Abstract
The aims of the present study were to assess the prevalence of Chlamydia suis in an Italian pig herd, determine the tetracycline susceptibility of C. suis isolates, and evaluate tet(C) and tetR(C) gene expression. Conjunctival swabs from 20 pigs were tested for C. suis by real-time polymerase chain reaction, and 55% (11) were positive. C. suis was then isolated from 11 conjunctival swabs resampled from the same herd. All positive samples and isolates were positive for the tet(C) resistance gene. The in vitro susceptibility to tetracycline of the C. suis isolates showed MIC values ranging from 0.5 to 4 μg/mL. Tet(C) and tetR(C) transcripts were found in all the isolates, cultured both in the absence and presence of tetracycline. This contrasts with other Gram-negative bacteria in which both genes are repressed in the absence of the drug. Further investigation into tet gene regulation in C. suis is needed.
Introduction
Chlamydiaceae are obligate intracellular Gram-negative bacteria that cause a broad range of diseases both in humans and animals. Chlamydia suis infects pigs and has been associated with conjunctivitis, rhinitis, pneumonia, enteritis and reproductive disorders. Subclinical chlamydial infections are highly prevalent among pigs and make them more susceptible to other infections [1]. Both animal and human chlamydial infections are primarily treated with tetracycline or its derivatives. A stable tetracycline resistance phenotype was described for the first time in C. suis isolates from diseased and apparently healthy pigs in the US [2]. The resistance pattern was subsequently associated with tet(C) genomic islands integrated into the chlamydial chromosome and probably acquired through horizontal gene transfer [3]. Each island contained genes encoding a tetracycline efflux pump and a regulatory repressor (tet[C] and tetR[C], respectively), a C. suis-specific insertion element (IScs605) and additional genes involved in plasmid replication and mobilization [4]. Neither tet(C) nor tetR(C) was detected in the tetracycline sensitive reference S45 C. suis strain [3]. Studies performed on C. suis isolates from intensively reared Italian pigs showed a tetracycline resistance phenotype associated with the genomic island carrying the tet(C) resistance gene in several C. suis isolates [5]. tet(C)-positive C. suis strains have been reported in Swiss pig herds [6], and in Belgian, Cypriot and Israeli pig farms, where in vitro testing to tetracycline was also performed [7].
The aims of the present study were to assess the C. suis prevalence in an Italian pig herd, test C. suis isolates for their susceptibility to tetracycline, and correlate findings to the expression of the tet(C) and tetR(C) genes.
Materials and Methods
Sample collection and bacterial isolation
In March 2014, conjunctival swabs were collected at from 20 asymptomatic pigs aged 2–6 months reared in a pig herd located in Central Italy (42°48´20´´N 13°7´21´´E). Clinical history in this herd included conjunctivitis in piglets that had resolved spontaneously. A tetracycline treatment was routinely applied in the herd after castration of boars. For this study, no specific permission was required as the investigation was undertaken on the request of the owner for diagnostic investigation. The pig herd and the accessed land are privately owned and the owner gave permission to conduct the study on this site. All efforts were made to minimize the discomfort of the animals during sampling. This field study did not involve endangered or protected species. The sampling was performed by a veterinarian, gently rubbing a disposable cotton swab on conjunctival mucosa of both eyes. Approval by an animal ethics committee was not required as that the sampling did not involve blood.
In May 2014, conjunctival swabs were collected from 23 pigs reared in the same herd but from a litter different from the first sampling, to attempt chlamydial isolation according to Donati et al. [8]. Conjunctival swabs were placed in sucrose phosphate transport medium, transported at 4°C and processed immediately. Specimens were vortexed, then inoculated in duplicate onto LLC-MK2 cells (a continuous cell line derived from Rhesus monkey kidney tissue, provided by IZSLER Brescia, Italy), seeded in individual vials containing a glass coverslip at the bottom. After centrifugation at 800×g for three hours, the infected cell monolayers were incubated at 37°C for 48 hours and then fixed in methanol before detection of intracellular chlamydial inclusions by immunofluorescence technique.
Bacterial DNA extraction
Genomic DNAs were extracted from the conjunctival swabs and the chlamydial positive cell cultures using the QIAamp DNA mini kit (Qiagen, Hilde, Germany), following the supplier's recommendations.
Genus and species identification
Genomic DNAs extracted from the conjunctival samples were screened by a Chlamydiaceae-specific real-time polymerase chain reaction (rt-PCR) targeting a region of the 23S rRNA gene conserved among all Chlamydiaceae [9]. Samples with Ct values < 40 were considered positive and reanalyzed by a rt-PCR assay targeting a C. suis-specific region 23S rRNA gene [10]. All DNAs extracted from the Chlamydia-positive cell cultures were tested with the C. suis-specific rt-PCR.
DNAs from C. suis-positive samples and isolates were used as a template for a 1050-bp ompA gene fragment amplification [11]. The ompA amplicons were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and both DNA strands were sequenced (Bio-Fab Research, Rome, Italy). The sequences obtained were compared with the public sequences available in GenBank using the BLAST server from the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi.).
MIC determination
The in vitro susceptibility to tetracycline of the C. suis isolates was tested according to Donati et al. [12]. Antimicrobial susceptibility testing was performed with LLC-MK2 cells grown in 24-well plates with Eagle's minimum essential medium. Each of the 24-well plates was inoculated with 5 × 103 inclusion-forming units (IFU) per milliliter. After centrifugation at 1,700 × g for 1 h, the medium was removed and replaced with medium containing different concentrations of antimicrobial drug. After incubation at 35°C for 48 h, infected monolayers were washed with phosphate-buffered saline (PBS), fixed with methanol, and stained for inclusions with a fluorescein-conjugated monoclonal antibody specific for the chlamydial lipopolysaccharide genus-specific antigen (Meridian Diagnostics, Inc., Cincinnati, OH, USA). The minimum inhibitory concentration (MIC) of tetracycline was defined as the lowest concentration preventing the detection of more than 90 per cent of the chlamydial inclusions compared with the drug-free control. All tests were run in triplicate.
Molecular analysis of tet genes
A PCR amplifying a 608 bp fragment including the tetR(C) region and a PCR amplifying a 457 bp fragment of the tetR(C)-tet(C) region were performed on the C. suis-positive samples and isolates. The first PCR used the new primer tetR-F (5’-TTGGGGCAACCATTTCTGGT-3’) and the primer CS38 (5’-CCAAGGGATGACGACGACTG-3’) [3]. The second PCR was performed using the new primer tetRC-F (5’-TGCGTCGAGCAACGCACGCT-3’) and the primer CS43 reverted (5’-CAAAGCGGTCGGACAGTGCT-3’) [3]. In the first PCR, cycling conditions were as follows: 5 min of denaturation at 95° C and 35 cycles each consisting of denaturation at 94° C for 1 min, annealing at 58° C for 1 min and extension at 72° C for 1 min. A final elongation step of 5 min at 72° C completed the reaction. In the second reaction, PCR conditions were as described above, except for a higher annealing temperature (63°C). All amplicons were purified and sequenced for both DNA strands, as described above.
Reverse transcription-PCR
Transcriptional analysis of tetR(C) and tet(C) genes was performed for all the C. suis isolates using a reverse transcriptase PCR (RT-PCR) performed on RNA templates extracted from C. suis-infected monolayers cultured in the absence or presence of tetracycline. A Swiss field C. suis isolate, named NB-1 [13] was included, as a negative control as it is tet(C)-PCR-negative and sensitive in vitro to tetracycline. RNA extraction was performed using the RNeasy Mini Kit (Qiagen, Hilden, Germany) following the supplier's recommendations. AMV Reverse Transcriptase (Promega, Madison, WI, USA) was used to catalyze the polymerization of DNA from RNA samples. PCRs for tetR(C) and tet(C) genes [3], targeting fragments of 450 bp and 525 bp, respectively, were performed on the complementary DNA (cDNA) obtained by the reverse transcription step.
Results and Discussion
In the first sampling, 11 out of the 20 (55%) sampled pigs were positive by the Chlamydiaceae-specific RT-PCR and all were identified as C. suis. The ompA-amplicons from samples showed 96% sequence similarity (GenBank accession number KU668376) to the corresponding sequence of Rogers 130 C. suis strain (ATCC VR1482, GenBank accession number AF269277). In the second sampling, C. suis was successfully isolated from 11 out of 23 conjunctival swabs. The ompA sequences of the isolates were identical to each other and to sequences previously obtained from the C. suis-positive conjunctival swabs.
In vitro testing of the isolates to tetracycline showed MIC values ranging from 0.5 to 4 μg/mL (Table 1). In particular, MIC values were 0.5 μg/mL for one C. suis isolate (Eu-21), 2 μg/mL for six isolates (Eu-10/15/17/18/22/23), and 4 μg/mL for four isolates (Eu-4/7/8/19).
Table 1. In vitro MICs of tetracycline against 11 Chlamydia suis isolates.
| No. isolates | MIC (μg/mL) |
|---|---|
| 1 | 0.5 |
| 6 | 2 |
| 4 | 4 |
TetR(C) and tetR(C)-tet(C) amplicons were detected in all positive samples and isolates. TetR(C) and tetR(C)-tet(C) sequences (GenBank accession numbers KU668377 and KU668378, respectively) were identical to each other, and all they differed from those of the tetracycline-resistant R19 and R27 C. suis strains (GenBank accession number AY428550 and AY428551, respectively) described by Dugan et al. [3] as they presented a sequence of eight nucleotides in the tetR(C)-tet(C) intergenic spacer (Fig 1).
Fig 1. TetR(C)-tet(C) region of the Eu-21 Chlamydia suis isolate compared with the same genomic region of the R19 and R27 C. suis strains.
The highlighted alignment shows the eight-nucleotide sequence deleted in R19 and R27 C. suis strains.
Tet(C) and TetR(C) transcripts were detected in all the C. suis isolates cultured both in the absence and in the presence of tetracycline (Figs 2 and 3).
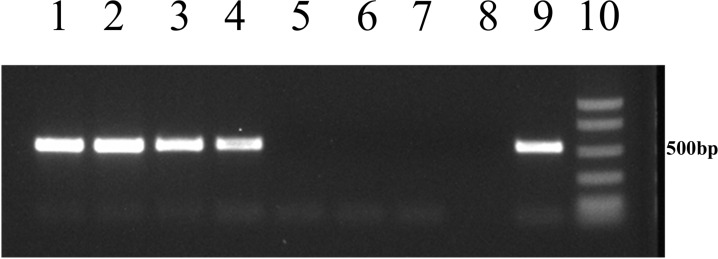
Fig 2. Analysis of tetR(C) transcription in Eu-22 and Eu-21 Chlamydia suis isolates.
Lanes 1 and 2: tetR(C) transcript from Eu-22 in the absence and presence of tetracycline at a concentration of 0.25 μg/mL. Lanes 3 and 4: tetR(C) transcript from Eu-21 in the absence and presence of tetracycline at a concentration of 0.25 μg/mL. Lane 5: negative control using NB-1 C. suis isolate cultured in the absence of tetracycline. Lane 6: blank control of RNA extraction. Lane 7: blank control of reverse transcription. Lane 8: blank control of PCR. Lane 9: positive control using bacterial genomic DNA as a template. Lane 10: BenchTop Markers (Promega, Madison, WI, USA).
Fig 3. Analysis of tet(C) transcription in Eu-22 and Eu-21 Chlamydia suis isolates.
Lanes 1 and 2: tet(C) transcript from Eu-22 in the absence and presence of tetracycline at a concentration of 0.25 μg/mL. Lanes 3 and 4: tet(C) transcript from Eu-21 in the absence and presence of tetracycline at a concentration of 0.25 μg/mL. Lane 5: negative control using NB-1 C. suis isolate cultured in the absence of tetracycline. Lane 6: blank control of RNA extraction. Lane 7: blank control of reverse transcription. Lane 8: blank control of PCR. Lane 9: positive control using bacterial genomic DNA as a template. Lane 10: BenchTop Markers (Promega, Madison, WI, USA).
In recent years, the emergence of tetracycline-resistant C. suis strains has attracted increasing interest. Suchland et al. [14] demonstrated the in vitro horizontal transfer of tetracycline resistance from C. suis to clinical strains of Chlamydia trachomatis, an important human pathogen. In view of the potential public health concerns resulting from in vivo tet(C) resistance gene transfer from porcine chlamydial strains to human chlamydial pathogens, the spread of tetracycline-resistant C. suis strains needs to be monitored.
The endemic trend of C. suis infection in Italian pig herds found in the present study is comparable to previous reports [15], and the presence of tet genes proved widespread. Comparison of the ompA sequences from samples and isolates suggests the circulation of a single C. suis strain in the sampled herd, but the isolates showed a different degree of susceptibility to tetracycline, with MIC values varying from 0.5 to 4 μg/mL. For one isolate (Eu-21) the MIC was lower than the MIC values detected in the other isolatesand in previous reports on C. suis strains [3,7]. Interestingly, an MIC value of 0.5 μg/mL was detected in our previous study in two tet(C) PCR-positive C. suis isolates [5].
Previous studies showed that overexpression of the tetracycline resistance protein is lethal for E. coli, probably due to collapse of the membrane potential [16]. Therefore, the efflux-encoding determinants are strictly regulated. Tetracycline-inducible Tet repressor proteins turn down transcription of the resistance genes and of their own genes in the absence of the drug and allow expression of both proteins only in the presence of tetracycline [17]. Dugan et al. [3] emphasized that the tet(C) transcript was found in C. suis R19 cultured both in the absence and presence of tetracycline, whereas it was not detected in E. coli (pSC101) when cultured in a medium lacking tetracycline. The comparison between the nucleotide sequences of C. suis and E. coli showed that tetR(C) in R19 strain was truncated and that the operator region had an octanucleotide deletion relative to the homologous sequence in pSC101. The authors suggested that these differences may affect the regulation of tet(C) gene, eliminating the tight control placed on tet(C) expression in the absence of tetracycline.
The present study found tet(C) and tetR(C) transcripts in all C. suis isolates cultured in the absence and presence of tetracycline. All C. suis isolates showed the truncation of tetR(C), whereas the eight-nucleotide deletion upstream of the tet(C) start site was not present. These findings suggest that the presence or absence of the eight-nucleotide sequence does not affect regulation of the tet(C) gene in the absence of tetracycline. Our data confirm the unusual features of tet gene regulation and expression in C. suis compared to other Gram-negative bacteria. Further investigation on tet gene regulation in C. suis is needed.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors have no support or funding to report.
References
- 1.Schautteet K, Vanrompay D. Chlamydiaceae infections in pig. Vet Res. 2011;42: 29 10.1186/1297-9716-42-29 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen AA, Rogers DG. Resistance to tetracycline and sulfadiazine in swine C trachomatis isolates, 1998, p. 313–316. In R. S. Stephens (ed.), Chlamydial infections. Proceedings of the Ninth International Symposium on Human Chlamydial Infection. International Chlamydia Symposium, San Francisco, CA.
- 3.Dugan J, Rockey DD, Jones L, Andersen AA. Tetracycline resistance inChlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob Agents Chemother. 2004;48(10): 3989–3995. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandoz KM, Rockey DD. Antibiotic resistance in Chlamydiae. Future Microbiol. 2010;5(9): 1427–1442. 10.2217/fmb.10.96 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Francesco A, Donati M, Rossi M, Pignanelli S, Shurdhi A, Baldelli R et al. (2008) Tetracycline resistant Chlamydia suis isolates, in Italy. Vet Rec. 2008;163(8): 251–252. . [DOI] [PubMed] [Google Scholar]
- 6.Borel N, Regenscheit N, Di Francesco A, Donati M, Markov J, Masserey Y, et al. Selection for tetracycline-resistant Chlamydia suis in treated pigs. Vet Microbiol. 2012;156(1–2): 143–146. 10.1016/j.vetmic.2011.10.011 . [DOI] [PubMed] [Google Scholar]
- 7.Schautteet K, De Clercq E, Miry C, Van Groenweghe F, Delava P, Kalmar I et al. Tetracycline-resistant Chlamydia suis in cases of reproductive failure on Belgian, Cypriote and Israeli pig production farms. J Med Microbiol. 2013;62(Pt 2): 331–334. 10.1099/jmm.0.042861-0 . [DOI] [PubMed] [Google Scholar]
- 8.Donati M, Pollini GM, Sparacino M, Fortugno MT, Laghi E, Cevenini R. Comparative in vitro activity of garenoxacin against Chlamydia spp. J Antimicrob Chemother. 2002;50(3): 407–410. . [DOI] [PubMed] [Google Scholar]
- 9.Ehricht R, Slickers P, Goellner S, Hotzel H, Sachse K. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol Cell Probes 2006;20(1): 60–63. . [DOI] [PubMed] [Google Scholar]
- 10.Pantchev A, Sting R, Bauerfeind R, Tyczka J, Sachse K. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp Immunol Microbiol Infect Dis. 2010;33(6): 473–484. 10.1016/j.cimid.2009.08.002 . [DOI] [PubMed] [Google Scholar]
- 11.Sayada C, Andersen AA, Storey C, Milon A, Eb F, Hashimoto N, et al. Usefulness of omp1 restriction mapping for avian Chlamydia psittaci isolate differentiation. Res Microbiol. 1995;146(2): 155–165. . [DOI] [PubMed] [Google Scholar]
- 12.Donati M, Di Francesco A, D’Antuono A, Delucca F, Shurdhi A, Moroni A, et al. In vitro activities of several antimicrobial agents against recently isolated and genotyped Chlamydia trachomatis urogenital serovars D through K. Antimicrob Agents Chemother. 2010; 54(12): 5379–5380. 10.1128/AAC.00553-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wanninger S, Hoffmann K, Donati M, Di Francesco A, Borel N. Tetracycline resistant Chlamydia suis in Swiss fattening pigs. In: Third European Meeting on animal Chlamydioses and Zoonotic Aspects, Anses, Maisons-alfort, France, September 24–25, 2015. Oral presentation.
- 14.Suchland RJ, Sandoz KM, Jeffrey BM, Stamm WE, Rockey DD. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob Agents Chemother. 2009;53(11): 4604–4611. 10.1128/AAC.00477-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Francesco A, Baldelli R, Cevenini R, Magnino S, Pignanelli S, Salvatore D, et al. Seroprevalence to chlamydiae in pigs in Italy. Vet Rec. 2006;159(25): 849–850. . [PubMed] [Google Scholar]
- 16.Eckert B, Beck CF. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J Bacteriol. 1989;171(6): 3557–3559. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillen W, Berens C. Mechanisms underlying expression of TN10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345‒369. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.