Abstract
The central role of the transcriptional template of the hepatitis B virus (HBV), covalently closed circular DNA (cccDNA), has been difficult to study in patients with chronic hepatitis B (CHB) infection. In this issue of the JCI, Zhang and colleagues reveal a mosaic distribution of viral antigens and nucleic acids and a mismatch between HBV cccDNA, RNA, and expression of the hepatitis B surface antigen (HBsAg). These unusual patterns varied over the natural history of CHB, prompting the authors to propose a new three-stage model of the HBV life cycle at the single-cell level.
Chronic HBV pathogenesis: limited data on the clinical pathogenesis
Hepatitis B virus (HBV) is a major human health problem, with more than 240 million people chronically infected with the virus worldwide and at least 780,000 people dying each year from HBV-related liver diseases such as cirrhosis and liver cancer (1). There is no known cure for chronic HBV (CHB) infection, and the preventative vaccines that are currently in use have no impact on existing infections. Knowledge about the molecular pathogenesis of CHB has come largely from animal model studies; however, matching clinical data on CHB etiology and progression are lacking.
In this issue, Zhang and colleagues used sensitive ISH technology to selectively detect HBV nucleic acids (DNA and RNA) and associated antigens in the liver tissue of Chinese patients with CHB (HBV genotype C) (2). Using this approach, the authors have confirmed and extended earlier seminal studies of duck HBV (DHBV) (3) and HBV (4, 5) that revealed an intracellular mismatch between hepadnavirus DNA replication and hepatitis B surface antigen (HBsAg) production. Zhang et al. developed novel molecular diagnostic tools for examining HBV DNA (covalently closed circular DNA [cccDNA] and protein-free relaxed circular DNA [rcDNA]), RNA, and protein in the same liver section. This study has demonstrated that the HBV nuclear genetic reservoir in humans consists of a population of protein-free cccDNA and rcDNA molecules, confirming the findings of previous studies done in hepatoma cell lines (6). In addition, the selective nature of the probe sets and the appropriate use of DNase and RNase to validate the nucleic acid population under investigation enabled Zhang and colleagues to comprehensively analyze HBV replicative intermediates and antigen production in the same cell (2). The observed disconnect between HBV DNA and RNA and HBsAg expression in the liver supports earlier studies showing that high levels of HBsAg were associated with very low levels of cccDNA (or RNA), and vice versa. This observation further supports the findings of Summers et al., who showed that higher levels of cccDNA are produced in the liver of primary duck hepatocytes infected with a DHBV mutant defective for envelope proteins (3).
A mismatch between HBV antigen production and transcription
The data presented by Zhang et al. (2) suggest that HBsAg regulates cccDNA levels via a negative feedback model that is similar to that first proposed by Tuttleman and colleagues in 1986 (7), whereby high levels of envelope antigen production blocks further cccDNA synthesis. However, if this model is correct, the threshold of HBsAg (or cccDNA) that is required to initiate and regulate such negative feedback is unclear. Additionally, Zhang and colleagues identified a similar mismatch between antigen production and HBV RNA, which is also important, as measuring the abundance of cccDNA alone cannot provide an accurate assessment of transcriptional activity. HBV cccDNA exists as a minichromosome, and the transcriptional activity of cccDNA is determined by histone acetylation and methylation status and is also regulated by the HBV antigen, HBx protein (8). A better understanding of the direct contribution of cccDNA (and the HBx protein) to HBV antigen production is important, as it is currently thought that rendering cccDNA transcriptionally inert (such as through locking up the minichromosome by hyperchromatination) may result in a functional cure (9). The development of such strategies to block cccDNA transcription is a major area of study; however, knowledge of the factors that regulate cccDNA transcription and the direct contribution of cccDNA to antigen production in human HBV infection is limited. Zhang et al. have shown that cccDNA persists in liver tissue, even in patients given a year-long course of antiviral therapy, which typically eliminates active viral replication. Interestingly, treatment did reduce cccDNA levels, most likely due to a decrease of cccDNA replenishment via inhibition of the intracellular conversion pathway (ref. 7 and Figure 1), as first demonstrated in the setting of adefovir treatment by Werle-Lapostolle et al. (10). Continued production of HBsAg during therapy is strongly associated with disease progression and liver cancer, indicating that a greater understanding of factors that regulate cccDNA transcription and antigen expression is warranted (9).
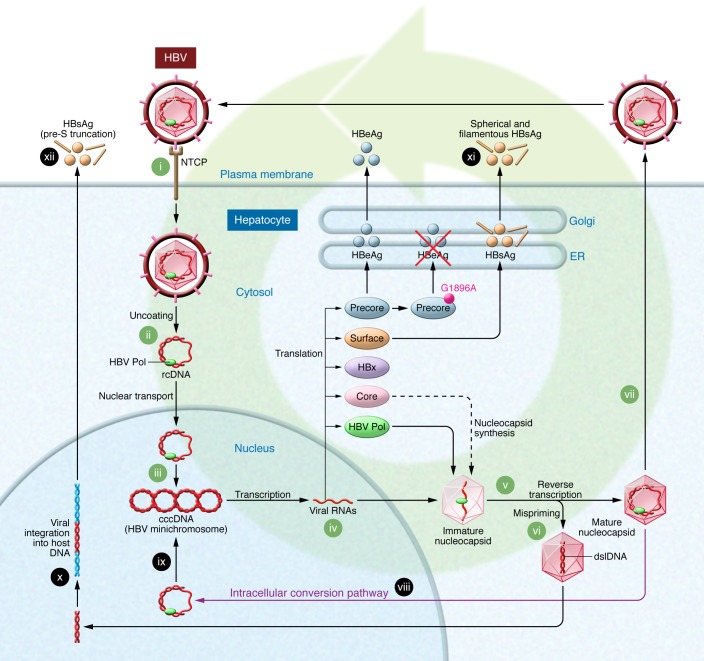
Figure 1. The life cycle of HBV.
HBV enters the hepatocyte via binding to the sodium taurocholate cotransporting polypeptide (NTCP) receptor (i). Following removal of the viral envelope and nucleocapsid (ii), viral rcDNA is transported to the nucleus, where it is converted to cccDNA (iii), which is the transcription template for all viral mRNAs (iv). pgRNA is transported to the cytoplasm, where it is encapsidated with HBV Pol and reverse transcribed to rcDNA (v). If mispriming occurs during this process, dslDNA is produced instead of rcDNA (vi). Virions encapsidating rcDNA become enveloped and are secreted from the cell as infectious virions (vii), or rcDNA and dslDNA may be transported to the nucleus (viii), where rcDNA is converted to cccDNA (ix) via the intracellular conversion pathway. The dslDNA may integrate into the host chromosome (x). HBsAg is synthesized from both the viral minichromosome–derived mRNA (xi) and from integrated HBV DNA (xii). The G1896A mutation introduces a premature stop codon and abrogates the production of HBeAg.
Zhang and colleagues have claimed that probe set 1 specifically detects pregenomic RNA (pgRNA). Probe set 1 will also detect the 3.5-kb precore mRNA (pcRNA), which is approximately 30 nucleotides longer than pgRNA at the N-terminus and shares the identical probe sequence. The pcRNA encodes the precore protein that is subsequently processed into intracellular p22 protein and then secreted as hepatitis B e antigen (HBeAg) (ref. 11 and Figure 1). While not required for productive HBV replication, the HBeAg is required for the establishment of persistent infection, and it is well recognized that mutations in the HBV genome associated with long-term persistence alter the balance between pgRNA and pcRNA. For example, mutations in the HBV basal core promoter (BCP) at positions 1762 and 1764 reduce transcription of pcRNA, leading to a relative increase in the proportion of the pgRNA replicative template (12). This may be important in the context of the study by Zhang et al., as BCP mutations are highly prevalent in genotype C infection and potentially impact the relative levels of pgRNA and pcRNA. The impact of the HBeAg and associated proteins on cccDNA, DNA, and RNA levels warrants further investigation. Specific quantification of pgRNA and pcRNA by real-time PCR in this context could also be considered.
Future directions
The study by Zhang et al. (2) was restricted to tissue collected from persons infected with HBV genotype C, and the authors have highlighted the need to design specific probe sets to enable similar studies of other HBV genotypes. The development of additional probe sets is important, as HBV’s natural history, mode of transmission, disease progression, and response to therapy differ markedly across genotypes and subtypes (13). HBV genotype C infection is almost exclusively transmitted at birth or shortly thereafter, meaning that the patients in the current study had been infected for most of their lives. Will there be a similar disconnect between cccDNA levels and antigen production in patients who acquired HBV in early childhood (African genotype A1), adolescence (Mediterranean genotype D3), or adulthood (North American genotype A2)?
It should be noted that, compared with HBeAg-positive patients, HBeAg-negative patients showed a trend toward higher levels of nuclear DNA than of cytoplasmic DNA (2). Although this difference failed to reach statistical significance, it is compelling when considered in the light of recent studies that have identified higher levels of integrated DNA in the setting of HBeAg-negative disease than in HBeAg-positive disease (14). The mechanisms that underlie these differences are unclear and may simply reflect a longer duration of infection. Zhang and colleagues acknowledge that their probe set for detecting nuclear DNA cannot distinguish cccDNA from integrated DNA, suggesting the possibility that some of the nuclear DNA may have been of integrant origin. A larger study of cccDNA and integrated DNA in the setting of HBeAg-negative HBV is warranted, perhaps using inverse PCR (15) to quantify integrated DNA in conjunction with the branched DNA (bDNA) assay for cccDNA developed by Zhang et al. This type of analysis should also include testing for double-stranded linear DNA (dslDNA), which is generated as a consequence of mis-priming during viral replication (16) and is the precursor to DNA integration (Figure 1). Additionally, the possible contribution of spliced HBV variants, which have also been shown to promote dslDNA formation (17), should be investigated.
A new model of CHB
Zhang and colleagues have proposed a new model for the phases of CHB based on the HBV life cycle at the single-cell level (2). This model suggests that HBV infection commences with an antigen-rich stage, followed by a DNA-rich stage, in which the relative proportion of DNA to viral antigens increases as virion production decreases. Decreased virion production then leads to what they term the “latent stage,” which is dominated by episomal DNA (cccDNA) that is not transcriptionally active or integrated and concurs with an almost complete absence of viral protein production. To date, the term “latent” has rarely been applied in the context of HBV infection. While a large pool of transcriptionally silent cccDNA (or integrated DNA) in the absence of productive viral replication would seem to be a reasonable definition of latency in the context of HBV infection, it is questionable whether HBV infection is ever truly latent. The term is relevant for persons who have cleared the acute infection, only to see the virus reemerge during immunosuppressive therapy (18, 19). Indeed, a major question for HBV investigators is whether HBV can ever be truly cured. Missing from the model proposed by Zhang et al. is the HBeAg-negative phase of chronic disease, which is characterized by productive viral replication in the absence of HBeAg. Indeed, the factors that trigger productive viral replication (and presumably cccDNA transcription) following the low-replicative phase of CHB’s natural history in this setting are unresolved. While an understanding of HBV replication at the single-cell level is important, this model should be refined further to account for the metabolic zones within the liver (20), where HBV replication levels and permissiveness are affected by oxygen and glucose gradients (periportal versus central vein). The metabolic state of the liver, host immune responses, and epigenetic host factors would also be considered relevant in this context.
The findings of Zhang and colleagues have provided new virological insights into the HBV life cycle within the infected hepatocyte and in the context of the natural history of CHB (2). Moreover, these results emphasize that there is still much to be learned about this important pathogen. Perhaps it is also time to reconsider the accepted definitions of the phases of CHB, namely the so-called “immune-tolerant,” “immune control,” “low-replicative,” and “high-replicative/HBeAg-negative” phases (21). These broad-brush definitions are largely meaningless from a virological perspective, and a greater understanding of the complete viral life cycle (Figure 1) at both the single-cell level and in the context of the whole liver should lead to more individualized management approaches that would be guided by the viral replicative state, including cccDNA levels, transcriptional activity, and viral antigen load, rather than serum alanine aminotransferase levels.
Acknowledgments
The authors thank Tania Candy for her assistance with the preparation of this Commentary. P.A. Revill is currently in receipt of a Melbourne Health Keir Research Fellowship.
Footnotes
Conflict of interest: P.A. Revill and S.A. Locarnini have received funding from Gilead Sciences Inc. for aspects of their research, and S.A. Locarnini has also received funding from Arrowhead Research Corporation.
Reference information:J Clin Invest. 2016;126(3):833–836. doi:10.1172/JCI86650.
See the related article beginning on page 1079.
References
- 1.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, et al. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J Clin Invest. 2016;126(3):1079–1092. doi: 10.1172/JCI83339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Summers J, Smith PM, Horwich AL. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol. 1990;64(6):2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrell CJ, Gowans EJ, Rowland R, Hall P, Jilbert AR, Marmion BP. Correlation between liver histology and markers of hepatitis B virus replication in infected patients: a study by in situ hybridization. Hepatology. 1984;4(1):20–24. doi: 10.1002/hep.1840040104. [DOI] [PubMed] [Google Scholar]
- 5.Tong WP, et al. An accurate quantitative method for screening effective siRNA probes targeting a hepatitis B virus transcript in single living cells. Biochem Biophys Res Commun. 2008;367(4):866–873. doi: 10.1016/j.bbrc.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol. 2007;81(22):12472–12484. doi: 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuttleman JS, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47(3):451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 8.Belloni L, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A. 2009;106(47):19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeisel MB, et al. Towards an HBV cure: state-of-the-art and unresolved questions — report of the ANRS workshop on HBV cure. Gut. 2015;64(8):1314–1326. doi: 10.1136/gutjnl-2014-308943. [DOI] [PubMed] [Google Scholar]
- 10.Werle-Lapostolle B, et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126(7):1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Messageot F, Salhi S, Eon P, Rossignol JM. Proteolytic processing of the hepatitis B virus e antigen precursor. J Biol Chem. 2003;278(2):891–895. doi: 10.1074/jbc.M207634200. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Buckwold VE, Hon MW, Ou JH. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J Virol. 1999;73(2):1239–1244. doi: 10.1128/jvi.73.2.1239-1244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim BK, Revill PA, Ahn SH. HBV genotypes: relevance to natural history, pathogenesis and treatment of chronic hepatitis B. Antivir Ther. 2011;16(8):1169–1186. doi: 10.3851/IMP1982. [DOI] [PubMed] [Google Scholar]
- 14.Wooddell C, et al. Reductions in cccDNA under NUC and ARC-520 therapy in chimpanzees with chronic hepatitis B virus infection implicate integrated DNA in maintaining circulating HBsAg. Hepatology. 2015;62(suppl 1): [Google Scholar]
- 15.Tu T, et al. Clonal expansion of hepatocytes with a selective advantage occurs during all stages of chronic hepatitis B virus infection. J Viral Hepat. 2015;22(9):737–753. doi: 10.1111/jvh.12380. [DOI] [PubMed] [Google Scholar]
- 16.Beck J, Nassal M. Hepatitis B virus replication. World J Gastroenterol. 2007;13(1):48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham TM, Lewellyn EB, Haines KM, Loeb DD. Characterization of the contribution of spliced RNAs of hepatitis B virus to DNA synthesis in transfected cultures of Huh7 and HepG2 cells. Virology. 2008;379(1):30–37. doi: 10.1016/j.virol.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kempinska A, Kwak EJ, Angel JB. Reactivation of hepatitis B infection following allogeneic bone marrow transplantation in a hepatitis B-immune patient: case report and review of the literature. Clin Infect Dis. 2005;41(9):1277–1282. doi: 10.1086/496924. [DOI] [PubMed] [Google Scholar]
- 19.Palmore TN, et al. Reactivation of hepatitis B with reappearance of hepatitis B surface antigen after chemotherapy and immunosuppression. Clin Gastroenterol Hepatol. 2009;7(10):1130–1137. doi: 10.1016/j.cgh.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanner MS. Mechanisms of liver injury relevant to pediatric hepatology. Crit Rev Clin Lab Sci. 2002;39(1):1–61. doi: 10.1080/10408360290795439. [DOI] [PubMed] [Google Scholar]
- 21.Dandri M, Locarnini S. New insight in the pathobiology of hepatitis B virus infection. Gut. 2012;61(suppl 1):i6–i17. doi: 10.1136/gutjnl-2012-302056. [DOI] [PubMed] [Google Scholar]