Summary
In this manuscript we consider the problem of jointly estimating multiple graphical models in high dimensions. We assume that the data are collected from n subjects, each of which consists of T possibly dependent observations. The graphical models of subjects vary, but are assumed to change smoothly corresponding to a measure of closeness between subjects. We propose a kernel based method for jointly estimating all graphical models. Theoretically, under a double asymptotic framework, where both (T, n) and the dimension d can increase, we provide the explicit rate of convergence in parameter estimation. It characterizes the strength one can borrow across different individuals and the impact of data dependence on parameter estimation. Empirically, experiments on both synthetic and real resting state functional magnetic resonance imaging (rs-fMRI) data illustrate the effectiveness of the proposed method.
Keywords: Graphical model, Conditional independence, High dimensional data, Time series, Rate of convergence
1. Introduction
Undirected graphical models encoding the conditional independence structure among the variables in a random vector have been heavily exploited in multivariate data analysis (Lauritzen, 1996). In particular, when X ~ Nd(0, Σ) is a d dimensional Gaussian vector, estimating such graphical models is equivalent to estimating the nonzero entries in the inverse covariance matrix Θ ≔ Σ−1 (Dempster, 1972). The undirected graphical model encoding the conditional independence structure for the Gaussian distribution is sometimes called a Gaussian graphical model.
There has been much work on estimating a single Gaussian graphical model, G, based on n independent observations. In low dimensional settings where the dimension, d, is fixed, Drton and Perlman (2007) and Drton and Perlman (2008) proposed to estimate G using multiple testing procedures. In settings where the dimension is much larger than the sample size, n, Meinshausen and Bühlmann (2006) proposed to estimate G by solving a collection of regression problems via the lasso. Yuan and Lin (2007), Banerjee et al. (2008), Friedman et al. (2008), Rothman et al. (2008), and Liu and Luo (2012) proposed to directly estimate Θ using the ℓ1 penalty (detailed definition provided later). More recently, Yuan (2010) and Cai et al. (2011) proposed to estimate Θ via linear programming. The above mentioned estimators are all consistent with regard to both parameter estimation and model selection, even when d is nearly exponentially larger than n.
This body of work is focused on estimating a single graph based on independent realizations of a common random vector. However, in many applications this simple model does not hold. For example, the data can be collected from multiple individuals that share the same set of variables, but differ with regard to the structures among variables. This situation is frequently encountered in the area of brain connectivity network estimation (Friston, 2011). Here brain connectivity networks corresponding to different subjects vary, but are expected to be more similar if the corresponding subjects share many common demographic, health or other covariate features. Under this setting, estimating the graphical models separately for each subject ignores the similarity between the adjacent graphical models. In contrast, estimating one population graphical model based on the data of all subjects ignores the differences between graphs and may lead to inconsistent estimates.
There has been a line of research in jointly estimating multiple Gaussian graphical models for independent data. On one hand, Guo et al. (2011) and Danaher et al. (2014) proposed methods via introducing new penalty terms, which encourage the sparsity of both the parameters in each subject and the differences between parameters in different subjects. On the other hand, Song et al. (2009a), Song et al. (2009b), Kolar and Xing (2009), Zhou et al. (2010), and Kolar et al. (2010) focused on independent data with time-varying networks. They proposed efficient algorithms for estimating and predicting the networks along the time line.
In this paper, we propose a new method for jointly estimating and predicting networks corresponding to multiple subjects. The method is based on a different model compared to the ones listed above. The motivation of this model arises from resting state functional magnetic resonance imaging (rs-fMRI) data, where there exist many natural orderings corresponding to measures of health status, demographics, and many other subject-specific covariates. Moreover, the observations of each subject are multiple brain scans with temporal dependence. Accordingly, different from the methods in estimating time varying networks, we need to handle the data where each subject has T, instead of one, observations. Different from the methods in Guo et al. (2011) and Danaher et al. (2014), it is assumed that there exists a natural ordering for the subjects, and the parameters of interest vary smoothly corresponding to this ordering. Moreover, we allow the observations to be dependent via a temporal dependence structure. Such a setting has not been studied in high dimensions until very recently (Loh and Wainwright, 2012; Han and Liu, 2013; Wang et al., 2013).
We exploit a similar kernel based approach as in Zhou et al. (2010). It is shown that our method can efficiently estimate and predict multiple networks while allowing the data to be dependent. Theoretically, under a double asymptotic framework, where both d and (T, n) may increase, we provide an explicit rate of convergence in parameter estimation. It sharply characterizes the strength one can borrow across different subjects and the impact of data dependence on the convergence rate. Empirically, we illustrate the effectiveness of the proposed method on both synthetic and real rs-fMRI data. In detail, we conduct comparisons of the proposed approach with several existing methods under three synthetic patterns of evolving graphs. In addition, we study the large scale ADHD-200 dataset to investigate the development of brain connectivity networks over age, as well as the effect of kernel bandwidth on estimation, where scientifically interesting results are unveiled.
We note that the proposed multiple time series model has analogous prototypes in spatial-temporal analysis. This line of work is focused on multiple times series indexed by a spatial variable. A common strategy models the spatial-temporal observations by a joint Gaussian process, and imposes a specific structure on the spatial-temporal covariance function (Jones and Zhang, 1997; Cressie and Huang, 1999). Another common strategy decomposes the temporal series into a latent spatial-temporal structure and a residual noise. Examples of the latent spatial- temporal structure include temporal autoregressive processes (Høst et al., 1995; Sølna and Switzer, 1996; Antunes and Rao, 2006; Rao, 2008) and mean processes (Storvik et al., 2002; Gelfand et al., 2003; Banerjee et al., 2004, 2008; Nobre et al., 2011). The residual noise is commonly modeled by a parametric process such as a Gaussian process. The aforementioned literature is restricted in three aspects. First, they only consider univariate or low dimensional multivariate spatial-temporal series. Secondly, they restrict the covariance structure of the observations to a specific form. Thirdly, none of this literature addresses the problem of estimating the conditional independence structure of the time series. In comparison, we consider estimating the conditional independence graph under high dimensional times series. Moreover, our model involves no assumption on the structure of the covariance matrix.
We organize the rest of the paper as follows. In Section 2, the problem setup is introduced and the proposed method is given. In Section 3, the main theoretical results are provided. In Section 4, the method is applied to both synthetic and rs-fMRI data to illustrate its empirical usefulness. A discussion is provided in the last section. Additional results and technical proofs are put in the online supplement.
2. The Model and Method
Let M = (Mjk) ∈ ℝd×d and υ = (υ1, …, υd)T ∈ ℝd. We denote υI to be the subvector of υ whose entries are indexed by a set I ⊂ {1, …, d}. We denote MI,J to be the submatrix of M whose rows are indexed by I and columns are indexed by J. Let MI,* be the submatrix of M whose rows are indexed by I and M*,J be the submatrix of M whose columns are indexed by J. For 0 < q < ∞, define the ℓ0, ℓq, and ℓ∞ vector norms as
where I(·) is the indicator function. For a matrix M, denote the matrix ℓq, ℓmax, and Frobenius norms to be
For any two sequences an, bn ∈ ℝ, we say that an ≍ bn if cbn ≤ an ≤ Cbn for some constants c, C.
2.1. Model
Let {Xu}u∈[0,1] be a series of d-dimensional random vectors indexed by the label u, which can represent any kind of ordering in subjects (e.g., any covariate or confounder of interest transformed to the space [0, 1]). For any u ∈ [0, 1], assume that Xu ~ Nd{0, Σ(u)}. Here is a function from [0, 1] to the d by d positive definite matrix set, . Let Ω(u) ≔ {Σ(u)−1 be the inverse covariance matrix of Xu and let G(u) ∈ {0, 1}d×d represent the conditional independence graph corresponding to Xu, satisfying that {G(u)}jk = 1 if and only if {Ω(u)}jk ≠ 0.
Suppose that data points in u = u1, …, un are observed. Let xi1, …, xiT ∈ ℝd be T observations of Xui, with a temporal dependence structure among them. In particular, for simplicity, in this manuscript we assume that follows a lag one stationary vector autoregressive (VAR) model, i.e.,
| (1) |
and xit ~ Nd{0, Σ(ui)} for t = 2, …, T. Here we note that extensions to vector autoregressive models with higher orders are also analyzable using the same techniques in Han and Liu (2013). But for simplicity, in this manuscript we only consider the lag one case. A(u) ∈ ℝd×d is referred to as the transition matrix. It is assumed that the Gaussian noise, εit ~ Nd{0, Ψ(ui)} is independent of {εit′}t′≠t and . Both A(·) and Ψ(·) are considered as functions on [0, 1]. Due to the stationary property, for any u ∈ [0, 1], taking the covariance on either side of Equation (1), we have Σ(u) = A(u)Σ(u){A(u)}⊤ + Ψ (u). For any i ≠ i′, it is assumed that are independent of . For i = 1, …, n and t = 1, …, T, denote xit = (xit1, …, xitd)⊤.
Of note, the function A(·) characterizes the temporal dependence in the time series. For each label u, A(u) represents the transition matrix of the VAR model specific to u. By allowing A(u) to depend on u, as u varies, the temporal dependence structure of the corresponding time series is allowed to vary, too.
As is noted in Section 1, the proposed model is motivated by brain network estimation using rs-fMRI data. For instance, the ADHD data considered in Section 4.3 consist of n subjects with ages (u) ranging from 7 to 22, while time series measurements within each subject are indexed by t varying from 1 to 200, say. That is, for each subject, a list of rs-fMRI images with temporal dependence are available. We model the list of images by a VAR process, as exploited in Equation (1). For a fixed age u, A(u) characterizes the temporal dependence structure of the time series corresponding to the subject with age u. As age varies, the temporal dependence structures of the images may vary, too. Allowing A(u) to change with u accommodates such changes. The VAR model is a common tool in modelling dependence for rs-fMRI data. Consider Harrison et al. (2003), Penny et al. (2005), Rogers et al. (2010), Chen et al. (2011), and Valdés-Sosa et al. (2005), for more details.
2.2. Method
We exploit the idea proposed in Zhou et al. (2010) and use a kernel based estimator for subject specific graph estimation. The proposed approach requires two main steps.. In the first step, a smoothed estimate of the covariance matrix Σ(u0), denoted as S(u0), is obtained for a target label u0. In the second step, Ω(u0) is estimated by plugging the covariance matrix estimate S(u0) into the CLIME algorithm (Cai et al., 2011).
More specifically, let K(·) : ℝ → ℝ be a symmetric nonnegative kernel function with support set [−1, 1]. Moreover, for some absolute constant C1, let K(·) satisfy that:
| (2) |
Equation (2) is satisfied by a number of commonly used kernel functions. Examples include:
Uniform kernel: K(s) = I(|s| ≤ 1)/2;
Triangular kernel: K(s) = (1 − |s|)I(|s| ≤ 1);
Epanechnikov kernel: K(s) = 3(1 − s2)I(|s| ≤ 1)/4;
Cosine kernel: K(s) = π cos(πs/2)I(|s| ≤ 1)/4.
For estimating any covariance matrix Σ(u0) with the label u0 ∈ [0, 1], the smoothed sample covariance matrix estimator S(u0) is calculated as follows:
| (3) |
where ωi(u0, h) is a weight function and Σ̂i is the sample covariance matrix of xi1, …, xiT:
| (4) |
Here c(u0) = 2I(u0 ∈, {0,1}) + I{u0 ∈ (0, 1)} is a constant depending on whether u0 is on the boundary or not, and h is the bandwidth parameter. We will discuss how to select h in the next section.
After obtaining the covariance matrix estimate, S(u0), we proceed to estimate Ω(u0) ≔ {Σ(u0)}−1. When a suitable sparsity assumption on the inverse covariance matrix Ω(u0) is available, we propose to estimate Ω(u0) by plugging S(u0) into the CLIME algorithm (Cai et al., 2011). In detail, the inverse covariance matrix estimator Ω̂(u0) of Ω(u0) is calculated via solving the following optimization problem:
| (5) |
where Id ∈ ℝd×d is the identity matrix and λ is a tuning parameter. Equation (5) can be further decomposed into d optimization subproblems (Cai et al., 2011). For j = 1, …, d, the j-th column of Ω̂(u0) can be solved as:
| (6) |
where ej is the j-th canonical vector. Equation (6) can be solved efficiently using a parametric simplex algorithm (Pang et al., 2013). Hence, the solution to Equation (5) can be computed in parallel.
Once Ω̂(u0) is obtained, we can apply an additional threshold step to estimate the Graph Ĝ(u0). We define a graph estimator G ∈ {0, 1}d×d to be:
| (7) |
Here γ is another tuning parameter.
Of note, although two tuning parameters, λ and γ are introduced, γ is introduced merely for theoretical soundness. Empirically, we found that setting γ to be 0 or a very small value (e.g., 10−5) has proven to work well. This is consistent with existing literature on graphical model estimation. We refer the readers to Cai et al. (2011), Liu et al. (2012), Liu et al. (2012), Xue and Zou (2012), and Han et al. (2013) for more discussion on this issue.
Procedures for choosing λ have also been well studied in the graphical model literature. On one hand, popular selection criteria, such as the stability approach based on subsampling (Meinshausen and Bühlmann, 2010; Liu et al., 2010), exist and have been well studied. On the other hand, when prior knowledge about the sparsity of the precision matrix is available, a common approach is trying a sequence of λ, and choosing one according to a desired sparsity level.
3. Theoretical Properties
In this section the theoretical properties of the proposed estimators in Equations (5) and (7) are provided. Under a double asymptotic framework, the rates of convergence in parameter estimation under the matrix ℓ1 and ℓmax norms are given.
Before establishing the theoretical result, we first pose an additional assumption on the function Σ(·). In detail, let Σjk(·) : u → {Σ(u)}jk be a real function. In the following, we assume that Σjk(·) is a smooth function with regard to any j, k ∈ {1, …, d}. Here and in the sequel, the derivatives at support boundaries are defined as one-sided derivatives.
-
(A1)There exists one absolute constant, C2, such that for all u ∈ [0, 1],
Under Assumption (A1), we propose the following lemma, which shows that when the subjects are sampled in u = u1, …, un with ui = i/n for i = 1, …, n, the estimator S(u0) approximates Σ(u0) at a fast rate for any u0 ∈ [0, 1]. The convergence rate delivered here characterizes both the strength one can borrow across different subjects and the impact of temporal dependence structure on estimation accuracy.
Lemma 1
Suppose that the data points are generated from the model discussed in Section 2.1 and Assumption (A1) holds. Moreover, suppose that the observed subjects are in ui = i/n for i = 1, …, n. Then, for any u0 ∈ [0, 1], if for some η > 0 we have
-
(A2)
and the bandwidth h is set as
where ξ ≔ supu∈[0,1] maxj[Σ(u)]jj/minj[Σ(u)]jj, then the smoothed sample covariance matrix estimator S(u0) defined in Equation (3) satisfies:(8) (9)
Assumption (A2) is a convolution between the smoothness of K(·) and Σjk(·), and is a weaker requirement than imposing smoothness individually. Assumption (A2) is satisfied by many commonly used kernel functions, including the aforementioned examples in Section 2.2. For example, with regard to the Epanechnikov kernel K(s) = 3(1 − s2)I(|s| ≤ 1)/4, it’s easy to check that
Therefore, as long as Σjk(u), , and are uniformly bounded, the Epanechnikov kernel satisfies Assumption (A2) with η ≤ 2.
There are several observations drawn from Lemma 1. First, the rate of convergence in parameter estimation is upper bounded by , which is due to the bias in estimating Σ(u0) from only n labels. This term is irrelevant to the sample size T in each subject and cannot be improved without adding stronger (potentially unrealistic) assumptions. For example, when none of ξ, supt ‖Σ(u)‖2, and supt‖A(u)‖2 scales with (n, T, d) and log d for some generic constant C, the estimator achieves a rate of convergence. Secondly, in the term {logd/(Tn)}1/4, n characterizes the strength one can borrow across different subjects, while T demonstrates the contribution from within a subject. When , the estimator achieves a {logd/(Tn)}1/4 rate of convergence. The first two points discussed above, together, quantify the settings where the proposed methods can beat the naive method which only exploits the data points in each subject itself for parameter estimation.
Finally, Lemma 1 also demonstrates how temporal dependence may affect the rate of convergence. Specifically, the spectral norm of the transition matrix, ‖A(u)‖2, characterizes the strength of temporal dependence. The term1/{1−supu∈[0,1]‖ A(u)‖2} in Equation (9) demonstrates the impact of the dependence strength on the rate of convergence. Further discussions on the effect of A(u) are collected in Section A of the online supplement.
Next, we consider the case where A(u) = 0 and hence are independent observations with no temporal dependence. In this case, following Zhou et al. (2010), the rate of convergence in parameter estimation can be improved.
Lemma 2
Under the assumptions in Lemma 1, if it is further assumed that
-
(B1)
are i.i.d. observations from Nd{0, Σ(u)};
-
(B2)
for all j, k ∈ {1, …, d};
-
(B3)There exists an absolute constant C3 such that
then, setting the bandwidth
we have(10)
We note again that the aforementioned kernel functions satisfy Assumptions (B2) for similar reasons. In detail, taking Epanechnikov kernel as an example, we have
So Assumption (B2) is satisfied as long as Σjk(u), , and are uniformly bounded.
Lemma 2 shows that the rate of convergence can be improved to {logd/(Tn)}1/3 when the data are independent. Of note, this rate matches the results in Zhou et al. (2010). However, the improved rate is valid only when a strong independence assumption holds, which is unrealistic in many applications, rs-fMRI data analysis for example.
After obtaining Lemmas 1 and 2, we proceed to the final result, which shows the theoretical performance of the estimators Ω̂ (u0) and Ĝ(u0) proposed in Equations (5) and (7). We show that under certain sparsity constraints, the proposed estimators are consistent, even when d is nearly exponentially larger than n and T.
We first introduce some additional notation. Let Md ∈ ℝ be a quantity which may scale with (n, T, d). We define the set of positive definite matrices in ℝd×d, denoted by ℳ(q, s, Md), as
For q = 0, the class ℳ(0, s, Md) contains all the matrices with the number of nonzero entries in each column less than s and bounded ℓ1 norm. We then let
| (11) |
| (12) |
Theorem 1 presents the parameter estimation and graph estimation consistency results for the estimators defined in Equations (5) and (7).
Theorem 1
Suppose that the conditions in Lemma 1 hold. Assume that Θ(u0) ≔ {Σ(u0)−1 ∈ ℳ(q, s, Md) with 0 ≤ q < 1. Let Θ̂(u0) be defined in Equation (5). Then there exists a constant C3 only depending on q, such that, whenever the tuning parameter
is chosen, one has that
Moreover, let Ĝ(u0) be the graph estimator defined in Equation (7) with the second step tuning parameter γ = 4Mdλ. If it is further assumed that Θ(u0) ∈ ℳ(0, s, Md) and
then
If the conditions in Lemma 2 hold, the above results are true with κ replaced by κ*.
Theorem 1 shows that the proposed method is theoretically guaranteed to be consistent in both parameter estimation and model selection, even when the dimension d is nearly exponentially larger than nT. Theorem 1 can be proved by following the proofs of Theorem 1 and Theorem 7 in Cai et al. (2011) and the proof is accordingly omitted.
4. Experiments
In this section, the empirical performance of the proposed method is investigated. This section consists of two parts. In the first, we demonstrate the performance using synthetic data, where the true generating models are known. On one hand, the proposed kernel based method is compared to several existing methods. The advantage of this new method is shown in both parameter estimation and model selection. On the other hand, implications of the theoretical results are also empirically verified. In the second part, the proposed method is applied to a large scale rs-fMRI data (the ADHD-200 data) and some potentially scientifically interesting results are explored. Additional experimental results are provided in Section B of the online supplement.
4.1. Synthetic Data
The performance of the proposed kernel-smoothing estimator (denoted as KSE) is compared to three existing methods: a naive estimator (donated as naive; details follow below), Danaher et al. (2014)’s group graphical lasso (denoted as GGL), and Guo et al. (2011)’s estimator (denoted as Guo). Throughout the simulation studies, it is assumed that the graphs are evolving from u = 0 to u = 1 continuously. Although there is one graphical model corresponding to each u ∈ [0, 1], it is assumed that data are observed at n equally spaced points u = 0, 1/(n − 1), 2/(n − 1), …, 1. For each u = 0, 1/(n − 1), 2/(n − 1), …, 1, T observations were generated from the corresponding graph under a stationary VAR(1) model discussed in Equation (1). To generate the transition matrix, A, the precision matrix was obtained using the R package Huge (Zhao et al., 2012) with graph structure “random“. Then it is divided by twice its largest eigenvalue to obtain A, so that ‖A‖2 = 0.5. The same transition matrix is used under every label u. Our main target is to estimate the graph at u0 = 0, as the endpoints represent the most difficult point for estimation. We also investigate one setting where the target label is u0 = 1/2, to demonstrate the performance at a non-extreme target label.
In the following, three existing methods for comparison are reviewed. naive is obtained by first calculating the sample covariance matrix at target label u0 using only the T observations under this label, and then plugged into the CLIME algorithm. Compared to KSE, GGL and Guo do not assume that there exists a smooth change among the graphs. Instead, they assume that the data come from n categories. That is, there are n corresponding underlying graphs that potentially share common edges, and observations are available within each category. Moreover, they assume that the observations are independent both between and within different categories. With regard to implementation, they solve the following optimization problem:
where Σ̂i is the sample covariance matrix calculated based on the data under label ui. GGL uses penalty
and Guo uses penalty
Here the regularity coefficients λ1, λ2, and λ control the sparsity level. Danaher et al. (2014) also proposed the fused graphical lasso that separately controls sparsity of and similarity between the graphs. However, this method is not scalable when the number of categories is large and therefore not included in our comparison.
After obtaining the estimated graph, Ĝ(u0), of the true traph G(u0), the model selection performance is further investigated by comparing the ROC curves of the four competing methods. Let Ê(u0) be the set of estimated edges corresponding to Ĝ(u0), and E(u0) the set of true edges corresponding to G(u0). The true positive rate (TPR) and false positive rate (FPR) are defined as
where for any set S, |S| denotes the cardinality of S. To obtain a series of TPRs and FPRs, for KSE, naive, and Guo, the values of λ are varied. For GGL, first λ2 is fixed and subsequently λ1 is tuned, and then the λ2 with the best overall performance is selected. More specifically, a series of λ2 are picked, and for each fixed λ2, λ1 is accordingly varied to produce an ROC curve. Of note, in the investigation, the ROC curves indexed by λ2 are generally parallel, thus motivating this strategy. Finally, the λ2 corresponding to the topleft most curve is selected.
4.1.1. Setting 1: Simultaneously Evolving Edges
In this section we investigate the performance of the four competing methods under one particular graphical model. In each simulation, nfix = 200 edges are randomly selected from d(d − 1)/2 potential edges and they do not change with regard to the label u. The strengths of these edges, i.e. the corresponding entries in the inverse covariance matrix, are generated from a uniform distribution taking values in [−0.3, −0.1] (denoted by Unif[−0.3, −0.1]) and do not change with u. We then randomly select ndecay and ngrow edges that will disappear and emerge over the evolution simultaneously. For each of the ndecay edges, the strength is generated from Unif[−0.3,−0.1] at u = 0 and will diminish to 0 linearly with regard to u. For each of the ngrow edges, the strength is set to be 0 at u = 0, and will linearly grow to a value generated from Unif[−0.3,−0.1]. The edges evolve simultaneously. For j ≠ k, when we subtract a value a from Ωjk and Ωkj, we increase Ωjj and Ωkk by a, and then further add 0.25 to the diagonal of the matrix to keep it positive definite.
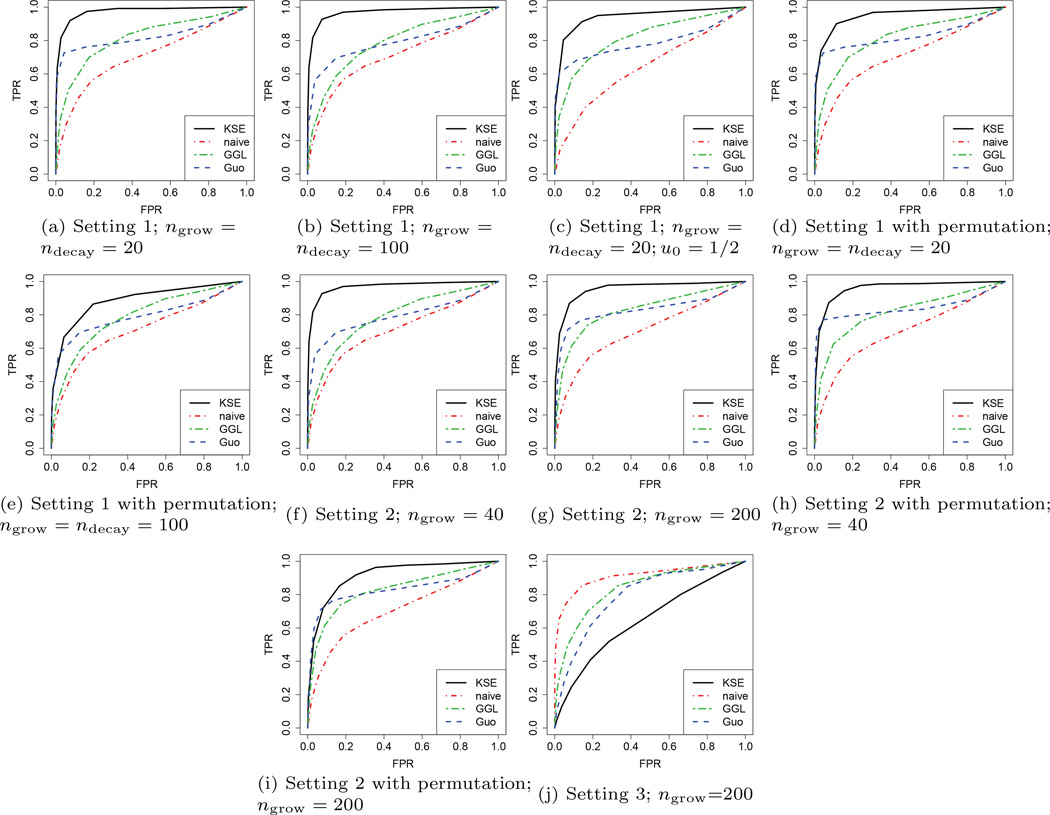
The ROC curves under this setting with different values of ngrow and ndecay are shown in Figures 1(a) and 1(b). We fix the number of labels n = 51, number of observations under each label T = 100, and dimension d = 50. The target label is u0 = 0. It can be observed that, under both cases, KSE outperforms the other three competing methods. Moreover, when we increase the values of ngrow and ndecay from 20 to 100, the ROC curve of KSE hardly changes, since the degree of smoothness in graphical model evolving hardly change. In contrast, the ROC curves of GGL and Guo drop, since the degree of similarity among the graphs is reduced. Finally, naive performances worst, which is expected because it does not borrow strength across labels in estimation. Figure 1(c) illustrates the performance under the same setting as in Figure 1(a) except u0 = 1/2. KSE still outperforms the other estimators.
Fig. 1.
ROC curves of four competing methods under three settings: simultaneous (a–e), sequential (f–i), and random (j). The target labels are u0 = 0 except for in (c), where u0 = 1/2. In each setting we set the dimension d = 50, the number of labels n = 51, the number of observations T = 100, and the result is obtained by 1,000 simulations.
Next, we exploit the same data, but permute the labels u = 1/50, 2/50, …, 1 so that the evolving pattern is much more opaque. Figures 1(d) and 1(e) illustrate the model selection result. We observe that under this setting, the ROC curves of the proposed method drop a little bit, but is still higher than the competing approaches. This is because the proposed method still benefits from the evolving graph structure (although more turbulent this time). The improvement over the naive method demonstrates exactly the strength borrowed across different labels. Note that the ROC curves of GGL, naive, and Guo shown in Figures 1(d) and 1(e) do not change compared to those in Figures 1(a) and 1(b), respectively, because they do not assume any ordering between the graphs.
4.1.2. Setting 2: Sequentially Growing Edges
Setting 2 is similar to Setting 1. The two differences are: (i) Here ndecay is set to be zero; (ii) The ngrow edges emerges sequentially instead of simultaneously. These ngrow edges are randomly selected, but there is no overlap with the existing 200 pre-fixed edges. The entries of the inverse covariance matrix for the ngrow edges each grow to a value generated from Unif[−0.3, −0.1], linearly in a length 1/ngrow interval in [0, 1], one after another. We note that there is possibility that n < ngrow, because n represents only the number of labels we observe. Under this setting, Figures 1(f) and 1(g) plot the ROC curves of the four competing methods. We also apply the four methods to the setting where the same permutation as in Setting 1 is exploited. We show the results in Figures 1(h) and 1(i). Here the same observations persist as in Setting 1.
4.1.3. Setting 3: Random Edges
In this setting, in contrast to the above two settings, we violate the smoothness assumption of KSE to the extreme. We demonstrate the limitedness of the proposed method in this setting. More specifically, in this setting, under every label u, ned edges are random selected with strengths from Unif[−0.3, −0.1]. In this case, the graphs do not evolve smoothly over the label u, and the data under the labels u ≠ 0 only contribute noises. We then apply the four competing methods to this setting and Figure 1(j) illustrates the result. Under this setting, we observe that naive beats all the other three methods. It is expected because naive is the only method that do not suffer from the noises. Here KSE performs worse than GGL and Guo, because there does not exist a natural ordering among the graphs.
Under the above three data generating settings, we further quantitatively com- pare the performance in parameter estimation of the inverse covariance matrix Ω(u0) for the four competing methods. Here the distances between the estimated and the true concentration matrices with regard to the matrix ℓ1, ℓ2, and Frobenius norms are shown in Table 1. It can be observed that KSE achieves the lowest estimation error in all settings except for the Setting 3. This coincides with the above model selection results. We omit the results for the label permutation cases and the case with u0 = 1/2, since they are again as expected from the model selection results above.
Table 1.
Comparison of inverse covariance matrix estimation errors in there data generating models. The parameter estimation error with regard to the matrix ℓ1, ℓ2, and Frobenius norms (denoted as ℓF here) is provided with standard deviations provided in the brackets. The results are obtained by 1,000 simulations.
| KSE | naive | ||||||
| Setting 1 | ngrow = ndecay | ℓ1 | ℓ2 | ℓF | ℓ1 | ℓ2 | ℓF |
| 20 | 3.25(0.232) | 1.53(0.104) | 4.42(0.220) | 5.02(0.287) | 2.68(0.132) | 8.30(0.412) | |
| 100 | 2.72(0.165) | 1.30(0.088) | 3.78(0.204) | 4.85(0.467) | 2.55(0.117) | 8.13(0.453) | |
| Setting 2 | ngrow | ||||||
| 40 | 3.39(0.553) | 1.56(0.213) | 4.47(0.302) | 5.26(0.740) | 2.73(0.313) | 8.24(0.386) | |
| 200 | 3.40(0.507) | 1.57(0.147) | 4.33(0.284) | 5.19(0.740) | 2.71(0.280) | 8.34(0.352) | |
| Setting 3 | ned | ||||||
| 50 | 2.21(0.194) | 1.37(0.120) | 3.20(0.104) | 1.60(0.249) | 0.84(0.113) | 3.09(0.185) | |
| GGL | Guo | ||||||
| Setting 1 | ngrow=ndecay | ℓ1 | ℓ2 | ℓF | ℓ1 | ℓ2 | ℓF |
| 20 | 3.28(0.298) | 1.45(0.112) | 4.13(0.190) | 3.22(0.418) | 1.42(0.259) | 4.04(0.280) | |
| 100 | 3.27(0.324) | 1.42(0.100) | 4.18(0.222) | 3.38(0.474) | 1.41(0.169) | 4.31(0.335) | |
| Setting 2 | ngrow | ||||||
| 40 | 3.47(0.580) | 1.47(0.163) | 4.22(0.153) | 3.06(0.417) | 1.40(0.274) | 4.00(0.205) | |
| 200 | 3.22(0.618) | 1.44(0.198) | 4.08(0.199) | 3.71(0.493) | 1.73(0.264) | 4.46(0.361) | |
| Setting 3 | ned | ||||||
| 50 | 1.52(0.224) | 0.85(0.105) | 2.04(0.104) | 1.48(0.263) | 0.67(0.116) | 1.81(0.150) | |
4.2. Impact of a Small Label Size n
As is shown in Lemma 1 and Theorem 1, the rates of convergence in parameter estimation and model selection crucially depend on the term . This is due to the bias in estimating Σ(u0) from n labels. This bias takes place as long as we include data under other labels into estimation, and cannot be removed by simply increasing the number of observations T under each label u. More specifically, Lemma C.1 of the online supplement shows that the rate of convergence for bias between the estimated and the true covariance matrix depends on n but not T.
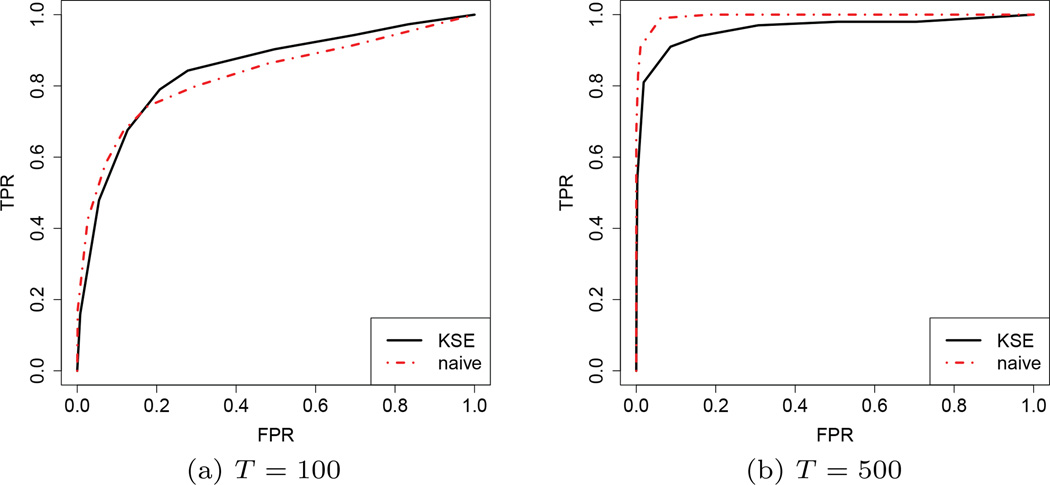
This section is devoted to illustrate this phenomenon empirically. We exploit Setting 2 in the last section with the number of labels n to be very small. Here we set n = 3. Moreover, we choose nfix = 100, ngrow = 500, and vary the number of observations T under each label. Figure 2 compares the ROC curves of KSE and naive corresponding to the settings when T = 100 or 500. There are two important observations we would like to emphasize: (i) When T = 100, KSE and naive have comparable performance. However, when T = 500, naive performs much better than KSE. (ii) The change of the ROC curves for KSE from T = 100 to T = 500 is less dramatic compared to the ROC curves for naive. These observations indicate the existence of bias in KSE that cannot be eliminated by only increasing T.
Fig. 2.
ROC curves of KSE and naive under Setting 1: sequentially evolving edges. We set dimension d = 50; number of labels n = 3; number of pre-fixed edges nfix=100; number of growing edges ngrow = 500.
4.3. ADHD-200 Data
As an example of real data application, we apply the proposed method to the ADHD-200 data (Biswal et al., 2010). The ADHD-200 data consist of rs-fMRI images of 973 subjects. Of them, 491 are healthy and 197 have been diagnosed with ADHD type 1,2, or 3. The remaining had their diagnosis withheld for the purpose of a prediction competition. The number of images for each subject ranges from 76 to 276. 264 seed regions of interest are used to define nodes for graphical model analysis (Power et al., 2011). A limited set of covariates including gender, age, handedness, IQ, are available.
4.3.1. Brain Development
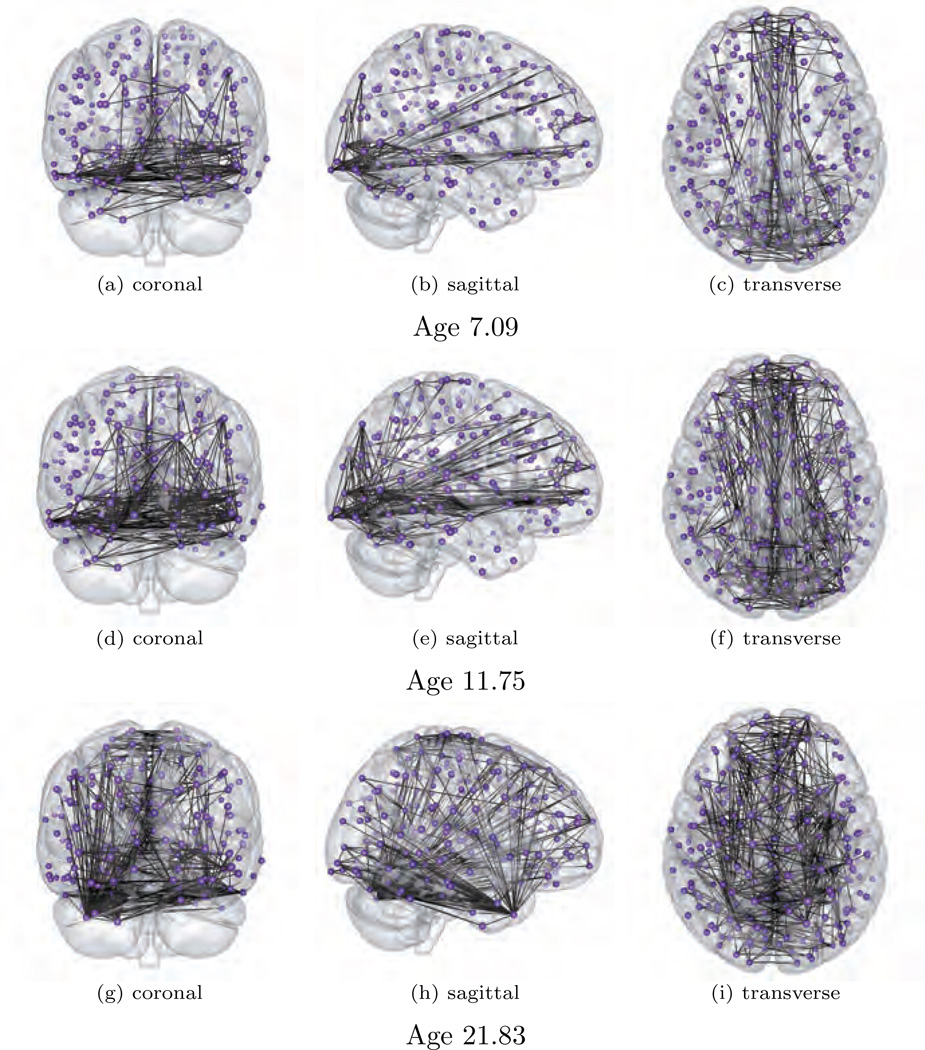
In this section, focus lies on investigating the development of brain connectivity network over age for control subjects. Here the subject ages are normalized to be in [0, 1], and the brain ROI measurements are centered to have sample means zero and scaled to have sample standard deviations 1. The bandwidth parameter is set at h = 0.5. The regularization parameter λ is manually chosen to induce high sparsity for better visualization and highlighting the dominating edges. Consider estimating the brain networks at ages 7.09, 11.75, and 21.83, which are the minimal, median, and maximal ages in the data. Figure 3 shows coronal, sagittal, and transverse snapshots of the estimated brain connectivity networks.
Fig. 3.
Estimated brain connectivity network at ages 7.09, 11.75, 21.83 in healthy subjects.
There are two main patterns worth noting in this experiment: (i) It is observed that the degree of complexity of the brain network at the occipital lobe is high compared to other regions by age seven. This is consistent with early maturation of visual and vision processing networks relative to others. We found that this conjecture is supported by several recent scientific results (Shaw et al., 2008; Blakemore, 2012). For example, Shaw et al. (2008) showed that occipital lobe is fully developed before other brain regions. Moreover, when considering structural development, the occipital lobe reaches its peak thickness by age nine. In comparison, portions of the parietal lob reaches their peak thickness as late as thirteen. (ii) Figure 3 also shows that dense connections in the temporal lobe only occur in the graph at age 21.83 among the ages shown. This is also supported by the scientific finding that grey matter in the temporal lobe doesn’t reach maximum volume untill age 16 (Bartzokis et al., 2001; Giedd et al., 1999). We also noticed that several confounding factors, such as scanner noise, subject motion, and coregistration, can have potential effects on inference (Braun et al., 2012; Van Dijk et al., 2012). In this manuscript, we rely on the standard data pre-processing techniques as described in Eloyan et al. (2012) for removing such confounders. The inuence of these confounders on our inference will be investigated in greater detail in the future.
5. Discussion
In this paper, we introduced a new kernel based estimator for jointly estimating multiple graphical models under the condition that the models smoothly vary according to a label. Methodologically, motivated by resting state functional brain connectivity analysis, we proposed a new model, taking both heterogeneity structure and dependence issues into consideration, and introduced a new kernel based method under this model. Theoretically, we provided the model selection and parameter estimation consistency result for the proposed method under both the independence and dependence assumptions. Empirically, we applied the proposed method to synthetic and real brain image data. We found that the proposed method is effective for both parameter estimation and model selection compared to several existing methods under various settings.
Supplementary Material
Acknowledgement
We would like to thank John Muschelli for providing the R tools to visualize the brain network. We would also like to thank one anonymous referee, the associate editor, and the editor for their helpful comments and suggestions. In addition, thanks also to Drs. Mladen Kolar, Derek Cummings, Martin Lindquist, Michelle Carlson, and Daniel Robinson for helpful discussions on this work.
Contributor Information
Huitong Qiu, Johns Hopkins University, Baltimore, USA..
Fang Han, Johns Hopkins University, Baltimore, USA..
Han Liu, Princeton University, Princeton, USA..
Brian Caffo, Johns Hopkins University, Baltimore, USA..
References
- Antunes AMC, Rao TS. On hypotheses testing for the selection of spatio-temporal models. Journal of Time Series Analysis. 2006;27(5):767–791. [Google Scholar]
- Banerjee O, El Ghaoui L, d'Aspremont A. Model selection through sparse maximum likelihood estimation for multivariate Gaussian or binary data. The Journal of Machine Learning Research. 2008;9:485–516. [Google Scholar]
- Banerjee S, Gelfand AE, Carlin BP. Hierarchical Modeling and Analysis for Spatial Data. CRC Press; 2004. [Google Scholar]
- Bartzokis G, Beckson M, Lu PH, Nuechterlein KH, Edwards N, Mintz J. Age-related changes in frontal and temporal lobe volumes in men: a magnetic resonance imaging study. Archives of General Psychiatry. 2001;58(5):461–465. doi: 10.1001/archpsyc.58.5.461. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo X-N, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, et al. Toward discovery science of human brain function. Proceedings of the National Academy of Sciences. 2010;107(10):4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakemore S-J. Imaging brain development: the adolescent brain. Neuroimage. 2012;61(2):397–406. doi: 10.1016/j.neuroimage.2011.11.080. [DOI] [PubMed] [Google Scholar]
- Braun U, Plichta MM, Esslinger C, Sauer C, Haddad L, Grimm O, Mier D, Mohnke S, Heinz A, Erk S, et al. Test–retest reliability of resting-state connectivity network characteristics using fMRI and graph theoretical measures. Neuroimage. 2012;59(2):1404–1412. doi: 10.1016/j.neuroimage.2011.08.044. [DOI] [PubMed] [Google Scholar]
- Cai T, Liu W, Luo X. A constrained L1 minimization approach to sparse precision matrix estimation. Journal of the American Statistical Association. 2011;106(494):594–607. [Google Scholar]
- Chen G, Glen DR, Saad ZS, Paul Hamilton J, Thomason ME, Gotlib IH, Cox RW. Vector autoregression, structural equation modeling, and their synthesis in neuroimaging data analysis. Computers in Biology and Medicine. 2011;41(12):1142–1155. doi: 10.1016/j.compbiomed.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressie N, Huang H-C. Classes of nonseparable, spatio-temporal stationary covariance functions. Journal of the American Statistical Association. 1999;94(448):1330–1339. [Google Scholar]
- Danaher P, Wang P, Witten DM. The joint graphical lasso for inverse covariance estimation across multiple classes. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2014;76(2):373–397. doi: 10.1111/rssb.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster AP. Covariance selection. Biometrics. 1972;28(1):157–175. [Google Scholar]
- Drton M, Perlman MD. Multiple testing and error control in Gaussian graphical model selection. Statistical Science. 2007;22(3):430–449. [Google Scholar]
- Drton M, Perlman MD. A SINful approach to Gaussian graphical model selection. Journal of Statistical Planning and Inference. 2008;138(4):1179–1200. [Google Scholar]
- Eloyan A, Muschelli J, Nebel MB, Liu H, Han F, Zhao T, Barber AD, Joel S, Pekar JJ, Mostofsky SH, et al. Automated diagnoses of attention deficit hyperactive disorder using magnetic resonance imaging. Frontiers in Systems Neuroscience. 2012;6(61):1–9. doi: 10.3389/fnsys.2012.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K. Functional and effective connectivity: a review. Brain Connectivity. 2011;1(1):13–36. doi: 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- Gelfand AE, Kim H-J, Sirmans C, Banerjee S. Spatial modeling with spatially varying coefficient processes. Journal of the American Statistical Association. 2003;98(462):387–396. doi: 10.1198/016214503000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL. Brain development during childhood and adolescence: a longitudinal MRI study. Nature Neuroscience. 1999;2(10):861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- Guo J, Levina E, Michailidis G, Zhu J. Joint estimation of multiple graphical models. Biometrika. 2011;98(1):1–15. doi: 10.1093/biomet/asq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Liu H. Transition matrix estimation in high dimensional time series; Proceedings of the 30th International Conference on Machine Learning; 2013. pp. 172–180. [Google Scholar]
- Han F, Liu H, Caffo B. Sparse median graphs estimation in a high dimensional semiparametric model. arXiv preprint arXiv:1310.3223. 2013 [Google Scholar]
- Harrison L, Penny WD, Friston K. Multivariate autoregressive modeling of fMRI time series. Neuroimage. 2003;19(4):1477–1491. doi: 10.1016/s1053-8119(03)00160-5. [DOI] [PubMed] [Google Scholar]
- Høst G, Omre H, Switzer P. Spatial interpolation errors for monitoring data. Journal of the American Statistical Association. 1995;90(431):853–861. [Google Scholar]
- Jones RH, Zhang Y. Modelling Longitudinal and Spatially Correlated Data. Springer; 1997. Models for continuous stationary space-time processes; pp. 289–298. [Google Scholar]
- Kolar M, Song L, Ahmed A, Xing EP. Estimating time-varying networks. The Annals of Applied Statistics. 2010;4(1):94–123. [Google Scholar]
- Kolar M, Xing EP. Sparsistent estimation of time-varying discrete Markov random fields. arXiv preprint arXiv:0907.2337. 2009 [Google Scholar]
- Lauritzen SL. Graphical Models. Vol. 17. Oxford University Press; 1996. [Google Scholar]
- Liu H, Han F, Yuan M, Lafferty J, Wasserman L. High-dimensional semiparametric gaussian copula graphical models. The Annals of Statistics. 2012;40(4):2293–2326. [Google Scholar]
- Liu H, Han F, Zhang C-H. Transelliptical graphical models. Advances in Neural Information Processing Systems. 2012:809–817. [PMC free article] [PubMed] [Google Scholar]
- Liu H, Roeder K, Wasserman L. Stability approach to regularization selection (StARS) for high dimensional graphical models. Advances in Neural Information Processing Systems. 2010:1432–1440. [PMC free article] [PubMed] [Google Scholar]
- Liu W, Luo X. High-dimensional sparse precision matrix estimation via sparse column inverse operator. arXiv preprint arXiv:1203.3896. 2012 [Google Scholar]
- Loh P-L, Wainwright MJ. High-dimensional regression with noisy and missing data: Provable guarantees with nonconvexity. The Annals of Statistics. 2012;40(3):1637–1664. [Google Scholar]
- Meinshausen N, Bühlmann P. High-dimensional graphs and variable selection with the lasso. The Annals of Statistics. 2006;34(3):1436–1462. [Google Scholar]
- Meinshausen N, Bühlmann P. Stability selection. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2010;72(4):417–473. [Google Scholar]
- Nobre AA, Sansó B, Schmidt AM. Spatially varying autoregressive processes. Technometrics. 2011;53(3):310–321. [Google Scholar]
- Pang H, Liu H, Vanderbei R. The fastclime package for linear programming and constrained L1-minimization approach to sparse precision matrix estimation in R. CRAN. 2013 [PMC free article] [PubMed] [Google Scholar]
- Penny W, Ghahramani Z, Friston K. Bilinear dynamical systems. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360(1457):983–993. doi: 10.1098/rstb.2005.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, et al. Functional network organization of the human brain. Neuron. 2011;72(4):665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS. Statistical analysis of a spatio-temporal model with location-dependent parameters and a test for spatial stationarity. Journal of Time Series Analysis. 2008;29(4):673–694. [Google Scholar]
- Rogers BP, Katwal SB, Morgan VL, Asplund CL, Gore JC. Functional MRI and multivariate autoregressive models. Magnetic Resonance Imaging. 2010;28(8):1058–1065. doi: 10.1016/j.mri.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman AJ, Bickel PJ, Levina E, Zhu J. Sparse permutation invariant covariance estimation. Electronic Journal of Statistics. 2008;2:494–515. [Google Scholar]
- Shaw P, Kabani NJ, Lerch JP, Eckstrand K, Lenroot R, Gogtay N, Greenstein D, Clasen L, Evans A, Rapoport JL, et al. Neurodevelopmental trajectories of the human cerebral cortex. The Journal of Neuroscience. 2008;28(14):3586–3594. doi: 10.1523/JNEUROSCI.5309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sølna K, Switzer P. Time trend estimation for a geographic region. Journal of the American Statistical Association. 1996;91(434):577–589. [Google Scholar]
- Song L, Kolar M, Xing EP. KELLER: estimating time-varying interactions between genes. Bioinformatics. 2009a;25(12):i128–i136. doi: 10.1093/bioinformatics/btp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Kolar M, Xing EP. Time-varying dynamic bayesian networks. Advances in Neural Information Processing Systems. 2009b:1732–1740. [Google Scholar]
- Storvik G, Frigessi A, Hirst D. Stationary space-time Gaussian fields and their time autoregressive representation. Statistical Modelling. 2002;2(2):139–161. [Google Scholar]
- Valdés-Sosa PA, Sánchez-Bornot JM, Lage-Castellanos A, Vega-Hernández M, Bosch-Bayard J, Melie-García L, Canales-Rodríguez E. Estimating brain functional connectivity with sparse multivariate autoregression. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360(1457):969–981. doi: 10.1098/rstb.2005.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Sabuncu MR, Buckner RL. The inuence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59(1):431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Han F, Liu H. Sparse principal component analysis for high dimensional multivariate time series; Proceedings of the 16th International Conference on Artificial Intelligence and Statistics; 2013. pp. 48–56. [Google Scholar]
- Xue L, Zou H. Regularized rank-based estimation of high-dimensional nonparanormal graphical models. The Annals of Statistics. 2012;40(5):2541–2571. [Google Scholar]
- Yuan M. High dimensional inverse covariance matrix estimation via linear programming. The Journal of Machine Learning Research. 2010;11:2261–2286. [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in the Gaussian graphical model. Biometrika. 2007;94(1):19–35. [Google Scholar]
- Zhao T, Liu H, Roeder K, Lafferty J, Wasserman L. The huge package for high-dimensional undirected graph estimation in R. The Journal of Machine Learning Research. 2012;13:1059–1062. [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Lafferty J, Wasserman L. Time varying undirected graphs. Machine Learning. 2010;80(2–3):295–319. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.