Abstract
Background
The cell adhesion molecule integrin α4β7 helps direct the migration of blood lymphocytes to the intestine and associated lymphoid tissues. We hypothesized that β7+ and β7- blood memory T helper cells differ in their expression of genes that play a role in the adhesion or migration of T cells.
Results
RNA was prepared from β7+ and β7- CD4+ CD45RA- blood T cells from nine normal human subjects and analyzed using oligonucleotide microarrays. Of 21357 genes represented on the arrays, 16 were more highly expressed in β7+ cells and 18 were more highly expressed in β7- cells (≥1.5 fold difference and adjusted P < 0.05). Several of the differentially expressed transcripts encode proteins with established or putative roles in lymphocyte adhesion and chemotaxis, including the chemokine receptors CCR9 and CCR10, the integrin α4 subunit, L-selectin, KLRB1 (CD161), NT5E (CD73), LGALS1 and LGALS2 (galectin-1 and -2), and RGS1. Flow cytometry was used to determine whether differences in levels of transcripts encoding cell surface proteins were associated with differential expression of those proteins. Using this approach, we found that surface expression of KLRB1, LAIR1, and NT5E proteins was higher on β7+ memory/effector T cells than on β7- cells.
Conclusions
Memory/effector T cells that express integrin β7 have a distinct pattern of expression of a set of gene transcripts. Several of these molecules can affect cell adhesion or chemotaxis and are therefore likely to modulate the complex multistep process that regulates trafficking of CD4+ memory T cell subsets with different homing behaviors.
Background
Lymphocyte migration is a multistep process that involves a complex interplay between adhesion molecules and chemokines and their G protein-coupled receptors [1,2]. Naïve T cells express the adhesion molecule L-selectin, the chemokine receptor CCR7 and other molecules that allow these cells to migrate preferentially to secondary lymphoid organs where they can encounter antigen-presenting cells. When presented with appropriate antigens, these T cells can differentiate into memory T cells. Some memory cells continue to express L-selectin and/or CCR7 and to migrate efficiently to secondary lymphoid organs, whereas others lose expression of these molecules and instead express other molecules that direct migration (or "homing") to other organs [1,2].
Extensive investigation has helped to define the role of some adhesion molecules and chemokine receptors in CD4+ memory T cell homing to the skin and the gut. The adhesion molecule cutaneous lymphocyte antigen (CLA) and the cutaneous T cell-attracting chemokine receptor CCR10 help control T cell homing to the skin [3-8]. The adhesion molecule integrin α4β7 and the chemokine receptor CCR9 play key roles in homing of lymphocytes to the intestine and Peyer's patches [9-13]. Integrin α4β7 is a receptor for mucosal addressin cell adhesion molecule-1 (MAdCAM-1), a glycoprotein that is expressed by gut endothelium. CCR9 is a receptor for the chemokine TECK (CCL25), which is expressed by endothelial cells and other cells in the small intestine [14,15]. The adhesion molecule and chemokine receptor expression pattern of memory/effector CD4+ T cells is strongly influenced by whether initial T cell activation takes place in cutaneous or intestinal lymph nodes [16-18]. Recent evidence suggests that T cell homing receptor expression patterns are "imprinted" by dendritic cells during antigen presentation [19-21]. Although many other molecules have been shown to help control lymphocyte adhesion or migration, it is not clear which if any of these are selective expressed in gut homing memory CD4+ T cells.
We used DNA microarrays to systematically compare RNA transcript expression in human blood β7+ and β7- CD4+ memory T cells. We identified a substantial number of differentially expressed genes, many of which have been previously shown to have effects on cell adhesion and migration. In addition, we showed that some of these transcript expression differences were reflected in differences in surface expression of the encoded proteins.
Results
Microarray analysis of differential gene expression by β7+ and β7- CD4+ memory T cells
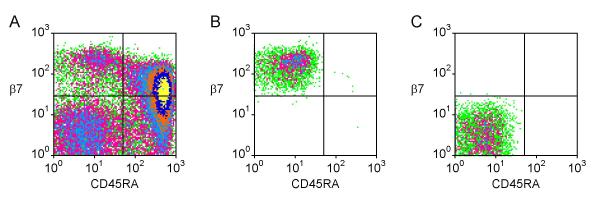
As previously reported [22], human blood CD4+ T cells could be divided into three distinct subsets based upon their surface expression of integrin β7 and the naïve cell marker CD45RA (Fig. 1A). CD45RA+ naïve cells expressed low levels of β7. CD45RA- memory T cells were divided into a larger population of β7- cells and a smaller population of β7+ cells. The integrin β7 subunit can combine with either integrin α4 (to form the gut homing receptor α4β7) or αE. Integrin αEβ7 is highly expressed on gut intraepithelial lymphocytes but is rarely expressed by blood T cells and its possible role in homing is less clear [23-25]. Consistent with previous reports [22,23,26], staining with antibodies specific for the α4β7 and αEβ7 complexes (Act-1 and HML-1 respectively) showed that the virtually all β7-expressing CD4+ T cells in the blood were α4β7+ , and that only 0.5 ± 0.3% (mean ± SD) of blood CD4+ T cells expressed αEβ7. We were able to obtain highly purified populations of β7+ and β7- CD4+ CD45RA- cells by flow cytometric sorting (Fig. 1B and 1C).
Figure 1.
Integrin β7 expression on blood T helper cells and on purified β7- and β7+ memory T helper cell subsets. Dot density plots depicting expression of the naïve cell marker CD45RA and integrin β7 on CD4+ blood T cells (A) and on purified β7+ (B) and β7- (C) memory cells in arbitrary fluorescence units. Sorted populations were ~99% pure. Quadrant markers indicate settings used to sort populations based on CD45RA and β7 expression. Colors indicate the relative frequency of cells (from lowest to highest density: green, purple, light blue, orange, dark blue, and yellow).
We used DNA microarrays to compare RNA transcript expression by β7+ and β7- cells from 9 healthy adult human subjects. Using this approach, we identified 16 genes that were expressed at higher levels in β7+ cells (at least 1.5-fold higher expression and P < 0.05 when adjusted for multiple comparisons). These are listed in Table 1. Our estimates of fold difference are likely to be conservative since the use of amplified RNA (as opposed to unamplified cDNA) tends to underestimate fold differences [27]. The most highly differentially expressed transcript encodes the chemokine receptor CCR9. CCR9 protein has previously been shown to be preferentially expressed on α4β7+ blood CD4+ T cells [13]. The ITGA4 transcript encodes the α subunit of integrin α4β7, and was expressed at 1.7-fold higher levels on β7+ cells. Other genes listed in Table 1 include genes encoding cell surface receptors (LRRN3, CD1C, KLRB1, LAIR1, IL18RAP, KLRG1), transcription factors (RAM2, SREBF1) and a transcriptional repressor known to play a role in T cell differentiation (GFI1). A complete listing of all gene expression results is provided in Appendix 2 [see additional file 2].
Table 1.
Gene transcripts with higher expression in β7+ versus β7- CD4+ CD45RA- T helper cells*
| Symbol | Name | Accession | Fold Difference | P value |
| CCR9 | chemokine (C-C motif) receptor 9 | NM_031200 | +3.0 | < 0.01 |
| CCL5 | chemokine (C-C motif) ligand 5 | NM_002985 | +2.4 | < 0.01 |
| RAM2 | transcription factor RAM2 | NM_018719 | +2.2 | < 0.01 |
| LRRN3 | leucine rich repeat neuronal 3 | AL442092 | +2.1 | < 0.01 |
| GFI1 | growth factor independent 1 | NM_005263 | +1.8 | < 0.01 |
| ITGA4 | integrin, alpha 4 (CD49D) | NM_000885 | +1.7 | < 0.01 |
| CD1C | CD1C antigen, c polypeptide | NM_001765 | +1.7 | < 0.01 |
| KLRB1 | killer cell lectin-like receptor subfamily B, member 1 | NM_002258 | +1.7 | < 0.01 |
| LAIR1 | leukocyte-associated Ig-like receptor 1 | NM_002287 | +1.7 | < 0.01 |
| RRM2 | ribonucleotide reductase M2 polypeptide | NM_001034 | +1.6 | < 0.01 |
| -- | Homo sapiens cDNA FLJ32290 fis, clone PROST2000463 | AK056852 | +1.6 | < 0.01 |
| HHL | expressed in hematopoietic cells, heart, liver | NM_014857 | +1.6 | 0.02 |
| IL18RAP | interleukin 18 receptor accessory protein | NM_003853 | +1.6 | < 0.01 |
| SREBF1 | sterol regulatory element binding transcription factor 1 | NM_004176 | +1.6 | < 0.01 |
| KLRG1 | killer cell lectin-like receptor subfamily G, member 1 | NM_005810 | +1.5 | < 0.01 |
| LGALS2 | lectin, galactoside-binding, soluble, 2 (galectin 2) | NM_006498 | +1.5 | 0.01 |
* Includes all transcripts with fold difference ≥+1.5 and adjusted P < 0.05. Positive fold difference values indicate higher expression on β7+ cells.
We also identified 18 gene transcripts that were expressed at significantly lower levels in β7+ cells compared with β7- cells (at least 1.5 times lower and adjusted P < 0.05). These are listed in Table 2. GPR2, the most highly differentially expressed transcript, encodes CCR10, the receptor for the cutaneous T cell attracting chemokine (CTACK/CCL27). Hudak et al. [8] found that CCR10 protein was present on a subset of CD4+ CD45RA- T cells that express the skin homing receptor CLA but do not express integrin α4β7. We also found that β7- cells expressed more transcript for SELL, which encodes L-selectin (CD62L), a non-integrin cell adhesion molecule that mediates lymphocyte adhesion to peripheral lymph node high endothelial venules. None of the other genes listed in Table 2 have established roles in organ-specific homing, although both RGS1 and LGALS1 have been reported to modulate lymphocyte adhesion (see Discussion). Given the importance of G protein-coupled receptors in regulating leukocyte migration, we note that in addition to GPR2/CCR10, one other G protein-coupled receptor, P2RY5, was expressed at higher levels on β7- cells. Although classified as a purinergic receptor, expression of P2RY5 did not result in nucleotide-evoked signaling responses, and the function of this receptor remains unknown [28].
Table 2.
Gene transcripts with higher expression in β7- versus β7+ CD4+ CD45RA- T helper cells*
| Symbol | Name | Accession | Fold Difference | P value |
| GPR2 | G protein-coupled receptor 2 (CCR10) | NM_016602 | -3.2 | < 0.01 |
| RGS1 | regulator of G-protein signalling 1 | NM_002922 | -2.0 | < 0.01 |
| TRIM2 | tripartite motif-containing 2 | NM_015271 | -1.9 | < 0.01 |
| C1orf24 | chromosome 1 open reading frame 24 | NM_052966 | -1.9 | < 0.01 |
| CAMTA1 | calmodulin binding transcription activator 1 | AB020640 | -1.9 | < 0.01 |
| LMNA | lamin A/C | NM_005572 | -1.9 | < 0.01 |
| LGALS1 | lectin, galactoside-binding, soluble, 1 (galectin 1) | NM_002305 | -1.9 | < 0.01 |
| P2RY5 | purinergic receptor P2Y, G-protein coupled, 5 | NM_005767 | -1.8 | < 0.01 |
| SELL | selectin L (lymphocyte adhesion molecule 1) | NM_000655 | -1.8 | 0.02 |
| -- | similar to RIKEN cDNA 1700007B22 | AK021437 | -1.8 | < 0.01 |
| ChGn | chondroitin beta1,4 N-acetylgalactosaminyltransferase | NM_018371 | -1.8 | < 0.01 |
| -- | similar to BcDNA:GH11415 gene product | AK056276 | -1.7 | < 0.01 |
| PTPLA | protein tyrosine phosphatase-like, member a | NM_014241 | -1.7 | < 0.01 |
| MRC2 | mannose receptor, C type 2 | NM_006039 | -1.6 | < 0.01 |
| PRNP | prion protein (p27-30) | NM_000311 | -1.6 | < 0.01 |
| PHLDA1 | pleckstrin homology-like domain, family A, member 1 | AK026181 | -1.6 | < 0.01 |
| KRT1 | keratin 1 | NM_006121 | -1.5 | < 0.01 |
| SEMA5A | semaphorin 5A | NM_003966 | -1.5 | < 0.01 |
* Includes all transcripts with fold difference ≤-1.5 and adjusted P < 0.05. Negative fold difference values indicate higher expression on β7- cells.
Comparison of surface protein expression on β7+ and β7- CD4+ memory T cells
We wished to determine whether differences in expression of selected transcripts were reflected in differences in expression of the cell surface proteins encoded by those transcripts. As previously discussed, the products of the two most highly differentially expressed genes (CCR9 and GPR2/CCR10) have already been shown to be highly selectively expressed on α4β7+ and α4β7- CD4+ T cells respectively [8,13]. We selected four other cell surface proteins encoded by transcripts that were expressed more highly on β7+ cells for further analysis based upon the availability of suitable antibodies. Three of these (LAIR1, KLRB1, and KLRG1) are listed in Table 1. A fourth, NT5E (ecto-5'-nucleotidase, also known as CD73), was also significantly higher in β7+ cells (adjusted P < 0.01), although the estimated magnitude of the difference was smaller (1.3-fold). We used flow cytometry to measure expression of each of these four molecules on CD4+ T cell subsets from human blood. For comparison, we also analyzed surface expression of one protein (SELL/CD62L) encoded by a transcript that was expressed at higher levels on β7- cells. We used a different population of study subjects from that used in the microarray studies to ensure that our findings could be validated in an independent sample. For four of the five proteins examined, the extent of differential surface expression was at least as great as predicted from the microarray data (Table 3). The other protein, KLRG1, was not consistently differentially expressed.
Table 3.
Surface expression of five gene products on β7+ versus β7- CD4+ T cells
| Mean surface protein expression* | |||||
| Gene | Fold difference: Transcript** | Fold difference: Surface protein*** | CD45RA- β7+ cells | CD45RA- β7- cells | CD45RA+ cells |
| LAIR1 | +1.7 | +2.5 (+1.6 to +3.9) | 34.6 | 15.3 | 37.3 |
| KLRB1 | +1.7 | +2.5 (+2.2 to +3.6) | 26.1 | 10.7 | 1.1 |
| KLRG1 | +1.5 | 1.0 (-3.2 to +2.1) | 17.7 | 15.6 | 9.1 |
| NT5E | +1.3 | +1.7 (+1.2 to +3.2) | 20.3 | 11.7 | 6.8 |
| SELL | -1.8 | -1.8 (-2.1 to -1.5) | 22.8 | 40.2 | 50.7 |
* Mean fluorescence intensity (MFI, in arbitrary units) was measured for 5–8 subjects.** As determined using DNA microarrays. *** Values represent mean (range) of the fold difference in MFI between CD45RA- β7+ cells and CD45RA- β7- cells for all subjects. Positive fold difference values indicate higher expression on β7+ cells and negative fold difference values indicate higher expression on β7- cells.
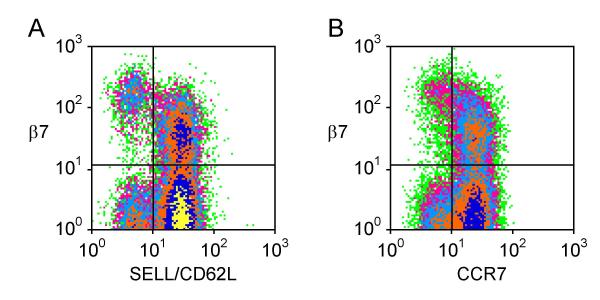
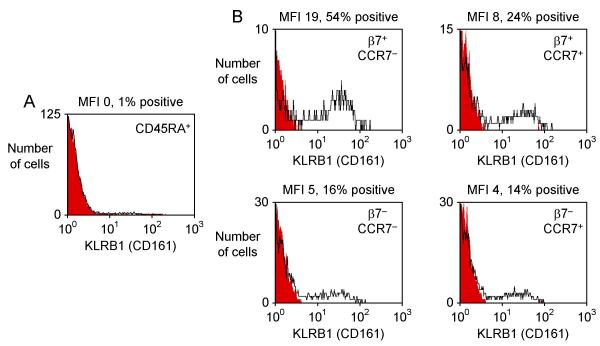
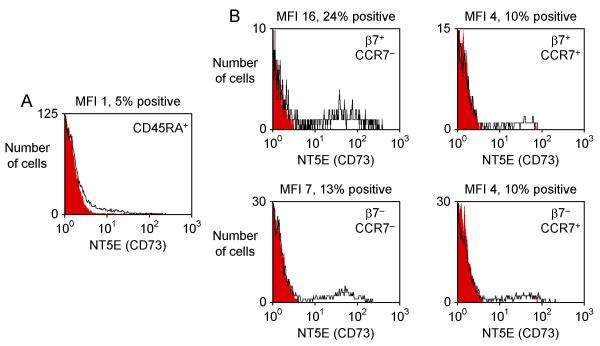
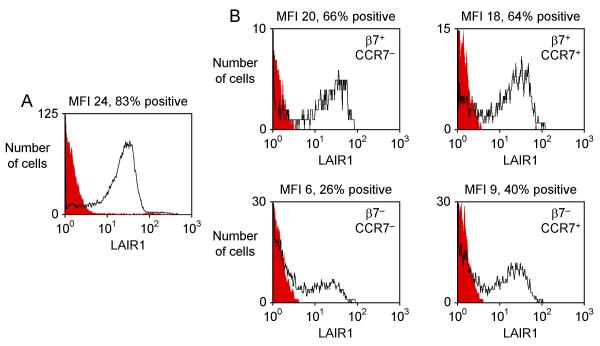
We used flow cytometry to conduct a more detailed analysis of the expression of KLRB1, NT5E and LAIR1 on T helper cell subsets with distinct homing properties. A small subset of memory cells expressed the skin homing receptor CLA (~4% of CD4+ CD45RA- T cells). These CLA+ cells had low expression of KLRB1, NT5E and LAIR1 (similar to levels found in other β7- CD45RA- cells, data not shown). The adhesion molecule L-selectin (SELL, CD62L) and the chemokine receptor CCR7 both play roles in homing to secondary lymphoid organs and each is expressed on subsets of memory/effector T cells. Memory/effector cells that lacked L-selectin and CCR7 included a subset with particularly high β7 expression and a larger subset with no β7 expression (Fig. 2). CD45RA+ cells that expressed L-selectin and CCR7 (central memory cells) also included β7+ and β7- subsets. We found that KLRB1, a member of the killer cell lectin-like receptor superfamily also known as CD161 or NKRP1A [29], was highly expressed on β7+ CCR7- memory effector T helper cells (Fig. 3). There was less expression on β7- CCR7- memory effector cells and on CCR7+ central memory cells. KLRB1 was essentially undetectable on CD45RA+ naïve cells. The expression pattern of NT5E/CD73 was similar, although NT5E expression was limited to a smaller fraction of cells than KLRB1 expression (Fig. 4). In contrast, LAIR1 had a different pattern of expression (Fig. 5). LAIR1 was most highly expressed on naïve cells, and was also highly expressed on β7+ CD45RA- cells (both CCR7- and CCR7+ subsets). LAIR1 was less highly expressed on β7- CD45RA- cells (especially the CCR7- subset). LAIR1 contains two ITIM domains and can transmit signals following ligation by antibody, however the nature of the LAIR1 ligand(s) and the biological function of LAIR1 on CD4+ T cells remains unknown [30].
Figure 2.
A β7high subset of memory/effector T helper cells lacks CD62L and CCR7. Dot density plots depicting expression of integrin β7 and (A) SELL (CD62L) or (B) CCR7 on CD4+ CD45RA- T cells. Quadrant lines were drawn based on staining obtained with control antibodies.
Figure 3.
Expression of KLRB1 (CD161) on T helper cell subsets. Histograms depict expression of KLRB1 (CD161) on (A) CD45RA+ naïve cells and (B) four subsets of CD45RA- memory/effector cells. These subsets were distinguished by their expression of β7 and CCR7, a marker for central memory cells, as shown in Fig. 2B. Black lines indicate KLRB1 staining and filled red lines indicate staining obtained with an isotype control antibody.
Figure 4.
Expression of NT5E (CD73) on T helper cell subsets. Histograms depict expression of NT5E (CD73) on (A) CD45RA+ naïve cells and (B) four subsets of CD45RA- memory/effector cells. These subsets were distinguished by their expression of β7 and CCR7, a marker for central memory cells, as shown in Fig. 2B. Black lines indicate NT5E staining and filled red lines indicate staining obtained with an isotype control antibody.
Figure 5.
Expression of LAIR1 on T helper cell subsets. Histograms depict expression of LAIR1 on (A) CD45RA+ naïve cells and (B) four subsets of CD45RA- memory/effector cells. These subsets were distinguished by their expression of β7 and CCR7, a marker for central memory cells, as shown in Fig. 2B. Black lines indicate LAIR1 staining and filled red lines indicate staining obtained with an isotype control antibody.
Discussion
We used DNA microarrays to identify gene transcripts that were differentially expressed on the subset of CD4+ memory/effector T cells that express integrin β7. Since the overwhelming majority of β7+ cells in blood express the gut homing receptor integrin α4β7, this approach was designed to identify genes that might play a role in organ-specific homing. We identified 16 transcripts that were more highly expressed in β7+ cells and 18 that were more highly expressed on β7- cells. We can relate transcript expression to expression of protein in seven cases (Table 3 and refs. [8,13]). In six of these cases (CCR9, GPR2/CCR10, KLRB1, LAIR1, NT5E, and SELL), differential transcript expression was accompanied by differential expression of the corresponding protein on the surface of β7+ versus β7- cells. In one case (KLRG1), we did not find evidence for a consistent difference in surface protein expression (Table 3). These data suggest that most of the differentially expressed transcripts correspond to differentially expressed proteins, although it will be important to confirm this directly when suitable reagents become available.
Some of the differentially expressed transcripts encode proteins with known roles in organ-specific homing. The two most highly differentially expressed genes we detected in our analysis encode chemokine receptors already shown to have important roles in gut and non-gut (skin) homing. These are CCR9, the TECK/CCL25 receptor which is expressed preferentially on α4β7+ gut homing cells [13], and GPR2/CCR10, the CTACK/CCL27 receptor which is expressed preferentially on CLA+ skin homing cells [8]. We found that transcripts encoding the α subunit (ITGA4) of integrin α4β7, was more highly expressed in β7+ cells. The microarray experiments indicated that the transcript encoding the β7 subunit itself (ITGB7) was not differentially expressed, consistent with previous results obtained using quantitative RT-PCR (M.W.R and D.J.E, unpublished results). This suggests that surface expression of the α4β7 heterodimer on CD4+ memory T cells may be determined primarily by the amount of α4 subunit available and not by the amount of β7 produced. The other differentially expressed transcript with a known role in homing is SELL, which encodes the peripheral lymph node homing receptor L-selectin (CD62L). We found that expression of both L-selectin transcript and surface protein was higher in β7- than β7+ memory/effector T cells. Although many β7-expressing memory cells also expressed L-selectin, cells expressing the highest levels of β7 did not express L-selectin (Fig. 2). This presumably contributes to the development of CD4+ T cell subsets with distinct homing properties.
We also found evidence that β7+ and β7- cells differ in their expression of other molecules that have been reported to modulate cell adhesion and migration. KLRB1 transcript and surface protein expression was higher on β7+ memory effector T cells. Overexpression of KLRB1 (also known as CD161 or NKRP1A) was previously found to increase transendothelial migration of CD4+ T cells, perhaps by modulating integrin activity [31]. The finding that KLRB1 was preferentially expressed on T cells from human intestine led to a recent suggestion that KLRB1 may help direct intestinal homing [32]. This hypothesis remains to be tested, but would be consistent with our finding of preferential expression of KLRB1 on β7+ blood memory CD4+ T cells. We also found higher expression of NT5E (CD73) transcripts and surface protein on β7+ memory effector T cells. NT5E is an ecto 5'-nucleotidase expressed on lymphocytes and on endothelial cells and has been reported to mediate lymphocyte-endothelial adhesion and transmigration by various mechanisms, including promotion of integrin-mediated binding via effects on integrin clustering [33-36]. Transcripts encoding two members of the galectin family were differentially expressed on β7+ versus β7- cells. Both LGALS1/galectin-1 (expressed at higher levels in β7- cells) and LGALS2/galectin-2 (expressed at higher levels in β7+ cells) have been shown to have several effects on cell adhesion in lymphocytes and other cells [37]. Transcripts for RGS1 were expressed at higher levels in β7- cells. RGS1 is a GTPase activating protein that regulates Gαi-stimulated pathways. Expression of RGS1 inhibits chemokine-induced cell migration and integrin-mediated adhesion in lymphocytes [38]. Our data suggest that there may be less RGS1-mediated inhibition of chemokine signaling in β7+ cells. We speculate that the differential expression of KLRB1, NT5E, LGALS1, LGALS2, and RGS1 on β7+ and β7- T helper cell subsets may contribute to homing specificity by modulating the complex multistep process of cell adhesion and migration.
The primary activation of T cells leads to changes in the expression of integrin α4β7 and other molecules that control homing. It has recently been shown that upregulation of integrin α4β7 expression and increased responsiveness to the CCR9 ligand TECK occur within two days after activation of murine naïve CD4+ T cells in intestinal, but not cutaneous, lymph nodes [18] and that activation by dendritic cells from Peyer's patches but not peripheral lymph node or spleen promotes murine CD8+ T cell α4β7 expression and TECK responsiveness [19]. We found that both KLRB1 and NT5E were virtually absent on CD45RA+ naïve CD4+ T cells and were expressed at low levels on CD45RA- CCR7+ central memory cells and on β7- CD45RA- non-gut homing memory effector cells but were expressed at higher levels in the β7high CCR7- CD45RA- memory effector subset. In contrast, LAIR1 was highly expressed on naïve T cells and on both CCR7- and CCR7+ β7+ CD45RA- cells but expression was lower on β7- CD45RA- cells. These results suggest that expression of α4β7 and expression of KLRB1, NT5E and LAIR1 may be coordinately regulated at the time of human T cell activation, as is apparently the case for α4β7 and CCR9 in mouse systems. Alternatively, it is possible that differences in expression arise from changes in gene expression that occur after the transition from naïve to memory effector cells or from the preferential retention of survival of particular memory effector cell subsets. Although we showed that expression of KLRB1, NT5E and LAIR1 was higher for the β7+ subset as a whole, many β7+ cells did not express these proteins and some β7- cells did. This suggests that the homing properties of individual T cells might be "fine tuned" by complex combinatorial regulation of molecules known to have major roles in determining organ-specific recruitment (e.g., integrin α4β7 and CCR9) and other molecules that can modulate adhesion and migration (e.g., KLRB1 and NT5E).
Conclusions
We used DNA microarrays to identify a set of gene transcripts that were differentially expressed on β7+ versus β7- blood memory CD4+ T cells. Some of these encode adhesion receptors (integrin α4 and L-selectin) and chemokine receptors (CCR9 and GPR2/CCR10) already known to be involved in organ-specific lymphocyte migration. Other differentially expressed transcripts encode additional molecules previously reported to affect cell adhesion and migration, including KLRB1, NT5E/CD73, LGALS1 and LGALS2 (galectin-1 and -2) and RGS1. As predicted from the transcript expression data, we found that KLRB1, NT5E and LAIR1 proteins were all preferentially expressed on the surface of β7+ versus β7- memory CD4+ T cells. Our results suggest that expression of several molecules that can modify T cell adhesion, endothelial transmigration and chemotaxis are also selectively regulated on CD4+ T cell subsets, perhaps during the "imprinting" process that takes place during initial T cell activation. These molecules are likely to contribute to the complex multistep process that regulates trafficking of CD4+ memory T cell subsets with different homing behaviors.
Methods
Isolation of β7+ and β7- lymphocytes from human blood
This protocol was approved by the Committee on Human Research at the University of California, San Francisco and informed consent was obtained from all subjects. Peripheral blood mononuclear cells (PBMC) were isolated from nine normal adult donors (six females and three males, ages 24–42 years) using sodium heparin coated Vacutainer Cell Preparation Tubes (Becton Dickinson) according to the manufacturer's instructions. PBMC were stained with CD4-FITC, CD45RA-Cy5 and anti-integrin β7-PE (all from BD Pharmingen). Anti-FITC microbeads and a MACS bead sorter (Miltenyi Biotec) were used to enrich for the CD4+ cells prior to flow cytometry. CD4+ memory (CD45RA- ) β7+ and β7- T cells were then purified using a FACS Vantage (Becton Dickinson).
Microarray analysis of transcript expression
Total RNA from β7+ and β7- CD4+ memory T cells was purified using the Mini RNA Isolation Kit (Zymo Research) and DNase treated using the DNA-free RNA Kit (Zymo Research) according to the manufacturer's instructions. RNA quality was evaluated using an Agilent Bioanalyzer. RNA amplification, labeling, and hybridization were performed as previously described with slight modifications [27]. In brief, two rounds of amplification were performed using T7 RNA polymerase (Ambion MessageAmp aRNA Kit). Cy3 or Cy5 was incorporated into the amplified RNA products using amino allyl-modified nucleotides. Fluorescently-labeled amplified RNAs were fragmented using Ambion RNA Fragmentation Reagents and hybridized to DNA microarrays using Ambion SlideHyb Glass Array Hybridization Buffer #1. Each hybridization involved β7+ cell RNA from a single subject (labeled with one dye) and β7- cell RNA from the same subject (labeled with the other dye). A total of 27 arrays were used to analyze samples from the 9 subjects (at least 2 arrays per subject, including a dye swap).
Arrays were produced in our microarray facility using the Operon version 2 set of 70-mer oligonucleotide probes supplemented with some custom-designed 70-mers as described [27]. Arrays included probes for 21357 genes plus control probes. After hybridization, arrays were scanned using an Axon GenePix 4000B scanner and images were processed using GenePix 5.0 software. The "print-tip loess" normalization was used to correct for within-array dye and spatial effects [39] and single channel quantile normalization was used to facilitate comparison between arrays [40]. We used functions in the library marrayNorm [41] of the R / Bioconductor package [42] to perform these normalizations. After normalization we determined a log ratio, log2 (β7+ sample intensity/β7- sample intensity), for each probe on each array. No background subtraction was performed. Complete information about the array platform and data from each of the individual arrays is available from GEO [43] (GEO accession GSE1039). Appendix 1 [see additional file 1] is a MIAME-compliant description of the array experiments.
Identification of differentially expressed gene transcripts
A fixed effects linear model was used to estimate differences in transcript expression between β7+ and β7- CD4+ T cells. For each individual gene (probe) on the array, the model can be described as
Y ij = μ + A i + εij ,
where Y ij represents the normalized log ratio determined on a single array, i represents the subject number and j represents the array number. The parameter μ represents the actual log ratio of gene expression between β7+ and β7- cells, which we wished to estimate. A i represents the difference between the log ratio for subject i and the average log ratio for all subjects and ε denotes the experimental error. We fit the model using a zero sum constraint (ΣA i = 0). Fold difference was calculated from the estimated log ratio. We computed moderated t-statistics [44], log-odds ratios of differential expression (based on empirical Bayes shrinkage of the standard errors towards a common value [45]), and adjusted p-values (obtained using the Bonferroni correction) using functions in the limma library of the Bioconductor software package [41,42]. Similar results were obtained under a random effects model (not shown).
Analysis of surface protein expression on blood CD4+ T cell subsets
Whole blood was collected from nine normal adult subjects (seven females and two males, ages 23–38 years, none of whom were included in the microarray experiments described above). Blood was stained with various antibody combinations and analyzed on a FACScalibur equipped for six color analysis. Antibodies recognizing CD4 (SK3), integrin β7 (FIB504), CD62L (DREG-56), CD73 (AD2), and CD161 (DX12) and isotype control antibodies were all from BD Biosciences. Other antibodies used were LAIR1 (DX26, generous gift of L. Lanier), KLRG1 (13A2, generous gift of D. Voehringer and H. Pircher), CD45RA (Immunotech), the anti-α4β7 mAb Act-1 [46], the anti-aEβ7 antibody HML-1 (Beckman Coulter), CCR7 (R&D Systems). Mean fluorescence intensity (MFI) was corrected by subtracting the MFI obtained with isotype control antibodies. The percentage of positive cells was determined by setting the threshold so that ≤1% of cells were considered positive after staining with control antibody.
Authors' contributions
MWR recruited the subjects, performed the experiments and participated in data analysis and drafting of the manuscript. ACP performed the statistical analysis of the microarray data. YHY supervised the statistical analysis of the microarray data. DJE conceived of the study and participated in design, analysis, and preparation of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Tab-delimited text file with calculated gene expression ratios for each probe on each array. Values represent normalized log2 (Cy5 intensity/Cy3 intensity).
MIAME-compliant description of the array experiments in rich text format.
Acknowledgments
Acknowledgements
Microarray studies were supported by Andrea Barczak, Michael Salazar, and the UCSF/San Francisco General Hospital General Clinical Research Center Core Genomics Laboratory (supported by grant M01-RR00083 from the National Center for Research Resources of the NIH). Flow cytometry was performed in the Flow Cytometry Core Laboratory of the J. David Gladstone Institutes. We thank Gordon Smyth and Mark Segal for helpful discussions regarding the microarray data analysis. The work was supported by NIH grant DK/AI54212.
Contributor Information
Madeleine W Rodriguez, Email: mwillkom@yahoo.com.
Agnés C Paquet, Email: apaquet@medsfgh.ucsf.edu.
Yee Hwa Yang, Email: jean@biostat.ucsf.edu.
David J Erle, Email: erle@itsa.ucsf.edu.
References
- von Andrian UH, Mackay CR. T-cell function and migration. Two sides of the same coin. N Engl J Med. 2000;343:1020–1034. doi: 10.1056/NEJM200010053431407. [DOI] [PubMed] [Google Scholar]
- Kunkel EJ, Butcher EC. Chemokines and the tissue-specific migration of lymphocytes. Immunity. 2002;16:1–4. doi: 10.1016/S1074-7613(01)00261-8. [DOI] [PubMed] [Google Scholar]
- Berg EL, Yoshino T, Rott LS, Robinson MK, Warnock RA, Kishimoto TK, Picker LJ, Butcher EC. The cutaneous lymphocyte antigen is a skin lymphocyte homing receptor for the vascular lectin endothelial cell-leukocyte adhesion molecule 1. J Exp Med. 1991;174:1461–1466. doi: 10.1084/jem.174.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietz W, Allemand Y, Borges E, von Laer D, Hallmann R, Vestweber D, Hamann A. CD4+ T cells migrate into inflamed skin only if they express ligands for E- and P-selectin. J Immunol. 1998;161:963–970. [PubMed] [Google Scholar]
- Homey B, Wang W, Soto H, Buchanan ME, Wiesenborn A, Catron D, Muller A, McClanahan TK, Dieu-Nosjean MC, Orozco R, Ruzicka T, Lehmann P, Oldham E, Zlotnik A. Cutting edge: the orphan chemokine receptor G protein-coupled receptor-2 (GPR-2, CCR10) binds the skin-associated chemokine CCL27 (CTACK/ALP/ILC) J Immunol. 2000;164:3465–3470. doi: 10.4049/jimmunol.164.7.3465. [DOI] [PubMed] [Google Scholar]
- Reiss Y, Proudfoot AE, Power CA, Campbell JJ, Butcher EC. CC chemokine receptor (CCR)4 and the CCR10 ligand cutaneous T cell-attracting chemokine (CTACK) in lymphocyte trafficking to inflamed skin. J Exp Med. 2001;194:1541–1547. doi: 10.1084/jem.194.10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homey B, Alenius H, Muller A, Soto H, Bowman EP, Yuan W, McEvoy L, Lauerma AI, Assmann T, Bunemann E, Lehto M, Wolff H, Yen D, Marxhausen H, To W, Sedgwick J, Ruzicka T, Lehmann P, Zlotnik A. CCL27-CCR10 interactions regulate T cell-mediated skin inflammation. Nat Med. 2002;8:157–165. doi: 10.1038/nm0202-157. [DOI] [PubMed] [Google Scholar]
- Hudak S, Hagen M, Liu Y, Catron D, Oldham E, McEvoy LM, Bowman EP. Immune surveillance and effector functions of CCR10(+) skin homing T cells. J Immunol. 2002;169:1189–1196. doi: 10.4049/jimmunol.169.3.1189. [DOI] [PubMed] [Google Scholar]
- Berlin C, Berg EL, Briskin MJ, Andrew DP, Kilshaw PJ, Holzmann B, Weissman IL, Hamann A, Butcher EC. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74:185–185. doi: 10.1016/0092-8674(93)90305-A. [DOI] [PubMed] [Google Scholar]
- Hamann A, Andrew DP, Jablonski-Westrich D, Holzmann B, Butcher EC. Role of alpha 4-integrins in lymphocyte homing to mucosal tissues in vivo. J Immunol. 1994;152:3282–3293. [PubMed] [Google Scholar]
- Abitorabi MA, Mackay CR, Jerome EH, Osorio O, Butcher EC, Erle DJ. Differential expression of homing molecules on recirculating lymphocytes from sheep gut, peripheral, and lung lymph. J Immunol. 1996;156:3111–3117. [PubMed] [Google Scholar]
- Williams MB, Butcher EC. Homing of naive and memory T lymphocyte subsets to Peyer's patches, lymph nodes, and spleen. J Immunol. 1997;159:1746–1752. [PubMed] [Google Scholar]
- Zabel BA, Agace WW, Campbell JJ, Heath HM, Parent D, Roberts AI, Ebert EC, Kassam N, Qin S, Zovko M, LaRosa GJ, Yang LL, Soler D, Butcher EC, Ponath PD, Parker CM, Andrew DP. Human G protein-coupled receptor GPR-9-6/CC chemokine receptor 9 is selectively expressed on intestinal homing T lymphocytes, mucosal lymphocytes, and thymocytes and is required for thymus-expressed chemokine-mediated chemotaxis. J Exp Med. 1999;190:1241–1256. doi: 10.1084/jem.190.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel EJ, Campbell JJ, Haraldsen G, Pan J, Boisvert J, Roberts AI, Ebert EC, Vierra MA, Goodman SB, Genovese MC, Wardlaw AJ, Greenberg HB, Parker CM, Butcher EC, Andrew DP, Agace WW. Lymphocyte CC chemokine receptor 9 and epithelial thymus-expressed chemokine (TECK) expression distinguish the small intestinal immune compartment: Epithelial expression of tissue-specific chemokines as an organizing principle in regional immunity. J Exp Med. 2000;192:761–768. doi: 10.1084/jem.192.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis KA, Landers C, Prehn J, Kouroumalis EA, Moreno ST, Gutierrez-Ramos JC, Hodge MR, Targan SR. CC chemokine receptor 9 expression defines a subset of peripheral blood lymphocytes with mucosal T cell phenotype and Th1 or T-regulatory 1 cytokine profile. J Immunol. 2003;171:159–165. doi: 10.4049/jimmunol.171.1.159. [DOI] [PubMed] [Google Scholar]
- Picker LJ, Treer JR, Ferguson-Darnell B, Collins PA, Buck D, Terstappen LW. Control of lymphocyte recirculation in man. I. Differential regulation of the peripheral lymph node homing receptor L-selectin on T cells during the virgin to memory cell transition. J Immunol. 1993;150:1105–1121. [PubMed] [Google Scholar]
- Picker LJ, Treer JR, Ferguson-Darnell B, Collins PA, Bergstresser PR, Terstappen LW. Control of lymphocyte recirculation in man. II. Differential regulation of the cutaneous lymphocyte-associated antigen, a tissue-selective homing receptor for skin-homing T cells. J Immunol. 1993;150:1122–1136. [PubMed] [Google Scholar]
- Campbell DJ, Butcher EC. Rapid acquisition of tissue-specific homing phenotypes by CD4(+) T cells activated in cutaneous or mucosal lymphoid tissues. J Exp Med. 2002;195:135–141. doi: 10.1084/jem.20011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora JR, Bono MR, Manjunath N, Weninger W, Cavanagh LL, Rosemblatt M, Von Andrian UH. Selective imprinting of gut-homing T cells by Peyer's patch dendritic cells. Nature. 2003;424:88–93. doi: 10.1038/nature01726. [DOI] [PubMed] [Google Scholar]
- Stagg AJ, Kamm MA, Knight SC. Intestinal dendritic cells increase T cell expression of alpha4beta7 integrin. Eur J Immunol. 2002;32:1445–1454. doi: 10.1002/1521-4141(200205)32:5<1445::AID-IMMU1445>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Dudda JC, Simon JC, Martin S. Dendritic cell immunization route determines CD8+ T cell trafficking to inflamed skin: role for tissue microenvironment and dendritic cells in establishment of T cell-homing subsets. J Immunol. 2004;172:857–863. doi: 10.4049/jimmunol.172.2.857. [DOI] [PubMed] [Google Scholar]
- Erle DJ, Briskin MJ, Butcher EC, Garcia-Pardo A, Lazarovits AI, Tidswell M. Expression and function of the MAdCAM-1 receptor, integrin alpha 4 beta 7, on human leukocytes. J Immunol. 1994;153:517–528. [PubMed] [Google Scholar]
- Cerf-Bensussan N, Jarry A, Brousse N, Lisowska-Grospierre B, Guy-Grand D, Griscelli C. A monoclonal antibody (HML-1) defining a novel membrane molecule present on human intestinal lymphocytes. Eur J Immunol. 1987;17:1279–1285. doi: 10.1002/eji.1830170910. [DOI] [PubMed] [Google Scholar]
- Cerf-Bensussan N, Begue B, Gagnon J, Meo T. The human intraepithelial lymphocyte marker HML-1 is an integrin consisting of a beta 7 subunit associated with a distinctive alpha chain. Eur J Immunol. 1992;22:273–277. doi: 10.1002/eji.1830220140. [DOI] [PubMed] [Google Scholar]
- Parker CM, Cepek KL, Russell GJ, Shaw SK, Posnett DN, Schwarting R, Brenner MB. A family of beta 7 integrins on human mucosal lymphocytes. Proc Natl Acad Sci U S A. 1992;89:1924–1928. doi: 10.1073/pnas.89.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erle DJ, Brown T, Christian D, Aris R. Lung epithelial lining fluid T cell subsets defined by distinct patterns of beta 7 and beta 1 integrin expression. Am J Respir Cell Mol Biol. 1994;10:237–244. doi: 10.1165/ajrcmb.10.3.7509610. [DOI] [PubMed] [Google Scholar]
- Barczak A, Rodriguez MW, Hanspers K, Koth LL, Tai YC, Bolstad BM, Speed TP, Erle DJ. Spotted long oligonucleotide arrays for human gene expression analysis. Genome Res. 2003;13:1775–1785. doi: 10.1101/gr.1048803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralevic V, Burnstock G. Receptors for purines and pyrimidines. Pharmacol Rev. 1998;50:413–492. [PubMed] [Google Scholar]
- Lanier LL, Chang C, Phillips JH. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol. 1994;153:2417–2428. [PubMed] [Google Scholar]
- Meyaard L, Adema GJ, Chang C, Woollatt E, Sutherland GR, Lanier LL, Phillips JH. LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity. 1997;7:283–290. doi: 10.1016/S1074-7613(00)80530-0. [DOI] [PubMed] [Google Scholar]
- Poggi A, Costa P, Zocchi MR, Moretta L. NKRP1A molecule is involved in transendothelial migration of CD4+ human T lymphocytes. Immunol Lett. 1997;57:121–123. doi: 10.1016/S0165-2478(97)00050-3. [DOI] [PubMed] [Google Scholar]
- Iiai T, Watanabe H, Suda T, Okamoto H, Abo T, Hatakeyama K. CD161+ T (NT) cells exist predominantly in human intestinal epithelium as well as in liver. Clin Exp Immunol. 2002;129:92–98. doi: 10.1046/j.1365-2249.2002.01886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henttinen T, Jalkanen S, Yegutkin GG. Adherent leukocytes prevent adenosine formation and impair endothelial barrier function by Ecto-5'-nucleotidase/CD73-dependent mechanism. J Biol Chem. 2003;278:24888–24895. doi: 10.1074/jbc.M300779200. [DOI] [PubMed] [Google Scholar]
- Airas L, Niemela J, Jalkanen S. CD73 engagement promotes lymphocyte binding to endothelial cells via a lymphocyte function-associated antigen-1-dependent mechanism. J Immunol. 2000;165:5411–5417. doi: 10.4049/jimmunol.165.10.5411. [DOI] [PubMed] [Google Scholar]
- Airas L, Hellman J, Salmi M, Bono P, Puurunen T, Smith DJ, Jalkanen S. CD73 is involved in lymphocyte binding to the endothelium: characterization of lymphocyte-vascular adhesion protein 2 identifies it as CD73. J Exp Med. 1995;182:1603–1608. doi: 10.1084/jem.182.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvilommi AM, Salmi M, Airas L, Kalimo K, Jalkanen S. CD73 mediates lymphocyte binding to vascular endothelium in inflamed human skin. Eur J Immunol. 1997;27:248–254. doi: 10.1002/eji.1830270137. [DOI] [PubMed] [Google Scholar]
- Hughes RC. Galectins as modulators of cell adhesion. Biochimie. 2001;83:667–676. doi: 10.1016/S0300-9084(01)01289-5. [DOI] [PubMed] [Google Scholar]
- Bowman EP, Campbell JJ, Druey KM, Scheschonka A, Kehrl JH, Butcher EC. Regulation of chemotactic and proadhesive responses to chemoattractant receptors by RGS (regulator of G-protein signaling) family members. J Biol Chem. 1998;273:28040–28048. doi: 10.1074/jbc.273.43.28040. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Thorne N. Normalization for Two-color cDNA Microarray Data. In: Goldstein D, editor. Science and Statistics: A Festschrift for Terry Speed. Vol. 40. 2003. pp. 403–418. [Google Scholar]
- Dudoit S, Yang YH. Bioconductor R packages for exploratory analysis and normalization of cDNA microarray data. In: Parmigiani G, Garrett ES, Irizarry RA and Zeger SL, editor. The Analysis of Gene Expression Data: Methods and Software. New York, Springer; 2002. [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. Journal of Computational and Graphical Statistics. 1996;5:299–314. [Google Scholar]
- Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Lönnstedt I, Speed TP. Replicated microarray data. Statistica Sinica. 2002;12:31–46. [Google Scholar]
- Schweighoffer T, Tanaka Y, Tidswell M, Erle DJ, Horgan KJ, Luce GE, Lazarovits AI, Buck D, Shaw S. Selective expression of integrin alpha 4 beta 7 on a subset of human CD4+ memory T cells with Hallmarks of gut-trophism. J Immunol. 1993;151:717–729. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tab-delimited text file with calculated gene expression ratios for each probe on each array. Values represent normalized log2 (Cy5 intensity/Cy3 intensity).
MIAME-compliant description of the array experiments in rich text format.