Abstract
Riboswitches are promising targets for the design of novel antibiotics and engineering of portable genetic regulatory elements. There is evidence that variability in riboswitch properties allows tuning of expression for genes involved in different stages of biosynthetic pathways by mechanisms that are not currently understood. Here we explore the mechanism for tuning of SAM-I riboswitch folding. Most SAM-I riboswitches function at the transcriptional level by sensing the cognate ligand— S-adenosyl methionine (SAM). SAM-I riboswitches orchestrate the biosynthetic pathways of cysteine, methionine and SAM, etc. We use base pair probability predictions to examine the secondary structure folding landscape of several SAM-I riboswitch sequences. We predict different folding behaviors for different SAM-I riboswitch sequences. We identify several “decoy” base pairing interactions involving 5’ riboswitch residues that can compete with the formation of a P1 helix, a component of the ligand-bound “transcription OFF” state, in the absence of SAM. We hypothesize that blockage of these interactions through SAM contacts contributes to stabilization of the OFF state in the presence of ligand. We also probe folding patterns for a SAM-I riboswitch RNA using constructs with different 3’ truncation points experimentally. Folding was monitored through fluorescence, susceptibility to base-catalyzed cleavage, nuclear magnetic resonance and indirectly through SAM binding. We identify key decision windows at which SAM can affect the folding pathway toward the OFF state. The presence of decoy conformations and differential sensitivities to SAM at different transcript lengths are crucial for SAM-I riboswitches to modulate gene expression in the context of global cellular metabolism.
Keywords: RNA, riboswitch, secondary structure prediction, dimmer switch, conformation ensemble, base pair probability, partition function
Introduction
Riboswitches illustrate the remarkable capacity of dynamic RNA folding to regulate gene expression. Riboswitches are folded elements within the 5’ untranslated regions (5’UTR) of messenger RNA (mRNA). Typically, downstream genes are involved in biosynthesis of small molecule metabolites. A riboswitch can form two or more alternative secondary structures in response to the cellular level of specific small molecules that are metabolically linked to products of the downstream genes. In this way, they provide a mechanism for feedback regulation of gene expression. These properties have sparked interest in the engineering of “designer riboswitches” for applications ranging from diagnostics to environmental clean up. Riboswitches have so far been identified in greatest abundance in bacteria and have been linked to the mode of action of several antibiotics, sparking interest in targeting of riboswitch RNA for drug design.
Bacterial riboswitches can be characterized as transcriptional or translational, depending on whether they control the synthesis of RNA or protein. Transcriptional riboswitch function is traditionally described in terms of a two-state (for example, ligand bound/transcription OFF and unbound/transcription ON) secondary structure model. These two state models facilitated the identification of riboswitch elements from genome sequencing data19, and guided the design of successful high resolution structural studies. For a number of riboswitches, X-ray and NMR structures provided insight into the basis for recognition of small molecule ligands by the so-called “aptamer” or ligand-sensing domain within the ligand-bound “OFF”state.
In most cases the unbound secondary structure (“ON”) state has not been solved to high resolution. Structural characterization of this ON state is linked to an understanding of the folding of a second riboswitch region, termed the “expression domain”, which actually controls gene expression. Typically the expression domain displays less sequence conservation than the aptamer region. So-called “rho-independent terminator” hairpin structures, or the Shine-Dalgarno ribosomal recognition sequence, are present in the expression domains of transcriptional and translational riboswitches, respectively. Aside from these general features, early models for the unliganded secondary structures of some riboswitches have been subject to revision.
Henkin and colleagues found variation in functional properties of a set of eleven S-adensoyl methionine (SAM)-I riboswitch sequences from B. subtilis27. A further indication of the complexity of riboswitch function arises from apparent discrepancies between levels of cognate ligand required for binding, in vitro function and cellular function. A mechanistic explanation of such functional variations requires an understanding of the role of the expression domain. In vitro structural studies are typically performed under equilibrium conditions with transcripts encompassing the full segment of the 5’UTR which is deemed to be functionally relevant. Co-transcriptional folding and ligand binding to incompletely transcribed RNA may modify the functional outcome from what would be predicted from in vitro equilibrium measurements that utilize full length transcripts.
Another contributing factor might be the presence of a broader distribution of riboswitch RNA secondary structures/conformations than acknowledged in the two-state picture. Recently the Free Energy Landscape approach, which assumes a population distribution of secondary structures, has been applied to predict riboswitch folding. This distribution can be calculated from a partition function.
Base pair probability (BPP) can also be calculated using the partition function implemented using McCaskill’s algorithm 38. BPP has been used to study the whole genome of an HIV-1 RNA virus 39 and the effects of Single Nucleotide Polymorphisms40 on the ensemble of RNA structures. It has also been used to evaluate multiple structures generated from free energy minimization 41 and to assess the quality of predicted secondary structures. An attractive aspect of BPP calculations is that they can be combined with experimental probing of secondary structure43
Riboswitches are a logical target for BPP calculations since their biological function precludes the formation of a single, strongly dominant secondary structure. The SAM-I riboswitch seems particularly suited to this analysis because a wealth of experimental binding and folding data is available particularly for sequence variants. The structural data and folding predictions suggest that relatively few non-canonical base pairs, (aside from tandem GA pairs in the well-characterized “kink-turn” domain51; 52) are present. One weakness of secondary structure predictions, the lack of parameters for non-canonical base pairing, is therefore less problematic than for some riboswitch systems.
In this study, we first chose to apply BPP calculations using a partition function to address a set of interrelated but unanswered questions regarding SAM-I riboswitch folding: What is the nature of the ON state? How might the need to “tune” the conformational distribution for varied expression of downstream genes be reflected in variations amongst riboswitch sequences? What mechanism allows a small molecular weight ligand to rearrange its folding so dramatically to form the OFF state? What switch point(s) during transcription are critical for this folding decision? We then found evidence for predicted switching transitions and “decoy” conformations from NMR measurements, monitoring of susceptibility to base-catalyzed cleavage, and other biophysical measurements, along with literature reports and sequence conservation patterns. Altogether, our findings suggest that riboswitch sequences undergo selection for metastable conformational states. We show that these results provide a potential explanation as to how specific SAM/riboswitch contacts53 stabilize formation of the OFF conformation in a series of events leading to transcription termination.
Results and Discussion
Potential Decision Points for SAM-I Riboswitch Folding
A. Alternative Models for Antiterminator Helices for two SAM-I Riboswitches
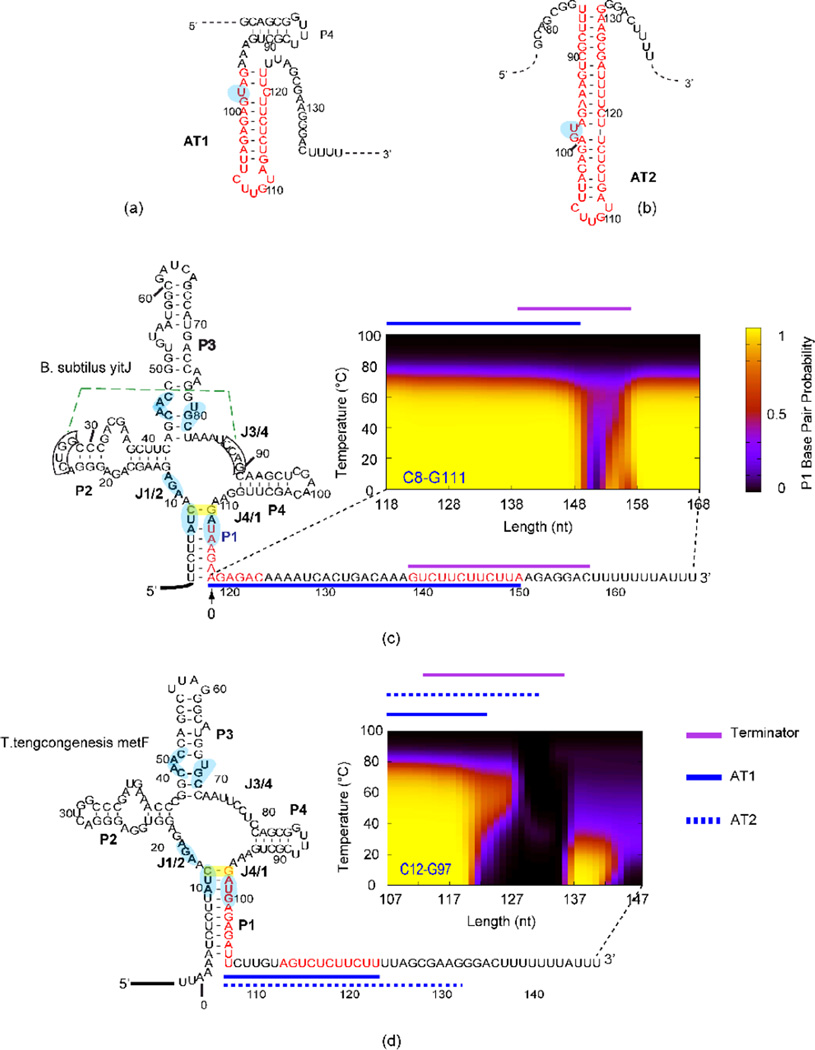
Atomic resolution X-ray structures have been reported for isolated OFF-state aptamers of SAM-I riboswitches from one thermophilic and one mesophilic50 organism. For the two sequences rather different secondary structure models for the ON state are presented in the literature. The model originally suggested for the AT helix in the T. tengcongensismetF sequence in ref56 (“AT1”-Figure 1a, compared to the OFF state in SI Figure 1a) differs from the model suggested in a recent biochemical study46 (Figure 1b) (which we call “AT2”). The model in Figure 1a (also shown in SI Figure 1b) is similar to that proposed for the B. subtilisyitJ sequence. In AT2, the AT helix intrudes into the P4 helix46. From these considerations, we suspected that a difference in distribution of conformer populations would be predicted for the two riboswitches based on BPP calculations.
Figure 1. Alternative AT helix models and results from SAM-I riboswitches.
(a) AT1 model as proposed in reference65, in which the P4 helical region can still form the secondary structure as observed in the crystal structure of the aptamer. (b) AT2 model as suggested in reference46 and this study. Note that the P4 helix of the aptamer is disrupted when this AT2 helix is formed.. Different patterns of secondary structure competition are predicted and observed for B. subtilisyitJ (c) and T. tengcongensismetF (d) SAM-1 riboswitches. Sequence of B. subtilisyitJ and T. tengcongensismetF SAM-I riboswitches in the secondary structure representation of the “OFF” state are shown at the left. The segments highlighted in red display the residues participating in the AT helix as proposed in the literature. BPP is plotted for the closing C8•G111 base pair of the P1 helix (highlighted with a yellow box) for transcripts with varying 3’ truncations (similar BPP patterns are predicted for other base pairs in the P1 helix-shown in SI Figure 2). The horizontals plots the length increment with 0 starting at the 3’ end of the aptamer, the vertical axis shows the temperature (0–100 °C). The color scale represents the magnitude of the BPP as indicated in the legend on the right of panel c. Lines above the plots are color coded for the different structural elements (Terminator, AT1 and AT2) as indicated in the legend next to panel d. Boxed residues participate in a pseudoknot interaction (dotted green line). Residues that contact SAM according to X-ray structures are highlighted in light blue. Side by side representations of models for ON and OFF state secondary structures for both riboswitches are presented in SI Figure 1a and b.
We input a series of RNA sequences to illuminate patterns of increasing transcript lengths for secondary structure prediction, incrementing the 3’ truncation point one nucleotide at a time. In this manner, we aim to predict folding intermediates as they may evolve during the synthesis of the riboswitch-containing region35. We call this type of calculation “co-transcriptional folding simulation” (CTFS). Then we monitored the BPP of representative base pairs from the P1 helix as a function of temperature for each transcript length. We incorporate temperature as a parameter to illuminate patterns of competitive folding. Results from CTFSs for the B. subtilis yitJ and T. tengcongensismetF SAM-I riboswitches (henceforth referred to as yitJ and metF, respectively), are shown in Figure 1c & d. The selected BPPs suggest the predicted population of P1 helix-forming riboswitches. Formation of this helix makes formation of the terminator helix, and thus transcription attenuation, more probable since the formation of the competing AT helix is blocked by P1 helix formation. The BPP patterns in Figure 1 are broadly representative of those predicted for most P1 helix base pairs within each riboswitch (SI Figure 2).
Comparison of predictions reported in Figure 1c & d for the two sequences reveals contrasting patterns of temperature dependence and transcript length dependent folding. For the metF SAM-I riboswitch there is a reduction of BPPs in the P1 helix near a length of 120 base pairs. At this length, the AT1 helix is nearly fully formed but the AT2 helix model cannot form completely, since the extension of the AT helix to displace J4/1 and the P4 helix is not yet possible. This AT1 helix participates in an overall secondary structure which appears partly analogous to that proposed for the yitJ system (a detailed comparison of these two helices and their competition with respective P1 helices is shown in SI Figure 3a and discussed in SI section “Contrasting P1 vs. AT helix competition patterns for the yitJ and metF SAM-I riboswitches”). For the yitJ SAM-I riboswitch, P1 helix BPPs also start to decrease significantly as the length of the RNA transcript reaches the position (length 32 nucleotides beyond the aptamer) that can form a full AT helix (corresponding to the AT1 helix for metF). On the other hand, some P1 pairing persists at high temperatures (40–80 °C) for the metF sequence in this range of transcript length, which is not observed for yitJ. In contrast to predictions for the metF sequence, P1 helix BPPs for yitJ are restored when the 3’ strand of the rho-independent terminator has been “synthesized”. The restoration of P1 helix BPPs for long metF transcripts is only predicted at low temperatures (0–40 °C) and when transcript length extends well beyond the AT2 helix.
The above calculations were performed with RNA sequences starting at the 5’ end of the riboswitch, omitting upstream residues. Calculations of P1 helix BPPs for riboswitch transcript sequences starting at the transcription start site (SI Figure 4) show similar trends, with slight increases in P1 helix BPPs across temperature and transcript lengths. Altogether, these results are consistent with suggestions that variability in the expression platform plays an important role in tuning the function of riboswitches.
A search for sequences with similar folding potential to that observed for the metF SAM-I riboswitch primarily yielded SAM-I riboswitches upstream of the metF gene homologs from closely related thermophilic organisms (SI Figure 5). Therefore the predicted co-transcriptional folding behavior in Figure 1d appears to be distinctive for SAM-I riboswitches located upstream of one particular set of genes.
B. Experimental Probing of the metF SAM-I Riboswitch Folding as a Function of Transcript Length
Aside from one recent study 46, there are few data on secondary structure dynamics for metF SAM-I riboswitches. Most SAM-I riboswitch experimental reports have utilized transcripts which were truncated at or near the 3’ end of the putative aptamer, or which contained the full length riboswitch. To determine whether the metF SAM-I riboswitch conformational folding follows the distinctive pattern predicted in Figure 1d, we probed conformations for transcripts of varying lengths using several experimental methods.
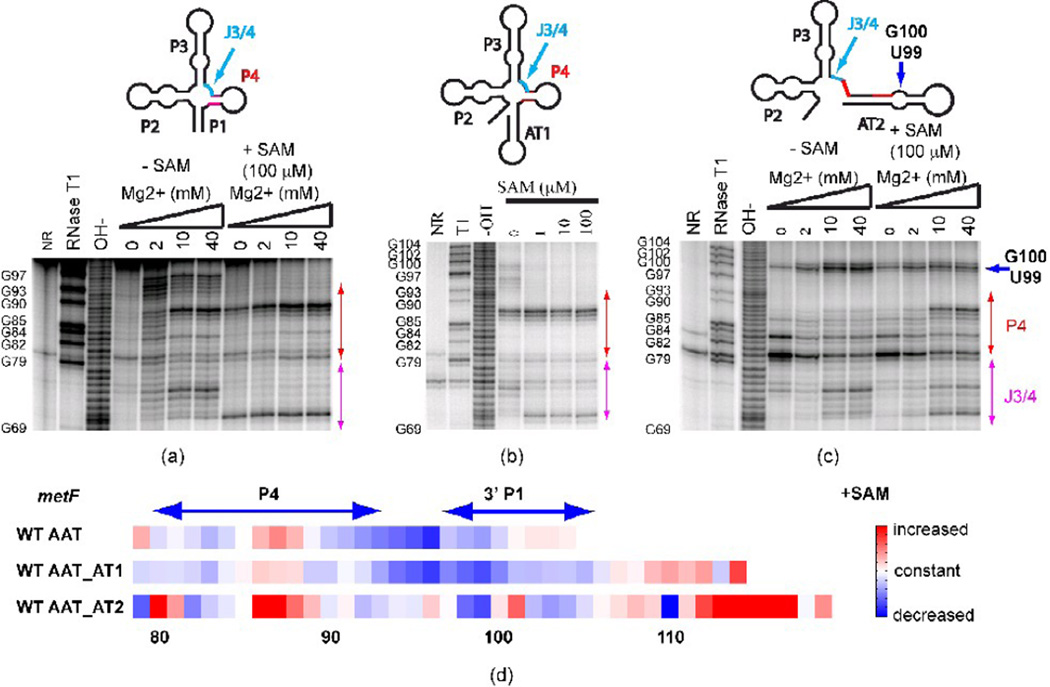
We synthesized a series of three metF SAM-I riboswitch transcripts with 3’ cutoff points at the aptamer, at the AT1 helix, or at the proposed AT2 helix. The folding of the three wild type RNAs is characterized using in-line probing (Figure 2). As expected, addition of SAM leads to a change in cleavage pattern for all three transcripts, with varying degrees of similarity to the SAM-induced pattern changes reported previously for full length yitJ riboswitches28. The trends are also consistent with other enzymatic and chemical probing reported for yitJ and metF SAM-I aptamers or full length riboswitches, in the presence and absence of SAM. In addition, the longest transcript probed (Figure 2c) shows a dramatic change in cleavage pattern in the P4 helix region (residues 80–85) in the presence or absence of SAM, as compared to shorter transcripts (Figure 2a, b). The observed pattern is consistent with the formation of an AT2 helix, as predicted for transcripts of this length. Also, the AT2 model predicts that a GU dinucleotide is bulged out of the helix (Figure 1b). Indeed, an enhanced cleavage in the absence of SAM is observed at these positions in Figure 2c (residues 99 and 100, blue arrow) as compared to Figure 2b.
Figure 2. In-line probing of secondary structure in metF SAM-I riboswitch-containing transcripts with varying 3’ truncation points.
(a) Truncation at the aptamer, (b) Truncation at the “switch point”, at which residues comprising AT1 have been transcribed (c) Truncation at the point of complete transcription of AT2 helix. Red arrows: change of cleavage pattern in the P4 helix. Blue arrow: the increased cleavage is likely to be due to the GU bulge in the alternative AT helix model. J3/4 region is indicated as the arrow in cyan. (d) Quantification of the effect of SAM on in-line probing experiments (40 mM Mg2+ lanes) for three metF SAM-I riboswitch RNA constructs starting from nucleotide 79. The color scale is shown on the right—red means increased cleavage rate, white denotes no change and blue indicates decreased cleavage rate in the presence of SAM. WT AAT—truncation at the aptamer; WT AAT_AT1— truncation at the “switch point”, at which residues comprising AT1 have been transcribed; WT AAT_AT2—truncation at the point of complete transcription of AT2 helix. Note that RNAse T1 cleavage bands for G100, G102, and G104 are not resolved from the full-length uncleaved band. Altogether, these results indicate that P1 helix and OFF state formation are inhibited in long metF SAM-I riboswitch transcripts by the formation of the AT2 helix.
Significantly, the cleavage patterns in regions that are most sensitive to SAM binding, such as J3/4 (highlighted in magenta in Figure 2), show a stronger residual of the minus SAM cleavage pattern when SAM is added for the longest transcript (Figure 2c, d). This observation and predictions in Figure 1 are consistent with the hypothesis that AT2 helix formation reduces OFF state formation and SAM binding, as proposed in reference 46. Gel mobility assays (SI Figure 6 and accompanying description in SI) for all constructs indicate that a fast migrating band increases in intensity and mobility when the RNA is pre-incubated with excess SAM, while mobility of slower moving bands are unaffected.
Figure 2a and Figure 2c also show that the cleavage pattern for the metF SAM-I riboswitch sequences is affected by magnesium concentrations in the absence of SAM. Some Mg2+-induced cleavage patterns, including that observed in residues in J3/4, parallel those associated with the addition of SAM (Figure 2a & c). This observation is consistent with fluorescence measurements that indicate that Mg2+ enhances SAM-dependent folding of the yitJ SAM-I riboswitch.
C. Evidence for the Shift of Conformational Equilibrium Upon SAM Binding from NMR Spectroscopy
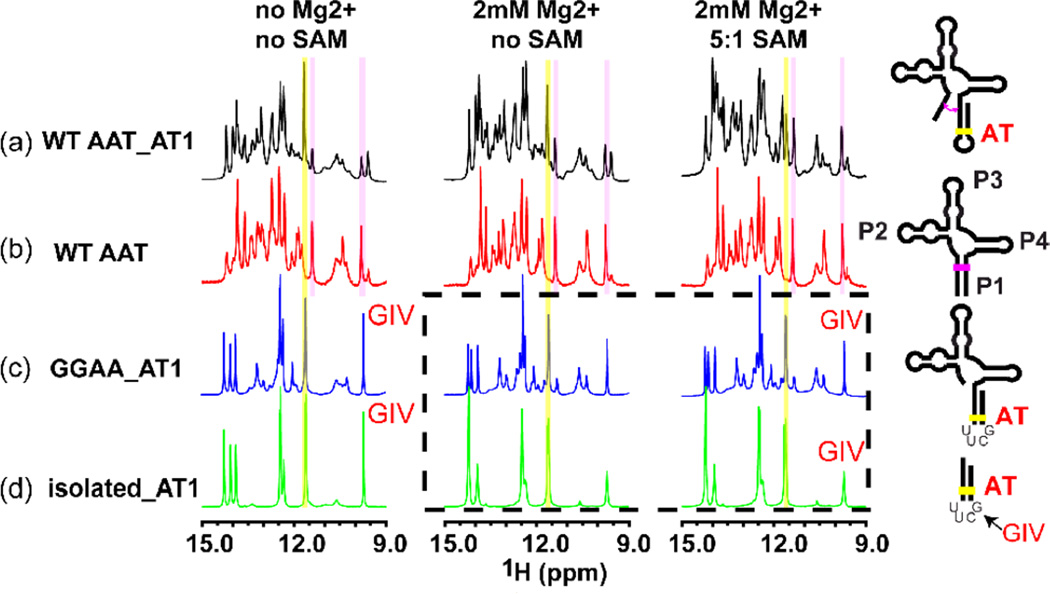
In favorable conditions, RNA base pairing can be monitored via the appearance of imino resonances in NMR spectra. To independently confirm the interpretation of AT helix formation from in-line probing measurements, we first obtained one and two-dimensional NMR spectra for a series of RNA constructs containing various segments of the metF SAM-I riboswitch. Combining these spectra confirmed the presence of P1 and AT1 helices within the longer RNAs utilized for in-line probing measurements in Figure 2a, and b. We then used these spectra to assign imino signals corresponding to AT1 helix base pairs. These assignments are shown in SI (SI Figures 7&8). Figure 3 displays the superposition of 1D imino proton spectra, which includes signals from protons involved in base pairing, for four different RNA constructs in low salt (left), with 2 mM MgCl2 and 100 mM KCl (center) and with SAM added to 5:1 stoichiometry (right). The strand switching mechanism leads to similar sequences with similar signals in the alternative P1/AT1 helices. The highlighted signals come from a GU base pair, one of the few positions which differ in the alternative helices.
Figure 3. Extension of the metF SAM-I riboswitch RNA transcripts to include AT1 is required to observe SAM-induced strand switching by NMR.
The 1d imino proton spectrum is shown at 35°C under three different conditions—no Mg2+, in the presence of 2 mM Mg2+, and with 5:1 ratio [SAM]/[RNA]. (a) SAM-I riboswitch RNA construct containing “strand switching” elements capable of forming P1 or AT1 helix (similar to AT1 helix but with 2 U residues truncated). (b) Truncated “aptamer” sequence capable of forming only P1-helix containing conformers. (c) Construct truncated at the 5’ end to prevent P1 helix conformation. (d) Isolated AT1 helix construct. For the AT1-containing RNA shown in (a), addition of SAM reduces intensity of signal associated with GU base pair within AT1 helix (yellow), while increasing intensity of another signal previously observed in truncated aptamer (pink). Note that whereas constructs used in (a) and (b) contain wild type sequences, RNAs used in panels c and d contain a UUCG tetraloop substitution in the AT helix, giving rise to a signal at 9.7 ppm89.
The WT AAT, (the isolated aptamer) displays no significant spectral change when SAM is added. For the WT AAT_AT1 construct that can form either the AT1 or P1 helix in a competition, we observed a number of changes in the spectrum upon addition of SAM. In particular, the signals from the GU base pair from within the AT and the P1 helix that are highlighted in Figure 3 demonstrate the most significant change. Upon the addition of SAM, the signals observed for the AT1 GU base pair (11.65 ppm, yellow) are reduced, while the intensities of the P1 GU base pair (9.8, 11.4 ppm, magenta) and other signals that line up with the WT AAT spectrum increase. These results suggest that the AT1 helix form is dominant in the absence of SAM, as predicted in Figure 1d and the equilibrium is shifted toward formation of the P1 helix when SAM is present.
These findings along with other one and two dimensional spectra of SAM-I riboswitch RNA segments demonstrate that 1) The truncated aptamer model (WT AAT) is not an adequate model for observation of ligand-induced conformational exchange 2) At least two conformational families exist in slow exchange for WT AAT_AT1, as indicated by the existence of separate signals. 3) Alignment between spectra from different constructs and comparison of spectra in the presence and in the absence of SAM are consistent with a conformational exchange between ON and OFF forms for WT AAT_AT1. 4) Assuming the interpretation described in 3), the SAM-I riboswitch is not well described by an “all or none” model of conformational switching. Rather, the ligand perturbs a delicately balanced equilibrium, allowing for fine tuning of the level of gene expression (a so-called “dimmer switch”).
D. SAM-Binding to Varying Length metF SAM-I Riboswitch Segments Parallels the Degree of OFF State Conformation
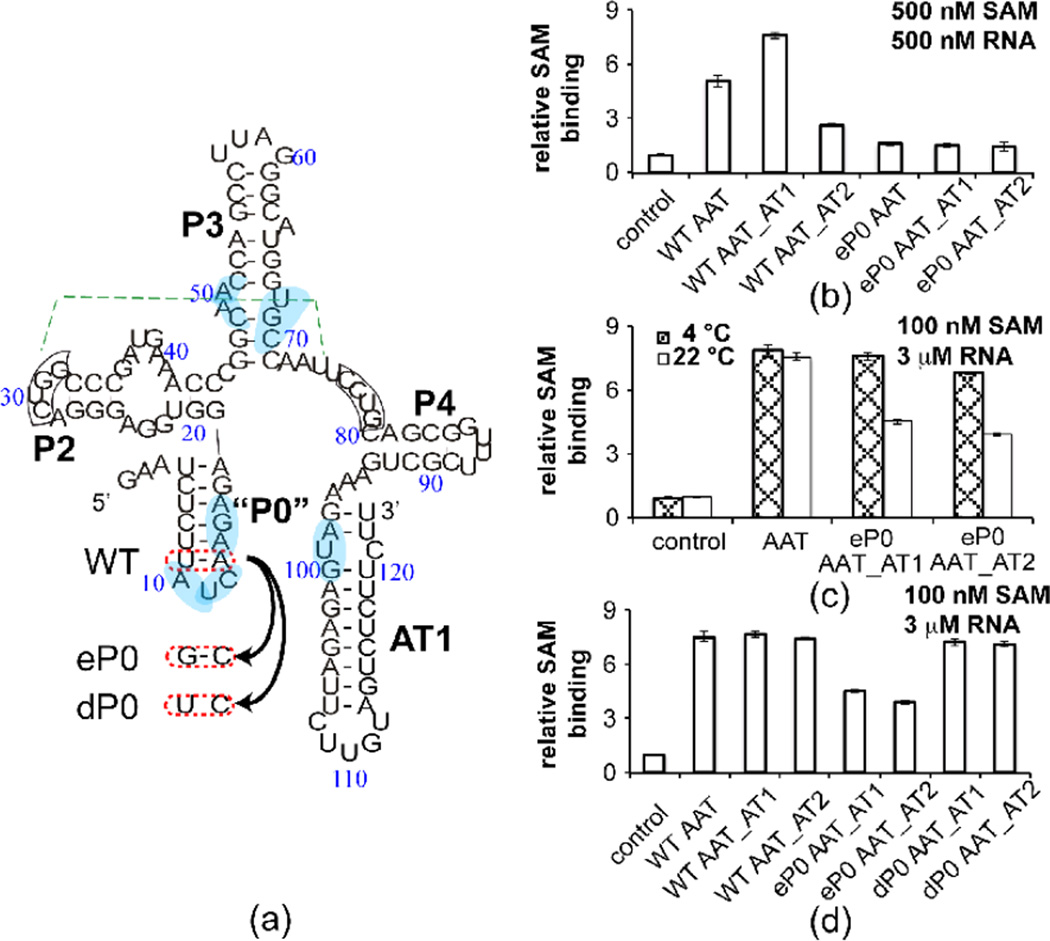
The reduction of SAM-induced conformational change observed in in-line probing and gel mobility experiments for AT2 helix-forming constructs led us to ask whether this helix inhibits SAM binding by perturbing the conformational equilibrium towards the ON state. Equilibrium dialysis has been used as a means of direct measurement of the binding of SAM and other ligands to riboswitches (Boyapati et al, manuscript in preparation). We utilized [3H]-SAM as a reporter in single titration point equilibrium dialysis measurements, in order to rank binding affinity of the ligand to the RNA constructs shown in Figure 2. Binding seems to correlate with the experimental and predicted degree of OFF state formation (Figure 4). The strongest binding affinity appears with the shortest and intermediate length wild type constructs (Figure 4b, “WT AAT”, “WT AAT_AT1”). The wild type transcript long enough to form an AT2 helix (“WT AAT_AT2”), however, shows the weakest SAM binding affinity, as would be expected if a high population of ON state conformer inhibits SAM binding.
Figure 4. SAM binding affinity examined by equilibrium dialysis partly correlates with predicted P1 helix BPP.
(a) Sequence information for eP0 and dP0 mutants. WT denotes wild-type, eP0 designates a double mutant that enhances the stability of the P0 helix by converting an AU base pair to a CG base pair and introducing a GA mismatch at the P1 helix, and dP0 contains a single mutant that restores full P1 helix pairing and destabilizes the P0 helix by changing an AU base pair to CU mismatch. The pseudoknot (which is still theoretically possible in the ON conformation) and base pairs that contact SAM are highlighted as in Figure 1. (b) Equilibrium dialysis (see Materials and Methods for details) performed with RNA: [3H] SAM is introduced in chamber a and RNA is added to chamber b, eachat the same concentration of 500 nM. A higher b/a ratio indicates tighter binding of SAM. The b/a ratio is therefore called “relative SAM binding affinity” in the Figure. The annotation of different 3’ truncation points is the same as Fig. 1b. (c) Equilibrium dialysis performed with 3 µM RNA and 100 nM [3H] SAM at 4 °C and 22 °C. The annotation is the same as (a). (d) Equilibrium dialysis performed with 100 nM [3H] SAM and 3 µM RNA at 22 °C.
Stabilization of ON State SAM-I Riboswitch Conformation via Sequestration of 5’P1 and J1/2 Residues
A. Residues in J1/2 Are Predicted to Interact with Decoy Regions in the Absence of SAM
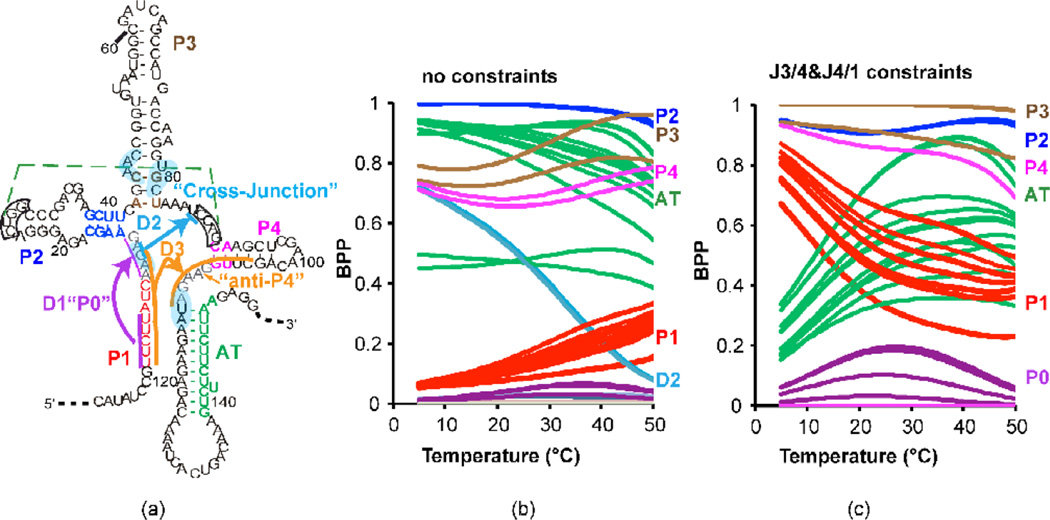
Figure 1 indicates that the transcript length at which either the P1 helix or the AT1 helix can form but the terminator is not yet fully transcribed could be an important decision point for SAM-I riboswitch regulated transcription. Since there was no obvious reason to suspect that an AT1 helix would melt at a lower temperature than a P1 helix, the temperature dependence for P1 BPPs predicted in Figure 1 caused us to suspect the involvement of additional secondary structural elements at low temperature. To detect where alternative interactions may be taking place within the BPP calculations, we monitored several possible alternative base pairings as a function of temperature for fixed lengths of the yitJ SAM-I riboswitch. Specifically we monitored base pairings involving residues in J1/2 and on the 5’ strand of the P1 helix. These alternative interactions are illustrated in Figure 5a, SI Figure 1c and listed in SI Figure 9. Figure 5b shows the selected BPPs as a function of temperature for a yitJ SAM-I riboswitch cut at length 151 (length 35 in Figure 1). As expected based upon Figure 1, base pairings in the AT helix (green) predominate over P1 helix pairings (red), with the probability of the latter increasing with increasing temperature. Note that two “cross-junction” base pairings (11/87 and 12/86) between the J1/2 and J3/4 (cyan) appear with a high BPP at low temperatures. The appearance of these base pairs is inversely correlated with the increase in P1 helix BPP with temperature. Note that monitors of P3 (brown) and P4 (magenta) helix formation indicate only 60–80% BPP, suggesting heterogeneity in secondary structure.
Figure 5. “Decoy” base pairings involving 5’ residues are predicted to favor AT formation in a fixed length yitJ SAM-I riboswitch fragment.
(a) Schematic illustration of alternative or “misfolded” conformations with in the context of an “ON” state secondary structure previously proposed for the yitJ SAM-I riboswitch. Residues participating in base pairing within the three alternative foldings D1, D2, and D3, are highlighted in purple, blue, and orange, respectively. J1/2 and J4/1 are highlighted in grey. Secondary structure schematics for the three alternative folds D1-D3 are illustrated explicitly in SI Figure 1c. The pseudoknot and base pairs that contact SAM are highlighted as in Figure 1. Predicted BPPs for individual base pairs for a 151 nucleotide length transcript are plotted as a function of temperature without (b) or with (c) the following residues constrained from base pairing: residues 87–90 (e. g. pseudoknot formation is blocked) and two residues in J4/1, blocking the formation of base pairings with 5’ residues involved in “decoy” helix D3. BPPs are color coded according to the secondary structure element that each base pair participates in as indicated on the right (P1, red, AT green, P2 dark blue, P3, brown, P4 magenta). Restricting formation of decoy base pairing increases the BPP for P1 helix formation at low temperatures. Note that all decoy pairings involve G11 in base pairing. D1 and D3 also inhibit P1 helix formation by incorporating corresponding nucleotides into competing base pairings.
The 11/87 and 12/86 cross-junction base pairings predicted in Figure 5b are inconsistent with the formation of a pseudoknot interaction between the apical loop of P2 and J3/4, as observed in the aptamer. RNA Fold does not take account of the possibility of pseudoknot formation. Since formation of the pseudoknot would preclude other base pairings involving residues 87–90, we repeated the BPP calculations shown in Figure 5b with those residues constrained from base pairing altogether. SI Figure 9 shows that the overall competition trend between P1 and AT helices changes slightly, while the probability of a fifth hairpin, termed “P0” (grey), is increased slightly. Base pairs between residues 5 and 7 and J4/1 are represented prominently at low temperature. We repeated the calculations constraining base pairing from residues 107 and 108 in J4/1, along with pseudoknot residues in J3/4 as before. Figure 5c shows that with these constraints the temperature trend for the P1/AT helix competition is reversed. P0 helix formation is correlated with AT helix formation at low temperatures. The P0 helix is formed from a complementarity between residues in J1/2, including G11, and the 5’ region of the P1 helix. An RNA segment containing this sequence in isolation forms a structure which melts above 50 degrees (SI Figure 10). In the higher temperature range, P1 and AT helix BPPs are relatively flat as a function of temperature (Figure 5c).
B. Potential for P0 (D1) Helix Formation and “anti-P4” (D3) Helix in Putative SAM-I Riboswitch Sequences
We term the P0 helix “D1”, for “decoy 1”, since it is one of three interactions which Figure 5 predicts could stabilize AT formation by sequestering J1/2 and possibly 5’P1 helix residues. We investigated whether a P0 helix could be predicted in other SAM-I riboswitches. SI Figure 11a shows that amongst the 2828 SAM-I riboswitch sequences identified through Rfam64, the vast majority (>95%) contain 3–5 potential base pairs between J1/2 and the 5’ strand of the P1 helix. Given the constraints in the alignment (requiring the conserved AUC motif to sit in a loop region) the odds of identifying a stretch of three base pairs within random sequences in this location is less than 14%. Formation of the P0 helix in the metF SAM-I riboswitch sequence may explain the observation in a previous study 46 that the J1/2 region is less solvent accessible in the absence of SAM.
In addition to P0 helix formation, BPP calculations for the yitJ aptamer suggest the potential for a cross-junction pairing which we term decoy 2 (D2). D2 involves only two base pairs. The D2 base pairs involve conserved residues, but this conservation may reflect the role of these residues in SAM contacts and pseudoknot formation-both known to favor P1 helix formation.
Figure 5 and SI Figure 9 suggest pairing of 5’ residues with residues in J4/1. This base pairing, which appears to disrupt the P4 helix by strand invasion, we term D3 (decoy 3), since, like the P0 helix, it sequestors 5’ residues and stabilizes AT helix formation. We performed a sequence analysis for this “anti-P4” (SI Figure 11b). Because this analysis is complicated by the requirement to align distal sequences, we have examined only the sequences used for the seed alignment by Rfam. D3 has the potential to form in a larger fraction of SAM-I riboswitch sequences than in shuffled sequences of similar length and base composition (SI Figure 11b). The distribution in SI Figure 11b is bimodal, suggesting that only a subset of SAM-I riboswitches (including yitJ but not metF) have the potential to form this interaction.
D3 helix formation can explain the cleavage of residues in one strand of the P4 helix by Winkler et al in the absence of SAM28. This interpretation is further strengthened by a contrasting pattern of protection for the corresponding segment from in-line probing measurements of a yitJ riboswitch in which five 5’ residues are truncated from the P1 helix (Boyapati et al, manuscript in preparation).
C. Experimental Probing for P0 (D1) Helix Formation in the metF SAM-I Riboswitch
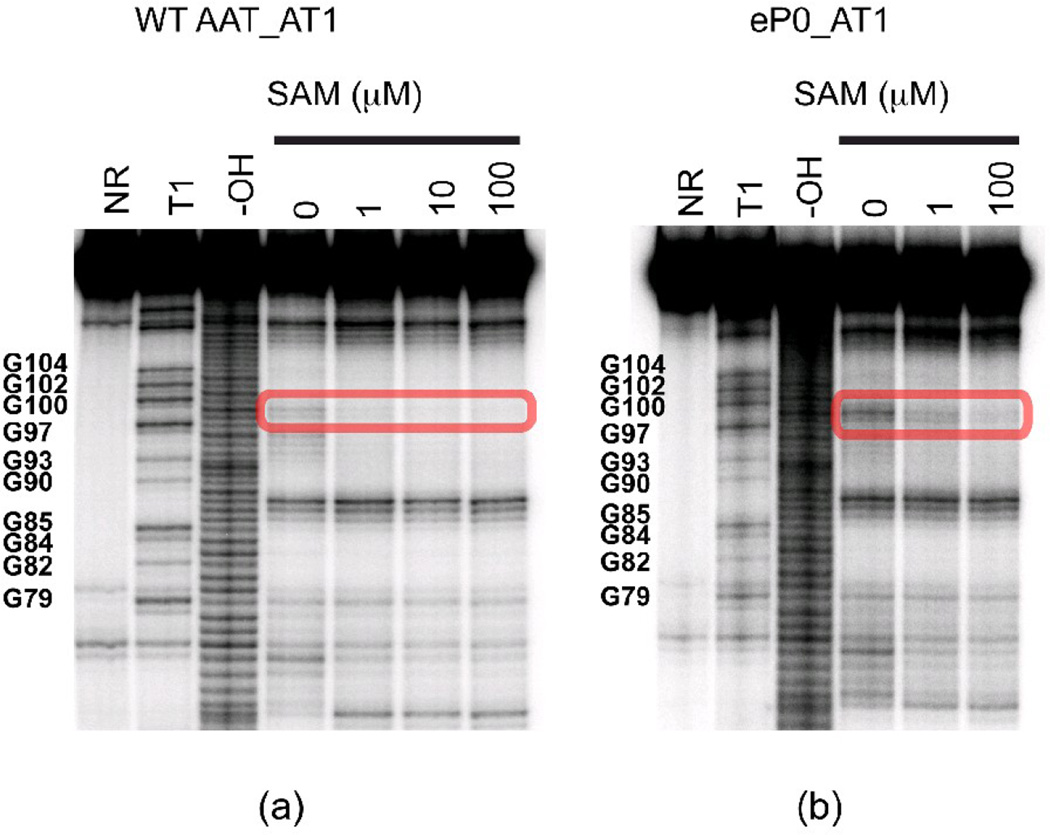
To test the possible impact of P0 (D1) helix formation on riboswitch conformation, we made similar transcripts to those shown in Figure 2 with different 3’ truncation points but containing a pair of point mutations in J1/2 and in the 5’ strand of the P1 helix (Figure 4a). This pair of mutations was designed to enhance P0 helix formation by converting an AU pair to a GC, while destabilizing the competing P1 helix by disrupting a single base pair. Figure 6 shows that introduction of the P0-helix-stabilizing/P1-helix-destabilizing mutation, as predicted, appears to increase the residual ON state cleavage pattern when SAM is added (positions 99 and 100, boxed). Gel mobility assays (SI Figure 6) also indicate that introducing the P0 mutation hinders formation of a fast-migrating SAM-induced conformation.
Figure 6. In-line probing of mutated metF SAM-I riboswitch RNA segments.
In-line probing experiment on wild-type (a) and eP0 mutant (b). The area highlighted in the red rectangle box is assigned to the GU highlighted by the blue arrow in Figure 2. In the context of sequences truncated at AT1, is opposite the fraying terminus of the AT1 helix. For the wild type sequence, this region has already been partially protected in the absence of SAM and fully protected in the presence of SAM. For the P0 mutant, the cleavage in this area is still persistent in the presence of low concentration SAM, indicating residual ON conformer formation.
RNA transcripts containing the P0 pair of mutations, still bind SAM but with lowered affinity (Figure 4b, “eP0” data points). Binding affinity is restored to P0 mutants by either lowering the temperature at which the dialysis incubation takes place (Figure 4c), or by introducing only a single mutation thus maintaining P1 helix formation and destabilizing P0 helix formation (Figure 4d, “dP0” data points).
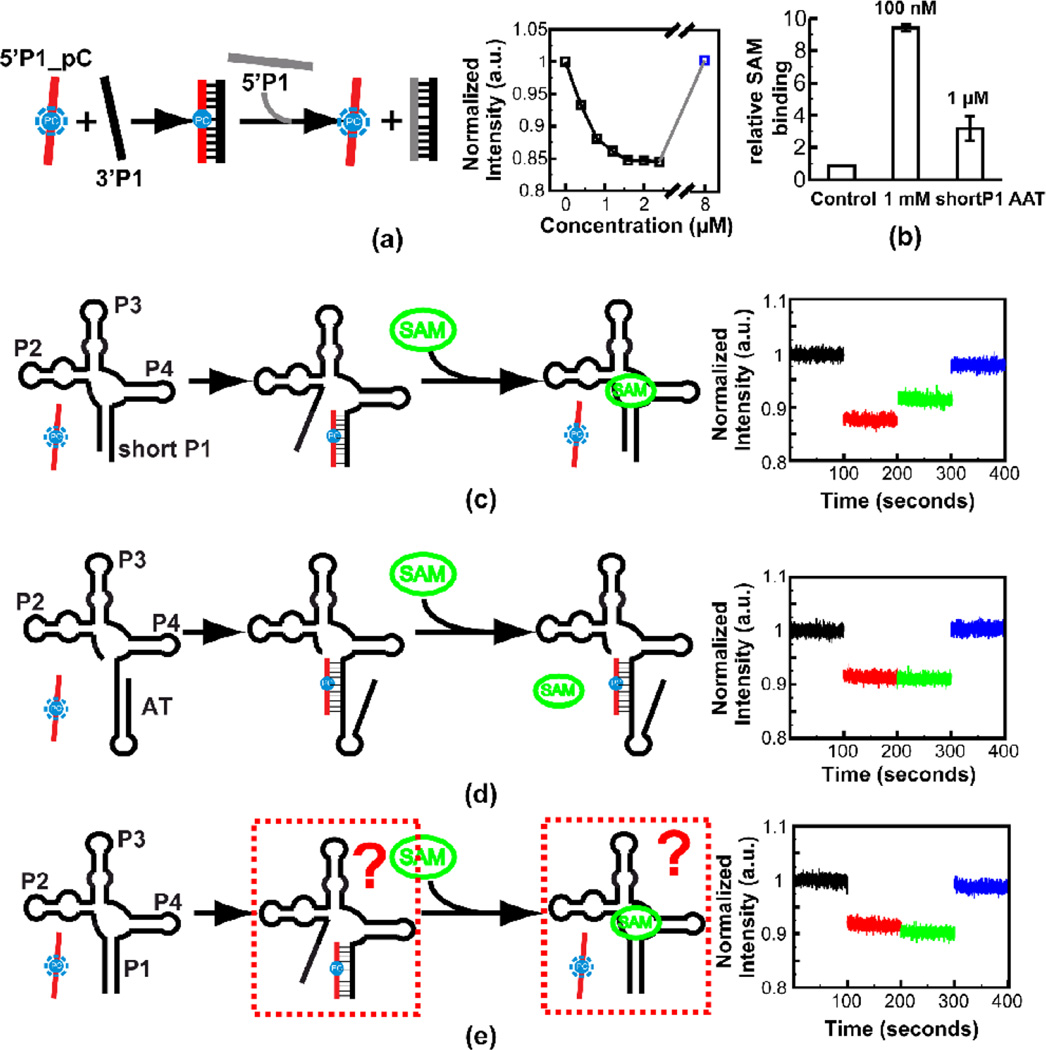
In-line probing measurements (Figure 6) indicated that the P0 mutation globally destabilizes OFF state formation. These measurements gave little indication, however, regarding the fate of residues near the 5’ ends of RNA constructs which were predicted to be involved in P0 helix formation. Pyrrolo C is a fluorescent analog of cytidine that can base pair with guanine 65. Upon incorporation within a base-paired A-form RNA helix, the fluorescence of pyrrolo C decreases 66. The reduced fluorescence signal is correlated with formation of hydrogen bonds as in a Watson-Crick GC base pair 67. We designed an assay (Figure 7) to utilize this molecular probe to compete with the 5’ segment of metF SAM-I riboswitch constructs for P1 helix formation. We reasoned that the capacity of this reporter to hybridize with riboswitch constructs would be an indicator of sequestration of these 5’ residues by decoy interactions.
Figure 7. Steady state fluorescence experiment on the metF SAM-I riboswitch RNA constructs.
(a) Titration of 3’ strand RNA of the P1 helix into solution containing Pyrrolo-C labeled 5’ strand RNA of the P1 helix. The fluorescence signal is recovered by adding excessive unlabeled 5’ strand RNA of the P1 helix (for the final (blue) data point, 8 µM unlabeled 5’ strand is added as a competitor). Schematic of the assay is shown at the left. (b) Equilibrium dialysis experiment to verify SAM binding of the WT AAT RNA construct with a short P1 helix (8 base pairs). The equilibrium dialysis experiment was performed with two different SAM concentrations—the RNA is in excess with 100 nM SAM or RNA to SAM is in 1:1 ratio with 1 µM SAM. (c) Experiment to mimic the effects of SAM on shifting the conformational state towards the “OFF” form. Addition of 20 µM SAM to complex of reporter with aptamer containing partial P1 helix, results in partial recovery of fluorescence due to displacement of the reporter. (d) Control experiment showing that SAM does not displace the reporter from hybridization to a construct that cannot bind SAM due to full truncation of 5’ P1 helix-forming residues. (e) Same experiment as (c) but with an RNA construct with full wild type P1 helix. In this case no recovery of fluorescence is observed when SAM is added.
First, we hybridized single strand RNAs analogous to an isolated P1 helix to verify the capacity of pyrrolo C as a conformation probe (Figure 7a). We observed a dose-dependent reduction in fluorescence upon titration of a complementary unlabeled single strand RNA to a pyrrolo C-containing analog of the 5’ strand of the P1 helix. Additionally, the fluorescence signal can be recovered upon addition of excess unlabeled competitor for the pyrrolo C labeled RNA oligo. This set of results confirms that pyrrolo C fluorescence is a sensitive indicator of duplex formation.
Next, to mimic the effect of intermolecular interaction in a single sequence RNA, a 1:1 ratio of pyrrolo C-labeled RNA oligo and SAM-I riboswitch RNA constructs were hybridized. We performed the experiment with two metF SAM-I riboswitch aptamer RNA constructs. In the first construct, 4 nucleotides at the 5’ end were truncated, which results in only 8 base pairs in the P1 helix, and prevents P0 helix formation. In this case, the equilibrium should favor the hybridization of pyrrolo C labeled RNA oligo to the 3’ strand region of the P1 helix. We used equilibrium dialysis to confirm the SAM binding capacity of the RNA construct with shortened P1 (Figure 7b).
The fluorescence assay on this RNA indeed displays the maximum fluorescence reduction (~12%) (Figure 7c, red). Moreover, the fluorescence signal can be partially recovered in the presence of excessive SAM (10:1 ratio) (Figure 7c, green), while no effect from SAM has been observed in the control experiment with an ON state construct (Figure 7d).
The same SAM effect, however, is not observed in the experiment with a second aptamer construct with a full length P1 helix (Figure 7e). This finding may be explained if the 5’ residues which were deleted in the first aptamer construct, participate in alternative base pairing, decreasing the likelihood of displacement of the hybridized pyrrolo-C. The lack of response to the presence of SAM in the full length P1 aptamer RNA construct may be due to P0 and/or cross-junction helix formation (Figure 5a).
G11 Base-Pairing Heterogeneity Is Predicted as a Key Factor for SAM-I Riboswitch Folding
A. BPP Predictions for Additional yitJ SAM-I Riboswitches
To extend our analysis to a larger SAM-I riboswitch sequence set, calculations similar to those shown in Figure 1 were performed for a series of ten other SAM-I riboswitch sequences in B. subtilis functionally characterized earlier by Tomsic et al27 (SI Figure 12). Variability in predicted P1 BPP as a function of temperature and transcript length is also observed amongst this group. Thus, the variability in predicted CTFS observed in Figure 1 is not solely due to the mesophilic or thermophilicorigin of the riboswitch sequence. Tuning of riboswitch folding characteristics is therefore predicted to vary amongst riboswitches within a single organism. One would then predict varied functional responses to the metabolite, as observed by Tomsic et al27, though this data does not prove a causal link between the two. Variability in predicted P1 helix BPP is greatest for transcripts which extend 12 nucleotides or more into the expression platform (SI Table 3).
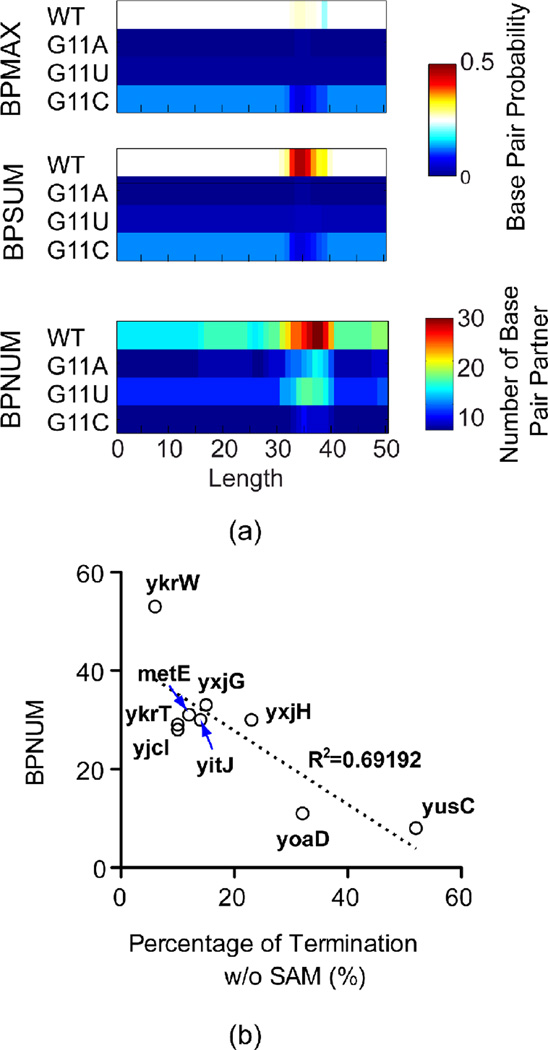
With the insights from BPP calculation, we revisited some experimental reports for SAM-I riboswitch function available in the literature. Lu and colleagues50 reported that three mutations at the G11 residue within J1/2 of the yitJ SAM-I riboswitch remove functional sensitivity to SAM. Paradoxically, termination for all three is enhanced even in the absence of SAM, though SAM binding is inhibited. The reported degree of enhancement from largest to smallest is G11C > G11U > G11A. Since P1 helix formation is believed to be coupled to terminator formation, we consider it to be a predictor of transcription termination. In the SI (SI Figure 13) we calculate changes in the BPP for P1 helix formation relative to wild type for each of the three mutants in question, as well as relative to each other. At the transcript length chosen in Figure 5 (which corresponds closely to the peak probability for AT helix formation) the mutations are predicted to increase the BPP for base pairs within the P1 helix, which would lead to increased transcription termination as observed 50. G11C has the greatest increase, followed by G11U and G11A. Therefore, the rank from our BPP calculation is consistent with the experimental result, assuming the correlation between constitutive P1 helix formation and constitutive transcription termination in the absence of SAM.
In Figure 8a, we observe that net probability of participation in any base pair (BPSUM) is much higher than the maximum probability for any one base pair (BPMAX) for residue 11 in the wild type yitJ sequence, indicating the pairing of G11 with alternate partners. Both BPSUM and BPMAX are very low for G11 in the mutants. The number of base pairing partners (BPNUM), is remarkably high for the WT sequence, and is reduced significantly for the mutants. The increase of base pair probability in the P1 helix in these mutants is therefore correlated with the elimination of alternative base pairing involving G11. Remarkably, BPNUM is also correlated with reported constitutive transcription termination for the set of B. subtilis SAM-I riboswitches characterized by Tomsic et al27 (Figure 8b). P3 helix mutations which were also reported to lead to constitutive transcription termination50, however, are predicted to diminish P1 helix BPPs or leave them unaffected, with little effect on G11 BPNUM (data not shown).
Figure 8. Conformatational heterogeneity for yitJ SAM-I riboswitch mutants at position G11 correlate, to some degree, with reported constitutive transcription termination.
(a) (Top) Maximum BPP (BPMAX) for any one of the possible base pairs involving position 11 as a function of transcript length, (Middle) The sum of BPP (BPSUM) for all base pairings which residue 11 participates in,90 The total number of predicted base pair partners with position 11 (BPNUM). In each panel the predicted values are plotted for the WT and for each of the three possible mutations at the same position. (b) Plot of possible base pair partners, BPNUM, for G11 versus the percentage of transcriptional termination without SAM for a set of SAM-I riboswitches from reference27.
B. Predicted Blockage of G11 Base Pairing Heterogeneity by SAM Binding
Above calculations, of course, have not taken account of the effect of ligand binding on SAM-I riboswitch secondary structure. It is well established that SAM binding favors formation of the OFF state secondary structure. From X-ray coordinates and studies of SAM binding to mutant riboswitches, we reasoned that we might derive some constraints on secondary structure base pairing as a consequence of SAM binding. We hypothesize that folding states that can bind SAM should satisfy the following minimal set of constraints: unpaired G11, no AU base pair in the AA-U internal loop in the P3 helix and 2 AU base pairs in the P1 helix. We calculated the BPP with the first and second constrained one at a time to test which constraint leads to the most significant enhancement of BPPs in the P1 helix (SI Figure 13b).
The constraint G11 unpaired causes the largest increase in BPPs in the P1 helix. The maximum difference is 0.663, which is observed at length 35nucleotides beyond the aptamer segment in the low temperature range (SI Figure 13b, left panel). SI Figure 13c displays BPPs for all base pairs in the P1 and the AT helix competition region at the same transcript length as in Figure 5a. The result indicates that the constraint G11 unpaired, which mimics one effect of SAM binding, increases the probability of P1 helix formation and reduces that of AT helix formation across the competing segment (solid vs. dotted lines). This result is consistent with the prediction that G11 mutants possess less heterogeneous base pairing patterns for residue 11, since it is suggested that AT helix formation is correlated with conformational heterogeneity in 5’ residues (Figure 5). Thus SAM may stabilize aptamer formation by preventing G11 from sampling those base pairings which stabilize the set of ON state secondary structures.
Summary of Evidence for Alternative SAM-1 Riboswitch ON State Secondary Structures
BPP calculations suggest a number of secondary structure folds for the yitJ and metF SAM-I riboswitches as shown above. At this stage, varying degrees of evidence can be cited for each of these alternative folds. For the AT2 and AT1 helices in the metF riboswitch, in-line probing data (Figure 2) provide strong evidence. For Decoy 3, there is strong evidence for its presence in the yitJ construct by comparing Figure 5 from Boyapati et al with Figure 1 from reference 28. Cleavage in one strand of the P4 helix observed in the latter (predicted to be displaced by D3) is suppressed when 5’ residues are truncated in the former. In addition, the D3 structure explains reduction in RNAse T1 cleavage of G108 upon addition of SAM within a full length yitJ SAM-I riboswitch construct44. Sequence analysis indicates that D3 will be present in only a subset of SAM-I riboswitches. Thus, between these two points, we have confirmed the hypothesis that individual SAM-I riboswitches have different conformer distributions in the ON state). A consequence of variability in ON conformation should be variable conformational response to SAM. Residues involved in Decoy 1 are at the 5’ end of the riboswitch, which is not readily characterized by the in-line probing data. Sequence conservation is strong and our flourescence data (figure 7) indicates that it may be induced by using a competing oligonucleotide. This element requires further study, perhaps incorporating tertiary structure in the analysis. Finally Decoy 2 is predicted in the calculations but at this stage not supported by experimental evidence.
A Schematic Model for Co-Transcriptional Folding of the metF and yitJ SAM-I Riboswitches
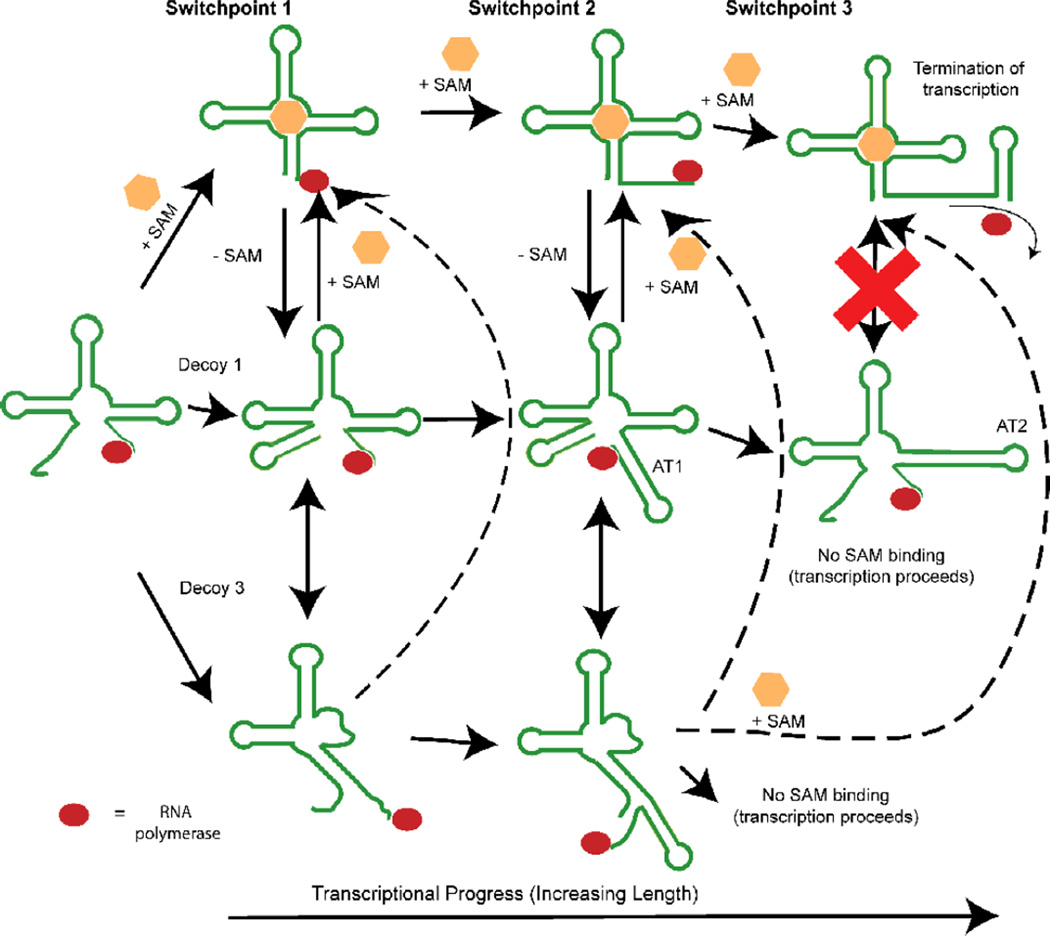
According to our BPP predictions, at least two, and, for one riboswitch, three switch points can be identified during the course of transcription. At each point, at least one conformational decision takes place that potentially biases the final transcriptional state of the riboswitch. Figure 9 illustrates the three decision points schematically.
Figure 9. Schematic illustration of three conformational decision points during the synthesis of the metF SAM-I riboswitch.
The first decision point determines whether 5’ residues engage in distal interactions, leading to formation of a P1 helix, or whether they are sequestered by “decoy” purine residues in junction regions. The second decision point involves a competition between P1 and AT1 helix formation. The final decision point, which is not apparent in the yitJ SAM-I riboswitch, converts the AT1 helix to an AT2 helix. Up to this final switch leading to the formation of the AT2 helix and the transcription ON state (lower right), it is proposed that SAM binding could readily perturb the equilibrium towards the transcription OFF state (upper right). Dotted arrows link decoy 3 to SAM-bound states, because no data is available to indicate the whether decoy 3 formation is reversible upon SAM binding. RNA polymerase is represented by a red oval.
First the decoy base pairings illustrated in Figure 5a may play a role in stabilizing the AT helix by occupying part of the switching region in the 5’ strand of the P1 helix. This competition is represented by switch point 1 in Figure 9. The ON state and OFF state secondary structures form the AT helix or the P1 helix, respectively. This choice represents the second structural switch (switch point 2 in Figure 9) as transcript length increases. Note that all of the decoy pairings illustrated in Figure 5a, involving 5’ pyrimidine residues, could take place early in transcription and hinder P1 helix formation. Note also that D3 will be present only in a subset of SAM-I riboswitches, as indicated in SI Figure 11.
In the third potential switch point, predicted only in the case of the metF SAM-I riboswitch, the AT helix can form two possible secondary structures. One, designated as AT1, resembling that proposed in reference56 may represent a switchable intermediate state (Figure 1a). The transcript length at which the BPP for P1 helix formation dips in Figure 1d ends with a stretch of Us for the metF SAM-I riboswitch sequence. A stretch of Us has been shown likely to be a transcriptional pausing site. This stretch of Us has the potential to form alternative base pairings resulting in either the AT1 model or the AT2 model in Figure 1. Hennelly and Sanbonmatsu46 found that a fluorescent reporter hybridization designed to mimic an AT2 structure could not be displaced by SAM binding. We found a similar result with a fluorescent reporter hybridized to the 3’ P1 helix-forming strand of a SAM-I riboswitch aptamer. Yet in-line probing and equilibrium dialysis measurements indicate that AT2-forming transcripts do bind SAM with lowered affinity at high ligand concentrations.
Here we observe that different alternative secondary structure elements are available in different SAM-I riboswitches (e. g. D3 in yitJ and AT2 in metF). One therefore predicts differing equilibrium folding behavior for each riboswitch as a function of transcript length, as calculated for the set of SAM-I riboswitches examined in reference27. The differing functional responses to SAM binding, and differing degrees of constitutive transcription termination in the absence of SAM observed by Tomsic and colleagues may be linked to a unique combination/configuration of conformational decoys within each individual riboswitch system.
Advantages and Limitations of BPP calculations and the FEL approach
Secondary structure prediction is the most popular computational application in RNA research 70. Multiple structures, such as suboptimal structures71, or a Boltzmann weighted ensemble72, can provide superior insight compared to a single secondary structure, especially for dynamic systems like riboswitches. Riboswitches typically act as “dimmer” switches allowing leaky expression, implying the simultaneous presence of permissive and non-permissive conformers. Overall, such behavior seems compatible with varying conformational population distributions, rather than a single lowest energy structure(MFE) on the one hand or an “all or none” picture on the other32.
The limitations of secondary structure predictions based upon partition function calculations are well documented42. (This point is discussed further in SI “Limitations of BPP calculations and the likely impact of pseudoknot formation”.) Nonetheless, several BPP predictions in this study are compatible with our experimental observations and literature reports, as discussed above and in SI.
Conclusions
We used BPP calculations, assuming a Boltzmann distribution of riboswitch conformers rather than a single MFE or a two-state “all or none” model, in order to understand the equilibrium behavior of SAM-I riboswitches. Following from Quart a et al’s35 study of the TPP riboswitch we carried out calculations as a function of transcript length. In our study we monitored BPP, and related parameters such as BPNUM, as readily interpretable metrics for the competition between P1 and AT helix formation.
A striking observation from the BPP predictions is the link between alternative base pairing configurations for residues in J1/2, and the overall equilibrium between the two dominant conformers associated with ON and OFF states respectively. From our observations, we can hypothesize that SAM binding perturbs the equilibrium through contacts with J1/2, and G11 in particular, thus blocking the aforementioned alternative or “decoy” pairings. Because the participation of the G11/J1/2 region in these decoy pairings is an important factor stabilizing the ON state, the ligand contact over a relatively small surface has a dramatic effect on global folding. In this way, consideration of the conformational heterogeneity of the ON state seems crucial for a full understanding of the mechanism of coupling between SAM binding and riboswitch folding.
The G11-mediated decoy base pairings play a critical role in the first of a set of tightly coupled “switches” which become active at different transcript lengths. In the case of the metF SAM-I riboswitch, we have identified at least three such switch points. We cannot say which of these represents the point at which SAM binding determines the ultimate transcriptional decision in vivo. This study has shown, however, how the switching decisions are linked through base pairing competitions, so that the effects of the early co-transcriptional folding decision can influence later stages.
Wild-type SAM-I riboswitches display variation in the regulatory response to SAM that cannot be solely explained by the binding affinity of the aptamer domain-this variability may be linked to precise genetic control on metabolic pathways73. We suggest that consideration of Boltzmann distributions of riboswitch conformers will be necessary to fully understand these tuning mechanisms. Functional SAM-I riboswitch sequences may need to maintain AT helix-stabilizing decoy interactions, just as adenine riboswitch sequences are selected to allow antitermination74. Additional factors such as cleavage of the 5’ segment 75, kinetic trapping30, tertiary contacts, other metabolites 76 etc., may be stabilizing the ON state conformation and maintaining expression of downstream genes in the absence of SAM under physiological conditions. Some of these factors may explain reports of constitutive termination in P3 mutants with reduced SAM binding50 which do not correlate with decreased predicted BPP for P1 helix formation.
It is believed that artificial riboswitches that fully mimic the natural riboswitches can be used as portable regulatory elements for synthetic biology. It has been shown that a tight control of the stoichiometry of different synthetic enzymes can greatly increase the yield of product79. New experimental techniques, such as SHAPE-seq, can be applied to study the differentiated regulatory mechanisms at a systematic level 80.
Materials and Methods
Sequence source and RNA preparation
The DNA templates for T. tengcongensismetF SAM-I riboswitches were amplified by overlapping PCR using oligos purchased from Integrated DNA Technologies, Inc.. The sequences of oligo templates and primers are listed in SI Table 1. RNA was transcribed from the PCR product by in vitro transcription using the following recipe: 100 µL PCR product, 50 µL 40 mM NTPs mix, 50 µL 400mg/mL PEG8000, 12.5 µL 20x transcription buffer (800 mM Tris-HCl, pH 8.1, 6 mg/mL spermidine, 0.2% Triton x-100, 15.6 mg/mL DTT), 11.5 µL 1M MgCl2, 2 µL 4 mg/mL in house T7 RNA polymerase, add d dH2O to 250 µL. Transcription and subsequent purification of the RNA was as described. The RNA sample was exchanged into 10 mM K Phosphate (pH 6.0), 10 mM KCl buffer and concentrated using Amicron. The RNA sample was stored at −20 °C.
Base pair probability calculation
BPP was calculated using McCaskill’s algorithm 38 implemented in Vienna RNA Package 1.8.5 85 with or without constraints. BPP values for specific base pairs were extracted from the dot plot files using in-house scripts. The net base pair probability for a single nucleotide was obtained by calculating the sum of base pair probabilities that the nucleotide was involved in (BPSUM). The number of possible base pairing partners (BPNUM) and the maximum BPP (BPMAX) among these base pairs were also extracted. These should be understood as predicted quantities throughout the manuscript. The scripts are available in the following link:http://dl.dropbox.com/u/24028147/scripts.tar.gz (refer to the README file in the tarball for more information).
Equilibrium Dialysis
Please see supplementary information for details of equilibrium dialysis measurements
In-line probing
In-line probing assays were performed following instructions in ref86. The procedures are briefly described here. The RNA was 5’ end labeled with γ-[32P] ATP and gel purified. The in-line probing assays were performed under the following buffer condition: 50 mM Tris-HCl, 100 mM KCl (pH 8.3).
The reactions were carried out with different Mg2+ concentration (0, 2 mM, 10 mM, 40 mM), in the absence or in the presence of 1, 10 and 100 µM SAM. The results are quantified using the software— SAFA 87. The intensity of bands in the presence of 100 µM SAM was normalized to that without SAM, as described in SI.
Steady State Fluorescence Experiment
The single strand of 5’P1 helix RNA with Pyrrolo C was obtained from the Keck Oligo Synthesis Lab in Yale University. The RNA is 2’ deprotected and desalted following the protocol from Glen Research (http://www.glenres.com). The steady-state fluorescence measurement is performed on a JASCO-FP6200 spectrofluorometer. The fluorescence intensity is recorded by exciting Pyrrolo C at 337 nm (5 nm band width) and monitoring the emission at 450 nm (10 nm band width) 66.
NMR Experiment
NMR samples were prepared by dialyzing RNA stock into NMR buffers using Amicon (Miillipore). NMR experiments were performed on a Varian 700 MHz spectrometer equipped with cryo-probe, or Inova 500 MHz. Experiments involving exchangeable protons were generally collected in 90% H2O/10% D2O at specified temperature. The buffer composition was in 10 mM potassium phosphate, 10 mM potassium chloride and 0.01 mM EDTA (pH 6.0) in 90% H2O/10% D2O, with additional components as specified in the result section. 1D spectra were processed in Mest ReNova LITE and multidimensional spectra were processed using NMR Pipe88.
Supplementary Material
Riboswitches control gene expression by coupling ligand binding to RNA folding
For the SAM-I riboswitch, a key residue engages in intramolecular decoy interactions.
SAM contacts with this key residue block decoy interactions, modifying the RNA fold.
The folding equilibrium is characterized at three potential decision points.
A small molecule can control RNA folding by blocking base-pairing promiscuity.
Acknowledgments
W.H. thanks Dr. Marcia Newcomer and Dr. Nathanial Gilbert for their help with using the JASCO-FP6200. W.H. also thanks Dr. Alain Laederach for his help with the SAFA software. We thank Alastair Murchie and Grover Waldrop for critical reading of the manuscript. This work has been supported in part by the Louisiana Experimental Program to Stimulate Competitive Research (EPSCOR), funded by the National Science Foundation and the Board of Regents Support Fund and by Award Number P20RR020159 from the National Center For Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Research Resources or the National Institutes of Health. W. H. was supported by Department of Biological Science, LSU.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith AM, Fuchs RT, Grundy FJ, Henkin T. Riboswitch RNAs: regulation of gene expression by direct monitoring of a physiological signal. RNA Biology. 2010;7:104–110. doi: 10.4161/rna.7.1.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annual Review of Biochemistry. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topp S, Gallivan JP. Emerging Applications of Riboswitches in Chemical Biology. ACS Chemical Biology. 2010;5:139–148. doi: 10.1021/cb900278x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muranaka N, Sharma V, Nomura Y, Yokobayashi Y. Efficient Design Strategy for Whole-Cell and Cell-Free Biosensors based on Engineered Riboswitches. Analytical Letters. 2009;42:108–122. [Google Scholar]
- 5.Wieland M, Hartig JS. Artificial Riboswitches: Synthetic mRNA-Based Regulators of Gene Expression. Chem Bio Chem. 2008;9:1873–1878. doi: 10.1002/cbic.200800154. [DOI] [PubMed] [Google Scholar]
- 6.Topp S, Gallivan JP. Guiding Bacteria with Small Molecules and RNA. Journal of the American Chemical Society. 2007;129:6807–6811. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biology. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chemical Biology. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 10.Anupam R, Nayek A, Green NJ, Grundy FJ, Henkin TM, Means JA, Bergmeier SC, Hines JV. 4,5-Disubstituted oxazolidinones: High affinity molecular effectors of RNA function. Bioorganic & Medicinal Chemistry Letters. 2008;18:3541–3544. doi: 10.1016/j.bmcl.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotech. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Cressina E, Leeper FJ, Smith AG, Abell C. A fragment-based approach to identifying ligands for riboswitches. ACS Chemical Biology. 2010;5:355–358. doi: 10.1021/cb9003139. [DOI] [PubMed] [Google Scholar]
- 13.Daldrop P, Reyes Francis E, Robinson David A, Hammond Colin M, Lilley David M, Batey Robert T, Brenk R. Novel Ligands for a Purine Riboswitch Discovered by RNA-Ligand Docking. Chemistry & Biology. 2011;18:324–335. doi: 10.1016/j.chembiol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 15.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci, USA. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription by bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 17.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 18.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 19.Barrick J, Breaker R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biology. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serganov A. The long and the short of riboswitches. Current Opinion in Structural Biology. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blouin S, Mulhbacher J, Penedo JC, Lafontaine DA. Riboswitches: Ancient and promising genetic regulators. ChemBioChem. 2009;10:400–416. doi: 10.1002/cbic.200800593. [DOI] [PubMed] [Google Scholar]
- 22.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annual Review of Biophysics. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 23.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rentmeister A, Mayer G, Kuhn N, Famulok M. Secondary structures and functional requirements for thiM riboswitches from Desulfovibrio vulgaris, Erwinia carotovora and Rhodobacter spheroides. Biological Chemistry. 2008;389:127–134. doi: 10.1515/BC.2008.012. [DOI] [PubMed] [Google Scholar]
- 25.Rentmeister A, Mayer G, Kuhn N, Famulok M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007;35:3713–3722. doi: 10.1093/nar/gkm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-Adenosylmethionine (SAM)-dependent riboswitches: S-Box elements in. Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. Journal of Bacteriology. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Struct. Biology. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 29.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 30.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 31.Cupal J, Flamm C, Renner A, Stadler PF. Proceedings International Conference On Intelligent Systems For Molecular Biology, 5Th, Halkidiki, Greece; 1997. [Google Scholar]
- 32.Hyeon C, Thirumalai D. Multiple Probes are Required to Explore and Control the Rugged Energy Landscape of RNA Hairpins. Journal of the American Chemical Society. 2008;130:1538–1539. doi: 10.1021/ja0771641. [DOI] [PubMed] [Google Scholar]
- 33.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–684. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Huang W, Maddineni S, Aboul-ela F, Jha S. Energy landscape analysis for regulatory RNA finding using scalable distributed cyberinfrastructure. Concurrency and Computation: Practice and Experience. 2011;23:2292–2304. [Google Scholar]
- 35.Quarta G, Kim N, Izzo JA, Schlick T. Analysis of riboswitch structure and function by an Energy Landscape framework. Journal of Molecular Biology. 2009;393:993–1003. doi: 10.1016/j.jmb.2009.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freyhult E, Moulton V, Clote P. Boltzmann probability of RNA structural neighbors and riboswitch detection. Bioinformatics. 2007;23:2054–2062. doi: 10.1093/bioinformatics/btm314. [DOI] [PubMed] [Google Scholar]
- 37.McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 38.McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–19. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 39.Huynen MA, Perelson A, Vieira WA, Stadler PF. Base pairing probabilities in a complete HIV-1 RNA. Journal of computational biology : a journal of computational molecular cell biology. 1996;3:253–274. doi: 10.1089/cmb.1996.3.253. [DOI] [PubMed] [Google Scholar]
- 40.Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 2010;6:e1001074. doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathews DH, Moss WN, Turner DH. Folding and Finding RNA Secondary Structure. Cold Spring Harbor Perspectives in Biology. 2010;2 doi: 10.1101/cshperspect.a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quarrier S, Martin JS, Davis-Neulander L, Beauregard A, Laederach A. Evaluation of the information content of RNA structure mapping data for secondary structure prediction. RNA. 2010;16:1108–1117. doi: 10.1261/rna.1988510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDaniel BA, Grundy FJ, Henkin TM. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Molecular Microbiology. 2005;57:1008–1021. doi: 10.1111/j.1365-2958.2005.04740.x. [DOI] [PubMed] [Google Scholar]
- 45.Montange RK, Mondragón E, van Tyne D, Garst AD, Ceres P, Batey RT. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. Journal of Molecular Biology. 2010;396:761–772. doi: 10.1016/j.jmb.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hennelly SP, Sanbonmatsu KY. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Research. 2011;39:2416–2431. doi: 10.1093/nar/gkq1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heppell B, Lafontaine DA. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry. 2008;47:1490–1499. doi: 10.1021/bi701164y. [DOI] [PubMed] [Google Scholar]
- 48.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci, USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler WC, Grundy FJ, Murphy BA, Henkin TM. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7:1165–1172. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu C, Ding F, Chowdhury A, Pradhan V, Tomsic J, Holmes WM, Henkin TM, Ke A. SAM recognition and conformational switching mechanism in the. Bacillus subtilis yitJ S Box/SAM-I Riboswitch. Journal of Molecular Biology. 2010;404:803–818. doi: 10.1016/j.jmb.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schroeder Kersten T, Daldrop P, Lilley David MJ. RNA Tertiary Interactions in a Riboswitch Stabilize the Structure of a Kink Turn. Structure. 2011;19:1233–1240. doi: 10.1016/j.str.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang W, Kim J, Jha S, Aboul-ela F. A mechanism for S-adenosyl methionine assisted formation of a riboswitch conformation: A small molecule with a strong arm. Nucleic Acids Res. 2009;37:6528–6539. doi: 10.1093/nar/gkp664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heppell B, Blouin S, Dussault A-M, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol. 2011;7:384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- 55.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 57.McDaniel BAM, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci, USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blouin S, Chinnappan R, Lafontaine DA. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic acids research. 2010 doi: 10.1093/nar/gkq1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY. Nonlocal helix formation is key to understanding S-Adenosylmethionine-1 riboswitch function. Biophysical Journal. 2009;96:L7–L9. doi: 10.1016/j.bpj.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baird NJ, Kulshina N, Ferré D’Amaré AR. Riboswitch function: Flipping the switch or tuning the dimmer? RNA Biology. 2010;7:328–332. doi: 10.4161/rna.7.3.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Batey RT. Recognition of S-adenosylmethionine by riboswitches. Wiley Interdisciplinary Reviews: RNA. 2011;2:299–311. doi: 10.1002/wrna.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim J, Winkler WC, Nakamura S, Scott V, Breaker RR. Molecular-recognition characteristics of SAM-binding riboswitches. Angewandte Chemie Int. Ed. 2006;45:964–968. doi: 10.1002/anie.200503198. [DOI] [PubMed] [Google Scholar]
- 63.Lu C, Smith AM, Ding F, Chowdhury A, Henkin TM, Ke A. Variable Sequences outside the Sam-Binding Core Critically Influence the Conformational Dynamics of the SAM-III/SMK Box Riboswitch. Journal of Molecular Biology. 2011;409:786–799. doi: 10.1016/j.jmb.2011.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic acids research. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berry DA, Jung KY, Wise DS, Sercel AD, Pearson WH, Mackie H, Randolph JB, Somers RL. Pyrrolo-dC and pyrrolo-C: fluorescent analogs of cytidine and 2 ‘-deoxycytidine for the study of oligonucleotides. Tetrahedron Letters. 2004;45:2457–2461. [Google Scholar]
- 66.Tinsley RA, Walter NG. Pyrrolo-C as a fluorescent probe for monitoring RNA secondary structure formation. RNA. 2006;12:522–529. doi: 10.1261/rna.2165806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hardman SJ, Botchway SW, Thompson KC. Evidence for a nonbase stacking effect for the environment-sensitive fluorescent base pyrrolo Cytosine--comparison with 2-aminopurine. Photochem Photobiol. 2008;84:1473–1479. doi: 10.1111/j.1751-1097.2008.00368.x. [DOI] [PubMed] [Google Scholar]
- 68.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An Allosteric Path to Transcription Termination. Molecular Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 69.Platt T. Transcription Termination and the Regulation of Gene Expression. Annual Review of Biochemistry. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 70.Mathews DH, Moss WN, Turner DH. Folding and finding RNA secondary structure. Cold Spring Harbor Perspectives in Biology. 2010;2:a003665. doi: 10.1101/cshperspect.a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuker M. Prediction of RNA secondary structure by energy minimization. Methods in Molecular Biology. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]
- 72.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Breaker RR. Riboswitches and the RNA World. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tremblay R, Lemay J-Fo, Blouin S, Mulhbacher Jrm, Bonneau Ãr, Legault P, Dupont P, Penedo JC, Lafontaine DA. Constitutive Regulatory Activity of an Evolutionarily Excluded Riboswitch Variant. Journal of Biological Chemistry. 2011;286:27406–27415. doi: 10.1074/jbc.M111.229047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seif E, Altman S. RNase P cleaves the adenine riboswitch and stabilizes pbuE mRNA in Bacillus subtilis. RNA. 2008;14:1237–1243. doi: 10.1261/rna.833408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watson PY, Fedor MJ. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nat Struct Mol Biol. 2011;18:359–363. doi: 10.1038/nsmb.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lucks JB, Qi L, Whitaker WR, Arkin AP. Toward scalable parts families for predictable design of biological circuits. Current opinion in microbiology. 2008;11:567–573. doi: 10.1016/j.mib.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 78.Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nature biotechnology. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 80.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yolov AA, Shabarova ZA. Constructing DNA by polymerase recombination. Nucleic acids research. 1990;18:3983–3986. doi: 10.1093/nar/18.13.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yon J, Fried M. Precise gene fusion by PCR. Nucleic acids research. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Varani G, Aboul-ela F, Allain FHT. NMR investigation of RNA structure. Progress in Nuclear Magnetic Resonance Spectroscopy. 1996;29:51–127. [Google Scholar]
- 84.Murchie AIH, Davis B, Isel C, Afshar M, Drysdale MJ, Bower J, Potter AJ, Starkey ID, Swarbrick TM, Mirza S, Prescott CD, Vaglio P, Aboul-ela F, Karn J. Structure-based Drug Design Targeting an Inactive RNA Conformation: Exploiting the Flexibility of HIV-1 TAR RNA. Journal of Molecular Biology. 2004;336:625–638. doi: 10.1016/j.jmb.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 85.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast Folding and Comparison of Rna Secondary Structures. Monatshefte Fur Chemie. 1994;125:167–188. [Google Scholar]
- 86.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods in Molecular Biology. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 87.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMR Pipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 89.Allain F, Varani G. Structure of the P1 helix from group I self-splicing introns. Journal Molecular Biology. 1995;250:333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- 90.Ley SVB, Bream IR, Jackson RN, Leach PS, Longbottom AG, Nesi DA, Scott M, Storer JS, Taylor RISJ. Multi-step organis synthesis using solid-supported reagents and scavengers: a new paradigm in chemical library generation. The Royal Society of Chemistry. 2000;2000:3815–4195. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.