Abstract
Artemisinin is highly effective against multidrug-resistant strains of Plasmodium falciparum, the etiological agent of the most severe form of malaria. However, a low level of accumulation of artemisinin in Artemisia annua is a major limitation for its production and delivery to malaria endemic areas of the world. While several strategies to enhance artemisinin have been extensively explored, enhancing storage capacity in trichome has not yet been considered. Therefore, trichome density was increased with the expression of β glucosidase (bgl1) gene in A. annua through Agrobacterium-mediated transformation. Transgene (bgl1) integration and transcript was confirmed by molecular analysis. Trichome density increased up to 20% in leaves and 66% in flowers of BGL1 transgenic plants than Artemisia control plants. High-performance liquid chromatography (HPLC, MS-TOF) data showed that artemisinin content increased up to 1.4% in leaf and 2.56% in flowers (g-1DW), similar to the highest yields achieved so far through metabolic engineering. Artemisinin was enhanced up to 5-fold in BGL1 transgenic flowers. The present study opens the possibility of increasing artemisinin content by manipulating trichomes density, which is a major reservoir of artemisinin. Combining biosynthetic pathway engineering with enhancing trichome density may further increase artemisinin yield in A. annua. Because oral feeding of Artemisia plant cells reduced parasitemia more efficiently than the purified drug, reduced drug resistance and cost of prohibitively expensive purification process, enhanced expression should play a key role in making this valuable drug affordable to treat malaria in a large global population that disproportionally impacts low-socioeconomic areas and underprivileged children.
Keywords: Genetic modification, artemisinin storage, malaria, global health, infectious disease
INTRODUCTION
Despite many advances in medical science, malaria is still a global issue with an estimated 198 million cases (uncertainty range: 124–283 million) occur every year worldwide (WHO, 2014), in which the majority of cases (82%) were in the African region followed by South-East Asia (12%) and Eastern Mediterranean Regions (4.5%, WHO, 2014). It is estimated that there were 584,000 malaria deaths globally in 2013 with about 82% of malaria cases and 90% of malaria deaths occurring in children under 5 years of age in the African region (WHO, 2014). Malaria is an entirely preventable and treatable disease, provided that the currently recommended interventions are properly implemented (WHO, 2014). The widely used antimalarial drug chloroquine, to control malaria, is now ineffective in most of falciparum malaria endemic areas, and is resistant to sulfadoxine-pyrimethamine (WHO, 2014).
Chinese researchers isolated artemisinin in the early 1970s from Artemisia annua, a herb of the Asteraceae family (Klayman, 1985). Although organic synthesis of artemisinin is possible, its low yield makes it economically nonviable as a mean of drug production (Avery et al. 1992). Thus, isolation of artemisinin from A. annua still is the best alternative (De Vries and Dien, 1996). Artemisinin is a sesquiterpenoid, produced in glandular trichomes of Artemisia annua (Covello 2008). Currently available commercial artemisinin is mainly extracted from wild plants which are subject to seasonal and environmental fluctuations. Thus, artemisinin concentration varies with genotype (Charles et al., 1990; Jain et al., 1996), plant tissue and time of harvesting (Laughlin, 1993, 1995; Morales et al., 1993; Ferreira and Janick, 1995), and is influenced by soil and climatological conditions (Van Geldre et al., 1997). Moreover, artemisinin content in A. annua is very low (0.01 - 1% dry weight, DW), and the demand for artemisinin is increasing along with the increasing number of people suffering from malaria (Qian et al., 2007). In an effort to increase artemisinin production, various approaches have been attempted including chemical synthesis (Avery et al., 1992) and genetic engineering of the pathway genes involved in artemisinin biosynthesis in A. annua (Vergauwe et al., 1996; Chen et al., 2000; Xie et al., 2001; Martin et al., 2003; Ro et al., 2006), but not much success has been achieved because of the complex nature of the gene regulation and expression in artemisinin biosynthesis. Thus, engineering of A. annua plants for increased artemisinin production still remains of great interest (Covello, 2008; Graham et al., 2010).
Monotherapy with artemisinin alone has failed to clear malarial parasites completely in some parts of the Asia (Sibley, 2015). Artemisinin-based combination treatments (ACTs) with other anti-malarial drugs are now commonly used and considered the best current treatment for uncomplicated falciparum malaria (WHO, 2014). Apart from malarial parasites, artemisinin also has antiviral activities (Romero et al., 2006) and can be used in treatment of hepatitis B (Romero et al., 2005) and for cancer treatment (Efferth et al., 2001). Artemisinin has been considered to be a relatively safe drug with no obvious adverse reactions or serious side effects, even for pregnant women (Dellicour et al., 2007). However, access of artemisinin based combination therapy to patients is inadequate in all surveyed countries (WHO, 2014).
The presence of glandular trichomes (GT) on the epidermis of the A. annua leaf is related to biosynthesis of several secondary metabolites and impacts accumulation of artemisinin (Kjær et al., 2012). However, molecular mechanism of artemisinin biosynthesis and the development of trichomes in A. annua remains poorly understood (Tan et al., 2015). A laser dissection study of GT of A. annua (Olsson et al. 2009) showed that key enzymes of artemisinin production were expressed exclusively in the two apical cells of GT. The initiation and development of GT in the genus Artemisia is completed at a very young primordial stage of the leaf development (Duke and Paul 1993), and density of GT of the fully developed leaf in A. annua is predetermined at an early primordial stage (Davies et al., 2009). The GT density observed highest at the maximum size of leaves; later density decreased rapidly, suggesting that some GT are ruptured after maturity (Lommen et al., 2006). In A. annua the number of GT increased as leaves reached full maturity and decreased thereafter (Arsenault et al. 2010). Duke and Paul (1993) showed that the cuticle covering the sub-cuticular storage area of the GT breaks in mature GTs and this influences density of intact trichomes. Further, a study on the floral morphology of A. annua has shown physiological maturity of GT in the inflorescence coincided with full bloom (Ferreira and Janick, 1995). In the past, attempts have been made to enhance GT to increase accumulation of artemisinin triggered by chemical or physical stress. However, such strategies have not been successful in A. annua ((Kjær et al., 2012). The presence of low density of GT in A. annua is responsible for the low accumulation of artemisinin (Kjær et al., 2012). Therefore, and there is a strong positive relationship between artemisinin content and GT densities. The relationship between GT densities, artemisinin content and key precursors was also confirmed by Graham et al. (2010).
Hormones play an important role in regulating plant growth and development (Davies, 2004). Plant vacuoles are known to be important reservoirs for storage of proteins, pigments, inactive hormones and cyanogenic glycosides, which play an important role in cellular homeostasis, plant defense mechanisms and detoxification (Taiz, 1992). Most of the phytohormones are synthesized within the cell and then transported to different cellular compartments, especially to the plant vacuoles. It has been reported that plant vacuoles are reservoirs for a lot of inactive hormone conjugates (Sembdner et al, 1994; Schneider et al, 1994). Hydrolytic enzymes like glucosidases releases active phytohormones by enzymatic cleavage of the glycosidic linkages in the glyco-conjugates (Jin et al 2011). Previously we reported that expression of β-glucosidase in tobacco chloroplasts increases biomass production as well as the number of trichomes (Jin et al, 2011). As pointed above, trichomes have been implicated in the biosynthesis of artemisinin; therefore, we explored the role of β-glucosidase in Artemisia annua. In the present study, bgl1 gene encoding β glucosidase enzyme from Trichoderma reesei tagged with vacuole targeting sequence chitinase at the C terminus was expressed in A. annua. The transgenic lines were characterized and transferred to greenhouse and evaluated for density of trichomes and artemisinin content in leaves and flowers.
RESULTS
Creation and characterization of Artemisia annua transgenic lines
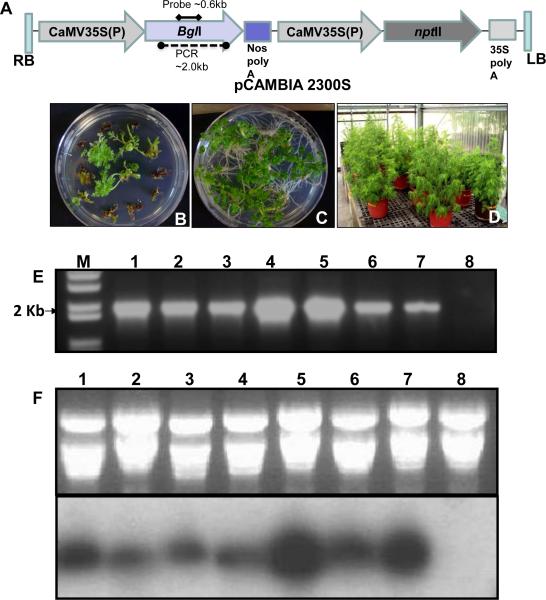
The binary transformation vector containing bgl1 gene with vacuolar targeting sequence (Fig. 1A) was used to transform Artemisia annua leaf explants using Agrobacterium tumefaciens LBA 4404. The transformation protocol was based on an earlier report with minor modifications (Han et al., 2005). After 4-6 weeks, shoots were regenerated from the leaf explants on selection medium (Fig. 1B) and resistant shoots were transferred to rooting medium containing kanamycin 50-mg/l (Fig. 1C). PCR analysis confirmed integration of the bgl1 gene into A. annua transgenic lines while no amplification was observed in the untransformed plants (Fig. 1E). These transgenic lines were transferred to a greenhouse for further growth and characterization. There was no phenotypic difference between transgenic and untransformed plants (Fig. 1D). RNA was extracted from the PCR positive transgenic lines and Bgl1 transcript was analyzed by northern blot with bgl1 gene specific probe (~0.6 Kb as shown in Fig. 1A). Bgl1 transcript was detected in all A. annua transgenic lines with various levels of expression, whereas no transcript was observed in untransformed plants (Fig. 1F).
Figure 1. Transformation and molecular characterization of A. annua transgenic plants.
(A) Schematic representation Agrobacterium transformation vector. (B) Putative Bgl1 transformants growing on regeneration selection medium containing kanamycin. (C) Bgl1 A. annua transgenic lines on rooting medium containing kanamycin. (D) Bgl1 transgenic lines grown in the greenhouse. (E) Confirmation of bgl1 gene integration by PCR. (F) Confirmation of Bgl1 transcript level of different transgenic lines by northern blot analysis using bgl1 gene probe (Lanes 1–7: Bgl1 transgenic lines; Lane 8: Untransformed plant). GOI: His Bgl1 Chitinase CTPP; Bgl1 Chitinase CTPP.
Beta-glucosidase activity in transgenic lines and transgene copy number
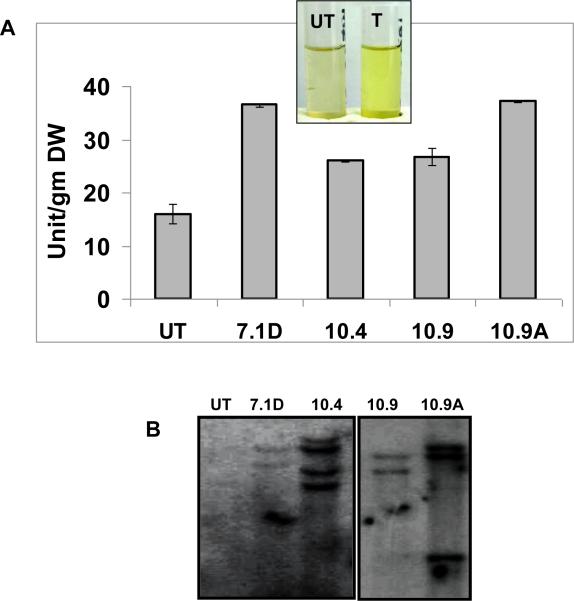
Leaf samples were collected from transgenic and untransformed plants. β-glucosidase activity was measured using p-nitrophenyl β-D-glucopyranoside (pNPG) as the substrate. One unit of β-glucosidase is defined as the amount of enzyme that released 1 μmol of p-nitrophenol from pNPG substrate under identical assay conditions. A. annua transgenic lines (26-33 units per gram fresh leaves) showed a 1.6-2 fold increase in β-glucosidase activity when compared to untransformed plants (16 units per gram mature fresh leaves) (Fig. 2A). A. annua plants showing higher activity of BGL1 enzyme activity were further evaluated for copy number of transgene Bgl1 by Southern blots. Transgenic lines showed multiple bgl1 transgene copies when probed using bgl1 gene specific probe (~0.6 kb) (Fig. 2B).
Figure 2. Evaluation of A. annua BGL-1 enzyme yield and transgene copy number.
One unit of BGL-1 enzyme is defined as the amount of enzyme that released 1 μmol of p-nitrophenol from pNPG substrate under the assay conditions described in “Materials and Methods. UT :untransformed plants; T: Bgl1 transgenic plant. (B) The plant showing higher activity of enzyme BGL1 in comparison to UT were further tested for copy number of Bgl1 by Southern blot using bgl1 gene specific probe (~0.6 kb) cut out from the vector using unique restriction sites EcoRI and BamHI. UT: untransformed; A. annua transgenic lines 7.1D, 10.4, 10.9, 10.9A.
Evaluation of trichome structure and density
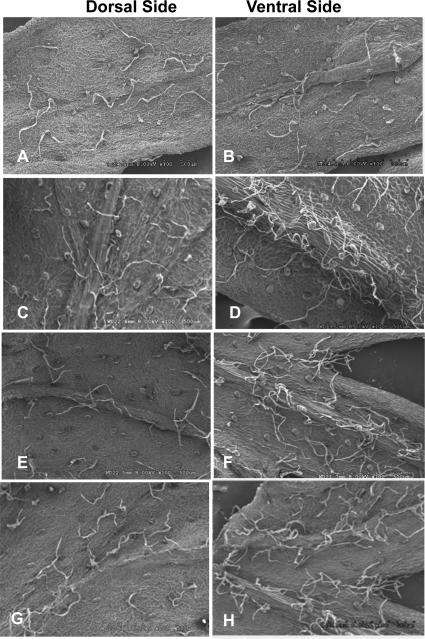
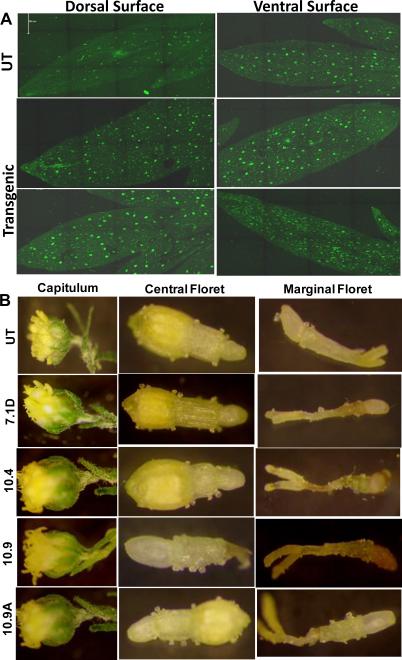
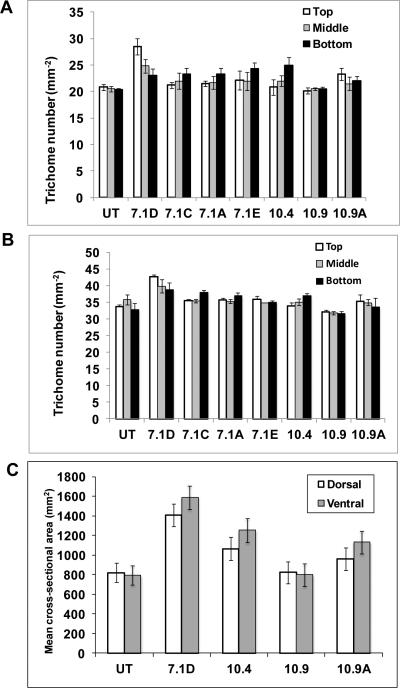
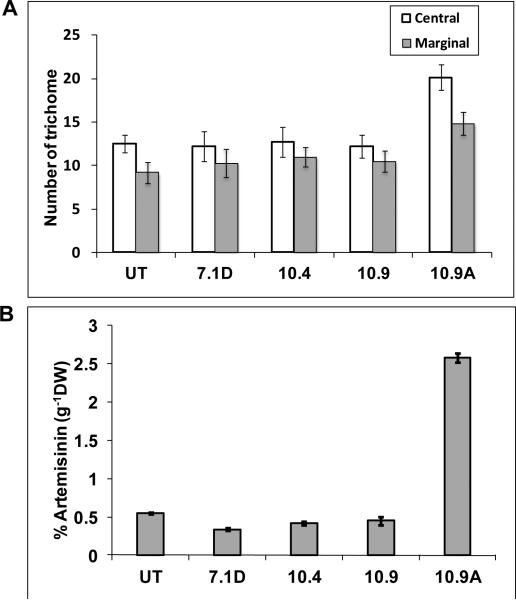
Trichomes in A. annua plants are epidermal outgrowth may have various functions depending on the plant species and organs. However, the function of the GTs of this species is unclear (Jessing et al., 2013). Scanning electron microscopy analysis revealed two types of trichomes, glandular and filamentous trichomes on both surfaces of leaves (Fig 3A-H). Similar results were obtained when leaf surface was observed under confocal laser microscope (Fig. 4A). In both leaf surfaces, density of glandular trichomes was 10-20% higher in Bgl1 transgenic lines when compared to untransformed plants (Fig. 5A & B). The elliptical cross-sectional area of the glandular trichomes was also measured. Transgenic lines showed up to 2-fold increase in cross-sectional area when compared to untransformed plants (Fig. 5C). The density of trichomes in flowers of Bgl1 transgenic lines was also observed under a stereomicroscope and compared with untransformed plants (Fig. 4B). One capitulum contains 10-14 central and 8-10 marginal florets. The number of trichomes was recorded for both florets. In A. annua transgenic lines, increase in trichome density in flowers was up to 66 % when compared to untransformed plants (Fig. 7A).
Figure 3. Evaluation of trichome density on leaf surface by scanning electron microscopy.
(A and B) Untransformed plant; (C to H) Bgl1 A. annua transgenic lines randomly selected for analysis showing higher trichome density.
Figure 4. Evaluation of trichome density in A. annua leaves and flowers.
(A) Density of trichomes on both the leaf surface observed under confocal microscope. (B) Density of trichomes in flowers. Three individual capitulum were collected and central and marginal florets were analyzed under light microscope. One capitulum contains 10-14 central and 8-10 marginal florets. The number of trichome was recorded for both florets. UT: untransformed plants; 7.1D, 10.4, 10.9 and 10.9A: Bgl1 A. annua transgenic plants.
Figure 5. Comparison of trichome density and cross-sectional area of leaf samples.
(A) Density of trichomes on dorsal surface (A) and ventral surface (B). Gland size (C). Numbers in the ordinate indicate the mean cross-sectional area of the gland. Elliptical cross-sectional areas of the glandular trichomes, the major (a) and minor (b) axes of the ellipse were measured with the program ImageJ (version 1.37; http://rsb.info.nih.gov/ij/). The area was calculated according to the formula, area = ABπ/4. UT: untransformed plants; 7.1A, 7.1C, 7.1D, 7.1E, 10.4, 10.9 and 10.9A: Bgl1 transgenic plants.
Figure 7. Number of trichome and quantification of artemisinin.
(A) Evaluation of trichome density in central and marginal florets. (B) Comparison of artemisinin content in Bgl1 transgenic and untransformed plants. UT: untransformed plants; 7.1D, 10.4, 10.9 and 10.9A: Bgl1 transgenic plants.
Evaluation of Artemisinin content
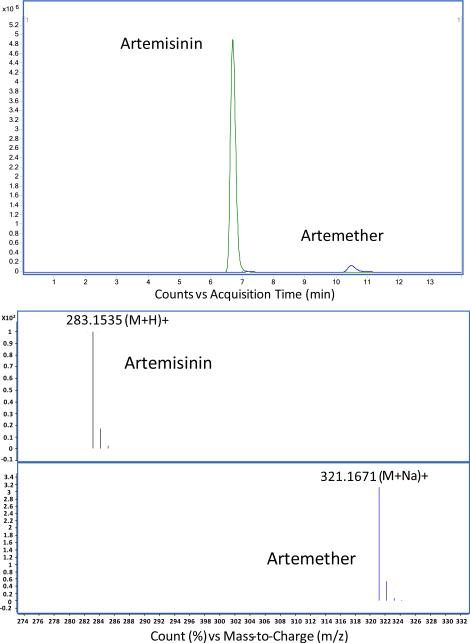
A high-performance liquid chromatography (HPLC) system equipped with a 6224A time of flight mass spectrometer (MS-TOF) was used to quantify artemisinin content in leaves harvested at different developmental changes and flowers. Artemether was used as an internal standard in all the samples (Fig. 6). The integrated extracted ion chromatograms (EIC) of artemisinin [M+H]+adduct and artemether [M+Na]+ adduct peaks were obtained. The artemisinin content was variable among in A. annua transgenic lines and also depended on the age and position of the harvested leaf samples (Table 1). The artemisinin content increased was up to 66.5% when compared to untransformed plants depending on the position and age of the leaf, while in some positions artemisinin content was lower than untransformed plants (Table 2). Highest artemisinin content was up to 1.4 % (g-1DW) in transgenic lines when compared to 1.22% (g-1DW) in untransformed plants. Artemisinin content was also quantified in flowers. In one particular transgenic line 10.9A, artemisinin content was 5 fold (2.56% g-1DW) more than untransformed flower (0.55% g-1DW, Fig. 7). Chromatogram of artemisinin and internal standard artemether is shown in Fig. 6.
Figure 6.
Chromatogram of artemisinin and internal standard artemether analyzed by high-performance liquid chromatography (HPLC) system equipped with a 6224A time of flight mass spectrometer (MS-TOF)
Table 1.
Quantification of Artemisinin from different leaf samples of Bgl1 transgenic and untransformed plants (% Artemisinin/gram DW). UT: Untransformed; 7.1D, 7.1A, 7.1C, 7.1E, 10.4, 10.9 and 10.9A: Bgl1 transgenic lines were grown under same conditions in the greenhouse. Quantification of artemisinin is provided as an average ± standard deviation of 3 replicates.
| Position | UT | 7.1D | 7.1A | 7.1C | 7.1E | 10.4 | 10.9 | 10.9A |
|---|---|---|---|---|---|---|---|---|
| Top (Young) | 0.84±0.02 | 1.21±0.04 | 0.87±0.03 | 0.33±0.22 | 0.87±0.03 | 0.78±0.02 | 0.81±0.01 | 1.40±0.02 |
| Top (Mature) | 1.15±0.02 | 1.37±0.06 | 0.68±0.07 | 0.42±0.12 | 0.87±0.01 | 0.72±0.09 | 1.11±0.01 | 0.93±0.02 |
| Top (Old) | 1.11±0.01 | 1.39±0.03 | 0.86±0.05 | 0.67±0.02 | 0.64±0.2 | 0.74±0.10 | 0.95±0.03 | 0.69±0.01 |
| Middle (Young) | 1.17±0.03 | 1.00±0.06 | 0.55±0.06 | 0.49±0.09 | 0.88±0.04 | 0.73±0.03 | 0.53±0.01 | 1.06±0.02 |
| Middle (Mature) | 0.97±0.03 | 1.31±0.03 | 0.82±0.03 | 0.64±0.05 | 1.00±0.1 | 0.77±0.10 | 0.88±0.02 | 0.63±0.03 |
| Middle (Old) | 1.02±0.04 | 1.11±0.01 | 1.12±0.06 | 1.03±0.07 | 1.26±0.05 | 0.85±0.01 | 0.88±0.07 | 0.98±0.01 |
| Bottom (Young) | 1.22±0.03 | 0.87±0.01 | 0.98±0.01 | 0.90±0.28 | 0.89±0.1 | 1.01±0.02 | 0.53±0.10 | 1.32±0.02 |
| Bottom (Mature) | 0.93±0.03 | 1.15±0.06 | 0.87±0.02 | 1.11±0.03 | 0.86±0.02 | 0.92±0.01 | 0.68±0.09 | 0.72±0.04 |
| Bottom (Old) | 0.82±0.02 | 0.90±0.06 | 1.17±0.02 | 1.24±0.01 | 1.10±0.35 | 1.18±0.03 | 0.69±0.01 | 0.84±0.01 |
Table 2.
Highest percentage increases of artemisinin content per gram DW in Bgl1 transgenic lines. Results were analyzed using t-test (www.graphpad.com) on artemisinin content in transgenic lines in leaves at different stages of development when compared to untransformed plants at identical stages of development and grown under similar conditions. A single asterisk represents significant difference at P< 0.01 and triple asterisk represents a highly significant difference at P< 0.0001 between transgenic and untransformed lines.
| 7.1D | 7.1A | 7.1C | 7.1E | 10.4 | 10.9A |
|---|---|---|---|---|---|
| 43% in top young leaf*** | 42% in bottom old leaf*** | 51% in bottom old leaf*** | 34% in bottom old leaf*** | 43% in bottom old leaf*** | 66.5% in top young leaf*** |
DISCUSSION
Although the World Health Organization (WHO) recommends ACT as first-line treatment for uncomplicated malaria caused by Plasmodium falciparum, there is great concern for limitation of supplying artemisinin over 100 million treatments per annum in order to meet the global requirement (Graham et al., 2010). It has been shown that the production of artemisinin occurs in specialized 10-celled biseriate glandular trichomes present on the leaves, stems and inflorescences of A. annua plants (Duke et al., 1994; Van Nieuwerburgh et al., 2006). Recently it has been shown that expression of β-glucosidase in tobacco plants releases active hormones, including GAs, from their conjugates stored within plastids and facilitate a 740% to 1,033% increase in trichome density in transplastomic lines and also increased leaf area (Jin et al., 2011). Therefore, substances that improve the leaf development and shoot growth are presumed to increase artemisinin yield. In an effort to increase the number of trichomes, Artemisia bgl1 transgenic lines were regenerated via Agrobacterium-mediated transformation. Many T0 transgenic lines were obtained and Bgl1 gene integration and copy number was confirmed by PCR and Southern blot, respectively. Northern blot analysis showed different levels of Bgl1 transcript between each individual line. This variation in transcript levels is not uncommon in nuclear transgenic lines and could be due to their site of integration (position effect), gene silencing, etc. However, Bgl1 enzyme unit per g/DW was 2 fold higher in transgenic plants than untransformed plants, which suggests that transgene expressed in Artemisia annua is functional.
Results of Van Nieuwerburgh's group (2006) supports the hypothesis that artemisinin and some of its structural analogs present in the leaves A. annua L. are localized entirely within the sub-cuticular space of glands on the leaf surface. The number of GT on leaves is directly proportional to artemisinin accumulation (Arsenault et al., 2010; Graham et al., 2010; Kjær et al., 2012). In our study, artemisinin content was higher in the sub-cuticular space of GTs of Bgl1-transformed plants than untransformed control plants. This suggests that larger size of the GTs may be associated with larger sub-cuticular space resulted due to BGL1 expression.
Scanning electron microscopic and confocal microscopic analysis have shown that density of glandular trichomes in leaf samples was 10-20% higher in most of the bgl1 transgenic lines when compared to untransformed plants (Fig. 5A & B). It has been reported that the application of phytohormones can increase cross sectional area of the trichome when compared to untreated Artemisia plants (Maes et al., 2011). In order to assess glandular trichome size, 4 transgenic lines were selected and a cross-sectional area of each individual glandular trichomes was measured. Notably, in addition to gland density, gland size also differed between each individual transgenic line (Fig. 5C). A. annua transgenic lines have larger glandular trichomes than untransformed plants. The presence and development of glandular trichomes in the inflorescences was also associated with artemisinin production based on extraction studies (Ferreira and Janick 1995). The glandular trichomes are greater in the corolla and receptacles florets than in leaves, stems, or bracts. Although these glands are present at the early stage of development on both leaves and inflorescences, artemisinin increases at anthesis, suggesting that it accumulates as the glands reach physiological maturity, a stage which coincides with the end of cell expansion in floret development (Ferreira and Janick 1995). Thus, mature flowers were dissected under stereomicroscope and trichome density was analyzed. Mature flowers of A. annua transgenic lines showed 11- 66% increase in trichome density when compared to untransformed plants. Large differences in artemisinin content have been reported, depending on genotype (variety), season (environmental changes), cultivation condition (soil nature) and plant developmental stage (Ferreira et al., 1995; Wallaart et al., 2000; Delabays et al., 2001; Yang et al., 2009). HPLC (MS-TOF) data shows variation in individual A. annua plants and different samples. This is in agreement with the various reports, which explains the influence of developmental stages of plants on artemisinin content. Some researchers believe that the artemisinin content reaches its peak before flowering (Abdin et al., 2003; Mauji Ram et al., 2010), while others have indicated the peak occurs at the full-flowering stage (Ferreira et al., 1995a). From the data presented here, it is clear that the accumulation of artemisinin is closely correlated with trichome density and stages of leaves vary between individual plants. This variation in trichome density among A. annua transgenic lines could be due to their site of integration (position effect), gene silencing, etc. However, considerable evidence suggests that the variation in artemisinin content in different organs is correlated with the density of trichomes. This is in agreement with our data although variations are also observed within individual lines. The association of artemisinin with glandular trichome sequestration explains why artemisinin was not detected in parts of the plant that do not bear glands, such as pollen or roots (Ferreira et al. 1995a) or in a glandless biotype (Duke et al. 1994). Recently, Jessing et al. (2013) reported that roots could produce very small amounts of artemisinin.
Several genes are involved in artemisinin biosynthetic pathways, which could be manipulated to increase the artemisinin content. Since artemisinin is stored in trichomes, there is need for storage room for large amounts of artemisinin synthesized from the over-expression of these genes. Thus the present study will open the possibility of increasing artemisinin content by manipulating trichome density. The present study is based on T0 transgenic lines, but further investigation is needed in T2/T3 homozygous transgenic lines where uniform trichome density and Bgl1 expression is expected. In order to achieve an even higher trichome density, chloroplast genome of A. annua should be transformed using the Bgl1 gene. However, chloroplast transformation has not yet been developed for this species.
Several questions should be addressed in further research. Why engineering of A. annua for increasing artemisinin yield has limited success so far? One answer is that it is due to a bottleneck of GT densities in foliar surface of A. annua as we observed in the present study. Thus, genetic modifications to the enzymatic pathway and enhancement of GT density would be important for enhanced artemisinin production. While several strategies to enhance artemisinin have been extensively explored, enhancing storage capacity has not yet been critically considered. This study shows that when GT density was increased artemisinin content was also increased in A. annua. In the wild type, densities of GT of the fully developed leaf in A. annua are predetermined at the early primordial stage (Davies et al., 2009), which may be a disadvantageous factor to enhance the yield. On an average A. annua produces ~ 0.1 to 1% artemisinin in the wild type depending on location and climatic conditions. Bgl1 transgenics produced artemisinin content on per gram dry weight (g-1DW) basis 1.4% (leaf) and 2.56% (flower), which is similar to the highest yield ~1.5% (15.1 mg/g DW) reported so far via overexpression of three (ADS, CYP71AV1 and CPR) transgenes (Lu et al. 2013). Combining these two approaches, an increase of GT densities via Bgl1 gene and introduction artemisinin biosynthetic pathway may be advantageous for further enhancement of artemisinin yield in A. annua. Artemisinin is lost during the rainy season due to disruption of trichomes. Artemisinin was measured up to 8 mgL-1 in rainy water suggesting that rain drops disrupt artemisinin-containing trichomes (Soetaert et al., 2013), again underscoring their importance. Relatively little is known about the regulation of artemisinin biosynthesis in A. annua plants, and the development of the specialized organ like the glandular trichome. A. annua transgenic lines created in this study could be useful to understand some of these important questions.
Despite progress made in understanding the malarial parasite, expression of malarial vaccine antigens (Davoodi-Semiromi et al 2010; Jones et al 2013), ability to produce low cost vaccines free of cold storage (Kwon et al 2013, Lakshmi et al 2013), developing an effective human vaccine against malaria is still elusive. Until then, drugs like artemisinin are the only hope to save lives. A more exciting new development in this field is oral delivery of dried A. annua leaves. When fed orally A. annua reduces parasitemia in rodent malaria more effectively than purified artemisinin drug (Elfawal et al., 2012). Most interestingly, feeding Artemisia annua plant cells to suitable animal models overcomes malarial parasite resistance more effectively than pure artemisinin. Stable resistance to this drug evolved three times slower than stable resistance to pure artemisinin drug (Elfawal et al., 2015).
Unfortunately, these reports on oral feeding of Artemisia (Elfawal et al 2012, 2015) did not explain the concept of bioencapsulation of this drug within plant cells but interpreted that this could be due to protection of flavonoids or other antioxidants present in plants or animal feed (which should be available for control animals fed with plant materials) or any other synergistic effect. However, we have previously shown that plant cells protect biopharmaceuticals from acids and enzymes in the stomach via bioencapsulation but gut microbes release these drugs in the gut lumen by the action of commensal microbes, where they are absorbed and delivered to the blood circulation system (Limaye et al., 2006; Kwon et al., 2013a,b; Sherman et al., 2014; Kohli et al., 2014; Shil et al., 2014). Human digestive enzymes are incapable of breaking down glycosidic bonds in carbohydrates that make up plant cell wall. However, when intact plant cells containing drugs reach the gut, commensal microbes digest the plant cell wall and release drugs. Gut microbes have evolved to breakdown every component of plant cell wall (Martens et al., 2011; Flint et al., 2008). Therefore, artemisinin bioencapsulated in plant cells provides higher efficacy than pure artemisinin drug delivered orally because it is degraded by gastric enzymes or acids minimizing, thereby reducing bioavailability. Most importantly, the primary cost of artemisinin is not in the growth of plants per se but the purification process is expensive. Elimination of this step should make artemisinin affordable to global population that disproportionally impacts those living in low-socioeconomic areas and underprivileged children. The demand for artemisinin is quite high (170 metric tons required annually for India). Although this drug is heavily subsidized, it still costs 10-40 times higher than other antimalarial drugs. Considering the fact that one third of global population earn <$2 per day, increasing artemisinin content and utilizing novel delivery methods are highly desirable.
MATERIALS AND METHODS
Plant materials and tissue culture conditions
Seeds of Artemisia annua L. were surface sterilized by soaking in 1.5% NaOCl for 10 min after immersing in 70% ethanol for 1 min. Seeds were rinsed 5 times with sterile distilled water and then germinated on MS medium (MS salt containing 2 mg/l glycine, 0.1 mg/l thiamine, 0.5 mg/l pyridoxin, 0.5 mg/l nicotinic acid, 100 mg/l myo-inositol, 3% (w/v) sucrose, 0.4 % (w/v) phytoblend, pH 6.1).
Construction of transformation vector
Full-length cDNA of bgl1 gene (U09580) was amplified from genomic DNA of T. reesei (American Type Culture Collection) as described previously (Jin et al., 2011) and fused with vacuole targeting sequence Chitinase (CTPP) at C terminus. The fusion gene was cloned into a pCR BluntII Topo vector (Invitrogen). After sequence confirmation, the fusion gene was digested with Sma1 and Xba1 and ligated into a pCAMBIA 2300S transformation vector. The neomycin phosphotransferase (nptII) was included in the construct to serve as a selectable marker. The nptII gene confers resistance to antibiotic kanamycin. Both genes were regulated by a 35S CaMV promoter. A polyA tail is included downstream to both genes. The complete expression cassette was then transformed into the Agrobacterium tumefaciens LBA 4404.
Transformation of A. annua
Agrobacterium tumefaciens cells were cultured in YEP agar medium supplemented with antibiotics rifamycin (50 mg/l) and Kanamycin (50 mg/l) and were incubated overnight at 25°C. A loop full of the inoculum was grown in YEP liquid medium supplemented with 50 mg/l of rifamycin/kanamycin and incubated overnight at 25°C. Transformed cells were collected by centrifugation. The pellet was then resuspended in Artemisia regeneration liquid medium and then diluted to obtain an OD of 0.1 at 600 nm. Leaf explants were harvested from 10 to 14 week old in vitro germinated Artemisia plants. Leaf pieces were inoculated with Agrobacterium suspension culture for 20 min and co-cultured for 2 days at 25°C in the dark. Then explants were washed with sterile distilled water 3-4 times until no bacterial turbidity was observed, followed by a final wash with water containing antibiotic cefotaxime (400 mg/l) to kill the bacteria. Following washing, explants were blotted dry and transferred on regeneration medium (MS medium + NAA 0.05 mg/l + BA 1.0 mg/l, pH 6.1) containing Kanamycin (35 mg/l) and cefotaxime (500 mg/l). Regenerated shoots were transferred to rooting medium (MS medium + NAA (0.05 mg/l) supplemented with kanamycin (15 mg/l) and cefotaxime (400 mg/l).
Evaluation of transgene integration
Genomic DNA was extracted form untransformed and transgenic A. annua lines using Qiagen DNAesy extraction kit (Qiagen). PCR was set up using bgl1 gene specific forward and reverse primers (F- 5′ GACACCATGCTGACTCTTGAATCAAGGTAGC 3′ and R 5′ GTCACTTGGCCAGAGTTTGCGATGTCAACG 3′) designed based on the bgl1 gene (accession No. U09580) to yield 2.0 kb PCR amplified product.
Evaluation of bgl1 transcript by northern blots
Total RNA was extracted from A. annua transgenic lines and untransformed plants using INTRON easy-BLUE plant RNA extraction kit following the manufacturer protocol (Boca Science). All apparatus and reagents were prepared and treated with diethyl pyrocarbonate (DEPC) treated water to inhibit RNase activities. Total RNA was separated on 1% agarose gel and transferred to nylon membrane. Further, the blot was hybridized with bgl1 gene specific probe as performed below for Southern blot.
Functional enzyme assay and copy number of transgene
Total soluble protein was extracted from 1 g of fresh leaf tissue ground in liquid nitrogen using 2 ml of ice-cold 100 mM citrate buffer (pH 5.2) which included protease inhibitor cocktail (Roche). Protein concentration was measured by Bradford method using the Bio-Rad protein assay kit. Beta-glucosidase activity was estimated as described by Berghem and Pettersson (1974). In brief, 100 μg of total soluble protein (TSP) was incubated with 4mM pNPG (Sigma) in 1ml of 100 mM citrate buffer, pH 5.2 at 50°C for 1 h. Addition of 2 ml of 1 M Na2CO3 terminated the reaction. Enzymatic release of nitrophenol was determined by spectrophotometer at 405 nm. The standard curve of p-nitrophenol was prepared under alkaline conditions using 1 M Na2CO3. One pNPG unit is defined as 1 μmol of p-nitrophenol release per min at 50 °C.
Transgenic lines that showed higher expression of BGL1 enzyme were evaluated for copy number of transgene using Southern blot. In brief, 10 μg of genomic DNA was digested with HindIII and separated onto 0.8% agarose gel. Further, the gel was transferred to nylon membrane and hybridized with 32P dCTP labeled bgl1 gene probe (~0.6kb, isolated from pCAMBIA 2300S with restriction enzymes EcoRI and BamHI) the Stratagene Quick-Hyb hybridization solution and protocol. Confirmed transgenic lines were transferred to jiffy pellets and after acclimatization in the growth room, plants were transferred to the greenhouse for further studies.
Evaluation of trichomes in leaves by SEM and confocal microscope
A. annua leaves were washed with distilled water. Drops of fixative (2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer) were added in a petri dish or on a glass plate and small pieces (2 mm) were dissected from mature leaves of transgenic and untransformed (UT) plants in the presence of the fixative and fixed for 3 to 4 h at room temperature. Tissues were washed with 0.1 M phosphate buffer four times for 15 min each and then rinsed with distilled water three times for 5 min each. Then, tissues were dehydrated with a graded series of ethanol: 30% ethanol for 10 min; 50% to 70% to 80% to 90% to 95% ethanol for 20 min each; and finally, 100% ethanol for 20 min three times. Leaf cross-sections were loaded into a gasket and placed into the critical point drier (Bomb; Electron Microscopy Sciences). After drying, samples were placed on carbon strips facing up. Gold- Palladium was coated with the Emitech K 550 Sputter Coater for 2 min to reach 200 A°. Pieces were excised and examined with a Hitachi S-3500N scanning electron microscope. The densities of trichomes were determined on both the upper and lower surfaces of leaves. A. annua leaf surfaces were also directly observed under confocal microscope (Leica). Images within the same section of a figure were taken using identical confocal settings.
Measurement of glandular trichome cross-sectional area
Elliptical cross-sectional area of the glandular trichome was calculated as described in Maes et al., 2011. In brief, the major (a) and minor (b) axes of the ellipse of the trichomes were measured using ImageJ program (version 1.37; http://rsb.info.nih.gov/ij/). Then cross-sectional area was calculated according to the formula, area = ABπ/4.
Evaluation of trichomes in floral structures
Mature flowers from untransformed and A. annua transgenic lines were harvested and three individual capitulum were dissected under the stereo microscope. Central and marginal florets were analyzed under the stereomicroscope. One capitulum contains 10-14 central and 8-10 marginal florets. The number of trichomes was recorded for both florets.
Evaluation of artemisinin content
Leaf samples were harvested from different positions of the individual plants and then dried for 24 h in an oven at 50°C. Flowers were also harvested from different A. annua transgenic lines. The dried plant material was ground to fine powder. Exactly 100 mg of powder was extracted in 20 ml hexane in an ultrasonic bath for 30 min. The supernatant was transferred to a 40 ml glass vial and evaporated to dryness under nitrogen gas. The residue was resuspended in 5 ml acetonitrile and filtered through a nylon filter (0.45 μm) attached to a 5-ml syringe used for artemisinin quantification.
Quantification of artemisinin by HPLC MS-TOF
Quantification was performed using high-resolution mass spectrometry with an Agilent Technologies 1200 series high-performance liquid chromatography (HPLC) system equipped with a 6224A time of flight mass spectrometer (MS-TOF) using electrospray ionization (ESI) operating in positive mode.. The mobile phases used were pump A, 1 mM ammonium acetate buffer, pH 5 (adjusted using acetic acid), and pump B, Methanol. With a flow gradient at 200 μl/min, the gradient mobile phase composition was 50% mobile phase B for 0.5 min, then linearly increased to 80% mobile phase B for 3.5 min and maintained 80% mobile phase B for 11 min. The total run time was 15 min. Separation was performed on a phenomenex Luna C18 (2) 2.00 × 100mm column with C18 2.00 × 4 mm guard column with peaks of interest eluting at 6.7 min for artemisinin and 10.5 min for artemether. The ESI capillary voltage was set to 3.5kV. The nitrogen nebulizing gas was set to 325°C, 12 l/min, and 40 psig. A standard curve was established over a range of five concentrations of artemisinin (1, 4, 16, 64, 256 ng/ml) while using a 5 μM artemether internal standard. The ratio of integrated extracted ion chromatograms (EIC) of artemisinin [M+H]+ adduct and artemether [M+Na]+ adduct peaks were used to construct a standard curve. Sample filtrate was serially diluted 1:100 twice using a diluent containing a 50:50 mixture of mobile phases A:B and 5μM artemether internal standard. An injection volume of 25 μl was used.
ACKNOWLEDGEMENT
This project was supported in part by NIH grants R01 GM 63879, R01 HL 109442, R01 HL 107904 to Henry Daniell. Dr. Dheeraj Verma is acknowledged for his contributions to this project in the Daniell lab at UCF.
References
- Abdin MZ, Israr M, Rehman RU, Jain SK. Artemisinin, a novel antimalarial drug: biochemical and molecular approaches for enhanced production. Planta Med. 2003;69:289–299. doi: 10.1055/s-2003-38871. [DOI] [PubMed] [Google Scholar]
- Arsenault PR, Vail D, Wobbe KK, Erickson K, Weathers PJ. Reproductive development modulates gene expression and metabolite levels with possible feedback inhibition of artemisinin in Artemisia annua. Plant Physiol. 2010;154:958–968. doi: 10.1104/pp.110.162552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellicour S, Hall S, Chandramohan D, Greenwood D. The safety of artemisinins during pregnancy: a pressing question. Malar. J. 2007;6:15. doi: 10.1186/1475-2875-6-15. doi: 10.1186/1475-2875-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery MA, Chong WKM, Jennings-White C. Stereo selective total synthesis of (+)-artemisinin, the antimalarial constituent of Artemisia annua L. J. Am. Chem. Soc. 1992;114:974–979. [Google Scholar]
- Berghem LE, Pettersson LG. The mechanism of enzymatic cellulose degradation: isolation and some properties of a beta-glucosidase from Trichoderma viride. Eur. J. Biochem. 1974;46:295–305. doi: 10.1111/j.1432-1033.1974.tb03621.x. [DOI] [PubMed] [Google Scholar]
- Charles DJ, Simon JE, Wood KV, Heinstein P. Germplasm variation in artemisinin content of Artemisia annua using an alternative method of artemisinin analysis from crude plant-extracts. J. Nat. Prod. 1990;53:157–160. [Google Scholar]
- Chen DH, Ye HC, Li GF. Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci. 2000;155:179–185. doi: 10.1016/s0168-9452(00)00217-x. [DOI] [PubMed] [Google Scholar]
- Covello PS. Making artemisinin. Phytochemistry. 2008;69:2881–2885. doi: 10.1016/j.phytochem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Davies PJ. Plant Hormones: Biosynthesis, Signal Transduction, Action! Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004. [Google Scholar]
- Davies MJ, Atkinson CJ, Burns C, Woolley JG, Hipps NA, Arroo RRJ, Dungey N, Robinson T, Brown P, Flockart I, Hill C, Smith L, Bentley S. Enhancement of artemisinin concentration and yield in response to optimization of nitrogen and potassium supply to Artemisia annua. Annals of Botany. 2009;104:315–323. doi: 10.1093/aob/mcp126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodi-Semiromi A, Schreiber M, Nallapali S, Verma D, Singh ND, Banks RK, Chakrabarti D, Daniell H. Chloroplast-derived vaccine antigens confer dual immunity against cholera and malaria by oral or injectable delivery. Plant Biotech. J. 2010;8:223–242. doi: 10.1111/j.1467-7652.2009.00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delabays N, Simonnet X, Gaudin MDAD. The genetics of artemisinin content in Artemisia annua L. and the breeding of high yielding cultivars. Curr. Med. Chem. 2001;8:1795–1801. doi: 10.2174/0929867013371635. [DOI] [PubMed] [Google Scholar]
- Dellicour S, Hall S, Chandramohan D, Greenwood BD. The safety of artemisinins during pregnancy: a pressing question. Malar. J. 2007;6:15. doi: 10.1186/1475-2875-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries PJ, Dien TK. Clinical pharmacology and therapeutic potential of artemisinin and its derivatives in the treatment of malaria. Drugs. 1996;52:818–836. doi: 10.2165/00003495-199652060-00004. [DOI] [PubMed] [Google Scholar]
- Duke MV, Paul RN, Elsohly HN, Sturtz G, Duke SO. Localization of artemisinin and artemisitene in foliar tissues of glanded and glandless biotypes of Artemisia annua L. Int. J. Plant Sci. 1994;155:365–372. [Google Scholar]
- Duke SO. Glandular trichomes - a focal point of chemical and structural interactions. International Journal of Plant Sciences. 1994;155:617–620. [Google Scholar]
- Duke SO, Paul RN. Development and fine structure of the glandular trichomes of Artemisia annua L. Internat. J. Plant Sci. 1993;154:107–118. [Google Scholar]
- Efferth T, Dunstan H, Sauerbrey A, Miyachi H, Chitambar CR. The antimalarial artesunate is also active against cancer. Int. J. Oncol. 2001;18:767–773. doi: 10.3892/ijo.18.4.767. [DOI] [PubMed] [Google Scholar]
- Elfawal MA, Towler MJ, Reich NG, Golenbock D, Weathers PJ, Rich SM. Dried Whole Plant Artemisia annua as an Antimalarial Therapy. PLoS ONE. 2012;7:e52746. doi: 10.1371/journal.pone.0052746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfawala MA, Towlerb MJ, Reich NG, Weathers PJ, Rich SM. Dried whole-plant Artemisia annua slows evolution of malaria drug resistance and overcomes resistance to artemisinin. Proc. Natl. Acad. Sci. USA. 2015;112:821–826. doi: 10.1073/pnas.1413127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JFS, Janick J. Floral morphology of Artemisia annua with special reference to trichomes. Int. J. Plant Sci. 1995;156:807–815. [Google Scholar]
- Ferreira JFS, Simon JE, Janick J. Relationship of artemisinin content of tissue-cultured, greenhouse-grown, and field-grown plants of Artemisia annua. Planta Med. 1995;61:351–355. doi: 10.1055/s-2006-958098. [DOI] [PubMed] [Google Scholar]
- Ferreira JFS, Simon JE, Janick J. Developmental studies of Artemisia annua: flowering and artemisinin production under greenhouse and field conditions. Planta Med. 1995a;61:167–170. doi: 10.1055/s-2006-958040. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121–31. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Graham IA, Besser K, Blumer S, Branigan CA, Czechowski T, Elias L, Guterman I, Harvey D, Isaac PG, Khan AM, Larson TR, Li Y, Pawson T, Penfield T, Rae AM, Rathbone DA, Reid S, Ross J, Smallwood MF, Segura V, Townsend T, Vyas D, Winzer T, Bowles D. The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science. 2010;327:328–331. doi: 10.1126/science.1182612. [DOI] [PubMed] [Google Scholar]
- Han J, Wang H, Ye H-C, Liu Y, Li Z-Q, Zhang Y, Zhang Y-S, Yan F, Li GF. High efficiency of genetic transformation and regeneration of Artemisia annua L. via Agrobacterium tumefaciens-mediated procedure. Plant Sci. 2005;168:73–80. [Google Scholar]
- Jain DC, Mathur AK, Gupta MN, Singh AK, Verma RK, Gupta AP, Kumar S. Isolation of high artemisinin-yielding clones of Artemisia annua. Phytochemistry. 1996;43:993e1001. [Google Scholar]
- Jessing K, Duke SO, Cedergreen N. Potential ecological roles of artemisinin produced by Artemisia anna L. J. Chem. Ecol. 2014;40:100–117. doi: 10.1007/s10886-014-0384-6. [DOI] [PubMed] [Google Scholar]
- Jessing K, Cedergreen N, Mayer P, Libous-Bailey L, Strobel BW, Rimando AM, Duke SO. Loss of artemisinin produced by Artemisia annua L. to the soil environment. J. Industrial Crops Products. 2013;43:132–140. [Google Scholar]
- Jin S, Kanagaraj A, Verma D, Lange T, Daniell H. Release of Hormones from Conjugates: Chloroplast Expression of β-Glucosidase Results in Elevated Phytohormone Levels Associated with Significant Increase in Biomass and Protection from Aphids or Whiteflies Conferred by Sucrose Esters. Plant Physiol. 2011;155:222–235. doi: 10.1104/pp.110.160754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Chichester JA, Mett V, Jaje J, Tottey S, Manceva S, Casta LJ, Gibbs SK, Musiychuk K, Shamloul M, Norikane J, Mett V, Streatfield SJ, van de Vegte-Bolmer M, Roeffen W, Sauerwein RW, Yusibov V. A plant-produced Pfs25 VLP malaria vaccine candidate induces persistent transmission blocking antibodies against Plasmodium falciparum in immunized mice. PLoS One. 2013;8(11):e79538. doi: 10.1371/journal.pone.0079538. doi: 10.1371/journal.pone.0079538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjær A, Grevsen K, Jensen M. Effect of external stress on density and size of glandular trichomes in full-grown Artemisia annua, the source of anti-malarial artemisinin. AoB PLANTS. 2012:pls018. doi: 10.1093/aobpla/pls018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klayman DL. Qinghaosu (artemisinin): an antimalarial drug from China. Science. 1985;228:1049–1055. doi: 10.1126/science.3887571. [DOI] [PubMed] [Google Scholar]
- Kohli N, Westerveld D, Ayache AC, Verma A, Shil P, Prasad T, Zhu P, Chan SC, Li Q, Daniell H. Oral delivery of bioencapsulated proteins across blood–brain and blood–retinal barriers. Molecular Therapy. 2014;22:535–546. doi: 10.1038/mt.2013.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K-C, Nityanandam R, New JS, Daniell H. Oral delivery of bioencapsulated exendin-4 expressed in chloroplasts lowers blood glucose level in mice and stimulates insulin secretion in beta-TC6 cells. Plant Biotech. J. 2013;11:77–86. doi: 10.1111/pbi.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon KC, Verma D, Singh ND, Herzog R, Daniell H. Oral delivery of human biopharmaceuticals, autoantigens and vaccine antigens bioencapsulated in plant cells. Adv. Drug Deliv. Rev. 2013;65:782–799. doi: 10.1016/j.addr.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmi PS, Verma D, Yang X, Lloyd B, Daniell H. Low cost tuberculosis vaccine antigens in capsules: expression in chloroplasts, bio-encapsulation, stability and functional evaluation in vitro. PLoS One. 2013;8:e54708. doi: 10.1371/journal.pone.0054708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin JC. Effect of agronomic practices on plant yield and antimalarial constituents of Artemisia annua L. Acta. Hortic. 1993;331:54–61. [Google Scholar]
- Laughlin JC. The influence of distribution of antimalarial constituents in Artemisia annua L. on time and method of harvest. Acta Hortic. 1995;390:67–73. [Google Scholar]
- Limaye A, Koya V, Samsam M, Daniell H. Receptor mediated oral delivery of a bioencapsulated green fluorescent protein expressed in transgenic chloroplasts into the mouse circulatory system. FASEB J. 2006;20:959–961. doi: 10.1096/fj.05-5134fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen WJM, Schenk E, Bouwmeester HJ, Verstappen FWA. Trichome dynamics and artemisinin accumulation during development and senescence of Artemisia annua leaves. Planta Medica. 2006;72:336–345. doi: 10.1055/s-2005-916202. [DOI] [PubMed] [Google Scholar]
- Lu X, Shen Q, Zhang L, Zhang F, Jiang W, Lv Z, Yan T, Fu X, Wang G, Tang K. Promotion of artemisinin biosynthesis in transgenic Artemisia annua by overexpressing ADS, CYP71AV1 and CPR genes. Industrial Crops and Products. 2013;49:380–385. [Google Scholar]
- Maes L, Van Nieuwerburgh FCW, Zhang Y, Reed DW, Pollier J, Vande Casteele SRF, Inzé D, Covello PS, Deforce DLDLD, Goossens A. Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytologist. 2011;189:176–189. doi: 10.1111/j.1469-8137.2010.03466.x. [DOI] [PubMed] [Google Scholar]
- Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in E. coli for production of terpenoids. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Martens EC, Loew EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Hanrissat B, Bolam DN, Gordon JI. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauji R, Khan MA, Jha P, Khan S, Kiran U, Ahmad MM, Javed S, Abdin MZ. HMG-CoA reductase limits artemisinin biosynthesis and accumulation in. 2010 [Google Scholar]
- Morales M, Charles DJ, Simon JE. Seasonal accumulation of artemisinin in Artemisia annua L. Acta Hortic. 1993;344:416–420. [Google Scholar]
- Olsson ME, Olofsson LM, Lindahl AL, Lundgren A, Brodelius M, Brodelius PE. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry. 2009;70:1123–1128. doi: 10.1016/j.phytochem.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Qian Z, Gong K, Zhang L, Lv J, Jing F, Wang Y, Guan S, Wang G, Tang KA. Simple and efficient procedure to enhance artemisinin content in Artemisia annua L. by seeding to salinity stress. African J. Biotech. 2007;6:1410–1413. [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Romero MR, Serrano MA, Vallejo M, Efferth T, Alvarez M, Marin JJG. Antiviral effect of artemisinin from Artemisia annua against a model member of the Flaviviridae family, the Bovine Viral Diarrhoea Virus (BVDV). Planta Med. 2006;72:1169–1174. doi: 10.1055/s-2006-947198. [DOI] [PubMed] [Google Scholar]
- Romero MR, Efferth T, Serrano MA, Castano B, Macias RIR, Briz O, Marin JJG. Effect of artemisinin/artesunate as inhibitors of hepatitis B virusproduction in an “in vitro” replicative system. Antiviral Res. 2005;68:75–83. doi: 10.1016/j.antiviral.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Schneider G, Schliemann W. Gibberellin conjugates: an overview. Plant GrowthRegulation. 1994;15:247–260. [Google Scholar]
- Sembdner G, Atzorn R, Schneider G. Plant hormone conjugation. Plant Molecular Biology. 1994;26:1459–1481. doi: 10.1007/BF00016485. [DOI] [PubMed] [Google Scholar]
- Sherman A, Su J, Lin S, Wang X, Herzog RW, Daniell H. Suppression of inhibitor formation against factor VIII in hemophilia A mice by oral delivery of antigens bioencapsulated in plant cells. Blood. 2014;124:1659–1668. doi: 10.1182/blood-2013-10-528737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shil PK, Kwon KC, Zhu P, Verma A, Daniell H, Li Q. Oral delivery of ACE2/Ang-(1-7) bioencapsulated in plant cells protects against experimental uveitis and autoimmune uveoretinitis. Molecular Therapy. 2014;22:2069–2082. doi: 10.1038/mt.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CH. Understanding artemisinin resistance. Science. 2015;347:373–374. doi: 10.1126/science.aaa4102. [DOI] [PubMed] [Google Scholar]
- Soetaert SSA, Van Neste CMF, Vandewoestyne ML, Head SR, Goossens A, Van Nieuwerburgh FCW, Deforce DLD. Differential transcriptome analysis of glandular and filamentous trichomes in Artemisia annua. BMC Plant Biol. 2013;13:220. doi: 10.1186/1471-2229-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L. The Plant Vacuole. J. exp. Biol. 1992;172:113–122. doi: 10.1242/jeb.172.1.113. [DOI] [PubMed] [Google Scholar]
- Tan H, Xiao L, Gao S, Li Q, Chen J, Xiao Y, Ji Q, Chen R, Chen W, Zhang L. Trichome and artemisinin regulator 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol. Plant. 2015 doi: 10.1016/j.molp.2015.04.002. doi: 10.1016/j.molp.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Van Geldre E, Vergauwe A, van den Eeckhout E. State of the production of the antimalarial compound artemisinin in plants. Plant Mol. Biol. 1997;33:199–209. doi: 10.1023/a:1005716600612. [DOI] [PubMed] [Google Scholar]
- Van Nieuwerburgh FCW, Vande Casteele SRF, Maes L, Goossens A, Inze D, Van Bocxlaer J, Deforce DLD. Quantitation of artemisinin and its biosynthetic precursors in Artemisia annua L. by high performance liquid chromatography-electrospray quadrupole time-offlight tandem mass spectrometry. J. Chromatography. 2006;1118:180–187. doi: 10.1016/j.chroma.2006.03.121. [DOI] [PubMed] [Google Scholar]
- Vergauwe A, Cammaert R, Vandenberghe D, Genetello C, Van Montagu M, Van den Eeckhout E. Agrobacterium tumefaciens-mediated transformation of Artemisia annua L.and regeneration of transgenic plant. Plant Cell Rep. 1996;15:929–937. doi: 10.1007/BF00231590. [DOI] [PubMed] [Google Scholar]
- Wallaart TE, Pras N, Beekman AC, Quax WJ. Seasonal variation of artemisinin and its biosynthetic precursors in plants of Artemisia annua of different geographical origin: Proof for the existence of chemotypes. Planta Med. 2000;2000:66, 57–62. doi: 10.1055/s-2000-11115. [DOI] [PubMed] [Google Scholar]
- WHO World malaria report. 2011 http://www.who.int/malaria/world_malaria_report_2011/en/
- WHO: Malaria. World Health Organization World Malaria Report. 2014 www.who.int/malaria.
- Xie DY, Zou ZR, Ye HC, Li GF, Guo ZC. Selection of hairy root clones of Artemisia annua L. for artemisinin production. Israel J. Plant Sci. 2001;49:129–134. [Google Scholar]
- Yang RY, Zeng XM, Lu YY, Lu WJ, Feng LL, Yang XQ, Zeng QP. Senescent leaves of Artemisia annua are one of the most active organs for overexpression of artemisinin biosynthesis responsible genes upon burst of singlet oxygen. Planta Medica. 2009;76:734–742. doi: 10.1055/s-0029-1240620. [DOI] [PubMed] [Google Scholar]