Abstract
Focal segmental glomerulosclerosis (FSGS) is a syndrome that involves kidney podocyte dysfunction and causes chronic kidney disease. Multiple factors including chemical toxicity, inflammation, and infection underlie FSGS; however, highly penetrant disease genes have been identified in a small fraction of patients with a family history of FSGS. Variants of apolipoprotein L1 (APOL1) have been linked to FSGS in African Americans with HIV or hypertension, supporting the proposal that genetic factors enhance FSGS susceptibility. Here, we used sequencing to investigate whether genetics plays a role in the majority of FSGS cases that are identified as primary or sporadic FSGS and have no known cause. Given the limited number of biopsy-proven cases with ethnically matched controls, we devised an analytic strategy to identify and rank potential candidate genes and used an animal model for validation. Nine candidate FSGS susceptibility genes were identified in our patient cohort, and three were validated using a high-throughput mouse method that we developed. Specifically, we introduced a podocyte-specific, doxycycline-inducible transactivator into a murine embryonic stem cell line with an FSGS-susceptible genetic background that allows shRNA-mediated targeting of candidate genes in the adult kidney. Our analysis supports a broader role for genetic susceptibility of both sporadic and familial cases of FSGS and provides a tool to rapidly evaluate candidate FSGS-associated genes.
Introduction
The glomerulus of the kidney is a specialized capillary bed that generates an ultrafiltrate that, after modification by the kidney tubule system, becomes urine. Diseases of the glomerulus often lead to chronic kidney disease, a major health care problem affecting between 5% and 10% of the adult population in developed countries (1). Treatment options are limited, in part owing to the poor understanding of the pathogenesis of glomerular disease. Better insights into the root cause of this disease will offer hope for improvement of this situation.
One of the most common glomerular syndromes is focal segmental glomerulosclerosis (FSGS). The pathologic change of FSGS is a scar that develops focally (in some but not all glomeruli) and segmentally (in only part of a glomerulus). While originally considered a disease, FSGS is now thought to consist of a variety of different syndromes. These include primary (idiopathic) FSGS, which is thought to be caused by a circulating factor, and secondary FSGS, which may be caused by viruses, medications, and genetic mutations. The most common form of secondary FSGS follows glomerular hyperfiltration arising from a mismatch between metabolic load and glomerular capacity and is associated with obesity, low birth weight, reduced renal mass, and other causes. Genetic mutations alone can be sufficient to cause disease (Mendelian) or may increase susceptibility to FSGS by potentiating the effects of environmental factors.
The glomerulus is composed of 3 different cell types: endothelial cells, mesangial cells, and epithelial cells known as podocytes. The podocyte is an unusual cell that covers the outside of the capillary wall and interdigitates with other podocytes to create small slits that allow the passage of fluid and small solutes into the urinary space. It is now clear that podocyte dysfunction is responsible for FSGS as well as other glomerular diseases such as minimal change disease, membranous glomerulopathy, and congenital nephrotic syndrome. Current models suggest that increased podocyte loss is the primary lesion in FSGS (2–5).
Over the past 10 years, various genetic approaches have identified mutations in over 20 podocyte genes as causative or leading to increased susceptibility to FSGS (6, 7). Mutations in these genes, however, explain only a small fraction of familial and sporadic FSGS cases (8–10). A larger fraction of cases may involve non-Mendelian forms of FSGS that could involve variants in multiple genes that interact together to generate susceptibility to podocyte injury and loss. Further gene discovery in oligogenic disease is challenged, however, by the fact that mutations will be distributed across many genes and be difficult to distinguish from numerous neutral gene variants (11, 12). A greater understanding of genetic causes of FSGS has the potential to elucidate molecular pathways that are involved in the disease.
In terms of the number of people affected, the most significant genetic contributor to FSGS susceptibility identified to date is APOL1. FSGS-associated alleles of APOL1, called G1 and G2, are common in West African populations, likely as a consequence of providing resistance to trypanosomiasis (13–15). The presence of 2 variant alleles significantly increases the risk of arterionephrosclerosis (hypertensive nephropathy) (odds ratio [OR] = 7), FSGS (OR = 17), or HIV-associated nephropathy (OR = 29) in African Americans (13, 16) and in South Africans (OR = 89) (17). Approximately 13% of African Americans carry 2 variant alleles and are at increased risk for chronic kidney disease. These variants by themselves largely explain the increased frequency of FSGS among African Americans. Despite this, the mechanisms by which APOL1 variants cause or predispose individuals to glomerular damage remain unknown. As these variants are absent from individuals lacking any African ancestry, they are not documented to play a role in FSGS susceptibility in individuals of other ancestries.
Here, we used high-throughput sequencing of DNA from FSGS patients of Northern European ancestry to identify genes that are potentially involved in susceptibility to the disease. The challenge of studying the genetics of sporadic FSGS is the possibility that a large number of genes may be involved and the likelihood that each gene contributes only a small amount of risk for the disease. In addition, the relatively low incidence of FSGS in adult and pediatric populations (~5/million/year) (1) and the even fewer number of cases that are confirmed by kidney biopsy preclude the assembly of a cohort of the size required for standard genetic approaches like GWAS, whole-genome sequencing, or exome sequencing (18). Thus, most genetic studies of FSGS have been family studies.
Here, we sequenced DNA from 214 patients of European ancestry with biopsy-confirmed FSGS and tested a variety of analytic approaches to mitigate our limited sample size. Since FSGS is considered a disease of podocytes, we focused our sequencing analysis on 2,500 genes that are highly and/or specifically expressed in podocytes. This approach significantly reduced the multitest penalty. We also developed a robust analytic pipeline permitting the use of individuals sequenced for other genetic studies as controls. Since there is no in vitro assay for FSGS, we developed a screening method using mice. Our system is based on a murine embryonic stem (ES) cell line with an FSGS-susceptible genetic background that allows for efficient, targeted delivery of shRNAs to generate mice that are nearly 100% derived from the ES cells, eliminating the need for subsequent breeding. This method allowed us to rapidly test 6 candidate genes and validate 3 new FSGS susceptibility genes. We expect that our system will allow for large numbers of candidate genes constituting the network of FSGS genes to be validated and that it will provide critical insight into the pathogenesis of this disease syndrome. In addition, our experimental approach should be broadly applicable to studying other uncommon diseases in which susceptibility genes are suspected.
Results
Sequencing of DNA from FSGS patients.
We conducted high-throughput DNA-sequencing studies focusing on 2,500 genes (~7 Mb) (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI82592DS1) that are highly expressed in podocytes, reasoning that the genetic susceptibility would be intrinsic to the podocyte (Supplemental Figure 1A). The list of genes that we sequenced included most genes currently implicated in familial FSGS (13, 19–23), approximately 200 genes that are functionally linked to these genes, 677 genes chosen on the basis of their high expression in microdissected human glomeruli (24), and 1,600 human orthologs of highly expressed genes identified by DNA microarrays of mouse podocytes (25–27).
We performed sequencing of DNA from 214 patients of European ancestry with biopsy-confirmed FSGS, including 192 patients with sporadic FSGS and 22 with familial FSGS. DNA samples were obtained from patients participating in a multicenter NIH study of biopsy-confirmed FSGS (16) and from patients diagnosed at Washington University. All subjects provided informed consent for the genetic studies. We focused on patients of European ancestry, because a well-characterized control set used for a genetic study of autism but unascertained for kidney disease was available that had a similar genetic ancestry (28). A similar control dataset for African or African admixture patients was not available at the time we performed this sequencing study, which prevented us from including these patients in our analysis.
To validate that our patient sequencing data were comparable to those of our control group, we processed data for both patients and controls in a single batch, with raw data aligned to the human genome (29–31). The depth of coverage was compared between patients and controls, and only those exons covered adequately (>20 times) and similarly in both patients and controls were advanced to the analysis stage (Supplemental Figure 1B). In summary, 16,784 exons and 2,769,942 bp were confidently covered in both patient and control cohorts, resulting in 16,008 SNPs and 1,724 genes analyzed in the final dataset. SNP calls were equally represented in patients and controls.
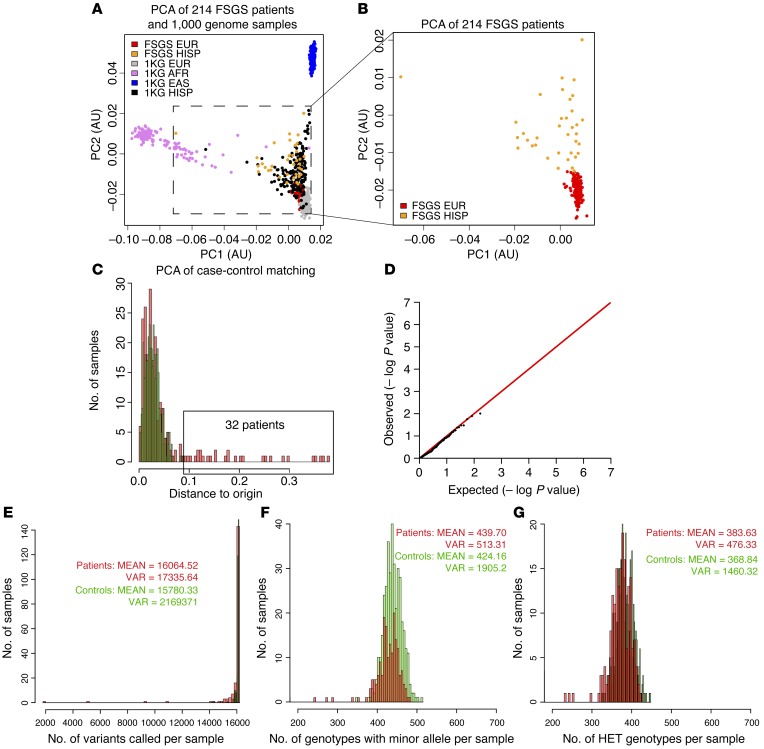
Thirty-two patients were removed from the study but reserved for follow-up because trace Hispanic ancestry was detected by principal component analysis (PCA) (Figure 1, A–C). Three patients were removed because the call rate of SNPs was less than 95%. The remaining FSGS patients (157 sporadic and 22 familial) had a similar number of SNPs, heterozygous genotypes, and genotypes containing an alternative allele per sample (Figure 1, E–G), allowing us to proceed to association analysis. Our final dataset contained 179 patients and 378 controls and included 157 sporadic and 22 familial FSGS patients. The accuracy of our analysis strategy was confirmed by resequencing key SNPs using Sanger sequencing and by showing that sequencing the same sample at both the Broad Institute and Washington University gave similar results.
Figure 1. Comparability of patients and controls.
(A) PCA plot of FSGS patients and 1,000 genome samples. The inset shows the distribution of putative Northern European FSGS patients in the PCA plot in relationship to 1,000 genome samples. (B) Magnified view of the inset area in A. (C) PCA analysis of patients and controls is depicted as the distance from the origin. Thirty-two patients with a highly similar variant profile but with a distance of more than 0.9 were removed and used as a follow-up group. (D) Fisher’s exact test of the common (MAF >5%) variants showed the absence of stratification and confirmed the validity and quality of our method for case-control matching. (E) Comparison of the total number of variants per sample showed that patients and controls were similar. (F) Comparison of the total number of heterozygous genotypes showed that patients and controls were similar. (G) Comparison of the total number of heterozygous and homozygous genotypes containing an alternative allele showed that patients and controls were similar. EUR, European; HISP, Hispanic; AFR, African; EAS, East Asian; VAR, variants; HET, heterozygous; PC1, principal component 1; PC2, principal component 2; KG, from 1000 Genomes Database.
No susceptibility genes were identified by single-variant analysis.
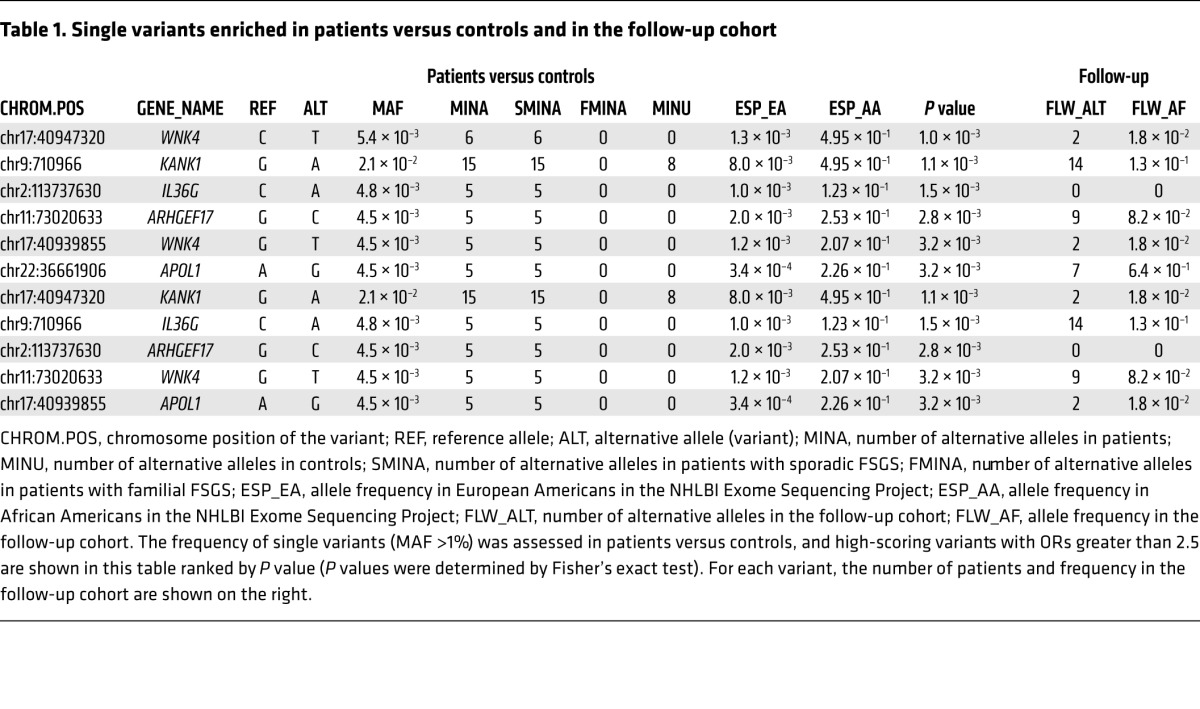
An association test examining single variants (minor allele frequency [MAF] >1%) was performed using Fisher’s exact test. No variants were detected with a P value below the multitest threshold (2 × 10–5) (Supplemental Table 2). The lack of significance is not surprising, given the relatively small size of our sample. This analysis did confirm that the distribution of synonymous and missense variants was similar between patients and controls (Figure 1D). Table 1 shows a list of the 10 highest-scoring variants. All the variants are missense variants. As a follow-up, we analyzed the 32 samples with Hispanic admixture, combined with 23 additional European ancestry samples that were sequenced from the Nephrotic Syndrome Study Network (NEPTUNE) cohort (32). This confirmed enrichment of 3 of the missense sequence variants in WNK4, KANK1, and ARHGEF17 (Table 1). Interestingly, KANK1 was recently identified as a susceptibility gene for familial nephrotic syndrome (33).
Table 1. Single variants enriched in patients versus controls and in the follow-up cohort.
Rare variant analysis identified 6 new potential FSGS susceptibility genes. We analyzed the rare variants (MAF <1%) using tests that compared the total numbers of rare variants between patients and controls. Currently, it is believed that there is an inverse correlation between the frequency of the allele and the potential risk. Thus, at each gene, we tested 2 distinct modes of inheritance for FSGS: the increased presence of extremely rare alleles that are predicted to be highly damaging and therefore highly penetrant (model 1), or the presence of low-frequency and less-damaging alleles with risk and protective variation intermingled with neutral variation (model 2). To discriminate between these 2 models, we analyzed 2 subsets of variants. For the first model, we used the Exome Aggregation Consortium (ExAC) browser (http://exac.broadinstitute.org) to identify 5,662 missense and loss-of-function variants in our dataset that are present at a frequency of less than 0.01% in the European population. We then tested the burden of these rare variants in FSGS patients versus controls (Supplemental Table 3 and ref. 34). Using this analysis, we found that no genes reached a level of statistical significance for rare, highly penetrant variants under this model.
To examine the second model involving low-frequency risk and protective variants, we selected all missense and loss-of-function variants with a MAF of less than 1% and compared their distribution between patients and controls using 2 different rare variant tests: the variable threshold (VT) (35) and the C-α test (36). Because the effect sizes of variants differ, the accuracy of each method can vary depending on the specific situation. Using a P value of less than 0.05 (Bonferroni-corrected P ≤ 0.00003) as a cutoff, no genes were identified that exceeded this value, but 8 genes (WNK4, APOL1, DLG5, GCC1, XYLT1, KAT2B, BPTF, and COL4A4) had P values close to the Bonferroni-corrected value (P < 0.00006 to P < 0.0008, Table 2, and Supplemental Table 4). Since the Bonferroni test tends to be conservative and APOL1 (13–15) and COL4A4 (37) are known FSGS genes, we selected these genes for further analysis as potential FSGS susceptibility genes.
Table 2. Top genes identified by rare variant analyses.

Our analysis of extremely rare variants (MAF <0.01%) in this set of 8 genes showed enrichment in 3 of these genes: GCC1, APOL1, and COL4A4. Examination of our follow-up group (55 samples) confirmed enrichment of a subset of the same rare variants found in our larger cohort in all of the genes except COL4A4 (Table 2), supporting the findings of our rare variant analysis. Since APOL1 and COL4A4 were already known (13–15, 37), the remainder of the identified genes (BPTF, DLG5, GCC1, KAT2B, WNK4, and XYLT1) could represent 6 new potential FSGS susceptibility genes. Notably, WNK4 was also identified by single-variant analysis.
Interestingly, 4 patients with sporadic FSGS carried the APOL1 G1 variant (G1), a known risk variant for FSGS that is present in 29% of African Americans but is a rare variant (0.03%) in European Americans. This finding was also seen in the follow-up set, in which 7 of 55 additional samples were identified with this mutation. Since the allele frequency of the APOL1 G1 allele in approximately 5,500 Hispanic samples in the ExAC dataset was only 0.5%, this represents significant enrichment, regardless of the Hispanic admixture.
Testing for the enrichment of rare variants in 20 previously known FSGS genes.
Family studies have identified nearly 30 genes that cause familial FSGS (6). To determine whether a set of 20 of these genes (Supplemental Table 5) are also involved in sporadic FSGS, we assessed the frequencies of predicted damaging, rare coding variants (missense and loss-of-function with a MAF <1%) in these genes in patients and controls. Approximately 36.9% of patients (66 of 179) had at least 1 predicted deleterious rare variant in these 20 genes compared with 3.4% of controls (13 of 378) (the list of variants is shown in Supplemental Tables 6 and 7). The distribution of variants between familial and sporadic cases was similar and consistent with previous studies showing that approximately 30% of steroid-resistant nephrotic syndrome patients presenting before the age of 25 have a variant in one of the known disease genes (38). There was also a difference in the total number of unique rare variants identified in patients (32.9%, 59 variants in 179 patients) versus controls (3.9%, 15 variants in 378 control subjects) (Supplemental Table 7). The significance of this finding was tested using a permutation analysis of differences in variants between patients and controls in all potential random groups of 20 genes chosen from our database of 1,724 genes. This showed, however, that 27% of the random sets of 20 genes had a similar or higher burden of rare variants compared with the set of 20 FSGS genes. This suggests that our patient dataset contains additional novel FSGS susceptibility genes with strong genetic effects.
Development of a high-throughput method to validate candidate genes in mice.
Since FSGS cannot be modeled in vitro and most confirmatory studies are performed today in zebrafish (33, 39–41), we developed a genetic system in mice to examine the function of candidate genes in vivo. Our strategy involved inhibiting the expression of candidate genes in podocytes from mice on a genetic background that is prone to develop FSGS. Mice that are heterozygous for 2 podocyte genes, Cd2ap and Synpo, develop FSGS with an incomplete penetrance (~25%–50%) and significant albuminuria occurring at about 6 months of age (42). Assuming that FSGS is an oligogenic disease, we reasoned that knocking down a bona fide disease gene in this background would accelerate disease onset.
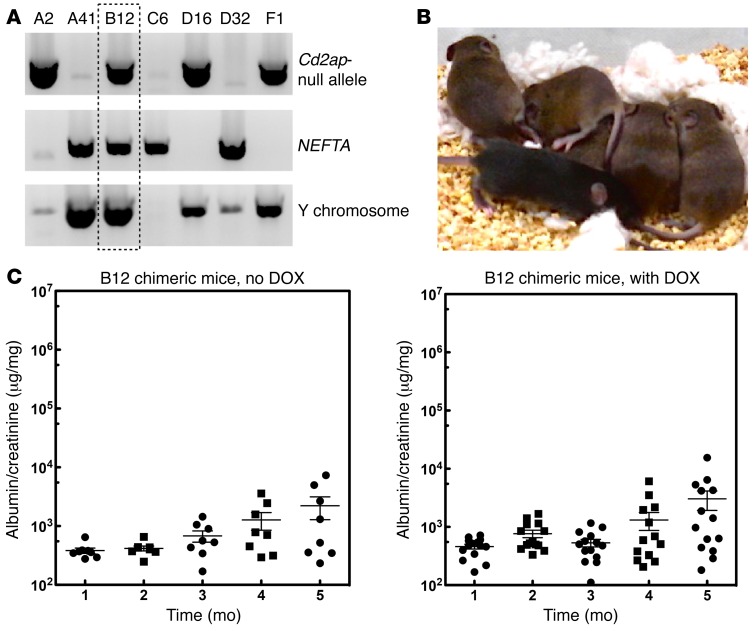
We generated a mouse ES cell line that was Cd2ap+/– and Synpo+/– using standard methods. The ES cell also expresses a podocyte-specific and doxycycline-inducible (DOX-inducible) transactivator (Nphs1-rtTA3G) that allows inducible expression of an shRNA (Figure 2A and ref. 43). We reasoned that inducible RNAi would allow us to study the role of a gene in the mature kidney without worrying about developmental effects. The new method using laser-assisted microinjection into 8-cell embryos (44) allowed us to generate mice that were nearly 100% derived from the ES cells without further breeding (Figure 2B). Consistent with mice generated by conventional breeding, approximately 50% of the mice generated from these ES cells developed mild proteinuria after 12 to 16 weeks of age (Figure 2C). To eliminate variability introduced by random integration of an RNAi transgene, we targeted a single copy of the RNAi transgene into the mouse Hprt1 locus (45) that allows the use of 6-thioguanine for efficient selection (Figure 3A).
Figure 2. Development of ES cells sensitized for FSGS.
(A) Identification of FSGS-sensitized ES cells. Our breeding strategy predicted that 1 of 8 embryos would have the correct genotype. ES cells were generated using standard approaches and genotyped for Cd2ap heterozygosity (upper panel), the NEFTA transgene (middle panel), and the Y chromosome (lower panel). (B) Laser-assisted injection generated mice with high chimerism. In the example shown, the ES cell line (agouti) was injected into 8-cell C57/BL6 (black) embryos. Compared with noninjected embryos (resulting in the 2 black mice shown on the bottom), all of the injected embryos generated pups that were close to being purely agouti. Injection of ES cells into C57/BL6 albino embryos resulted in completely agouti animals (not shown). (C) Mice generated from ES cells developed mild proteinuria after 4 months, with no DOX treatment. Fifteen mice were generated from the sensitized ES cells and treated with or without DOX in the drinking water. Urine was tested every month by measuring the albumin/creatinine ratios. Mice developed low-level proteinuria at 4 months of age, but the level of proteinuria was not affected by DOX treatment.
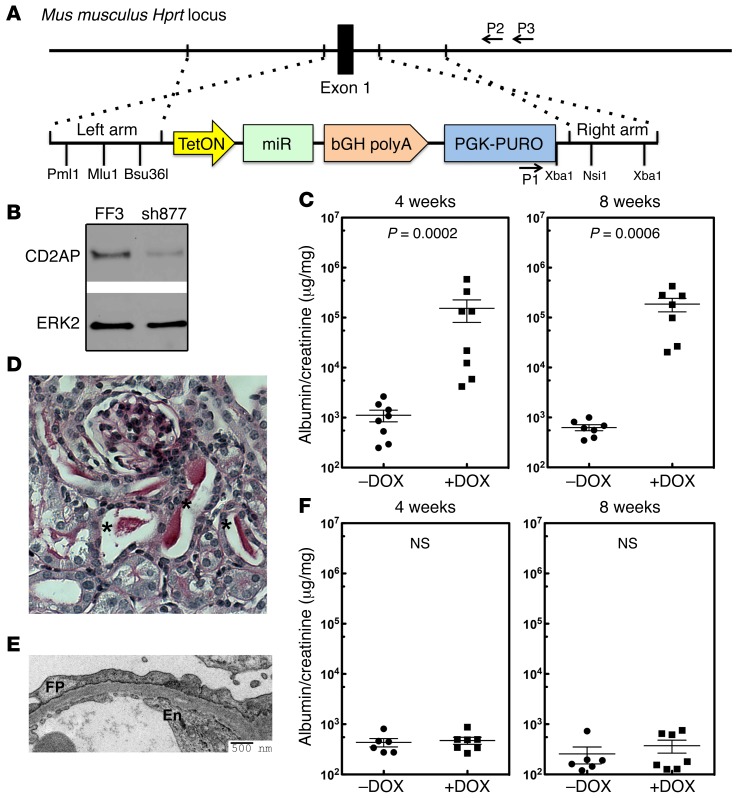
Figure 3. Validation of the system using CD2AP knockdown.
(A) Targeting strategy used to integrate a miR30-shRNA transgene into the Hprt1 locus. (B) Knockdown efficiency of a miR30-shRNA for Cd2ap (sh877). Immunoblot shows endogenous CD2AP in NIH3T3 cells stably transduced with FF3 (control shRNA) or sh877. The validation of sh877 is shown in Supplemental Figure 2A. Panel B represents multiple experiments (n = 3) conducted to test the efficiency of the RNAi. (C) Sixteen mice generated with ES cells with the Cd2ap shRNA that was targeted to the Hprt1 locus were treated with or without DOX, and urine was analyzed by measuring the urine albumin/creatinine ratio at 4 and 8 weeks. (D) Histology from a representative Cd2ap RNAi mouse treated with DOX showing protein casts (indicated with asterisks; n >5). (E) Representative electron microscopic image from a Cd2ap RNAi mouse treated with DOX shows podocyte foot process (FP) effacement. En, endothelial cells (n = 9). (F) Thirteen control mice were generated with a control luciferase RNAi targeted to the Hgprt locus. Mice were treated with (n = 6) or without (n = 7) DOX, and urine was analyzed by measuring the albumin/creatinine ratio at 4 and 8 weeks. A 2-tailed Mann-Whitney U test was used to calculate the P values in C and F. A P value of less than 0.05 was considered statistically significant.
Since Cd2ap is an FSGS disease gene (46) and knockout (KO) mice develop severe proteinuria (47), we validated our system by generating Cd2ap RNAi mice. Multiple Cd2ap-specific RNAis were tested for their ability to inhibit Cd2ap expression (Figure 3B and Supplemental Figure 2A), and the RNAi showing the greatest inhibition (sh877) was embedded into a miR30 sequence that allows for DOX-inducible expression (Supplemental Figure 2, B and C, and ref. 48). An RNAi for the firefly luciferase gene (FF3) was used as a control. Half of the founder (F0) animals were treated with DOX at 2 weeks of age to induce shRNA transgene expression. All of the DOX-treated mice developed sustained proteinuria that was over 150-fold higher than that seen in the control animals (Figure 3C). Histological analysis of the kidneys revealed protein casts in the tubules (Figure 3D). Electron microscopic examination of the kidney showed widespread foot process effacement, a marker of proteinuria (Figure 3E), validating that our RNAi strategy could be used to test candidate FSGS genes. Interestingly, the proteinuric mice recovered after removal of DOX treatment (Supplemental Figure 2D). In contrast, FF3-RNAi mice did not show proteinuria after treatment with DOX for 8 weeks (Figure 3F), and no abnormalities were detected by electron microscopy or histology (Supplemental Figure 2E).
Validation of WNK4, KANK1, and ARHGEF17 as potential FSGS susceptibility genes.
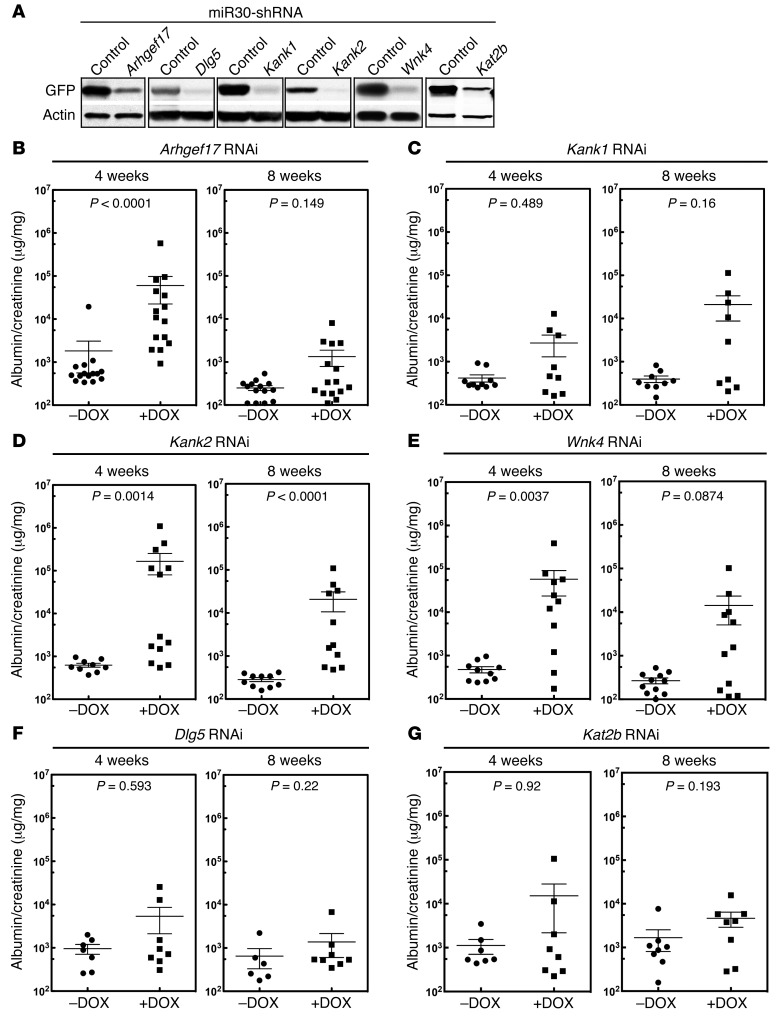
Three of the six genes, WNK4, DLG5, and KAT2B, identified by rare variant analysis were chosen for testing. We also chose the 3 single-variant candidates, KANK1, WNK4, and ARHGEF17. Since WNK4 was present on both lists, a total of 5 genes were selected for analysis. Because the exact mouse ortholog for human KANK1 is unknown, because Kank2 is more highly expressed in mouse podocytes (49), and because Kank1 and Kank2 were recently identified as susceptibility genes for nephrotic syndrome, we targeted both Kank1 and Kank2. Multiple shRNAs were generated for the 6 candidate genes. Their efficacy was validated in vitro, and the best one was targeted to the Hprt1 locus (Figure 4A and Supplemental Figure 3, A and B). Two independent clones for each candidate gene were selected, and 15–30 mice were generated by laser-assisted microinjection.
Figure 4. Validation of 5 candidate FSGS disease genes.
(A) Validation of shRNAs for Arhgef17, Dlg5, Kank1, Kank2, Wnk4, and Kat2b. As described in Methods, shRNAs were tested for the ability to inhibit a target sequence fused to GFP in 293 cells. GFP immunoblotting was used to measure the degree of inhibition. Each immunoblot is representative of at least 3 independent experiments measuring RNAi efficiency. (B–G) Mouse validation screening for candidate FSGS genes. ES cells were generated with the specific RNAis targeted to the Hgprt locus. Essentially pure chimeric mice were generated by laser-assisted microinjection of ES cells into C57BL6 8-cell embryos. Injections generally resulted in cohorts of 14 to 30 animals; smaller cohorts of animals were not used. Mice were divided into 2 groups and treated with or without DOX to induce expression of the RNAi transgene. Urine albumin/creatinine ratios were measured 4 and 8 weeks after DOX treatment. Albumin/creatinine ratios are shown for each cohort of mice at the indicated time points. A 2-tailed Mann-Whitney U test was used to calculate the P values for B–G. A P value of less than 0.0083 was considered statistically significant (multitest penalty was used).
Half of each cohort was given DOX, and proteinuria (albumin/creatinine), an indicator of podocyte function, was assessed at 4 and 8 weeks after DOX treatment (Figure 4, B–G). All 3 RNAi transgenes, Wnk4, Arhgef17, and Kank2, induced substantial proteinuria, with a level of proteinuria that was significantly higher than that seen in the control mice (Figure 4, B–E). In contrast, the Dlg5, Kat2b, and Kank1 RNAi mice did not show statistically significant elevations of proteinuria after 4 or 8 weeks of DOX treatment. Because there was a slight trend toward increased proteinuria in the Kat2b and Kank1 mice, we followed the proteinuria levels for an additional 4 weeks. After 12 weeks, Kank1 mice had a clear proteinuric phenotype, while the proteinuria present in Kat2b mice was still not significant (Supplemental Figure 4A). Thus, Wnk4, Arhgef17, Kank1, and Kank2 mice were positive for proteinuria, while Dlg5 and Kat2b mice were negative for proteinuria.
We confirmed the Dlg5 result by obtaining Dlg5-KO mice (50) and generating Dlg5, Cd2ap, and Synpo triple-heterozygous mice using conventional breeding. No kidney dysfunction was detected (Supplemental Figure 4B), confirming our RNAi result. As expected, electron microscopic examination of the kidneys showed podocyte foot process effacement in Arhgef17, Kank1 (12 week-time point), Kank2, and Wnk4 mice, but not in Dlg5 RNAi mice (Supplemental Figure 4A). While the overall morphology was normal, some focal areas of mild foot process effacement could be seen in the Kat2b mice.
Testing for the enrichment of rare variants in patients versus controls in 20 previously known FSGS genes plus 3 new genes.
We added KANK1, WNK4, and ARHGEF17 to the list of the 20 known FSGS genes and reanalyzed differences in the number of rare variants between patients and controls. Approximately 53.5% of patients (84 of 179) had at least 1 predicted deleterious rare variant in these 23 genes compared with 5.6% of controls (21 of 378) (Supplemental Table 7). We separated the patients by sporadic and familial FSGS and found a similar distribution, with 50% of sporadic FSGS patients and 64% of familial FSGS patients having variants in these 23 genes.
We tested random sets of 23 genes by permutation analysis of patients and controls, which showed that only 0.67% (P < 1.6 × 10–27) of random sets of 23 genes chosen from the controls equaled or matched the burden of rare variants seen in the patients for these 23 genes. This supports the idea that genetic variants in these 23 genes account for most of the disparity between patients and controls in the numbers of rare variants. This also supports the idea that a specific subset of genes may function more broadly to create a susceptible background for the development of sporadic FSGS.
Discussion
FSGS is a syndrome of diverse etiology that shares a common histologic pattern of focal and segmental glomerular scarring, together with glomerular proteinuria and progressive loss of renal function. The majority of FSGS cases involve primary FSGS, adaptive FSGS, or APOL1 FSGS; less common are viral FSGS, Mendelian FSGS, and medication-associated FSGS. As there are no validated methods to specifically distinguish sporadic (nonfamilial) FSGS, the present study included subjects with both primary and adaptive FSGS as well as subjects with familial FSGS. Because of the strong predictive power of family history, and because only a small percentage of individuals affected by known etiological factors develop FSGS, the genetic background of the individual is thought to play an important role (7).
The critical locus of injury in FSGS is now thought to be the podocyte (5), a terminal-differentiated cell that has limited replication potential (51). In the normal kidney, small numbers of podocytes are continuously lost over time (3), and when podocyte numbers drop below a critical level, kidney failure inexorably ensues (2, 5, 52). Environmental insults and genetic susceptibility are thought to enhance the rate of podocyte loss, and this increases the probability of developing FSGS. Interpreted this way, the FSGS “lesion” likely represents the common outcome of a wide variety of pathogenetic causes.
In validating genetic susceptibility in sporadic FSGS, a significant challenge is the likelihood that a large number of genes may be involved and that each gene contributes only a small amount of risk for the disease. Additionally, the role of mutations in a specific gene may affect only a small number of patients. This substantially increases the challenge of gene identification in any large genetic study. An additional complication is that sporadic FSGS is relatively uncommon, and most patients do not have a biopsy-confirmed diagnosis. This currently precludes the assembly of a large enough cohort for strong statistical analysis. Because of this, most of the FSGS disease genes identified to date are from family studies, from the sequencing of candidate genes based on the phenotype of mouse models, or from admixture linkage studies of African Americans (13, 19–23, 25, 46, 53–56).
Here, we used next-generation sequencing to identify FSGS susceptibility genes. Because of our relatively modest sample size, we adjusted our analytic approach to maximize our ability to identify candidate genes. As both rare and common variants have allele frequencies that are determined by ancestry, well-matched controls for ancestry are required. Since large control datasets for individuals of European ancestry are already available, we focused on FSGS subjects of European ancestry. Because DNA variant calling can be different between institutions and between platforms, we established a pipeline to validate that the datasets were comparable. FSGS is more common in African Americans, but the complex genetic admixture in this population will require a large and complex control dataset that is currently not available. Our focus on genes expressed in podocytes allowed us to focus on higher-likelihood genes and minimized the multitest penalty.
Genetic analysis of FSGS is challenging because of the potentially broad genetic heterogeneity of the disease and the relatively small number of subjects available for analysis when the subjects’ ancestry needs to be controlled. Rare variant analysis in ethnically admixed populations such as those found in the United States will require new statistical approaches and the development of large, ancestrally matched control datasets. Nonetheless, our work shows that current statistical approaches, combined with focused sequence analysis, can identify candidate genes from a relatively small sample for a syndrome like FSGS that has widely divergent etiologies. While our sample size was sufficient to extract a list of candidate genes using rare variant analysis, a sample size of at least one order of magnitude larger would be necessary to generate statistically significant data for the single variants (18). Our sample size, however, allowed for a candidate susceptibility list to be generated from both rare and single variants that we were able to confirm using a small confirmatory or follow-up dataset. Although large-scale whole-exome and whole-genome–sequencing efforts are becoming more commonplace, focused approaches on subsets of genes may have biological and statistical advantages in diseases for which samples are limited.
True validation of some of these genes in humans will probably require identification of high-penetrant familial mutations in some of these genes and/or analysis of a much larger group of patients. As shown by our analysis of 20 known familial variants in this population, their assessment individually would not have implicated them in the pathogenesis of disease in this population of subjects. However, clustering genes together allowed us to generate greater statistical power and arrive at a better understanding of this oligogenic disease. Given that some of our subjects had rare variants in more than 1 candidate gene, it is tempting to speculate that possessing more than 1 risk allele may increase susceptibility. In the future, when most, if not all, susceptibility genes are identified, a risk index could be generated on the basis of the number of gene variants present and their predicted deleterious effect.
Sequence analysis.
An innovation of our approach was the development of a robust pipeline that allowed the use of data on individuals sequenced for other studies as controls. The ability to combine datasets generated at different institutions for different types of studies will become increasingly important and powerful as sequencing becomes more widespread. In our initial studies, we found that batch effects caused by different approaches used for sequencing among different institutions could be a confounding factor precluding the use of analyzed data generated at 2 different institutions. However, by applying the same sequencing read alignment and variant calling pipelines to the primary sequencing data from both patients and controls, we were able to eliminate this variable. We validated our approach by establishing a method for case-control genotype matching and removal of any stratification as well as verifying that primary sequencing data from 2 different institutions using the same control DNA sample gave similar results.
Using a P value of less than 0.05, no genes were identified by rare or single-variant analysis that reached genome-wide significance because of Bonferroni’s multiple test correction. Because the Bonferroni test tends to be conservative, we assembled a list of the top 8 genes identified by rare variant analysis and the top 3 genes identified by single-variant analysis with P values that were close to the Bonferroni corrected P value. Supporting the veracity of this analysis, 3 of the genes, APOL1, COL4A4, and KANK1, were already known FSGS susceptibility genes (13, 16, 33, 37), and WNK4 was identified on both lists.
We initially sequenced over 700 biopsy-confirmed FSGS samples, but most of these samples were genetically admixed, preventing further analysis because of the lack of matched controls. We therefore focused only on the patients of European ancestry as defined by PCA. Because the number of patients of European ancestry with biopsy-confirmed FSGS is extremely small, it is not possible to assemble a true replication study. Also, because of cost, case-control studies with replication using whole-exome or whole-genome sequencing have, in general, been extremely limited and are not yet commonplace in the literature. As a confirmatory approach, we used the 33 samples that we had eliminated because of Hispanic admixture and 23 additional European ancestry samples that we had sequenced subsequent to the original analysis as a confirmatory or follow-up dataset. Our analysis of this second dataset confirmed an increased burden of rare variants in the 6 listed rare variants as well as an increase in 3 common variants (Table 1). Since WNK4 was identified by both processes and APOL1, COL4A4, and KANK1 are known genes, at least 7 new candidate genes were identified by our sequencing analysis. While the groups were small, the distribution of variants did not seem to differ significantly between the sporadic and familial FSGS cases. A complete summary of our results is provided in Table 1.
We were surprised to identify the APOL1 G1 variant in 4 of our subjects and in 7 of the subjects in our follow-up set, as it is rare in non-African populations. The enrichment of this variant in 11 of 208 of our non-African subjects suggests that this particular allele may interact with other variants, leading to susceptibility to FSGS. This is supported by the enrichment of rare, predicted deleterious APOL1 variants in our subjects. The enrichment in our European American subjects with variants that are common in African Americans, but rare in European Americans, was also found in WNK4, KANK1, and ARHGEF17. The absence of neighboring African SNPs suggests that these are ancestral variants and not due to admixture.
Validation method.
Currently, there are few approaches to validate potential susceptibility genes using animal models. While some important FSGS susceptibility genes have been identified and validated using genetic KO studies in mice (47, 57), such studies are labor intensive and not suitable for high-throughput screening. More recently, zebrafish and Drosophila are being used, given the presence of podocyte-like cells in these organisms and the convenience of knocking down gene expression (33). Differences in gene expression and in gene orthologs for the human gene limit the physiologic significance of such approaches for a human disease such as FSGS.
We therefore wanted to develop an efficient mouse model that could be used to mimic FSGS as well as validate candidate genes. Since FSGS is probably oligogenic (8), we started with an FSGS mouse model we had generated previously that was based on bigenic heterozygosity of Cd2ap and Synpo. Mice with this genetic background develop FSGS and exhibit proteinuria with an incomplete penetrance (~25%) after they are over 6 months of age (42). Because of the slow onset of disease in only a fraction of animals, we reasoned that this could be a good background on which to test candidate FSGS genes. We generated ES cells from this background and confirmed that nearly pure mice generated from ES cells from this background developed a mild proteinuria that was first detectable in approximately half of our animals at around 4 months of age and slowly worsened after that (42). We reasoned that partial inhibition of gene expression by RNAi, combined with the heterozygosity of Cd2ap and Synpo, would constitute a good model of oligogenic FSGS. Since we did not want to confound our results with RNAi effects on renal development, we designed our RNAi expression system to be podocyte specific and inducible after birth.
We targeted shRNAs for candidate genes to the Hprt1 locus by homologous recombination using a strategy we have described previously (45). To achieve podocyte-specific expression, our ES cells also expressed a DOX-inducible transactivator molecule (rtTA3G) under the control of a podocyte-specific promoter (nephrin) (43). shRNA oligos were embedded into a miR30 transgene (48). Using the method of laser-assisted injection of ES cells into 8-cell embryos, we could generate large cohorts (10–20 mice) of almost identical animals in a single day of injections (44). We believe our study is the first to use this method to generate large numbers of F0 animals for genetic screening. Coat color confirmed that our animals were close to being completely derived from the ES cells. That proteinuria was only seen in some, but not all, animals probably reflects variability in the intrinsic rate of disease in our mouse model and the short time window we used to measure the effects.
Our animal screening system is limited, because it relies on epistasis with Cd2ap and Synpo. Because of this, it is possible that on a different genetic background, Dlg5 might be an FSGS susceptibility gene. If all susceptibility genes are components of a single network of genes involved in disease pathogenesis, a strategy such as ours could be very useful. Conversely, our strategy could be used to determine whether one or more networks of genes are involved by using ES cells with different genetic backgrounds.
Without knowing how many different networks of pathways are involved in FSGS, our animal screening strategy might not be sensitive to testing all susceptibility genes. While less of a problem in mice compared with zebrafish or Drosophila, differences between mice and humans in the expression of gene paralogs might be important. The differences we observed between Kank1 and Kank2 RNAi cohorts could be due to differences in their expression in murine versus human podocytes. Alternatively, it could be differences in epistatic interactions between Cd2ap/Synpo and Kank1 versus Kank2. As we showed with Kank1 RNAi mice, our system is potentially more sensitive if a longer screening period was used. As proteinuria and FSGS begin to be manifest in mice around 4 to 6 months of age, we purposefully designed the screen to conclude after 8 weeks of RNAi expression.
Our method is similar in principle to one described by Premsrirut et al. that targeted shRNAs to the Col1a1 locus using ES cells developed by Jaenisch and coworkers. These ES cells expressed the rtTA ubiquitously from the Rosa locus (58, 59). We originally tried this method and found that transgenes targeted to the Col1a1 locus were weakly expressed in the podocyte (data not shown). In addition, our method used an ES cell line from a sensitized background, allowing us to customize our screen for FSGS. Our method is distinct from that of Premsrirut et al., who used tetraploid complementation, which is much more labor intensive and produces far fewer transgenic animals (59). The ability to derive ES cells from various disease-susceptible backgrounds should make our approach suitable for a wide variety of disease models.
With the availability of large-scale DNA sequencing of human populations, the identification of disease candidate genes and potential disease-associated variants will become more common. How these candidate genes and variants will be validated is unclear. Here, we demonstrate a pipeline that uses common and rare variant association analyses to identify candidate genes from sporadically affected unrelated individuals and control sequence data that were previously generated. We then developed a method to allow these candidates to be tested in vivo. Our method relied on generating an ES cell line that was sensitized for the development of FSGS. It should be possible to generate ES cells on other disease-specific backgrounds to allow for validation studies that will be required to facilitate the discovery of genetic variants associated with both rare and common diseases.
Methods
Exon capture and Illumina sequencing.
Sample preparation and sequencing were carried out using standard protocols for targeted capture and Illumina sequencing. In brief, genomic DNA was fragmented to 150 to 200 bp using a Covaris E220 Focused Ultrasonicator. The ends of the fragmented DNA were repaired using a mixture of T4 DNA polymerase, Klenow polymerase, and T4 polynucleotide kinase. Subsequently, adapters for Illumina sequencing were ligated onto the fragments. These libraries were then hybridized to biotinylated DNA probes from regions of interest (manufactured by MyGenostics). After washing away DNA libraries that bound nonspecifically to the probes, DNA of interest was recovered using Dynabeads MyOne Streptavidin T1 (Life Technologies). Resulting DNA libraries were amplified, if needed, for sequencing on an Illumina HiSeq 2500.
Variant calling and data quality control.
We performed alignment of the raw sequencing data and variant calling according to GATK best practices with the BWA/Picard/GATK software pipeline of the Broad Institute. To insure that gene loci were equally covered in both patients and controls, we performed quality control on patients’ and controls’ genotypes separately, applying the following filters: (a) retention of only variants that PASS all GATK quality filters; (b) retention of genotypes with DP>10,GQ>30,AB for hets 0.3<AB<0.7, for homozygous alternative AB<0.3; and (c) retention of all variants with less than 5% missing genotypes. After applying these filters, variants were combined from patients and controls, and only those variants with less than 5% missing genotypes in both patients and controls were kept for further analysis. Our final dataset contained 16,108 SNPs in 1,874 genes. The sequencing data were deposited in the NCBI’s Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/), under accession number SRP067711.
PCA and case-control matching.
PCA was performed with Eigenstrat software using the common (MAF >5%) variants found in autosomes only. We computed a Euclidean distance from each point on the PCA plot to the origin and plotted distributions of this parameter for both patients and controls. Using the 3-sigma rule, 30 samples of mixed Hispanic ancestry were identified as outliers and removed from the dataset.
Sample statistics and case-control–matching metrics were computed using PLINK/SEQ analysis software. We used the number of variants called per sample, the number of heterozygous genotypes per sample, and the number of genotypes with minor allele per sample as a metric representing the genetic background of the cohort. The similarity between the genetic background of patients and controls was established by matching the mean and variance of patient and control distributions for every metric. We tested the validity of this approach by running Fisher’s exact test on the common variation and QQ-plot of the P values. This showed no inflation, confirming the absence of any population stratification in the case-control dataset (Supplemental Figure 1).
Mouse strains.
Cd2ap+/– mice were generated previously (47). Synpo–/– mice were obtained from Peter Mundel’s laboratory (60). The Nphs1-rtTA3G (NEFTA) strain was a gift from Jeffrey Miner’s laboratory (43).The Dlg5+/– mouse strain was a gift from Valeri Vasioukhin’s laboratory (50). All mouse strains were genotyped by established methods.
Generation of a male Cd2ap+/–, Synpo+/–, NEFTA+ ES cell line.
To generate a male ES cell line that was sensitized to FSGS, we bred Cd2ap+/– Synpo–/– males with NEFTA+ females. The females were superovulated using standard methods. After mating, the embryos were isolated at the 8-cell stage (morulae) and cultured overnight in EmbryoMax KSOM medium (MR-121-D; EMD Millipore) microdrops overlaid with mineral oil at 5% CO2 and 37°C. Blastocysts were transferred, 1 per well, into 48-well plates with γ-irradiated mouse embryonic fibroblast (MEF) feeders and standard ES cell media containing 15% ES-qualified FBS (SH30070.03E; Hyclone). The inner cell mass (ICM) was allowed to grow out and was trypsinized after 5 to 7 days, depending on the size and shape of the outgrowth. Cells were cultured until ES colonies were identified. The colonies were expanded and genotyped using standard methods.
Generation of miR30-shRNA knockin transgenic mice.
Integration of a single-copy transgene into the Hprt1 locus using 6-thioguanine was performed as we previously described (45) and was modified by the addition of a puromycin resistance cassette to increase the efficiency of selection of a positive ES clone. A PGK-Puro cassette was inserted between the left and right arm of the pHPRT targeting vector. The miR30-based shRNA-expressing transgene that was driven by the tetracycline-responsive promoter (TRE) was inserted between the left arm and the PGK-Puro cassette. The linearized targeting vector was transfected into ES cells. Twenty-fours hours after transfection, the ES cells were treated with 1 μg/ml puromycin for 48 hours. After passaging once, the ES cells were treated with 6-thioguanine (5 μg/ml) for an additional 48 hours. Surviving ES cell colonies were selected, expanded, and examined by genomic PCR across the right arm (forward primer: 5′-CAAGCCCGGTGCCTGATCTAGATCATAATC-3′; reverse primer: 5′-CTGTAAAGGTCTCTGAACTACCAATTGCAC-3′). Positive ES cells were then stocked for injection.
Laser-assisted microinjection.
The ES cells were maintained at the expansion phase before injection. Eight ES cells were injected into a recipient embryo at the 8-cell stage by following a protocol published previously (44). Since the ES cell line produces mice with agouti coat color, albino B6 (C57BL/6J-Tyrc–2J) mice were used as hosts to allow for direct evaluation of chimerism by coat color.
Cell culture and lentiviral infection.
Immortalized murine podocytes were maintained and differentiated as described previously (25). To examine the knockdown efficiency of CD2AP-sh877, podocytes were infected with lentiviral vectors encoding miR30-sh877. A control lentiviral vector encoding miR30-FF3 that targets firefly luciferase cDNA was used as a control. CD2AP expression was examined by immunoblot analysis of the whole-cell lysates.
Design and validation of the miR30-shRNA constructs for genes of interest.
The shRNA oligo sequences were chosen using an online algorism (http://katahdin.cshl.org/siRNA/RNAi.cgi?type=shRNA) as described previously (48). The miR30-shRNA backbone was subcloned by PCR from pPRIME-CMV-GFP-FF3 (https://www.addgene.org/11663/) and inserted into a pcDNA3.1-Zeo(+) vector to generate the pcMIR vector. To examine knockdown efficiency, the miR30-shRNA construct and its artificial target (Supplemental Figure 3B) were cotransfected into HEK293T cells at a molar ratio of 5:1. The expression of EGFP in whole-cell lysates was examined by immunoblot analysis.
Abs.
The Abs used for immunoblotting were mouse anti-XFP (632381; 1:10,000 dilution; Clontech); rabbit anti-ERK2 (sc-154; 1:5,000 dilution; Santa Cruz Biotechnology Inc.); mouse anti–β-actin (A2228; 1:10,000 dilution; Sigma-Aldrich); and rabbit anti-CD2AP (generated in our previous study; 1:10,000 dilution).
Albumin-creatinine assay.
Mouse urine samples were collected at the time points indicated in the figures, and urinary albumin (E90-134; Bethyl Laboratories Inc.) and creatinine (DICT-500; BioAssay Systems) were quantified by ELISA according to the manufacturers’ protocols.
Transmission electron microscopy.
Portions of kidney cortex were fixed with 2% paraformaldehyde and 2% glutaraldehyde. Specimen processing, ultrathin sectioning, and imaging were performed by the Electron Microscopy (EM) Core Facility at Washington University.
Statistics.
P values of all albumin/creatinine ratio plots (Figure 3, C and F, and Figure 4, B–G) were calculated using a 2-tailed Mann-Whitney U test. For Figure 3, C and F, a P value of less than 0.05 was considered statistically significant. For Figures 4, B–G, a P value of less than 0.0083 was considered statistically significant (the multitest penalty was applied). All error bars represent the mean ± SEM.
Study approval.
All animal experiments were conducted with the approval of the Washington University Animal Studies Committee. Because all of the patient samples were deidentified, the Washington University IRB deemed these studies exempt from IRB approval.
Author contributions
The general experimental scheme was conceived by HY and ASS. The list of genes to be sequenced was determined by MGS, MK, JBK, and ASS. Methods for exome capture and sequencing were piloted by ASS, RDM, MCS, and SJ and largely completed by MCS and GS. FSGS patient samples and their selection for this study were provided by CAW, JBK, and SJ. Sequencing analysis was performed by MA and MJD. HY and JMW generated the ES cells, and the laser-assisted microinjection was performed by JMW. HY devised the RNAi validation screen and the ES cell–targeting strategy with the help of JHM. Validation of the candidate genes by testing RNAis, generating ES cells, and phenotyping mice was performed by HY and SB.
Supplementary Material
Acknowledgments
This project was supported by grants from the NIDDK (to A.S. Shaw, J.H. Miner, M. Kretzler, and M.G. Sampson); the HHMI (to A.S. Shaw); and the Intramural Research Programs of the NIDDK (to J.B. Kopp), NCI (to C.A. Winkler), and the Deutsche Forschungsgemeinschaft (DFG) (to S. Brähler). We thank V. Vasioukhin for sharing Dlg5+/– mice, overexpression and RNAi lentiviral plasmids, and histology stainings of Dlg5–/– kidney. We also thank C. Der for sharing the Arhgef17 cDNA construct. We also thank R. Kopan and members of the Shaw laboratory for critical discussions.
Footnotes
Haiyang Yu’s present address is: Ludwig Institute for Cancer Research, La Jolla, California, USA.
Andrey S. Shaw’s present address is: Genentech, South San Francisco, California, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(3):1067–1078. doi:10.1172/JCI82592.
Contributor Information
Haiyang Yu, Email: yuhaiyangsam@gmail.com.
Mykyta Artomov, Email: artomov@fas.harvard.edu.
Sebastian Brähler, Email: sbraehler@path.wustl.edu.
M. Christine Stander, Email: cstander@WUSTL.EDU.
Ghaidan Shamsan, Email: shams015@umn.edu.
Matthew G. Sampson, Email: mgsamps@med.umich.edu.
Matthias Kretzler, Email: kretzler@umich.edu.
Jeffrey H. Miner, Email: minerj@wustl.edu.
Sanjay Jain, Email: sjain22@wustl.edu.
Jeffrey B. Kopp, Email: jeffreyk@intra.niddk.nih.gov.
Mark J. Daly, Email: mjdaly@atgu.mgh.harvard.edu.
References
- 1.D’Agati VD, Kaskel FJ, Falk RJ. Focal segmental glomerulosclerosis. N Engl J Med. 2011;365(25):2398–2411. doi: 10.1056/NEJMra1106556. [DOI] [PubMed] [Google Scholar]
- 2.Kim YH, et al. Podocyte depletion and glomerulosclerosis have a direct relationship in the PAN-treated rat. Kidney Int. 2001;60(3):957–968. doi: 10.1046/j.1523-1755.2001.060003957.x. [DOI] [PubMed] [Google Scholar]
- 3.Wharram BL, et al. Podocyte depletion causes glomerulosclerosis: diphtheria toxin-induced podocyte depletion in rats expressing human diphtheria toxin receptor transgene. J Am Soc Nephrol. 2005;16(10):2941–2952. doi: 10.1681/ASN.2005010055. [DOI] [PubMed] [Google Scholar]
- 4.Wickman L, et al. Urine podocyte mRNAs, proteinuria, and progression in human glomerular diseases. J Am Soc Nephrol. 2013;24(12):2081–2095. doi: 10.1681/ASN.2013020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiggins RC. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney Int. 2007;71(12):1205–1214. doi: 10.1038/sj.ki.5002222. [DOI] [PubMed] [Google Scholar]
- 6.Rood IM, Deegens JK, Wetzels JF. Genetic causes of focal segmental glomerulosclerosis: implications for clinical practice. Nephrol Dial Transplant. 2012;27(3):882–890. doi: 10.1093/ndt/gfr771. [DOI] [PubMed] [Google Scholar]
- 7.Pollak MR. The genetic basis of FSGS and steroid-resistant nephrosis. Semin Nephrol. 2003;23(2):141–146. doi: 10.1053/snep.2003.50014. [DOI] [PubMed] [Google Scholar]
- 8.Pollak MR. Inherited podocytopathies: FSGS and nephrotic syndrome from a genetic viewpoint. J Am Soc Nephrol. 2002;13(12):3016–3023. doi: 10.1097/01.ASN.0000039569.34360.5E. [DOI] [PubMed] [Google Scholar]
- 9.Barua M, Brown EJ, Charoonratana VT, Genovese G, Sun H, Pollak MR. Mutations in the INF2 gene account for a significant proportion of familial but not sporadic focal and segmental glomerulosclerosis. Kidney Int. 2013;83(2):316–322. doi: 10.1038/ki.2012.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurin LP, Lu M, Mottl AK, Blyth ER, Poulton CJ, Weck KE. Podocyte-associated gene mutation screening in a heterogeneous cohort of patients with sporadic focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2014;29(11):2062–2069. doi: 10.1093/ndt/gft532. [DOI] [PubMed] [Google Scholar]
- 11.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11(6):415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 12.Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59(1):5–15. doi: 10.1038/jhg.2013.114. [DOI] [PubMed] [Google Scholar]
- 13.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22(11):2098–2105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomson R, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A. 2014;111(20):E2130–E2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopp JB, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22(11):2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasembeli AN, et al. APOL1 risk variants are strongly associated with HIV-associated nephropathy in black South Africans. J Am Soc Nephrol. 2015;26(11):2882–2890. doi: 10.1681/ASN.2014050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322(5903):881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiser J, et al. TRPC6 is a glomerular slit diaphragm-associated channel required for normal renal function. Nat Genet. 2005;37(7):739–744. doi: 10.1038/ng1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winn MP, et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science. 2005;308(5729):1801–1804. doi: 10.1126/science.1106215. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan JM, et al. Mutations in ACTN4, encoding α-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24(3):251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 22.Brown EJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42(1):72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JM, et al. CD2-associated protein haploinsufficiency is linked to glomerular disease susceptibility. Science. 2003;300(5623):1298–1300. doi: 10.1126/science.1081068. [DOI] [PubMed] [Google Scholar]
- 24.Lindenmeyer MT, et al. Systematic analysis of a novel human renal glomerulus-enriched gene expression dataset. PLoS One. 2010;5(7): doi: 10.1371/journal.pone.0011545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akilesh S, et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest. 2011;121(10):4127–4137. doi: 10.1172/JCI46458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boerries M, et al. Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int. 2013;83(6):1052–1064. doi: 10.1038/ki.2012.487. [DOI] [PubMed] [Google Scholar]
- 27.Brunskill EW, Georgas K, Rumballe B, Little MH, Potter SS. Defining the molecular character of the developing and adult kidney podocyte. PLoS One. 2011;6(9): doi: 10.1371/journal.pone.0024640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;11(1110):11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampson MG, Hodgin JB, Kretzler M. Defining nephrotic syndrome from an integrative genomics perspective. Pediatr Nephrol. 2015;30(1):51–63. doi: 10.1007/s00467-014-2857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gee HY, et al. KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest. 2015;125(6):2375–2384. doi: 10.1172/JCI79504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Leal SM. Discovery of rare variants via sequencing: implications for the design of complex trait association studies. PLoS Genet. 2009;5(5): doi: 10.1371/journal.pgen.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, et al. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86(6):832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neale BM, et al. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7(3): doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voskarides K, et al. COL4A3/COL4A4 mutations producing focal segmental glomerulosclerosis and renal failure in thin basement membrane nephropathy. J Am Soc Nephrol. 2007;18(11):3004–3016. doi: 10.1681/ASN.2007040444. [DOI] [PubMed] [Google Scholar]
- 38.Sadowski CE, et al. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2015;26(6):1279–1289. doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gee HY, et al. ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest. 2013;123(8):3243–3253. doi: 10.1172/JCI69134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou W, Hildebrandt F. Inducible podocyte injury and proteinuria in transgenic zebrafish. J Am Soc Nephrol. 2012;23(6):1039–1047. doi: 10.1681/ASN.2011080776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashraf S, et al. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest. 2013;123(12):5179–5189. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huber TB, et al. Bigenic mouse models of focal segmental glomerulosclerosis involving pairwise interaction of CD2AP, Fyn, and synaptopodin. J Clin Invest. 2006;116(5):1337–1345. doi: 10.1172/JCI27400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin X, Suh JH, Go G, Miner JH. Feasibility of repairing glomerular basement membrane defects in Alport syndrome. J Am Soc Nephrol. 2014;25(4):687–692. doi: 10.1681/ASN.2013070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poueymirou WT, et al. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat Biotechnol. 2007;25(1):91–99. doi: 10.1038/nbt1263. [DOI] [PubMed] [Google Scholar]
- 45.Yu H, et al. Rac1 activation in podocytes induces rapid foot process effacement and proteinuria. Mol Cell Biol. 2013;33(23):4755–4764. doi: 10.1128/MCB.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowik MM, et al. Focal segmental glomerulosclerosis in a patient homozygous for a CD2AP mutation. Kidney Int. 2007;72(10):1198–1203. doi: 10.1038/sj.ki.5002469. [DOI] [PubMed] [Google Scholar]
- 47.Shih NY, et al. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science. 1999;286(5438):312–315. doi: 10.1126/science.286.5438.312. [DOI] [PubMed] [Google Scholar]
- 48.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102(37):13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu X, et al. Expression of novel podocyte-associated proteins sult1b1 and ankrd25. Nephron Exp Nephrol. 2011;117(2):e39–e46. doi: 10.1159/000320049. [DOI] [PubMed] [Google Scholar]
- 50.Nechiporuk T, Fernandez TE, Vasioukhin V. Failure of epithelial tube maintenance causes hydrocephalus and renal cysts in Dlg5–/– mice. Dev Cell. 2007;13(3):338–350. doi: 10.1016/j.devcel.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greka A, Mundel P. Cell biology and pathology of podocytes. Annu Rev Physiol. 2012;74:299–323. doi: 10.1146/annurev-physiol-020911-153238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Agati VD. Podocyte injury in focal segmental glomerulosclerosis: lessons from animal models (a play in five acts) Kidney Int. 2008;73(4):399–406. doi: 10.1038/sj.ki.5002655. [DOI] [PubMed] [Google Scholar]
- 53.Boute N, et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24(4):349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 54.Gigante M, et al. CD2AP mutations are associated with sporadic nephrotic syndrome and focal segmental glomerulosclerosis (FSGS) Nephrol Dial Transplant. 2009;24(6):1858–1864. doi: 10.1093/ndt/gfn712. [DOI] [PubMed] [Google Scholar]
- 55.Kao WH, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40(10):1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kopp JB, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40(10):1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krendel M, et al. Disruption of Myosin 1e promotes podocyte injury. J Am Soc Nephrol. 2009;20(1):86–94. doi: 10.1681/ASN.2007111172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44(1):23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 59.Premsrirut PK, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145(1):145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deller T, et al. Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity. Proc Natl Acad Sci U S A. 2003;100(18):10494–10499. doi: 10.1073/pnas.1832384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.