Abstract
Cancer stem cells (CSCs) are associated with cancer recurrence and metastasis. Prostate cancer cells often metastasize to the bone with a complex microenvironment of cytokines favoring cell survival. In this study, the cell stemness influence of a group of interleukins including IL-3, 6, 10, 11 and 24 on human prostate cancer cell lines LNCaP and PC-3 was explored in vitro. Sulforhodamine B(SRB) and 5-ethynyl-2′-deoxyuridine (EdU) assays were applied to examine the effect on cell proliferation, and wound healing and transwell assays were used for migration and invasion studies, in addition to colony formation, Western blotting and flowcytometry for the expression of stemness factors and chemotherapy sensitivity. We observed that ILs-3, 6 and 11 stimulated while ILs-10 and 24 inhibited the growth, invasion and migration of both cell lines. Interestingly, ILs-3, 6 and 11 significantly promoted colony formation and increased the expression of SOX2, CD44 and ABCG2 in both prostate cancer cell lines. However, ILs-10 and 24 showed the opposite effect on the expression of these factors. In line with the above findings, treatment with either IL-3 or IL-6 or IL-11 decreased the chemosensitivity to docetaxel while treatment with either IL-10 or IL-24 increased the sensitivity of docetaxel chemotherapy. In conclusion, our results suggest that ILs-3, 6 and 11 function as tumor promoters while ILs-10 and 24 function as tumor suppressors in the prostate cancer cell lines PC-3 and LNCaP in vitro, and such differences may attribute to their different effect on the stemness of PCa cells.
Keywords: interleukin, cytokines, cancer stem cells, chemotherapy, CD44 and ABCG2
INTRODUCTION
Prostate cancer(PCa) is the second leading cause of cancer death among men in the European and American, followed by lung cancer [1], and the morbidity and mortality are rapidly increasing in China [2, 3]. Prostate specific antigen (PSA) is a specific marker for PCa, especially in the early stage. The androgen deprivation and radical prostectomy has been the gold standard therapy. Often PCa patients have a progression without clinical symptoms and the disease is detected in advanced stages.
The standard treatment for patients with primary hormone-dependent prostate cancer is endocrine therapy, and the treatment originally inhibits tumor growth. However, nearly all patients who received this treatment will ultimately relapse and develop bone metastases [4]. Recently, increasing evidence indicates that there are cancer stem cells (CSCs) survived in circulating system or bone marrow of prostate cancer patients, which attribute to therapy-resistant and relapse [5–7]. The most notable features of these multipotent and self-renewing cells are their high survival capability and high resistance to chemotherapy.
CSCs are driving force for the onset, development and progression of various cancers. Recent studies point to the possibility that a number of cytokines in the tumor microenviroment [8, 9] influence the stemness of cancer cells and CSC properties, and the growth and metastasis of tumors [10–12].
It is believed that conventional treatment options eradicate the bulk of more differentiated tumor cell clones. However, CSCs survive most of the classical chemotherapy regimens and contribute to the tumor progression and recurrence [13–15].
Interleukins (ILs) are a subgroup of cytokines being primarily reported in leukocytes. IL-3 is produced in activated T cells and mast cells, and influences the production, differentiation and function of granulocytes and macrophages [16, 17]. It has been reported that IL-3 receptor(IL-3R) overexpression on leukaemia stem cell populations is a common occurrence, and therapeutic IL-3-IL-3R interference option already shown promising [18]. And importantly, the IL-3R(CD123) had been considered as a biomarker of leukemia stem cells [19, 20], although contradictory results exist in the literature that IL-3 has not direct relationship with the hematopoietic stem cells (HSCs) [21], or breast cancer [22].
IL-6, also called B-cell stimulatory factor-2 and interferon beta-2, is involved in different biological functions, including B lymph cell differentiation and myeloma and plasmacytoma growth [23, 24]. Recently, it was observed that IL-6 is secreted from tumor-associated macrophages (TAMs) in hepatocellular carcinoma (HCC) and involved in the HCC tumorigenesis and also the CSC expansion [25]. Similar findings are also reported in human colon cancer [26] and malignant transformation of rat mesenchymal stem cells (MSCs) [27].
IL-10, another immunomodulatory cytokine, had been used as vehicle of the neural stem/progenitor cells (NSPCs) into the central nervous system (CNS) to replace damaged cells and cure inflammation [28]. IL-10 is implicated in enhancing the ability of MSCs in anti-inflammatory application [29] and hematopoietic stem cell transplantation (HSCT) in treatment of the inflammatory bowel disease (IBD) [30]. Other studies have shown that IL-10 may inhibit the growth and differentiation of neural stem cells (NSCs) in normal adult brain [31], and help the MSCs to inhibit the mature of dendritic cells (DCs) by JAK1 and STAT3 signaling pathway [32].
IL-11 is also a secreted cytokine and involves in megakaryocytopoiesis, platelet production, osteoclast activation and inhibition of epithelial cell proliferation and apoptosis [33]. IL-11 is active in peripheral blood stem cell(PBSC) mobilisation [35]. Clinically, it has been shown that IL-11 can decrease the occurrence of the development of graft versus host disease (GVHD), which is induced by allogeneic hematopoietic stem cell transplantation (allo-HSCT) [34].
Increasing evidence indicates that IL-24 may suppress the growth of cancer stem cells. Further studies revealed that IL-24 effectively inhibited proliferation and angiogenesis of cancer cells, even reduce the percentage of breast cancer-initiating/stem cells [36] and mesenchymal stem cells (MSCs) [37] in vivo.
We have previously reported that granulocyte-macrophage colony stimulating factor (GM-CSF) and colony stimulating factor(CSF) stimulate the stemness of PCa cells [38]. In our present study, we have examined the effect of different interleukins (IL-3,6,10,11 and 24) on the cell growth, migration, invasion, apoptosis, colony formation capability and chemotherapy resistance of the androgen-dependent LNCaP and the androgen-independent cell line PC-3 PCa cell lines, aiming to explore whether these cytokines could influence the cell stemness of the PCa cells in vitro.
RESULTS
The effect on cell growth
In order to study the proliferation effect of the ILs on LNCaP and PC-3 cells, dose-dependent growth curves were made. As shown in Figure 1A, dose-dependent growth curves are not always lineage for all of these ILs. However, it is apparently from this figure that 5 ng/ml treatment is rather representative for all the ILs, and 5 ng/ml was therefore chosen to use for the following experiments. Representative images of fluorescence microscopy using the EdU incorporation and Hoechest 33342 for the cells treated for 24 hrs are shown in Figure 1B. Histograms of the fluorescence microscopy results are shown in Figure 1C and 1D for LNCaP and PC-3 cells, respectively. It was found that 24 hrs treatment with IL-3, IL-6 and IL-11 in 5 ng/ml significantly increased the cell numbers in both cells lines, with p-values of 0.02, 0.037 and 0.032 in LNCaP cells, and p-values of 0.043, 0.029 and 0.029 in PC-3 cells, respectively. However, 24 hrs treatment of IL-10 and IL-24 in 5 ng/ml significantly decreased the number of cells in both cell lines, with p-values of 0.023 and 0.018 in LNCaP cells, and p-values of 0.027and 0.029 in PC-3 cells, respectively. These results indicate that IL-3, IL-6 and IL-11 significantly stimulate the cell growth, while IL-10 and IL24 significantly inhibit the cell growth in both cell lines.
Figure 1. Growth influence of ILs on LNCaP and PC-3 cells.
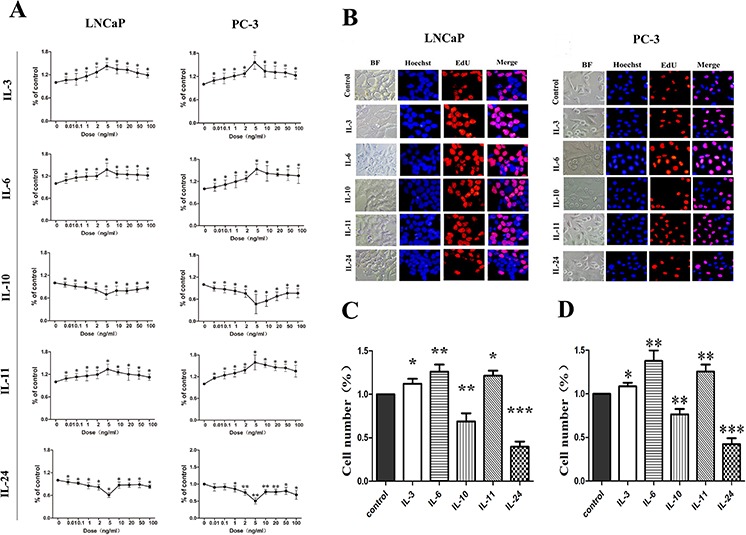
A. shows growth curves of the dose dependent effect of the interleukins. There is no linear dose-dependent effect. Representative fluorescence microscopic images of the EdU and Hoechest 33342 staining are shown in B. for LNCaP and PC-3 cells, respectively. Both cells are stained with EdU (red) for DNA synthesis and Hoechest 33342(blue) for nuclear staining. C. and D. show histograms of the living cell numbers after treatments of the interleukins for 48 hrs for LNCaP and PC-3 cells, respectively. Data are presented as mean ± SD of three separate experiments. * means p < 0.05, ** means p < 0.01 and *** means p < 0.001, in comparison to the control groups, respectively.
The effect on cell mobility
The motility of human PCa cells lines LNCaP and PC-3 cells were examined by wound healing assay when treated with different ILs. Confluent monolayers of cells were scratched to be wounded and cultured for 18 hrs (Figure 2). Compared with the control cells, treatment with IL-3, IL-6 and IL-11 demonstrated significantly higher mobility in both cell lines, and the rates of wound healing increased 10.2%(p = 0.046), 21.1%(p = 0.004) and 11.9% (p = 0.047) in LNCaP cells and 13.6%(p = 0.049) 30.4%(p = 0.045) and 16.1%(p = 0.040) in PC-3 cells, respectively. But treatment with IL-10 and IL-24 showed an inhibition effect on the wound healing in comparison to the control cells, and the rates of wound healing decreased with 20.8%(p = 0.008) and 39.3%(p = 0.031) in LNCaP cells and 26.2%(p < 0.001) and 48.5%(p = 0.002) in PC-3 cells, respectively.
Figure 2. Results of wound healing assay.
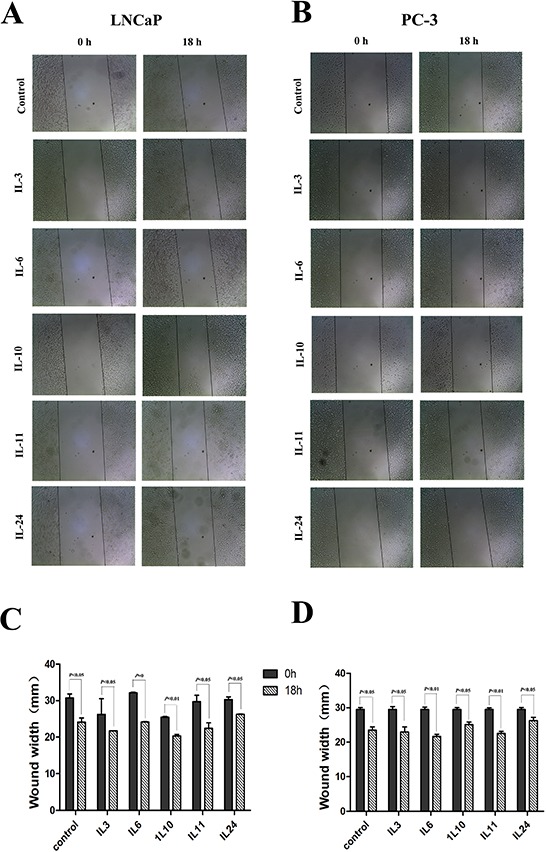
A. and C. show representative images and histograms of the effect of different interleukins on LNCaP cell line, respectively. B. and D. show representative images and histograms of the effect of different interleukins on PC-3 cell line, respectively. Data are presented as mean ± SD of three separate experiments, n = 3. * means p < 0.05, ** means p < 0.01, and *** means p < 0.001, in comparison to the control groups, respectively.
Migration and invasion effect
A transwell chamber system was employed to measure the migration and invasion effect of different ILs on LNCaP and PC-3 cells. In general, migration and invasion ability of both cell lines was increased when treated with IL-3, IL-6 and IL-11, but decreased when treated with IL-10 and IL-24 (Figure 3A and 3B). When cell migratory ability was examined with the non-treated cells as controls in LNCaP cells, 24 hrs of IL-3, IL-6 and IL-11 treatment significantly increased the number of cells migrated through the membrane, with increased rates of 13.2% (p = 0.014), 65.3%(p = 0.014) and 55.4%(p < 0.001), respectively. However, 24 hrs of IL-10 and IL-24 treatment significantly decreased the number of cells migrated through the membrane, and the migration rates declined 25.3% and 40.0% with p = 0.002 and p < 0.001, respectively. The migratory effect on PC-3 cells was similar. Compared to the non-treated cells, 24 hrs of IL-3, IL-6 and IL-11 treatment significantly increased the number of cells migrated through the membrane with increased rates of 10.7% (p = 0.002), 50.5% (p = 0.004) and 41.2%(p = 0.002), respectively, while 24 hrs treatment of IL-10 and IL-24 significantly decreased the number of cells migrated through the membrane with decreased rates of 22.4% (p = 0.007) and 24.7% (p = 0.002), respectively(Figure 3C).
Figure 3. Migratory and invasion influence of ILs on LNCaP and PC-3 cells.
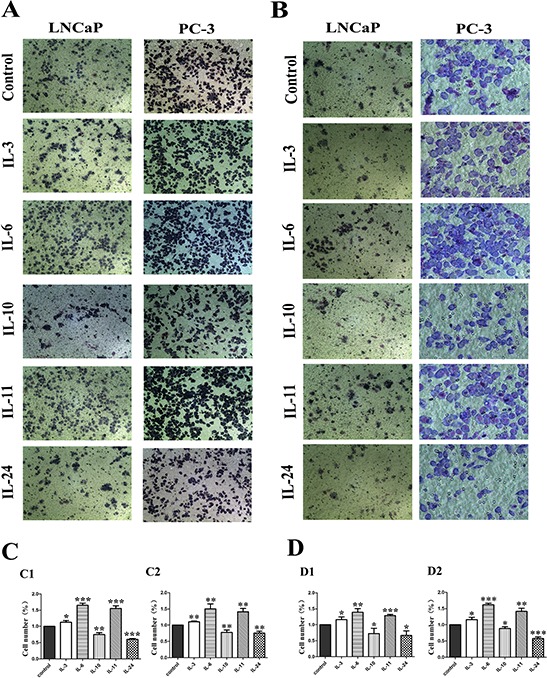
A. shows representative photographs of the cells migrated through the polycarbonate membrane stained by Gimsa. B. shows representative photographs of the invasive cells. C. shows histograms of the migration assay results and D. shows histograms of invasion assay results for both cell lines, respectively. While IL-3, IL-6 and IL-11 stimulate the migration and invasion of both cell lines, IL-10 and IL-24 significantly inhibit the migration and invasion of the cells as shown in C and D. All data represent means from three independent experiments. * means p < 0.05, ** means p < 0.01, and *** means p < 0.001.
For cell invasion examination where the membrane was coated with 60 μL of matrigel, 24 hrs of IL-3, IL-6 and IL-11 treatment significantly increased the number of invasive cells. Compared with the control cells, the invasion rate increased 16.6% (p = 0.026), 39.5% (p = 0.004) and 28.9% (p < 0.001) in the IL-3, IL-6 and IL-11 treated LNCaP groups, and 16.3% (p = 0.017), 61.2% (p < 0.001) and 41.7% (p = 0.002) in the IL-3, IL-6 and IL-11 treated PC-3 groups, respectively. While 24 hrs of IL-10 and IL-24 treatment significantly decreased the number of cells penetrated through the membrane in both cell lines. Comparatively, the decreased invasion rates were 27.7% (p = 0.044) and 33.6% (p = 0.015) in the IL-10 and IL-24 treated LNCaP groups, and 27.7% (p = 0.023) and 42.3% (p < 0.001) in the IL-10 and IL-24 treated PC-3 groups, respectively (Figure 3D).
The effect on chemotherapy resistance
The apoptotic effect of the ILs was firstly examined by flow cytometry.
Compared with the control cells, significantly lower numbers of apoptotic cells were seen in the cells treated with IL-3, IL-6 and IL-11 for 24 hrs, with p-values of 0.049, 0.003 and 0.011 in LNCaP cells, and p-values of 0.012, 0.001 and 0.002 in PC-3 cells, respectively. But treatment with IL-10 and IL-24 significantly increased the number of apoptotic cells, with p-values of 0.045 and 0.001 in LNCaP cells, and p-values of 0.040 and 0.009 in PC-3 cells, respectively (Figure 4).
Figure 4. Docetaxel treatment sensitivity influence.
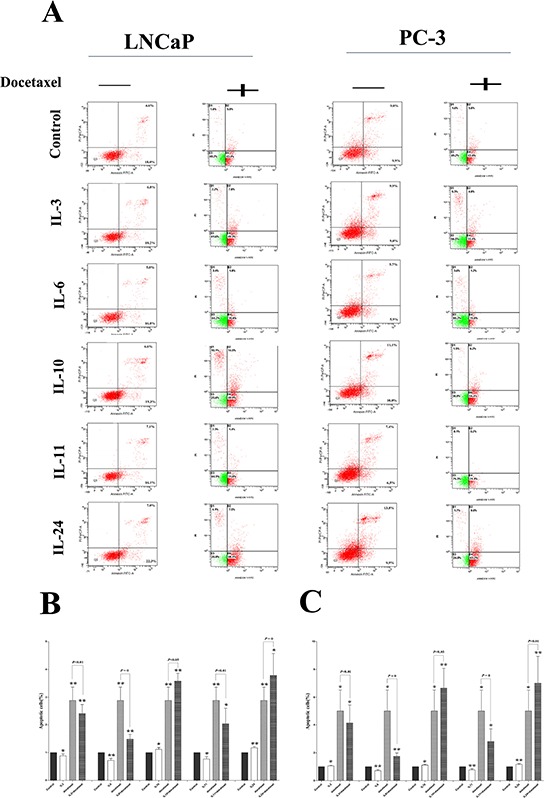
A. shows representative flowcytometry figures for LNCaP and PC-3 cell lines treated with different ILs with and without docetaxel application. Corresponding histogram results are shown in B. and C. for LNCaP and PC-3 cells, respectively. While docetaxel significantly induces apoptosis in both cell lines, application of either IL-3, or IL-6 or IL-11 slightly inhibits the apoptosis in the both cell lines. On the contrary, application of either IL-10 or IL-24 slightly increases the apoptosis already induced by the docetaxel. * means p < 0.05, ** means p < 0.01, and *** means p < 0.001, in comparison to the control groups, respectively.
The apoptotic effect of docetaxel on these cells was further examined. After optimization of the dose, 10nmol/L concentration of docetaxel was applied in this study. As shown in Figure 4, 24 hrs of docetaxel treatment alone significantly increased the number of apoptotic cells in these cell lines, with a p-value of 0.002 in LNCaP cells, and a p-value of 0.010 in PC-3 cells.
Then we asked whether the application of such ILs could influence the apoptosis influenced by docetaxel in these cells by joint application of the ILs and docetaxel. It was discovered that combination of docetaxel application with either IL-3 or IL-6 or IL-11 significantly reduced the numbers of apoptotic cells than those with docetaxel application alone, with p-values of 0.001, 0.007 and 0.017 in LNCaP cells, and p-values of 0.014, 0.002 and 0.026 in PC-3 cells, respectively. Also, the combination of docetaxel and either IL10 or IL-24 could significantly increase the number of apoptotic cells, with p-values of 0.001 and 0.024 in LNCaP cells, and p-values of 0.007 and 0.005 in PC-3 cells, respectively, indicating a chemo-sensitizing role of IL-10 and IL-24 in docetaxel treatment in prostate cancer cells.
The effect on SOX2 expression
To explore how ILs influenced the expression of SOX2, the expression of mRNA level of SOX2 was examines by RT-PCR(Figure 5A). Compared with the control groups, the expression of SOX2 mRNA was increased when treated with IL-3, IL-6 and IL-11 in both cells, while the expression of SOX2 mRNA when treated with IL-10 and IL-24, was decreased. All the mRNA level changes were consistent with the SOX2 protein expression alterations revealed with Western blotting as shown in Figure 5B. To further evaluate the expression of SOX2 in both cells, especially the cellular localization of SOX2, immunofluorescence microscopy of SOX2 was performed (Figure 5C–5F). All positive staining of the SOX2 in these cells was confined in the nuclei in both cell lines. Quantification of the positive cell numbers revealed similar results as demonstrated by RT-PCR (Figure 5A) and Western blotting (Figure 5B).
Figure 5. Influence of ILs on the RNA and protein expression of SOX2 in LNCaP and PC-3 cells by RT-PCR, Western blotting and immunofluorescence.
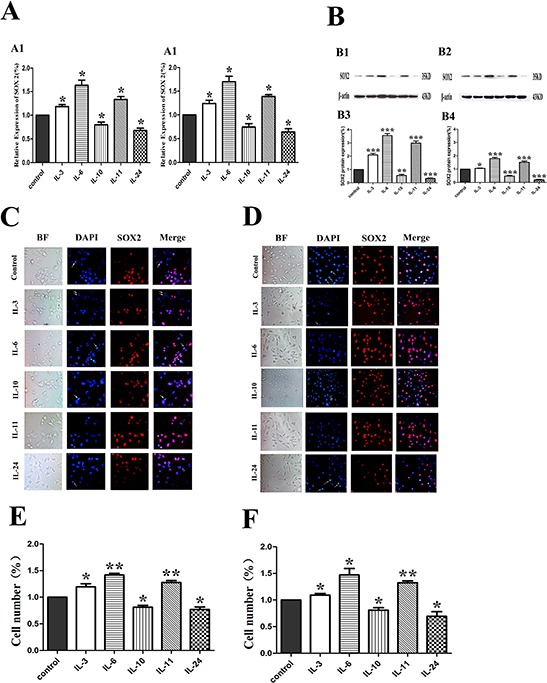
A. shows the relative gene expression of SOX2 figures for LNCaP and PC-3, respectively. B. shows the expression of SOX2 in LNCaP and PC-3 cells by Western blotting in protein level (upper) and the corresponding histograms (lower) in both cells. C. and D. show representative images of immunofluorescence microscopy of SOX2 in both cell lines. E. and F. show corresponding histograms of the SOX2 immunofluorescence microscopy for LNCaP and PC-3 cell lines, respectively. Cells are stained with DAPI to visualize the nuclei (blue). SOX2 (red) is localized in the nuclei. BF stands for bright field. All photographs were originally taken at 200 × .* means p < 0.05, and ** means p < 0.01.
Clonogenicity effect
The colony formation assay was carried out to examine the clonogenicity effect of these cytokines. Representative photos of the colony formation assay for both cell lines are shown on Figure 6A, and corresponding histograms of the results are shown on Figure 6B for LNCaP cells, and Figure 6C for PC-3 cells. Compared to the control cells, treatment of IL-3, IL-6 and IL-11 increased the number of clones by 18.6% (p = 0.005), 45.3% (p = 0.045) and 31.3% (p = 0.042) in LNCaP cells as shown on Figure 6B, and by 22.6% (p = 0.021), 57.0% (p = 0.027) and 43.3% (p = 0.001) in PC-3 cells as shown on Figure 6C. However, treatment with IL10 and IL24 resulted in significantly fewer clones by 16.9% (p = 0.032) and 31.4% (p = 0.030) as shown on Figure 6B for LNCaP cells and 19.0% (p = 0.027) and 42.4% (p = 0.002) as shown on Figure 6C for PC-3 cells, respectively.
Figure 6. Results of colony formation assay.
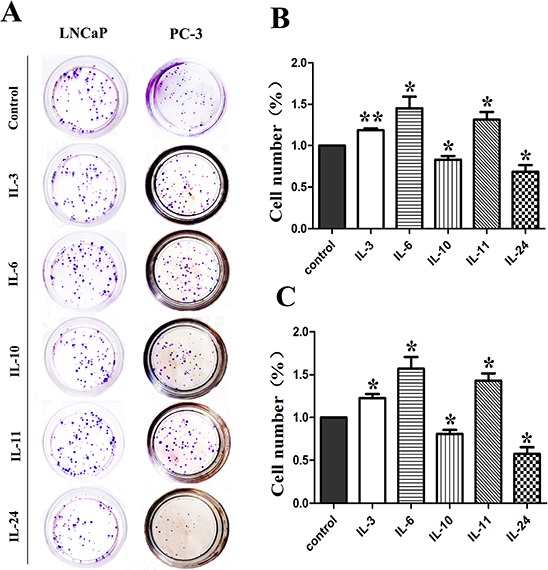
Representative colony formation assay plates are shown in A. for both cell lines. Corresponding histograms for the effect of interleukins on LNCaP and PC-3 are shown in B. and C. respectively. * means p < 0.05 and ** means p < 0.01.
The effect on the expression of CD44 and ABCG2
The effect of the interleukins on the expression of CD44 and ABCG2 in LNCaP and PC-3 cells was examined by flowcytometry. Typical photos of the flowcytometry and corresponding histograms are shown on Figure 7A. Compared with the control groups, IL-3, IL-6 and IL-11 application in LNCaP cells significantly increased the expression of CD44 and ABCG2 with p-values of 0.042, 0.037 and 0.015 for CD44 expression and 0.001, < 0.001, and 0.001 for ABCG2, respectively, while the IL-10 and IL-24 application significantly decreased the expression of CD 44 and ABCG2 with p-values of 0.003 and 0.015 for CD44, and 0.010 and 0.027 for ABCG2 as shown on Figure 7B, 7C and 7D. Similar results for PC-3 cells were obtained. Compared to the control PC-3 cells, IL-3, IL-6 and IL-11 application in PC-3 cells significantly activated the expression of CD44 and ABCG2 with p-values of 0.017, 0.004 and 0.011 for CD44, and 0.001, 0.006 and 0.008 for ABCG2, respectively. Again, the application of IL-10 and IL-24 resulted in significantly lower levels of CD44 and ABCG2 expression in PC-3 cells, with p-values of 0.002 and 0.021 for CD44 and 0.004 and 0.009 for ABCG2 as shown on Figure 7E, 7F and 7G, respectively.
Figure 7. The effect of ILs on the expression of CD44 and ABCG2.
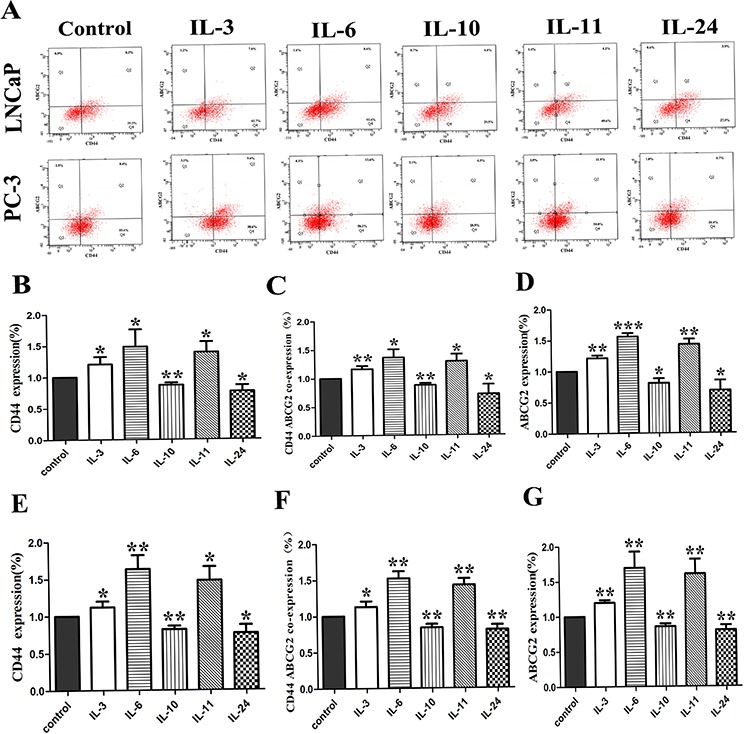
A. shows representative flowcytometry of CD44 and ABCG2 in both cell lines. B, C and D. show corresponding histograms of CD44 alone, CD44 and ABCG2, and ABCG2 alone for LNCaP cells, respectively. E, F and G. show corresponding histograms of CD44 alone, CD44 and ABCG2, and ABCG2 alone for PC-3 cell lines, respectively. * means p < 0.05, and ** means p < 0.01.
DISCUSSION
It is known the ILs, a subset of cytokines, actively influence molecule signaling and cellular behavior [41], such as cell proliferation, migration, invasion and adhesion. Increasing evidence points to the important roles of ILs in the microenvironment of tumors. Tumors may rapidly progress when the microenvironment is favorable, and the tumor cells may develop dormancy when the microenvironment allows for such a status. Tumor cell dormancy and tumor cell micrometastasis are two major issues for cancer treatment failure. Although it is known that increasing tumor cell stemness is associated with the features of tumor cell dormancy and micrometastases, molecular mechanisms are still obsolete.
In our current study, the effect of ILs on cell proliferation was examined by a SRB assay, and the results show that IL-3, IL-6 and IL-11 all have similar role in stimulating cell growth in the LNCaP and PC-3 cells. Wound healing and transwell assays also show that these ILs promote migration and invasion of these cells. The cells treated with these ILs show significantly higher colony formation ability and lower number of apoptotic cells, an indication of higher cell stemness in these cells after ILs treatment. In deed, these cells show increasing expression of stemness factors SOX2, and CD44, in addition to the drug-resistant gene ABCG2.
Interestingly, it is already reported that IL-3 is one of the primary factors capable of supporting the growth and acting on early progenitors [42]. Injection of IL-3 in the peripheral blood can maintain more CD34(+) /CD36(+) double-positive erythroid progenitors, which can significantly mobilize the peripheral blood (MPB) CD34(+) cells and increase the repopulating cells in marrow [43]. Presence of IL-3 in cell cultures can support the mesenchymal stem cells expansion of the irradiated CD34(+) cells in vitro [44]. It has been suggested to combine IL-3 and G-CSF injection in peripheral blood for peripheral blood stem cell propagation and allogenic stem cell transplantation [45, 46]. Recently, IL-3 hypersecretion has been reported to be associated with cutaneous B-lymphoblastic lymphoma [47]. Our study reveals another aspect of IL-3 functions, namely stimulating the stemness of prostate cancer cells.
IL-6 has been shown to be a major contributing factor in growth and progression of ovarian cancer [48], celiac disease [49] and neck squamous cell carcinoma [50]. It has been reported that the early growth response 3(EGR3) directly activates the excessive production of IL-6 in prostate cancer and promoted the progression [51]. Autophagy, a critical process for breast cancer stem cells (CSC), maintains the cell stemness of breast CSC by regulating the secretion of IL-6 [52]. Chang and co-workers [53] reported that IL-6 induces the expression of OCT4/NANOG and then activates the IGFIR to promote the progression of HBV [53]. Several reports show that IL-11 stimulate and accelerate the development of ulcerative colitis[54], promote the tumor progression by activating the STAT3 and suppress the antitumor immune response [55]. Furthermore, higher levels of IL-11 expression have been reported in distant metastatic gastric cancer cells [56]. The results are in line with our present study showing that IL-6 and IL-11 stimulated the stemness of prostate cancer cells in vitro.
On the contrary to the above findings for ILs-3, 6 and 11, we have demonstrated that IL-10 and IL-24 inhibit the proliferation capability of LNCaP and PC-3 cells. All other experiments for migration and invasion, colony formation, chemotherapeutic effect and the expression of stemness factors etc. show opposite results than using ILs-3, 6, and 11, indicating that these two factors suppressed the stemness of these cancer cells in vitro.
Contradictory observations for the role of IL-10 in cancer have been reported. Beguelin and associates [57] reported that IL-10 promotes tumor cell proliferation and survival by STAT3 signal pathway, and blocking of IL-10 receptor was suggested as a novel therapeutic target in diffused large B-cell lymphoma [57]. Others have reported that absence of IL-10 to increase the risk of acute lymphoblastic leukaemia occurrence (ALL) [58], and the polymorphisms of IL-10 may be associated with an increasing risk of colorectal cancer [59]. Nevertheless, accumulating evidence indicates that IL-10 is involved in the prostate cancer progression [60, 61], as well as breast [62] and non-small cell lung cancers [63]. We have shown that IL-10 significantly inhibit the growth, migration and invasion, most probably by down regulating the cell stemness in these two prostate cancer cell lines in vitro.
IL-24 is a member of the IL-10 family [41]. Increasing number of studies has pointed out the possibility of IL-24 as a promising therapeutic target for tumors. IL-24 inhibits the growth of breast [64] and lung [65]cancers. It is also reported that IL-24 induces apoptosis in melanoma cells [66], and enhances antitumor activities when applied in combination with paclitaxel in breast [67] and prostate [68] cancers. Recombinant human IL-24 (rhIL-24) reverses the chemoresistance in human breast cancer cell line MCF-7 cells [69] and significantly suppresses the growth of ovarian cancer cell [70]. It is also reported that IL-24 plays a critical antitumor role in oral [71], rectal [72] and pancreatic cancers [73]. To our best knowledge, this is the first report exploring the effect of IL-24 on the stemness of prostate cancer cells in vitro.
In summary, we have discovered in our present study that while ILs-3, 6 and 11 have similar tumor promotion and stemness stimulation effects, IL-10 and IL-24 reveal opposite effects on prostate cancer cells in vitro, underlining a complex role of ILs in vivo, which merit further studies.
MATERIALS AND METHODS
Cell culture
Human PCa cell lines LNCaP and PC3 cells were obtained from the Chinese Academy of Sciences (ATCC, USA). All cells were cultivated in RPMI 1640 (Gibco/Invitrogen, USA) medium supplemented with 10% fetal bovine serum (FBS) (Hyclone, USA), 1% penicillin/streptomycin (Sigma-Aldrich, USA) and L-glutamine (Sigma-Aldrich, USA) in a humidified 5% CO2 incubator at 37°C.
Sulforhodamine B assay
Human PCa LNCaP and PC3 cells were plated at 6,000 per well in 96-well plates and incubated overnight at 37°C in a humidified incubator containing 5% CO2. On the following day, IL-3, 6, 10 and 11(Invitrogen, USA) and IL-24(R&D, USA) up to 100 ng/ml (0.01, 0.1, 1, 2,5,10, 50, 100 ng/ml) in complete medium were added to different wells and cultivated for additional 48 hrs. Control wells were added with complete medium. Cell viability was determined using the Sulforhodamine B assay (SRB) (Sigma, USA) according to the manufacturer's instructions. Briefly, culture medium was aspirated and the cells were fixed by addition of 100 μl cold 10% trichloroacetic acid (TCA) at 4°C for 1 h, washed five times with deionized water and left to dry at room temperature. The cells were then stained with 100 μl SRB dye 0.4% (w/v) dissolved in 1% acetic acid (v/v) for at least 15 minutes, washed four times with 1% acetic acid to remove unbound dye and left to dry at room temperature. The dye bound protein was solubilized with 150 μl 10 mM unbuffered tris base and examined with Multi-Mode Microplate Reader (Biotek Synergy2, USA) for optical density reading at 560nm.
5-ethynyl-2′-deoxyuridine assay
Cells were plated at 5 × 103 per well in 96-well microtiter plates, treated with different cytokines and incubated for 24 hrs. Then 100 μl 50 μM 5-ethynyl-2′-deoxyuridine (EdU) (CellLight EdU DNA imaging Kit, Guangzhou RiboBio, China) were added into each well and the cells were cultured for an additional 2 hrs. The cells were stained with EdU according to the manufacturer's protocol. EdU medium was discarded and 4% paraformaldehyde was added to fix the cells at room temperature for 30 min. The cells were washed with glycine (2 mg/ml) for 5 min in a shaker and treated with 0.5% Trion X-100 for 10 min before washed with PBS for five minutes. The cells were then incubated with 1× Apollo® click stain reaction buffer for 30 min while protecting from light, washed with 0.5% Triton X-100 for three times to permeate the cells, stained with Hoechst33342 (10ug/ml) for 30 min at room temperature, washed with PBS for three times. The cells were examined with an inverted florescence microscope(Olympus, Japan) immediately.
In Vitro wound healing assay
Cells were seeded into a 6-well plate and allowed to grow to 60% confluent in complete medium. Cells were then wounded by a sterile pipette tip (1 mm), washed with PBS for several times to remove cell debris and incubated for additional 18 h with serum-free RPMI 1640 medium with or without corresponding interleukins. During the incubation at 37°C, cells migrated into wound surface which was considered as a process of in vitro healing. The healing process was recorded by inverted fluorescence microscopy. The rate of wound healing=[(the wound width of 0 h- the wound width 18 h)/the wound width 0 h wound width] × 100%.
Transwell migration and invasion assay
Transwell 24-well filters (Corning, USA) with 8.0 μM pores were used for the migration and invasion assays, according to the protocols recommended by the manufacturer. Briefly, for invasion assay, transwell membranes were coated with 60 μL of matrigel (BD, USA) at a final concentration of 0.1 mg/mL and dried 1 × 105 cells in 100 μL with serum-free RPMI 1640 medium. The membranes were added to the upper chamber triplicate wells and allowed to migrate through matrigel overnight at 37°C with 5% CO2 in a humidified incubator. The lower compartment of the transwell chamber was filled with 600 μL RPMI 1640 with 10% FBS. The migration assay was performed with exactly the same procedure except that the membranes were not coated with matrigel. After incubated for 24 hrs(migration assay was 6 h), the cells on the upper surface of filter were removed with a cotton swab, fixed with 4% formaldehyde and stained with Giemsa solution. The cells on the lower surface, which were the migrated/invaded cells, were photographed under the high-power microscopic field(HPF)(200 ×). The rate of migration/invasion inhibition = [(OD values of the control group - OD values of the transfection group)/OD values of the control group] × 100%.
Flow cytometry analysis
All cells were harvested at logarithmic growth phase before analyzed by a flow cytometer (FCM, FACSCalibur, BD, USA) within 1 h.
Apoptotic cells were detected using Annexin V-FITC/PI kit(KeyGen Biotech, China) according to manufacturer's instructions. Briefly, after treated with interleukin that combined with and without docetaxel(10nmol/L), cells were centrifuged at 1000 rpm for 5 min at the concentration of 1 × 106 cells/ml. Then the pellets were washed twice with ice-cold phosphate buffered saline (PBS) and resuspended in 500 μl binding buffer before 10 μl Annexin V-FITC and 5 μl of PI were added into each of the solution, and the cells were gently vortexed and incubated for 15 min at room temperature in the dark before flowcytometry.
For cell surface stem cell markers ABCG2 and CD44, an anti-ABCG2 monoclonal antibody directly conjugated with phycoerythrin(PE) and an anti-CD44 monoclonal antibody directly conjugated with APC, purchased from BD Pharmingen Company, were applied in this study. The antibodies were used at optimized dilutions, and the cells were prepared with a similar procedure as described above before flowcytometry. PE Mouse IgG2b (eBioscience, USA) and APC Mouse IgG2b (eBioscience, USA) isotype controls were used as negative controls. Viable and single cells were gated for each sample before examination
FQ-RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) by following the manufacturer's instructions. Briefly, after cells were treated with or without IL-3, 6, 10, 11 and 24 at 48 hrs, the cells (1 × 106) were harvested and washed twice with cold phosphate-buffered saline (PBS). For each well, 1.0 mL TRIzol reagent was added, and then RNA was precipitated by isopropanol, washed with 75% ethanol, dissolved with 20 μL DEPC (0.1%), and quantified using a UV spectrophotometer.
Reverse transcription was carried out with the PrimeScript™RT reagent Kit (TaKaRa, Janpan) according to the standard protocol. In brief, in a 20 μL reaction mixture containing 2 μg RNA and oligo primers, at 42°C for 1 h, and the synthesized cDNA was used for PCR by using SYBR®Premix Ex Taq™II (TaKaRa, Dalian, China) with the primers as shown in Table 1. PCR was performed as follows: 30 sec of pre-degeneration at 95°C; 5 sec at 95°C and 20 sec at 60°C for 40 cycles with Roche instrument- Lightcycler 480 (Roche, USA). SOX2 gene and β-actin gene were amplified in the same reaction where β-actin gene was applied as an internal loading control. The amplification specificity was confirmed by the melting curves, and the fluorescence was collected at 60°C (n = 3), and the relative quantitative results were analyzed by the 2−ΔΔCt values.
Table 1. Sequences of the primers used for RT-PCR.
Western blotting
Cells ready for Western blotting analysis were harvested and washed with cold PBS twice, then lysed on ice in RIPA buffer (1 × PBS, 1% NP-40, 0.1% sodium dodecylsulfate (SDS), 5 mM EDTA, 0.5% sodium deoxycholate, and 1 mM sodium orthovanadate) that contained 100 μg/mL phenylmethylsul-fonylsuoride and protease inhibitors (KeyGen, Nanjing, China).
Approximately 50 μg of protein from each sample was separated using a 10% SDS-polyacrylamide gel, electrotransfered to polyvinylidene fluoride(PVDF) membranes and blocked in 5% nonfat dry milk in Tris-buffered saline, pH 7.5 (100 mM NaCl, 50 mM Tris, and 0.1% Tween-20). The transferred membranes were incubated with anti-SOX2 (Cell Signaling Technology, USA) and anti-β-actin primary antibodies (Beyotime, Jiangsu, China) overnight at 4°C, followed by incubation with horseradish peroxidase(HRP) conjugated IgG(JacksonImmunoResearch, USA). Proteins were detected by Quantity-one software (Bio-Rad, Laboratories, Inc, USA) using Immobilon ECL Chemiluminescence HRP Substrate (Millipore, Merck, USA).
Immunofluorescence microscopy
2 × 105 cells were plated on coverslips, which were placed on the bottom in 6-well plates, and treated with different cytokines. Before examination, the cells were fixed with 4% paraformaldehyde in PBS(pH 7.5) for 30 min and permeated with 0.5% Trion X-100 for 15 min at room temperature. The coverslips were first immersed for 1 h in blocking solution that contained 5% bovine serum albumin (BSA) in PBS, and the cells were then incubated for overnight at 4°C with rabbit antibodies against SOX2 (CST, USA). DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI, 5ug/ml) and observed under inverted fluorescence microscope.
Colony formation assay
Single LNCaP and PC3 cells (200 cells/well) were planted in 35 mm well plates for overnight incubation to allow for cells attachment. Different interleukins were then added into the medium for culturing another two weeks. The cells were then fixed with 4% paraformaldehyde for 30 min, and stained with Giemsa for 30 min at room temperature. The plates were gently washed with PBS and evaluated under microscope. Cell cluster with more than 30 cells was considered as a colony. Colony formation efficiency was estimated as follows: Colony formation efficiency = colonies/input cells × 100%.
Statistical analysis
All data are expressed as mean ± the standard deviation (SD) and subjected to one-way analysis of variance (ANOVA). Differences between groups were examined by Student t test unless otherwise noted. Statistical significance was accepted at the level of p-value less than 0.05 by using SPSS 17.0 software.
ACKNOWLEDGEMENTS AND FUNDING
This study was financially supported by National Natural Science Foundation of China (81272824) and Special Research Fund for the Doctoral Program of Higher Education (20121107120021) as well.
Footnotes
CONFLICTS OF INTERESTS
All authors declare that they have no conflicts of interests to state
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, He J. Report of incidence and mortality in China cancer registries, 2009. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2013;25:10–21. doi: 10.3978/j.issn.1000-9604.2012.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zhang S, Zhao P, Zeng H, Zou X, He J. Annual report on status of cancer in China, 2010. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2014;26:48–58. doi: 10.3978/j.issn.1000-9604.2014.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sieber PR. Emerging Therapeutic for the Treatment of Skeletal-related Events Associated With Metastatic Castrate-resistant Prostate Cancer. Reviews in urology. 2014;16:10–20. [PMC free article] [PubMed] [Google Scholar]
- 5.Ni J, Cozzi P, Hao J, Duan W, Graham P, Kearsley J, Li Y. Cancer stem cells in prostate cancer chemoresistance. Current cancer drug targets. 2014;14:225–240. doi: 10.2174/1568009614666140328152459. [DOI] [PubMed] [Google Scholar]
- 6.Frame FM, Maitland NJ. Cancer stem cells, models of study and implications of therapy resistance mechanisms. Advances in experimental medicine and biology. 2011;720:105–118. doi: 10.1007/978-1-4614-0254-1_9. [DOI] [PubMed] [Google Scholar]
- 7.Ni C, Huang J. Dynamic regulation of cancer stem cells and clinical challenges. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2013;15:253–258. doi: 10.1007/s12094-012-0927-7. [DOI] [PubMed] [Google Scholar]
- 8.Chen W, Wang GM, Guo JM, Sun LA, Wang H. NGF/gamma-IFN inhibits androgen-independent prostate cancer and reverses androgen receptor function through downregulation of FGFR2 and decrease in cancer stem cells. Stem cells and development. 2012;21:3372–3380. doi: 10.1089/scd.2012.0121. [DOI] [PubMed] [Google Scholar]
- 9.Pasquier J, Rafii A. Role of the microenvironment in ovarian cancer stem cell maintenance. BioMed research international. 2013;2013:630782. doi: 10.1155/2013/630782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Wang L, Pappan L, Galliher-Beckley A, Shi J. IL-1beta promotes stemness and invasiveness of colon cancer cells through Zeb1 activation. Molecular cancer. 2012;11:87. doi: 10.1186/1476-4598-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang T, Long H, He L, Han X, Lin K, Liang Z, Zhuo W, Xie R, Zhu B. Interleukin-17 produced by tumor microenvironment promotes self-renewal of CD133 cancer stem-like cells in ovarian cancer. Oncogene. 2013 doi: 10.1038/onc.2013.537. [DOI] [PubMed] [Google Scholar]
- 12.Waldner MJ, Foersch S, Neurath MF. Interleukin-6-a key regulator of colorectal cancer development. International journal of biological sciences. 2012;8:1248–1253. doi: 10.7150/ijbs.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selbo PK, Bostad M, Olsen CE, Edwards VT, Hogset A, Weyergang A, Berg K. Photochemical internalisation, a minimally invasive strategy for light-controlled endosomal escape of cancer stem cell-targeting therapeutics. Photochemical & photobiological sciences : Official journal of the European Photochemistry Association and the European Society for Photobiology. 2015 doi: 10.1039/c5pp00027k. [DOI] [PubMed] [Google Scholar]
- 14.Lin M, Yuan Y, Xu J, Cai X, Liu S, Niu L, Chen J, Li Q, Xu K. Safety and efficacy study of nasopharyngeal cancer stem cell vaccine. Immunology letters. 2015 doi: 10.1016/j.imlet.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Choi GH, Kim GI, Yoo JE, Na DC, Han DH, Roh YH, Park YN, Choi JS. Increased Expression of Circulating Cancer Stem Cell Markers During the Perioperative Period Predicts Early Recurrence After Curative Resection of Hepatocellular Carcinoma. Annals of surgical oncology. 2015 doi: 10.1245/s10434-015-4480-9. [DOI] [PubMed] [Google Scholar]
- 16.Ymer S, Tucker WQ, Sanderson CJ, Hapel AJ, Campbell HD, Young IG. Constitutive synthesis of interleukin-3 by leukaemia cell line WEHI-3B is due to retroviral insertion near the gene. Nature. 1985;317:255–258. doi: 10.1038/317255a0. [DOI] [PubMed] [Google Scholar]
- 17.Ihle JN, Pepersack L, Rebar L. Regulation of T cell differentiation: in vitro induction of 20 alpha-hydroxysteroid dehydrogenase in splenic lymphocytes from athymic mice by a unique lymphokine. Journal of immunology (Baltimore, Md : 1950) 1981;126:2184–2189. [PubMed] [Google Scholar]
- 18.Frolova O, Benito J, Brooks C, Wang RY, Korchin B, Rowinsky EK, Cortes J, Kantarjian H, Andreeff M, Frankel AE, et al. SL-401 and SL-501, targeted therapeutics directed at the interleukin-3 receptor, inhibit the growth of leukaemic cells and stem cells in advanced phase chronic myeloid leukaemia. British journal of haematology. 2014;166:862–874. doi: 10.1111/bjh.12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li LJ, Tao JL, Fu R, Wang HQ, Jiang HJ, Yue LZ, Zhang W, Liu H, Shao ZH. Increased CD34+CD38 -CD123 + cells in myelodysplastic syndrome displaying malignant features similar to those in AML. International journal of hematology. 2014;100:60–69. doi: 10.1007/s12185-014-1590-2. [DOI] [PubMed] [Google Scholar]
- 20.Nievergall E, Ramshaw HS, Yong AS, Biondo M, Busfield SJ, Vairo G, Lopez AF, Hughes TP, White DL, Hiwase DK. Monoclonal antibody targeting of IL-3 receptor alpha with CSL362 effectively depletes CML progenitor and stem cells. Blood. 2014;123:1218–1228. doi: 10.1182/blood-2012-12-475194. [DOI] [PubMed] [Google Scholar]
- 21.Rybtsov S, Batsivari A, Bilotkach K, Paruzina D, Senserrich J, Nerushev O, Medvinsky A. Tracing the Origin of the HSC Hierarchy Reveals an SCF-Dependent, IL-3-Independent CD43(−) Embryonic Precursor. Stem cell reports. 2014;3:489–501. doi: 10.1016/j.stemcr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawicki S, Bedkowska GE, Wojtukiewicz M, Szmitkowski M. Hematopoietic cytokines as tumor markers in breast malignancies. A multivariate analysis with ROC curve in breast cancer patients. Advances in medical sciences. 2013;58:207–215. doi: 10.2478/ams-2013-0023. [DOI] [PubMed] [Google Scholar]
- 23.Gabler J, Wittmann J, Porstner M, Renz H, Jack HM, Abram M, Zemlin M. Contribution of microRNA 24-3p and Erk1/2 to interleukin-6-mediated plasma cell survival. European journal of immunology. 2013;43:3028–3037. doi: 10.1002/eji.201243271. [DOI] [PubMed] [Google Scholar]
- 24.Tormo AJ, Meliani Y, Beaupre LA, Sharma M, Fritz JH, Elson G, Crabe S, Gauchat JF. The composite cytokine p28/cytokine-like factor 1 sustains B cell proliferation and promotes plasma cell differentiation. Journal of immunology (Baltimore, Md : 1950) 2013;191:1657–1665. doi: 10.4049/jimmunol.1201595. [DOI] [PubMed] [Google Scholar]
- 25.Wan S, Zhao E, Kryczek I, Vatan L, Sadovskaya A, Ludema G, Simeone DM, Zou W, Welling TH. Tumor-Associated Macrophages Produce Interleukin 6 and Signal via STAT3 to Promote Expansion of Human Hepatocellular Carcinoma Stem Cells. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JT, Wang JY, Chen MK, Chen HC, Chang TH, Su BW, Chang PJ. Colon cancer mesenchymal stem cells modulate the tumorigenicity of colon cancer through interleukin 6. Experimental cell research. 2013;319:2216–2229. doi: 10.1016/j.yexcr.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Cui X, Liu J, Bai L, Tian J, Zhu J. Interleukin-6 induces malignant transformation of rat mesenchymal stem cells in association with enhanced signaling of signal transducer and activator of transcription 3. Cancer science. 2014;105:64–71. doi: 10.1111/cas.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klose J, Schmidt NO, Melms A, Dohi M, Miyazaki J, Bischof F, Greve B. Suppression of experimental autoimmune encephalomyelitis by interleukin-10 transduced neural stem/progenitor cells. Journal of neuroinflammation. 2013;10:117. doi: 10.1186/1742-2094-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy O, Zhao W, Mortensen LJ, Leblanc S, Tsang K, Fu M, Phillips JA, Sagar V, Anandakumaran P, Ngai J, et al. mRNA-engineered mesenchymal stem cells for targeted delivery of interleukin-10 to sites of inflammation. Blood. 2013;122:e23–32. doi: 10.1182/blood-2013-04-495119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah N, Kammermeier J, Elawad M, Glocker EO. Interleukin-10 and interleukin-10-receptor defects in inflammatory bowel disease. Current allergy and asthma reports. 2012;12:373–379. doi: 10.1007/s11882-012-0286-z. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Asensio FJ, Perpina U, Planas AM, Pozas E. Interleukin-10 regulates progenitor differentiation and modulates neurogenesis in adult brain. Journal of cell science. 2013;126:4208–4219. doi: 10.1242/jcs.127803. [DOI] [PubMed] [Google Scholar]
- 32.Liu WH, Liu JJ, Wu J, Zhang LL, Liu F, Yin L, Zhang MM, Yu B. Novel mechanism of inhibition of dendritic cells maturation by mesenchymal stem cells via interleukin-10 and the JAK1/STAT3 signaling pathway. PloS one. 2013;8:e55487. doi: 10.1371/journal.pone.0055487. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Lu H, Jiang T, Li R, Wang S, Zhang Q, Zhao S. Bone marrow stromal cells transduced with a thrombopoietin, interleukin-6, and interleukin-11 syncretic gene induce cord mononuclear cells to generate platelets in vitro. Transfusion. 2015;55:176–186. doi: 10.1111/trf.12800. [DOI] [PubMed] [Google Scholar]
- 34.Xu XJ, Niu XM, Guo ZW, He HQ, Qiu DF, Liu C, Lin SH, Song K, Ren ZJ, Li WC, et al. [Effects of recombinant human interleukin 11 on hematological malignancy after allogeneic hematopoietic cell transplantation] Zhonghua yi xue za zhi. 2011;91:100–102. [PubMed] [Google Scholar]
- 35.Zhu HY, Da WM, Gao CJ, Wang FF, Han XP, Li HH, Huang WR, Zhang YZ, Wang SH, Bo J, et al. [Effects of recombinant human interleukin 11 and granulocyte colony stimulating factor in mobilization for autologous peripheral blood stem cell transplantation] Zhongguo shi yan xue ye xue za zhi / Zhongguo bing li sheng li xue hui = Journal of experimental hematology / Chinese Association of Pathophysiology. 2008;16:345–349. [PubMed] [Google Scholar]
- 36.Bhutia SK, Das SK, Azab B, Menezes ME, Dent P, Wang XY, Sarkar D, Fisher PB. Targeting breast cancer-initiating/stem cells with melanoma differentiation-associated gene-7/interleukin-24. International journal of cancer Journal international du cancer. 2013;133:2726–2736. doi: 10.1002/ijc.28289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Zhang L, Xu W, Qian H, Ye S, Zhu W, Cao H, Yan Y, Li W, Wang M, et al. Experimental therapy for lung cancer: umbilical cord-derived mesenchymal stem cell-mediated interleukin-24 delivery. Current cancer drug targets. 2013;13:92–102. [PubMed] [Google Scholar]
- 38.Ma Y, Liang D, Liu J, Axcrona K, Kvalheim G, Giercksky KE, Nesland JM, Suo Z. Synergistic effect of SCF and G-CSF on stem-like properties in prostate cancer cell lines. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33:967–978. doi: 10.1007/s13277-012-0325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croy A, Brodowski L, Burlakov J, Myerski AC, von Kaisenberg CS, Grundmann M, Hubel CA, von Versen-Höynck F. Vitamin D Prevents Endothelial Progenitor Cell Dysfunction Induced by Sera from Women with Preeclampsia or Conditioned Media from Hypoxic Placenta. PloS one. 2014;9:e98527. doi: 10.1371/journal.pone.0098527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forget MA, Voorhees JL, Cole SL, Dakhlallah D, Patterson IL, Gross AC, Moldovan L, Mo X, Evans R, Marsh CB, et al. Macrophage colony-stimulating factor augments tie2-expressing monocyte differentiation, angiogenic function, and recruitment in a mouse model of breast cancer. PloS one. 2014;9:e98623. doi: 10.1371/journal.pone.0098623. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Brocker C, Thompson D, Matsumoto A, Nebert DW, Vasiliou V. Evolutionary divergence and functions of the human interleukin (IL) gene family. Human genomics. 2010;5:30–55. doi: 10.1186/1479-7364-5-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang YC, Clark SC. Interleukin-3: molecular biology and biologic activities. Hematology/oncology clinics of North America. 1989;3:441–452. [PubMed] [Google Scholar]
- 43.Liu J, Samuel K, Turner ML, Gallagher RC. Use of IL3 and chromatin-modifying reagents valproic acid and 5-aza-2′-deoxycytidine to affect mobilized peripheral blood CD34+ cell fate decisions. Vox sanguinis. 2014;107:83–89. doi: 10.1111/vox.12124. [DOI] [PubMed] [Google Scholar]
- 44.Mourcin F, Grenier N, Mayol JF, Lataillade JJ, Sotto JJ, Herodin F, Drouet M. Mesenchymal stem cells support expansion of in vitro irradiated CD34(+) cells in the presence of SCF, FLT3 ligand, TPO and IL3: potential application to autologous cell therapy in accidentally irradiated victims. Radiation research. 2005;164:1–9. doi: 10.1667/rr3384. [DOI] [PubMed] [Google Scholar]
- 45.Ohi S, Sakamaki S, Matsunaga T, Kuga T, Hirayama Y, Kohgo Y, Niitsu Y. Co-administration of IL3 with G-CSF increases the CFU-S mobilization into peripheral blood. International journal of hematology. 1995;62:75–82. doi: 10.1016/0925-5710(95)00386-7. [DOI] [PubMed] [Google Scholar]
- 46.Ihle JN, Morishita K, Parker DS, Bartholomew C, Askew D, Buchberg A, Jenkins NA, Copeland N, Weinstein Y. Mechanisms in the transformation of IL3-dependent hematopoietic stem cells. Current topics in microbiology and immunology. 1989;149:59–69. doi: 10.1007/978-3-642-74623-9_5. [DOI] [PubMed] [Google Scholar]
- 47.Bomken S, Haigh S, Bown N, Carey P, Wood K, Windebank K. Cutaneous B-lymphoblastic lymphoma with IL3/IgH translocation presenting with hypereosinophilia and acute endocarditis. Pediatric blood & cancer. 2014 doi: 10.1002/pbc.25318. [DOI] [PubMed] [Google Scholar]
- 48.Ataie-Kachoie P, Morris DL, Pourgholami MH. Minocycline suppresses interleukine-6, its receptor system and signaling pathways and impairs migration, invasion and adhesion capacity of ovarian cancer cells: in vitro and in vivo studies. PloS one. 2013;8:e60817. doi: 10.1371/journal.pone.0060817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fekih M, Sahli H, Ben Mustapha N, Mestiri I, Fekih M, Boubaker J, Kaabachi N, Sellami M, Kallel L, Filali A. [Bone metabolism, biochemical markers of bone resorption and formation processes and interleukine 6 cytokin level during coeliac disease] La Tunisie medicale. 2013;91:59–65. [PubMed] [Google Scholar]
- 50.Mojtahedi Z, Khademi B, Hashemi SB, Abtahi SM, Ghasemi MA, Fattahi MJ, Ghaderi A. Serum interleukine-6 concentration, but not interleukine-18, is associated with head and neck squamous cell carcinoma progression. Pathology oncology research : POR. 2011;17:7–10. doi: 10.1007/s12253-010-9261-y. [DOI] [PubMed] [Google Scholar]
- 51.Baron VT, Pio R, Jia Z, Mercola D. Early Growth Response 3 regulates genes of inflammation and directly activates IL6 and IL8 expression in prostate cancer. British journal of cancer. 2015;112:755–764. doi: 10.1038/bjc.2014.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maycotte P, Jones KL, Goodall ML, Thorburn J, Thorburn A. Autophagy Supports Breast Cancer Stem Cell Maintenance by Regulating IL6 Secretion. Molecular cancer research : MCR. 2015 doi: 10.1158/1541-7786.MCR-14-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang TS, Wu YC, Chi CC, Su WC, Chang PJ, Lee KF, Tung TH, Wang J, Liu JJ, Tung SY, et al. Activation of IL6/IGFIR confers poor prognosis of HBV-related hepatocellular carcinoma through induction of OCT4/NANOG expression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:201–210. doi: 10.1158/1078-0432.CCR-13-3274. [DOI] [PubMed] [Google Scholar]
- 54.Klein W, Tromm A, Griga T, Fricke H, Folwaczny C, Hocke M, Eitner K, Marx M, Duerig N, Epplen JT. A polymorphism in the IL11 gene is associated with ulcerative colitis. Genes and immunity. 2002;3:494–496. doi: 10.1038/sj.gene.6363897. [DOI] [PubMed] [Google Scholar]
- 55.Ernst M, Putoczki TL. Molecular pathways: IL11 as a tumor-promoting cytokine-translational implications for cancers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:5579–5588. doi: 10.1158/1078-0432.CCR-13-2492. [DOI] [PubMed] [Google Scholar]
- 56.Shen Z, Seppanen H, Vainionpaa S, Ye Y, Wang S, Mustonen H, Puolakkainen P. IL10, IL11, IL18 are differently expressed in CD14+ TAMs and play different role in regulating the invasion of gastric cancer cells under hypoxia. Cytokine. 2012;59:352–357. doi: 10.1016/j.cyto.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 57.Beguelin W, Sawh S, Chambwe N, Chun Chan F, Jiang Y, Choo JW, Scott DW, Chalmers A, Geng H, Tsikitas L, et al. IL10 receptor is a novel therapeutic target in DLBCLs. Leukemia. 2015 doi: 10.1038/leu.2015.57. [DOI] [PubMed] [Google Scholar]
- 58.Winkler B, Taschik J, Haubitz I, Eyrich M, Schlegel PG, Wiegering V. TGFbeta and IL10 have an impact on risk group and prognosis in childhood ALL. Pediatric blood & cancer. 2015;62:72–79. doi: 10.1002/pbc.25142. [DOI] [PubMed] [Google Scholar]
- 59.Andersen V, Holst R, Kopp TI, Tjonneland A, Vogel U. Interactions between diet, lifestyle and IL10, IL1B, and PTGS2/COX-2 gene polymorphisms in relation to risk of colorectal cancer in a prospective Danish case-cohort study. PloS one. 2013;8:e78366. doi: 10.1371/journal.pone.0078366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dluzniewski PJ, Wang MH, Zheng SL, De Marzo AM, Drake CG, Fedor HL, Partin AW, Han M, Fallin MD, Xu J, et al. Variation in IL10 and other genes involved in the immune response and in oxidation and prostate cancer recurrence. Cancer epidemiology, biomarkers &prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:1774–1782. doi: 10.1158/1055-9965.EPI-12-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang MH, Helzlsouer KJ, Smith MW, Hoffman-Bolton JA, Clipp SL, Grinberg V, De Marzo AM, Isaacs WB, Drake CG, Shugart YY, et al. Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. The Prostate. 2009;69:874–885. doi: 10.1002/pros.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang X, Zhang J, Zhu Y, Lu Y, Zhou X, Wang Z, Yu J, Yan Y, Di L, Che L, et al. Specific genetic polymorphisms of IL10-592 AA and IL10-819 TT genotypes lead to the key role for inducing docetaxel-induced liver injury in breast cancer patients. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2013;15:331–334. doi: 10.1007/s12094-012-0936-6. [DOI] [PubMed] [Google Scholar]
- 63.Hart K, Landvik NE, Lind H, Skaug V, Haugen A, Zienolddiny S. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung cancer (Amsterdam, Netherlands) 2011;71:123–129. doi: 10.1016/j.lungcan.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 64.Wei S, Cao H, Zhou X, Wu H, Yang J. Prokaryotically and eukaryotically expressed interleukin-24 induces breast cancer growth suppression via activation of apoptosis and inhibition of tumor angiogenesis. Molecular medicine reports. 2015;11:3673–3681. doi: 10.3892/mmr.2014.3136. [DOI] [PubMed] [Google Scholar]
- 65.Xu M, Li M, Yang J, Guo J, Sun W. [Adenovirus-mediated interleukin-24 enhances the inhibitory effect of paclitaxel on the growth of lung cancer A549 cells] Xi bao yu fen zi mian yi xue za zhi = Chinese journal of cellular and molecular immunology. 2014;30:1150–1153. [PubMed] [Google Scholar]
- 66.Tian H, Zhang DF, Zhang BF, Li HZ, Zhang Q, Li LT, Pei DS, Zheng JN. Melanoma differentiation associated gene-7/interleukin-24 induces caspase-3 denitrosylation to facilitate the activation of cancer cell apoptosis. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2015;35:157–167. doi: 10.1089/jir.2014.0061. [DOI] [PubMed] [Google Scholar]
- 67.Fang L, Cheng Q, Bai J, Qi YD, Liu JJ, Li LT, Zheng JN. An oncolytic adenovirus expressing interleukin-24 enhances antitumor activities in combination with paclitaxel in breast cancer cells. Molecular medicine reports. 2013;8:1416–1424. doi: 10.3892/mmr.2013.1680. [DOI] [PubMed] [Google Scholar]
- 68.Fan JK, Wei N, Ding M, Gu JF, Liu XR, Li BH, Qi R, Huang WD, Li YH, Xiong XQ, et al. Targeting Gene-ViroTherapy for prostate cancer by DD3-driven oncolytic virus-harboring interleukin-24 gene. International journal of cancer Journal international du cancer. 2010;127:707–717. doi: 10.1002/ijc.25069. [DOI] [PubMed] [Google Scholar]
- 69.Amirzada MI, Ma X, Gong X, Chen Y, Bashir S, Jin J. Recombinant human interleukin 24 reverses Adriamycin resistance in a human breast cancer cell line. Pharmacological reports : PR. 2014;66:915–919. doi: 10.1016/j.pharep.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 70.Wang S, Guo J, Tang Y, Zheng R, Song M, Sun W. [Effects of recombinant human interleukin-24 alone and in combination with cisplatin on the growth of ovarian cancer cells in vitro] Xi bao yu fen zi mian yi xue za zhi = Chinese journal of cellular and molecular immunology. 2014;30:33–36. [PubMed] [Google Scholar]
- 71.Kim JS, Yu SK, Lee MH, Park MG, Park E, Kim SG, Lee SY, Kim CS, Kim HJ, Chun HS, et al. MicroRNA-205 directly regulates the tumor suppressor, interleukin-24, in human KB oral cancer cells. Molecules and cells. 2013;35:17–24. doi: 10.1007/s10059-013-2154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi Y, Roh MS, Hong YS, Lee HS, Hur WJ. Interleukin-24 is correlated with differentiation and lymph node numbers in rectal cancer. World journal of gastroenterology : WJG. 2011;17:1167–1173. doi: 10.3748/wjg.v17.i9.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lebedeva IV, Su ZZ, Vozhilla N, Chatman L, Sarkar D, Dent P, Athar M, Fisher PB. Mechanism of in vitro pancreatic cancer cell growth inhibition by melanoma differentiation-associated gene-7/interleukin-24 and perillyl alcohol. Cancer research. 2008;68:7439–7447. doi: 10.1158/0008-5472.CAN-08-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]


