic lesions. These CCR10+CD4+ T cells harbored a Th9/Th2/Tr1 phenotype, and high levels were associated with a bad prognosis. A recent study reported that CCR10 expression is preferentially induced on regulatory memory skin-resident T cells, but not on effector cells during an inflammatory skin process (55). Other explanations for the high numbers of recirculating CCR10+CD4+ T cells during distant metastases and the lack of chemoattraction toward CCL27-producing keratinocytes could be the autocrine production of CCL27 by melanoma cells themselves (56) or the high serum levels of CCL27, as observed in mycosis fungoides (57). Since CCR10-expressing T cells do not home to tumors and are endowed with a suppressive cytokine–release pattern, it may be useful to interfere with the CCR10/CCL27 axis with neutralizing Abs.
Immune checkpoint blockade has come of age in the management of MMel. However, uncoupling efficacy from toxicity (skin, gut, and liver) remains an unmet medical need for the administration of anti-CTLA4 Abs, alone or in combination with PD1/PD-L1 blockade. The dynamic of the skin-homing receptors may be useful for identifying patients prone to respond to ipilimumab. Indeed, in a cohort of 47 patients with MMel, the induction of CLA expression on CD8+ TEMs after 1 injection was associated with disease stabilization or objective responses. It is intriguing to note that CLA+ TEMs do not have a natural propensity to accumulate in skin-dLNs in the absence of therapy. However, it remains to be investigated whether this would be the case in patients responding to ipilimumab. Moreover, these findings warrant a prospective validation in the context of new-generation protocols (such as those combining anti-CTLA4 with anti-PD1 or anti-KIR Abs).
Certain homeostatic chemokine receptors play key roles in the homing of various stem cells including tumor cells themselves (58). Indeed, CXCR4 is involved in bone, LN, and lung metastases in breast cancer as well as in melanomagenesis (58). CXCR4 upregulation also correlated with BRAF mutations in other cancers (59). CCR7 and its corresponding ligands CCL19 and CCL21 have been associated with LN metastases, the CCR10/CCL27 axis with melanoma skin survival and metastases (29), and the CCR9-CCL25 interaction in metastases in the small intestine (28). Finally, CXCR3 expression in cutaneous melanoma cells negatively correlates with lymphocyte infiltration (60). Hence, it is conceivable that there could be a competition for ligand access between malignant cells and host leukocytes. This notion is particularly important with regard to therapeutic targeting. Indeed, an impairment of CXCR4/CXCL12-driven chemotaxis by AMD3100 or AMD11070 could inhibit skin tumor development (by mast cell suppression) and block liver melanoma metastases (61) on the one hand, but could prevent trafficking of T cells into lesions on the other hand. Thus, by inhibiting antigen-mediated T cell accumulation in the small intestine (62), the CCR9 antagonist CCX8037 could interfere with the natural immunosurveillance against melanoma. Therefore, our data should prompt detailed investigations into how these drugs affect lymphocyte trafficking (53).
In conclusion, specific CCR/CXCR expression patterns on circulating T lymphocytes may guide novel diagnostic and therapeutic approaches in the context of MMel. In the future, it will be important to investigate the dynamic expression of CCR/CXCR ligands as well as expression of CCR/CXCR by melanoma cells during immunotherapy or targeted therapy, with the aim of gaining even more clinically relevant information on the dialog between malignant cells and their microenvironment.
Methods
Characteristics of patients and cohorts
SORAFTEM.
The SORAFTEM (European Union Drug Regulating Authorities clinical trial EudraCT 2007-000527-18) cohort has been described previously (63). Patients over 18 years of age with histologically confirmed metastatic or unresectable melanoma, measurable disease, an Eastern Cooperative Oncology Group performance status below 2, and adequate hematologic, renal, hepatic, and coagulopathic functions were included in the SORAFTEM phase I/II investigator-sponsored study. T cell analyses were performed on peripheral blood mononuclear cells (PBMCs) from patients’ blood samples drawn prior to temozolomide and sorafenib treatment.
LUDWIG: MAGE3 protein–based vaccines.
Details on patient characteristics, protocol description, and survival have been previously reported by Kruit et al. (64). T cells were analyzed at the start of the study, prior to vaccine inoculation (1 month after relapse with conventional therapies), and correlated with OS.
IMAIL-2 study.
Adult patients with solid malignancy or measurable or evaluable disease and who were refractory to standard therapy were eligible for the study (IMAIL-2 phase I trial). The IMAIL-2 trial and its participating patients have been previously described, and T cell analyses were performed prior to cyclophosphamide, IL-2, and imatinib treatment (65). The clinical characteristics of these 3 cohorts are described in detail in Supplemental Table 2.
The ipilimumab-treated cohorts included 47 patients with MMel from 4 centers
Prospective French cohort of 22 patients.
All patients had stage IIIB/IV MMel and were treated at the Gustave Roussy Cancer Campus between March 2013 and October 2014. Blood samples were drawn from patients before and after 1, 2, 3, and 4 cycles of ipilimumab treatment. Patients’ samples were provided by C. Robert, N. Chaput, C. Coutzac, and S. Cotteret (Gustave Roussy Institute, Villejuif, France).
Retrospective German cohort of 4 patients.
Blood was collected, before and after 2 ipilimumab injections, from 4 patients participating in a phase II study evaluating the safety and efficacy of combined ipilimumab and intratumoral IL-2 treatment in pretreated patients with stage IV melanoma (ClinicalTrials.gov identifier: NCT01480323). Chemokine receptors were assessed in patients at baseline and after 2 injections of ipilimumab with concomitant treatment consisting of 7 or 8 injections of intratumoral IL-2 (9 mIU/injection).
Retrospective Italian cohort of 10 patients.
Blood samples were collected from patients before and after 1 and 2 injections of ipilimumab at the University Hospital of Siena between July 2011 and June 2015. The chemokine receptor profile was assessed after the samples were thawed.
Retrospective American cohort of 11 patients.
Blood samples were collected from patients before and after 2 injections of ipilimumab (ClinicalTrials.gov identifier: NCT00495066). The chemokine receptor profile was assessed after the samples were thawed. Clinical characteristics of these 4 cohorts are described in detail in Supplemental Table 3.
Prospective cohort of 20 patients with stage III/IV metastatic melanoma
Blood and metastatic LNs were collected at the time of surgery at the Gustave Roussy Cancer Center or at the CHU (Lyon, France) between May 2013 and March 2015 and were processed as mentioned below. Eighty-six percent of patients presented with stage III disease at sampling. Two stage IV patients presented with pulmonary metastases, and a third patient had abdominal metastases. Five patients (23%) were treated before sampling with DTIC (dacarbazine), IFN-α, or tyrosine kinase inhibitors.
Mice
C57Bl/6 mice expressing the human RET transgene in melanocytes under control of the murine metallothionein I promoter enhancer were provided by I. Nakashima (Chubu University, Aichi, Japan). Animals were crossed and kept under specific pathogen–free (SPF) conditions in the animal facility at the German Cancer Research Center (Heidelberg, Germany). All mice studied were between 5 and 7 weeks of age and had visible macroscopic tumors of different sizes. For s.c. tumors in the MCA205 model, mice between 7 and 14 weeks of age were used. WT SPF C57BL/6J mice were obtained from Harlan Laboratories and were kept in SPF conditions at the Gustave Roussy Institute. The murine fibrosarcoma MCA205 cell line was cultured at 37°C in 5% CO2 in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, 2 mM L-glutamine, 1 mM sodium pyruvate, and nonessential amino acids (all reagents from Gibco, Invitrogen). Mice were s.c. injected into the right flank with 0.8 million cells and i.p. with anti-CCL25 Ab (100 μg/mouse; clone 89818; R&D Systems); anti-CXCR3 (200 μg/mouse; clone CXCR3-173; BioXcell); and their respective isotype controls the same day but several hours before tumor inoculation and every 3 days for 5 injections. Organs were collected, digested, counted, and stained with the appropriate Abs, as described below.
PBMC preparations
PBMCs were thawed and washed in culture medium (SORAFTEM, IMAIL-2, LUDWIG, German, Italian, and American cohorts), i.e., RPMI 1640 (Gibco, Invitrogen), 10% human AB+ serum (Institut de Biotechnologies Jacques Boy), 1% penicillin/streptomycin (PEST; Gibco, Invitrogen), and 2 mM L-glutamine (Gibco, Invitrogen). Peripheral blood samples from patients (prospective cohorts) were carefully layered on top of a Ficoll-Hypaque density gradient (PAA Laboratories). PBMCs were collected and washed twice in Dulbecco’s PBS (Gibco, Invitrogen), followed by resuspension in PBS. Cells were then stained for flow cytometric analyses or resuspended in CryoMaxx medium (PAA Laboratories) for storage in liquid nitrogen. No significant differences between handling of cells from HVs versus those from patients could be detected in these processes.
TIL preparations
Resected LN specimens from 20 patients with MMel were analyzed for infiltrating lymphocyte phenotyping in parallel with paired-blood specimens. Tissue samples were cut and placed into dissociation medium, which consisted of RPMI 1640, 1% penicillin-streptomycin, type IV collagenase (50 IU/ml), hyaluronidase (280 IU/ml), and DNAse I (30 IU/ml) (all from Sigma-Aldrich) and were run on a gentleMACS Dissociator (Miltenyi Biotec). The cell samples were diluted in PBS and then passed through a cell strainer and centrifuged for 5 minutes at 450 g. Next, cells were resuspended in PBS and stained for flow cytometric analysis.
Flow cytometric analyses
Human study.
PBMCs and TILs were stained with fluorochrome-coupled mAbs (detailed in Supplemental Table 1), incubated for 20 minutes at 4°C, and then washed. Cell samples were acquired with a multicolor Cyan ADP 9 Color Flow Cytometer (Beckman Coulter). For compensations, single-stained Ab-capturing beads were used (Compbeads; BD Biosciences). Data were analyzed with FlowJo software, version 7.6.5. For dead-cell discrimination and to exclude B cells, NK cells, and macrophages or neutrophils, a dump channel with a LIVE/DEAD Cell-Staining Kit (Invitrogen, Life Technologies) and CD14, CD15, CD16, and CD19 mAbs were used. To discriminate between the different subsets of lymphocytes, T cells were stained with CD4, CD8, CCR7, CD45RA, CRTH2, CCR10, CXCR3, CCR6, CXCR5, CD103, CCR9, CLA, and CXCR4 mAbs (see Supplemental Figure 1 for the gating strategy). Of note, 2 tubes containing the same CCR10 Ab assessing different additional markers in each tube of flow cytometric staining were featured on the graphs as CCR10 and CCR10.2 (but significant correlations between the 2 tubes for CCR10 expression levels were found).
Mouse study.
The following fluorescence-labeled mAbs were used: anti–CD3-PerCP-Cy5.5, anti–CD4-PE-Cy7, anti–CD8-APC-Cy7, and anti–CD62L-APC (all from BD Biosciences); and anti–CD45RB-APC-Cy7, anti–CD183-PE, anti–CD199-FITC, and anti–CD44-PE-Cy7 (all from BioLegend). Single-cell suspensions were treated with FcR Blocking Reagent (Miltenyi Biotec) for 15 minutes at 4°C, followed by incubation with mAbs for 30 minutes at 4°C. Acquisition was performed by 6-color flow cytometry using the FACSCanto II with FACSDiva version 6.0 software, which includes the Cytometer Setup and Tracking System (both from BD Biosciences). Dead-cell exclusion was based on scatter profile or 7-AAD (BioLegend). The compensation control was performed with a BD CompBeads set (BD Biosciences) following the manufacturer’s instructions. FlowJo software was used to analyze samples.
Cell-sorting and T cell polarization analyses
Frozen PMBCs and/or TILs were quickly thawed in culture media, washed in sorting media (PBS supplemented with 10% FCS; PAA Laboratories) and 1 mM EDTA (Research Organics Inc.) and resuspended after filtration through a cell strainer. Cells were stained with the appropriate mAbs (see Supplemental Table 1) and sorted by flow cytometry on a 3-laser FACSAria III Cell Sorter (BD) or on a 3-laser MoFlo XDP Cell Sorter (Beckman Coulter). Sorted positive and negative cell populations were washed and resuspended in culture media and seeded in a 96-well plate with 20 UI/ml IL-2 (Proleukin) and stimulated with Dynabeads Human T-Activator CD3/CD28 (Gibco, Life Technologies) according to the manufacturer’s instructions. After 40 hours, supernatants were collected and frozen at –20°C until cytokine dosage with a Human Th1/Th2/Th9/Th17/Th22 13-plex RTU FlowCytomix Kit (eBioscience) according to the manufacturer’s instructions and acquired on a multicolor CyAn ADP 9 Color Flow Cytometer (Beckman Coulter). Analyses were performed using FlowCytomix Pro 3.0 Software (eBioscience).
Protein dosage
CCL25 dosage was done using a commercially available ELISA kit (R&D Systems) following the manufacturer’s recommendations. This concentration was normalized to the total protein content as measured by the DC protein assay (Bio-Rad) following the manufacturer’s protocol.
Statistics
Data were analyzed and presentations were performed within the R statistical environment (66). Flow cytometric parameters were converted to Z-scores prior to any correlation analyses. The estimation of metastatic group effect (human data) and organ distributions (mouse data) on FACS parameters was performed by beta regression modeling, with the dispersion allowed to differ between groups. For the other continuous parameters, generalized least-squares regression was applied to log-transformed data and contrasts of interest tested by Wald tests. Parameter distributions between the blood and tumor were assessed by mixed-effects linear modeling. OS, determined from the date of diagnosis, and progression-free survival (PFS), determined from the date of sampling, were used as the primary endpoints. Survival curves were estimated by the Kaplan-Meier product-limit method. Survival distributions were compared by Firth’s penalized likelihood Cox regression after adjusting for metastatic location or stage. The predictive abilities of individual markers for the response of patients to ipilimumab treatment were evaluated by logistic regression after adjusting for investigation center differences. Significance levels were determined by P values with up to 4 digits and adjusted according to the Benjamini-Hochberg (expressed as the FDR) or Holm (denoted by symbols in the graphs) method. Unless otherwise stated, P values are 2-sided, and 95% CIs for the statistic of interest are reported.
Study approval
Human studies.
IRB approval was granted by the University of Kremlin Bicêtre and the Institut Gustave Roussy (for the SORAFTEM and IMAIL-2 studies); the Ludwig Institute for Cancer Research (for the LUDWIG study); and the University of Kremlin Bicêtre, the University of Tuebingen, Memorial Sloan Kettering Cancer Center, and the University Hospital of Siena (for the ipilimumab-treated cohorts). The human study protocols were in accordance with the Declaration of Helsinki principles, and all patients provided informed consent before enrollment in the study.
Mouse studies.
All mouse experiments were approved by the DKFZ (Heidelberg, Germany) and the Comité d’éthique en expérimentation animale no. 26 (CEEA) (Villejuif, France).
Author contributions
LZ, AE, GK, and VU designed the study. NJ, CF, NV, CB, JMP, TY, MPR, RD, MV, VPC, MS, and SR conducted experiments. NJ, CF, DPE, CB, and LZ analyzed the data. DPE performed statistical evaluations. A Caignard, CLS, FS, and AM provided technical and scientific advice. A Cavalcanti, BW, HK, SD, AMDG, MM, PW, JY, and JW provided patients’ samples and clinical information. LZ wrote the paper, with input from NJ, DPE, and GK.
Supplementary Material
Acknowledgments
The authors thank C. Robert, N. Chaput, C. Coutzac, and S. Cotteret (Gustave Roussy Cancer Center) for patients’ specimens. We are indebted to P. Rameau and Y. Lecluse from the flow cytometry and imaging platform; to the Department of Biology and Pathology (Gustave Roussy and CHU of Lyon); and to the Plateforme d’Evaluation Préclinique of Gustave Roussy. The Institute for Research in Biomedicine is supported by the Helmut Horten Foundation. This work was supported by the Institut National du Cancer, the Agence Nationale pour la Recherche, the Ligue contre le Cancer (équipe labellisée de L. Zitvogel), the European Union (Inflammation and Cancer Research in Europe [INFLACARE]), the Site de Recherche Intégrée sur le Cancer (SOCRATES), LABEX OncoImmunology, the Direction Générale de l’Offre de Soins (Université Paris-Sud), and the Paris Alliance for Cancer Research Institutes. J.M. Pitt received a fellowship from the Association pour la Recherche sur le Cancer (ARC). N. Jacquelot received a fellowship from Cancéropole Ile-de-France.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(3):921–937. doi:10.1172/JCI80071.
Contributor Information
Nicolas Jacquelot, Email: jacquelot.nicolas@gmail.com.
Caroline Flament, Email: caroline.flament@igr.fr.
Carolin Blattner, Email: c.blattner@dkfz-heidelberg.de.
Jonathan M. Pitt, Email: dr.jonathan.pitt@gmail.com.
Takahiro Yamazaki, Email: tak.yamazak@gmail.com.
María Paula Roberti, Email: mpaula.roberti@gmail.com.
Romain Daillère, Email: romaindaillere@gmail.com.
Marie Vétizou, Email: marie.vetizou@gmail.com.
Vichnou Poirier-Colame, Email: vichnou.poiriercolame@gustaveroussy.fr.
Anne Caignard, Email: anne.caignard@inserm.fr.
Federica Sallusto, Email: federica.sallusto@irb.usi.ch.
Sylvie Rusakiewicz, Email: s_rusakiewicz@yahoo.com.
Benjamin Weide, Email: benjamin.weide@med.uni-tuebingen.de.
Aurélien Marabelle, Email: aurelien.marabelle@gustaveroussy.fr.
Holbrook Kohrt, Email: kohrt@stanford.edu.
Stéphane Dalle, Email: stephane.dalle@chu-lyon.fr.
Andréa Cavalcanti, Email: Andrea.CAVALCANTI@gustaveroussy.fr.
Guido Kroemer, Email: kroemer@orange.fr.
Anna Maria Di Giacomo, Email: a.digiacomo@ao-siena.toscana.it.
Michele Maio, Email: mmaiocro@gmail.com.
Phillip Wong, Email: wongp@mskcc.org.
Jianda Yuan, Email: yuanj@mskcc.org.
Jedd Wolchok, Email: wolchokj@mskcc.org.
Alexander Eggermont, Email: Alexander.EGGERMONT@gustaveroussy.fr.
Laurence Zitvogel, Email: laurence.zitvogel@orange.fr.
References
- 1.Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36(5):705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zweemer AJ, Toraskar J, Heitman LH, IJzerman AP. Bias in chemokine receptor signalling. Trends Immunol. 2014;35(6):243–252. doi: 10.1016/j.it.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Reiter E, Lefkowitz RJ. GRKs and β-arrestins: roles in receptor silencing, trafficking and signaling. Trends Endocrinol Metab. 2006;17(4):159–165. doi: 10.1016/j.tem.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 4. [May 10, 2015];IUPHAR/BPS Guide to Pharmacology, http://www.guidetopharmacology.org
- 5.Harlin H, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69(7):3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mabuchi T, Chang TW, Quinter S, Hwang ST. Chemokine receptors in the pathogenesis and therapy of psoriasis. J Dermatol Sci. 2012;65(1):4–11. doi: 10.1016/j.jdermsci.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Rivino L, Messi M, Jarrossay D, Lanzavecchia A, Sallusto F, Geginat J. Chemokine receptor expression identifies Pre-T helper (Th)1, Pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J Exp Med. 2004;200(6):725–735. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10(8):857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 9.Chevalier N, et al. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. J Immunol. 2011;186(10):5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- 10.Erdag G, et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 2012;72(5):1070–1080. doi: 10.1158/0008-5472.CAN-11-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 13.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 14.Messina JL, et al. 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci Rep. 2012;2: doi: 10.1038/srep00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salerno EP, Olson WC, McSkimming C, Shea S, Slingluff CL., Jr T cells in the human metastatic melanoma microenvironment express site-specific homing receptors and retention integrins. Int J Cancer. 2014;134(3):563–574. doi: 10.1002/ijc.28391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clancy-Thompson E, King LK, Nunnley LD, Mullins IM, Slingluff CL, Jr, Mullins DW. Peptide vaccination in Montanide adjuvant induces and GM-CSF increases CXCR3 and cutaneous lymphocyte antigen expression by tumor antigen-specific CD8 T cells. Cancer Immunol Res. 2013;1(5):332–339. doi: 10.1158/2326-6066.CIR-13-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salerno EP, et al. Activation, dysfunction and retention of T cells in vaccine sites after injection of incomplete Freund’s adjuvant, with or without peptide. Cancer Immunol Immunother. 2013;62(7):1149–1159. doi: 10.1007/s00262-013-1435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullins IM, et al. CXC chemokine receptor 3 expression by activated CD8+ T cells is associated with survival in melanoma patients with stage III disease. Cancer Res. 2004;64(21):7697–7701. doi: 10.1158/0008-5472.CAN-04-2059. [DOI] [PubMed] [Google Scholar]
- 19.Hannani D, et al. Anticancer immunotherapy by CTLA4 blockade: obligatory contribution of IL-2 receptors and negative prognostic impact of soluble CD25. Cell Res. 2015;25(2):208–224. doi: 10.1038/cr.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robert C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364(26):2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 21.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umansky V, et al. Melanoma-specific memory T cells are functionally active in Ret transgenic mice without macroscopic tumors. Cancer Res. 2008;68(22):9451–9458. doi: 10.1158/0008-5472.CAN-08-1464. [DOI] [PubMed] [Google Scholar]
- 23.Sevko A, et al. Antitumor effect of paclitaxel is mediated by inhibition of myeloid-derived suppressor cells and chronic inflammation in the spontaneous melanoma model. J Immunol. 2013;190(5):2464–2471. doi: 10.4049/jimmunol.1202781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umansky V, Sevko A. Ret transgenic mouse model of spontaneous skin melanoma: focus on regulatory T cells. Pigment Cell Melanoma Res. 2013;26(4):457–463. doi: 10.1111/pcmr.12104. [DOI] [PubMed] [Google Scholar]
- 25.Meyer C, et al. Chronic inflammation promotes myeloid-derived suppressor cell activation blocking antitumor immunity in transgenic mouse melanoma model. Proc Natl Acad Sci U S A. 2011;108(41):17111–17116. doi: 10.1073/pnas.1108121108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sevko A, et al. Cyclophosphamide promotes chronic inflammation-dependent immunosuppression and prevents antitumor response in melanoma. J Invest Dermatol. 2013;133(6):1610–1619. doi: 10.1038/jid.2012.444. [DOI] [PubMed] [Google Scholar]
- 27.Hong M, et al. Chemotherapy induces intratumoral expression of chemokines in cutaneous melanoma, favoring T-cell infiltration and tumor control. Cancer Res. 2011;71(22):6997–7009. doi: 10.1158/0008-5472.CAN-11-1466. [DOI] [PubMed] [Google Scholar]
- 28.Amersi FF, et al. Activation of CCR9/CCL25 in cutaneous melanoma mediates preferential metastasis to the small intestine. Clin Cancer Res. 2008;14(3):638–645. doi: 10.1158/1078-0432.CCR-07-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Baruch A. Organ selectivity in metastasis: regulation by chemokines and their receptors. Clin Exp Metastasis. 2008;25(4):345–356. doi: 10.1007/s10585-007-9097-3. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto J, et al. Differential expression of the chemokine receptors by the Th1- and Th2-type effector populations within circulating CD4+ T cells. J Leukoc Biol. 2000;68(4):568–574. [PubMed] [Google Scholar]
- 31.Dengel LT, et al. Interferons induce CXCR3-cognate chemokine production by human metastatic melanoma. J Immunother. 2010;33(9):965–974. doi: 10.1097/CJI.0b013e3181fb045d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antonicelli F, et al. CXCL10 reduces melanoma proliferation and invasiveness in vitro and in vivo. Br J Dermatol. 2011;164(4):720–728. doi: 10.1111/j.1365-2133.2010.10176.x. [DOI] [PubMed] [Google Scholar]
- 33.Nardin A , et al. Dacarbazine promotes stromal remodeling and lymphocyte infiltration in cutaneous melanoma lesions. J Invest Dermatol. 2011;131(9):1896–1905. doi: 10.1038/jid.2011.128. [DOI] [PubMed] [Google Scholar]
- 34.Zou L, et al. Bone marrow is a reservoir for CD4+CD25+ regulatory T cells that traffic through CXCL12/CXCR4 signals. Cancer Res. 2004;64(22):8451–8455. doi: 10.1158/0008-5472.CAN-04-1987. [DOI] [PubMed] [Google Scholar]
- 35.Roy LD, Sahraei M, Schettini JL, Gruber HE, Besmer DM, Mukherjee P. Systemic neutralization of IL-17A significantly reduces breast cancer associated metastasis in arthritic mice by reducing CXCL12/SDF-1 expression in the metastatic niches. BMC Cancer. 2014;14: doi: 10.1186/1471-2407-14-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13(5):587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 37.Schiraldi M, et al. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J Exp Med. 2012;209(3):551–563. doi: 10.1084/jem.20111739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguilar-Cazares D, et al. Relationship of dendritic cell density, HMGB1 expression, and tumor-infiltrating lymphocytes in non-small cell lung carcinomas. Appl Immunohistochem Mol Morphol. 2014;22(2):105–113. doi: 10.1097/PAI.0b013e3182849808. [DOI] [PubMed] [Google Scholar]
- 39.Stenstad H, Svensson M, Cucak H, Kotarsky K, Agace WW. Differential homing mechanisms regulate regionalized effector CD8αβ+ T cell accumulation within the small intestine. Proc Natl Acad Sci U S A. 2007;104(24):10122–10127. doi: 10.1073/pnas.0700269104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 41.Wurbel M-A, Malissen M, Guy-Grand D, Malissen B, Campbell JJ. Impaired accumulation of antigen-specific CD8 lymphocytes in chemokine CCL25-deficient intestinal epithelium and lamina propria. J Immunol. 2007;178(12):7598–7606. doi: 10.4049/jimmunol.178.12.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans-Marin HL, et al. Unexpected regulatory role of CCR9 in regulatory T cell development. PLoS One. 2015;10(7): doi: 10.1371/journal.pone.0134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Li F, Wei H, Lian ZX, Sun R, Tian Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J Exp Med. 2014;211(12):2397–2410. doi: 10.1084/jem.20140625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamoto N, et al. CCR9+ macrophages are required for acute liver inflammation in mouse models of hepatitis. Gastroenterology. 2012;142(2):366–376. doi: 10.1053/j.gastro.2011.10.039. [DOI] [PubMed] [Google Scholar]
- 45.Mavigner M, et al. Altered CD4+ T cell homing to the gut impairs mucosal immune reconstitution in treated HIV-infected individuals. J Clin Invest. 2012;122(1):62–69. doi: 10.1172/JCI59011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson-Holiday C, et al. CCL25 mediates migration, invasion and matrix metalloproteinase expression by breast cancer cells in a CCR9-dependent fashion. Int J Oncol. 2011;38(5):1279–1285. doi: 10.3892/ijo.2011.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.et al. CCR9 mediates PI3K/AKT-dependent antiapoptotic signals in prostate cancer cells and inhibition of CCR9-CCL25 interaction enhances the cytotoxic effects of etoposide. Int J Cancer. 2010;127(9):2020–2030. doi: 10.1002/ijc.25219. [DOI] [PubMed] [Google Scholar]
- 48.Johnson-Holiday C, Singh R, Johnson EL, Grizzle WE, Lillard JW, Jr, Singh S. CCR9-CCL25 interactions promote cisplatin resistance in breast cancer cell through Akt activation in a PI3K-dependent and FAK-independent fashion. World J Surg Oncol. 2011;9: doi: 10.1186/1477-7819-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta P, et al. CCR9/CCL25 expression in non-small cell lung cancer correlates with aggressive disease and mediates key steps of metastasis. Oncotarget. 2014;5(20):10170–10179. doi: 10.18632/oncotarget.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye LF, Huang J, Zhang LP, Chen Z. Intracellular expression profile and clinical significance of the CCR9-CCL25 chemokine receptor complex in nasopharyngeal carcinoma. J Laryngol Otol. 2015;129(10):1013–1019. doi: 10.1017/S0022215115002108. [DOI] [PubMed] [Google Scholar]
- 51.Zhong Y, et al. Expression of CC chemokine receptor 9 predicts poor prognosis in patients with lung adenocarcinoma. Diagn Pathol. 2015;10: doi: 10.1186/s13000-015-0341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen HJ, et al. Comprehensive models of human primary and metastatic colorectal tumors in immunodeficient and immunocompetent mice by chemokine targeting. Nat Biotechnol. 2015;33(6):656–660. doi: 10.1038/nbt.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vela M, Aris M, Llorente M, Garcia-Sanz JA, Kremer L. Chemokine receptor-specific antibodies in cancer immunotherapy: achievements and challenges. Front Immunol. 2015;6: doi: 10.3389/fimmu.2015.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faaij CM, et al. A possible role for CCL27/CTACK-CCR10 interaction in recruiting CD4 T cells to skin in human graft-versus-host disease. Br J Haematol. 2006;133(5):538–549. doi: 10.1111/j.1365-2141.2006.06058.x. [DOI] [PubMed] [Google Scholar]
- 55.Xia M, et al. CCR10 regulates balanced maintenance and function of resident regulatory and effector T cells to promote immune homeostasis in the skin. J Allergy Clin Immunol. 2014;134(3):634–644. doi: 10.1016/j.jaci.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monteagudo C, et al. CCL27-CCR10 and CXCL12-CXCR4 chemokine ligand-receptor mRNA expression ratio: new predictive factors of tumor progression in cutaneous malignant melanoma. Clin Exp Metastasis. 2012;29(6):625–637. doi: 10.1007/s10585-012-9476-2. [DOI] [PubMed] [Google Scholar]
- 57.Fujita Y, et al. Presence of circulating CCR10+ T cells and elevated serum CTACK/CCL27 in the early stage of mycosis fungoides. Clin Cancer Res. 2006;12(9):2670–2675. doi: 10.1158/1078-0432.CCR-05-1513. [DOI] [PubMed] [Google Scholar]
- 58.Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol. 2011;11(9):597–606. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]
- 59.Torregrossa L, et al. CXCR4 expression correlates with the degree of tumor infiltration and BRAF status in papillary thyroid carcinomas. Mod Pathol. 2012;25(1):46–55. doi: 10.1038/modpathol.2011.140. [DOI] [PubMed] [Google Scholar]
- 60.Monteagudo C, Martin JM, Jorda E, Llombart-Bosch A. CXCR3 chemokine receptor immunoreactivity in primary cutaneous malignant melanoma: correlation with clinicopathological prognostic factors. J Clin Pathol. 2007;60(6):596–599. doi: 10.1136/jcp.2005.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Boyle G, et al. Inhibition of CXCR4-CXCL12 chemotaxis in melanoma by AMD11070. Br J Cancer. 2013;108(8):1634–1640. doi: 10.1038/bjc.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tubo NJ, Wurbel MA, Charvat TT, Schall TJ, Walters MJ, Campbell JJ. A systemically-administered small molecule antagonist of CCR9 acts as a tissue-selective inhibitor of lymphocyte trafficking. PLoS One. 2012;7(11): doi: 10.1371/journal.pone.0050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romero AI, et al. Regulation of CD4(+)NKG2D(+) Th1 cells in patients with metastatic melanoma treated with sorafenib: role of IL-15Rα and NKG2D triggering. Cancer Res. 2014;74(1):68–80. doi: 10.1158/0008-5472.CAN-13-1186. [DOI] [PubMed] [Google Scholar]
- 64.Kruit WH, et al. Phase 1/2 study of subcutaneous and intradermal immunization with a recombinant MAGE-3 protein in patients with detectable metastatic melanoma. Int J Cancer. 2005;117(4):596–604. doi: 10.1002/ijc.21264. [DOI] [PubMed] [Google Scholar]
- 65.Pautier P, et al. Phase I clinical trial combining imatinib mesylate and IL-2 in refractory cancer patients: IL-2 interferes with the pharmacokinetics of imatinib mesylate. Oncoimmunology. 2013;2(2): doi: 10.4161/onci.23079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R Development Core Team . R: A Language And Environment For Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.