Abstract
Colonization resistance by the commensal microbiota is a key defense against infectious pathogens in the gastrointestinal tract. The microbiota directly competes with incoming pathogens by occupying the colonization niche, depleting nutrients in the gut lumen as well as indirectly inhibiting the growth of pathogens through activation of host immunity. Enteric pathogens have evolved strategies to cope with microbiota-mediated colonization resistance. Pathogens utilize a wide array of virulence factors to outcompete their commensal rivals in the gut. However, since the expression of virulence factors is costly to maintain and reduces bacterial fitness, pathogens need to regulate their virulence properly in order to maximize their fitness. To this end, most pathogens use environmental cues to regulate their virulence gene expression. Thus, a dynamic regulation of virulence factor expression is a key invasion strategy utilized by enteric pathogens. On the other hand, host immunity selectively targets virulent pathogens in order to counter infection in the gut. The host immune system is generally tolerant of harmless microorganisms, such as the commensal microbiota. Moreover, the host relies on its commensal microbiota to contribute, in concert with its immune system, to the elimination of pathogens. Collectively, regulation of virulence determines the fate of enteric pathogens, from the establishment of infection to the eventual elimination. Here, we will review the dynamics of virulence and its role in infection.
Keywords: enteric infection, microbiota, host immunity, competition
Introduction
The human gastrointestinal (GI) tract harbors trillions of harmless and/or beneficial microorganisms. On average there are more than a thousand different species and collectively they represent the most densely populated habitat in the human body (1). The communities of endogenous microorganisms, referred to as the commensal microbiota, confer a wide variety of benefits on the host. For instance, the commensal microbiota provides enzymes that are not encoded by the host genome, thereby helping with the digestion of complex carbohydrates as well as the generation of essential nutritional factors, such as vitamins (2). The commensal microbiota also contributes to the differentiation and maturation of the mucosal immune system, including Th17 cells, regulatory T cells and IgA production (3–5). Furthermore, commensal bacteria play a crucial role in host resistance against infectious pathogens, so-called colonization resistance (6, 7). The commensal microbiota is capable of competing with incoming enteric pathogens, such as Salmonella enterica serovar Typhimurium (S. typhimurium), Vibrio cholerae (V. cholerae), Shigella flexneri (S. flexneri), or pathogenic Escherichia coli, and prevent their colonization and growth in the gut. In contrast, once colonization resistance is breached, the abnormally expanded pathogens often cause serious infectious disease and often death. Indeed, enteric infections are a major cause of morbidity and mortality in the developing world. Thus, a better understanding of the competition mechanisms between commensal and pathogenic microbes will lead to the development of novel therapeutic strategies for the treatment of these life-threatening infections.
Competition between commensal and pathogenic bacteria
Colonization resistance conferred by the commensal microbiota is a key host defense mechanism against enteric infection. Colonization resistance is achieved, in part, through niche competition. Commensal microbes occupy the luminal niche and expend available nutrients, thereby limiting the growth of newcomers. Certain environmental factors, i.e. inflammation, dietary changes, and antibiotics, lead to the disruption of commensal microbial communities, therefore significantly increasing the risk of colonization and expansion of incoming pathogens. For example, antibiotic treatment leads to an expansion of enteric pathogens, including Clostridium difficile (C. difficile), vancomycin-resistant Enterococcus faecalis (E. faecalis), and S. typymurium, in mice and humans (8). Moreover, germ-free (GF) mice are highly susceptible to Citrobacter rodentium (C. rodentium), a murine enteric pathogen that is a model for A/E lesion forming human pathogens, such as enterohemorragic Escherichia coli (EHEC) or enteropathogenic Escherichia coli (EPEC). Importantly, unlike conventional SPF mice, GF mice are unable to eradicate C. rodentium from the gut. Taken together, this suggests that the commensal microbiota is essential for prevention of colonization and proliferation of enteric pathogens in the gut as well as their elimination from the gut (9). Recent accumulating evidence indicates that nutritional competition is a key mechanism that the commensal microbiota uses to prevent the proliferation of pathogens in the gut lumen (6). In other words, the resident microbes consume available nutrients in the gut lumen, effectively preventing any incoming pathogens from accessing luminal nutrients and starving them. For example, it has been shown that C. rodentium is outcompeted by commensal E. coli, a bacterium with similar metabolic preferences (e.g., monosaccharides) (9). Likewise, microbes use metabolites to regulate their environment. This is demonstrated by the metabolic exclusion of certain pathogens by fermentation end products of Bacteroidetes and Firmicutes, the most dominant phyla in the colon. Complex polysaccharides are fermented into short-chain fatty acids (SCFAs) by these bacteria in the gut (10); SCFAs in turn can act as potent growth inhibitors of certain pathogens (e.g. C. difficile, S. typhimurium) (11) (12).
Although the use of commensal microbes is an effective defense mechanism employed by the host against invading pathogens, pathogens have evolved strategies to evade colonization resistance mediated by the microbiota. Mounting evidence suggests that pathogens exploit host inflammatory responses or host-derived metabolic byproducts, which cannot be utilized by the commensal microbiota, for their own growth in the gut. Intestinal inflammation is known to provide a growth advantage to various enteric pathogens (e.g., S. Typhimurium, C. rodentium, EHEC, and Campylobacter jejuni), resulting in the overgrowth of pathogens at the expense of commensal microbes (13, 14). For example, ethanolamine, released from the intestinal epithelial cells due to inflammation-caused tissue damage, selectively fuels the growth of EHEC. Commensal E. coli, unlike EHEC, lacks the eut operon, which is required for the utilization of ethanolamine (15, 16). Likewise, during gut inflammation caused by S. typhimurium infection, reactive oxygen species (ROS) are released from the inflamed tissue. ROS convert host-derived thiosulfate (S2O32−) into tetrathionate (S4O62−), a respiratory electron acceptor that provides a growth advantage selectively to S. typhimurium in the presence of other competing microbes (17). In addition to being a source of alternative nutrients and electron donors important for the growth of pathogens, intestinal inflammation also increases the niche and availability of common nutrients utilized for the growth of pathogens through inflammation-triggered perturbations of microbial configurations (i.e. reduced bacterial diversity and richness)(18). These findings clearly signify that pathogen-induced inflammation and microbiota perturbations can be plausible mechanisms employed by pathogens to enhance their ability to colonize and replicate in the gut. Although intestinal inflammation further facilitates the ability of pathogens to thrive in the gut, pathogens need to breach microbiota-mediated resistance and colonize the gut to initiate inflammation. In this context, there has been more evidence indicating that pathogens express a wide variety of factors, namely virulence factors, which allow them to overcome colonization resistance mediated by the commensal microbiota and survive in the harsh environment present in the gut. Virulence factors comprise a myriad of molecules including toxins, molecules associated with attachment to and invasion of host cells and factors required for modulation of the host environment. Notably, virulence factors are not essential for in vitro growth and survival or in vivo growth of pathogens in the absence of competing bacteria (e.g. mono-colonization in GF mice). In contrast, virulence factors are essential for colonization and growth in the gut in the presence of commensal microbes (9). For example, C. rodentium uses its virulence factors to localize near/at the intestinal epithelium, so-called pathogen-specific niche, where it may utilize niche-specific nutrients while at the same time escaping from the nutritional competition exerted by commensal microbes. Since mutant C. rodentium strains, which lack virulence factors, are incapable of residing in the pathogen-specific niche, these mutants fail to colonize and proliferate in SPF mice. Thus, virulence factor expression is a key requisite for successful pathogen colonization of the gut. Once successfully established in the gut, pathogens can initiate inflammation and further shape the luminal microenvironment so that it better accommodates their needs for growth.
Environmental cues regulate virulence factor expression
Although virulence factors are essential for the ability of pathogens to overcome commensal-mediated colonization resistance and establish infection, pathogens do not always express them because of the associated fitness cost. It has been reported that mutant strains deficient in virulence factors display increased fitness compared to their virulent counterparts, due to the cost of virulence (19). Therefore, it does not seem to benefit pathogens to express virulence factors constitutively. It is ideal for pathogens to stay avirulent before reaching their colonization sites and/or in the absence of competitors, and only to express virulence factors upon reaching the destination sites and/or for competition with resident microbes. In other words, pathogens should not express virulence factors until they sense that the required timing and location are right to maximize their fitness during infection. The next question is how do pathogens determine the right timing and location? Mounting evidence suggests that pathogens are able to sense the microenvironment in the GI tract during the course of infection (20, 21). Even in the same anatomical site the environment can be highly variable (e.g. luminal space, mucus layer, epithelial surface and inside the tissue and cells). Successful colonization requires that pathogens efficiently adapt to this highly variable environment through the appropriate and coordinate expression of virulence genes. To this end, pathogens will respond to various environmental cues that may differentially regulate their virulence genes. Here, we review the key environmental cues that elicit or inhibit pathogen virulence factors expression (Table 1).
Table 1.
Intestinal environmental cues utilized by virulence gene regulation of pathogens.
| Related-Pathway | |||||||
|---|---|---|---|---|---|---|---|
| Stimulus | Putative Site | Pathogen | Regulator | Virulence gene | Pathological meanings | Ref. | |
| Physico-chemical sensing | pH | stomach | EPEC | GadX |
gadA↑ ler↓ |
Adaptation to acidic environment -Resistance to acid↑ -Virulence factors↓ |
22 |
| Bicarbonate | duodenum-colon | C. rodentium | AraC/XylS-like proteins (RegA) |
adcA↑ kfcC↑ |
Increased chance to find niches -Motility (fimbriae)↑ -Carbon metabolism↑ |
25 | |
| Osmolarity | ileum | S. typhimurium | EnvZ/OmpR-TviA |
flhC↓ viaB↑ |
Adaptation to host tissue -Vi capsule↑ -Motility↓ |
32 | |
| Oxygen | colon | S. flexneri | FNR | T3SS↑ | Entry to the niche -Invasion ability↑ |
34 | |
| Metabolic sensing | Bile | duodenum- | V. cholerae | ToxT |
flrA↑ ctxAB, tcpA ↓ |
Increased chance to find niches -Motility (fimbriae)↑ -Cholera toxin↓ |
36 |
| SCFAs | ileum | S. typhimurium | BarA-SirA | hilA↑ | Entry to the niches (cells) -Invasion ability↑ |
42 | |
| Fucose | colon | EHEC | FusKR | LEE↓ | Prevention of Energy expenditure -Repression of virulence genes ↓ |
47 | |
| Mechano sensing |
Attachment Shear stress |
colon | C. rodentium | GrlA | LEE↑ | Entry to the niches -Invasion ability↑ |
49 |
| Viscosity | colon | V. cholerae | flagella |
ctxAB↑ tcpA ↑ |
Increased chance to find niches -Invasion ability↑ |
52 | |
| Quorum sensing | Cell density | ileum | S. aureus | AgrA/AgrC |
hla↑ spa↓ |
Adaptation to host tissue -Endosomal escape↑ -Surface proteins↓ |
54 |
| - | Unknown | unknown | S. typhimurium | unknown | TTSS-1↑↓ | Stabilization of cooperative virulence -Bistable virulence expression |
57 |
Sensing of physical/chemical stimuli
pH
Successful intestinal colonization depends on the ability of the pathogen to tolerate a dramatic shift in pH associated with the various compartments of the GI tract. Certain pathogens, such as EPEC, express GadX in response to acid stimulation, an activator of genes involved in acid tolerance. GadX is a member of the XylR/AraC family of transcriptional regulators, also known to negatively regulate the expression of perA, the indirect activator of the virulence regulatory gene Ler. This evidence suggests that GadX is simultaneously a positive regulator of acid tolerance genes and a suppressor of virulence genes, which are not required in the acidic environment (22).
Bicarbonate
In mammals, bicarbonate (HCO3−) plays a central role in whole body homeostasis and is known to be abundant in the gut. Bicarbonate acts to maintain intestinal homeostasis by controlling the pH levels in the intestine. It is released from the pancreas in response to the hormone secretin and neutralizes the acidic chyme, a thick semifluid mass of partially digested food entering the duodenum from the stomach. The large intestine also secretes bicarbonate to neutralize any increases in acidity resulting from the formation of several by-products of microbial fermentation. In this bicarbonate-rich host environment, several pathogens, such as Streptococcus pyogenes and EHEC, have evolved a bicarbonate associated signal transduction system to control the virulence factors involved in bacterial colonization of the host surfaces (23, 24), For example, the virulence regulator regA is known to play an essential role in C. rodentium pathogenesis through the regulation of more than 60 operons, including the locus of enterocyte effacement (LEE) virulence genes (25, 26). It has been reported that transcriptional regulation of the regA gene itself is governed by two putative virulence loci, adcA and kfc, both of which are responsive to environmental bicarbonate (27).
Osmolality
In S. typhimurium infection, the pathogen expresses flagellin when in the intestinal lumen. However, once the pathogen migrates into the tissue, it ceases to express flagellin and, in turn, starts to produce the Vi capsule, which prevents host recognition of the pathogen through TLR4 (28–30). This reciprocal expression of virulence related genes is thought to be of crucial importance to the pathogen as it allows it to evade innate immune surveillance at the intestinal mucosa and contributes to the increased systemic dissemination of S. typhimurium. As the bacterium traverses the mucosa, it is able to detect the difference in osmolality between the lumen and the mucosal tissue. The osmolality of the intestinal lumen is known to be higher than that of the tissue.
Certain pathogens express the transmembrane histidine kinase EnvZ, a sensor of environmental osmolality. During conditions of high osmolality, EnvZ autophosphorylates and transfers the phosphoryl group to the response regulator OmpR, resulting in the formation of phosphorylated OmpR (31). In response to a relative decrease in osmolarity, EnvZ/OmpR induce the expression of tviA gene. In concert with Rcs-B, TviA represses the biosynthesis of flagellin (32), and enhances Vi capsule production through the activation of the viaB locus. Collectively, these mechanisms enable the pathogen to rapidly cease flagellin expression when crossing the epithelial lining and prevent the induction of host immune responses that would limit systemic dissemination.
Oxygen
Oxygen tension is low in the intestinal lumen, but increases adjacent to the mucosal surface. In response to the oxygen gradient, certain pathogens control the expression of virulence factors during the initial step of host cell invasion. During S. typhimurium infection, the anaerobic response regulator fumarate and nitrate reductase (FNR) is activated in response to oxygen and enhances the expression of several loci related to flagellar biosynthesis, chemotaxis, anaerobic carbon utilization and the Salmonella pathogenicity island (SPI)-1 genes (33). Likewise, S. flexneri utilizes FNR-mediated oxygen tension sensing for effective colonization of the gut. Under microaerobic conditions in the intestinal lumen, the activation of FNR leads to elongation of type 3 secretion system (T3SS) needles. However, at the same time, FNR represses the secretion of effector proteins, such as invasion plasmid antigens (ipas), leaving the T3SS needles extended, but not competent for secretion. However, upon reaching the aerobic zone adjacent to the mucosal surface, FNR is inactivated and, in turn, the T3SS is fully activated, reversing the anaerobic block of ipa secretion into the host. Thus, S. flexneri is primed in the anaerobic environment of the intestinal lumen and host entry is triggered by the aerobic conditions at the intestinal cell surface. This finely tuned strategy allows the pathogen to precisely control the timing of activation of the T3SS at the intended site of action, maximizing its invasion ability and virulence (34). Notably, FNR is known to be widely conserved among enteric pathogens, suggesting the importance of oxygen in the regulation of virulence.
Sensing of metabolites
Bile
Bile is a digestive liquid produced by the liver and secreted into the duodenum through the bile duct. Enteric pathogens encounter bile in the early stages of infection. Although bile has potent antimicrobial properties, is a natural detergent and plays an important role in host defense, it is evident that certain pathogens are able to tolerate it, or even use it to their advantage (35). The concentration of bile is known to decrease gradually from the intestinal lumen to the epithelial surface. By utilizing the luminal bile gradient, certain pathogens, such as V. cholera, selectively control their virulence genes. In the pathogenesis of V. cholerae infection, the major virulence factors of V. cholera are cholera toxin (CT, encoded as ctxAB) and toxin-coregulated pilus (TCP, encoded as tcpA), both positively regulated by the dimerized transcription activator ToxT. It has been shown that unsaturated fatty acids (UFAs) present in bile directly bind ToxT, preventing its homo-dimerization and repressing ctxAB and tcpA genes (36, 37). In contrast, flagellar genes are known to be up-regulated by bile stimulation. In fact, in V. cholerae infection, CT and TCP production is down-regulated by ToxT in the intestinal lumen where bile concentration is high and the expression of flagellar genes is enhanced (38). However, once the pathogen reaches the epithelial surface where bile concentration is low, CT and TCP repression is removed. Since it is known that CT-induced fluid secretions from epithelial cells could flush away the pathogen from the cellular surface, the bile-mediated regulatory mechanism allows the pathogen to successfully approach the epithelial barrier. Also, TCP expression carries a fitness cost and as long as the bacterium remains in the luminal space it is counterproductive. On the other hand, it would be more reasonable to think that the pathogen benefits from increased motility in the lumen that enables it to penetrate the thick mucus layer and adhere to the underlying epithelial cells. Then, once it reaches the epithelial layer where bile concentration is lower, the pathogen ceases to express motility genes and virulence genes are up-regulated, allowing the bacterium to firmly colonize the epithelium.
Short-chain fatty acids (SCFAs)
Short-chain fatty acids (SCFAs) are the end products of microbial fermentation. Dietary fibers resist digestion and absorption in the GI tract and can only be fermented by the gut commensals, such as Bacteroidetes and Furmicutes - the most abundant phyla in the human colon. SCFAs are highly concentrated in the colon, reaching concentrations of up to 140 mM (39). SCFAs mainly consist of acetate (C2), propionate (C3), and butyrate (C4); in the human gut the approximate molar ratio is 60:20:20 (40). The Bacteroidetes phylum mainly produces acetate and propionate, whereas the Firmicutes phylum includes a large population of butyrate-producing bacteria (41). SCFAs are known to have many beneficial effects, such as their role in immune-modulatory functions that contribute to the maintenance of gut homeostasis. Additionally, SCFAs can also control bacterial fitness in the gut. Although the effects of SCFAs on enteric pathogens have not been fully elucidated, some pathogens are capable of sensing SCFAs. This sensory ability is often connected to regulation of virulence gene expression. For example, S. typhimurium uses SCFA concentration and composition to regulate its invasion genes encoded on the Salmonella pathogenicity island 1 (SPI-1) via the BarA/SirA two-component regulatory system (42). The conversion of acetate to acetyl-phosphate in the bacterial cytoplasm activates the BarA/SirA regulatory system, resulting in the upregulation of SPI-1 genes, such as hilA. Interestingly, it has been shown that low total SCFAs (~30 mM) with a predominance of acetate (~25mM), like the conditions found in the distal ileum, induce the expression of SPI-1 genes. On the other hand, high total SCFAs (~200mM) with greater proportions of propionate and butyrate, like the conditions found in the colon, suppress invasion genes. Although the detailed mechanism remains unclear, this evidence suggests that S. typhimurium is able to use the SCFA composition in the terminal ileum as a signal for invasion.
Fucose
Fucose is one of the major components of mucin and is expressed on the surface of host epithelial cells (43, 44). Certain commensals, such as Bacteroides thetaiotaomicron (B. thetaiotaomicron), cleave fucose from host mucus by using several fucosidases and control free fucose availability in the gut lumen (10, 45, 46). Luminal fucose represses the expression of the LEE virulence genes in EHEC through the activation of a two-component signal transduction system named FusKR (47). As long as the bacterium remains in the lumen, the LEE virulence genes do not provide EHEC any competitive advantage and remain repressed (47). In contrast, when EHEC is localized in proximity to the epithelium, an area with low availability of fucose, host-derived adrenergic signals begin to repress FusKR (47). Consequently, the expression of the LEE genes is no longer repressed, freeing the pathogen to up-regulate its virulence genes. These findings suggest that EHEC is able to use fucose to modulate its virulence and metabolism (47).
Sensing of mechanical stimuli
Attachment
Pathogen attachment to host cells is a crucial step in the development of infection or disease. In fact, a recent study clearly demonstrated that bacterial attachment (e.g., C. rodentium and EHEC) is critical for triggering the Th17-induced gene expression program in the epithelium (48). It has also been shown that pathogen attachment to host cells triggers expression of the LEE genes in a GrlA-dependent manner. GrlA is a cytoplasmic regulator of virulence genes, including the LEE associated virulence genes. GrlA dependent induction of the LEE genes is observed in the mode of attachment based on either electrostatic interactions or specific receptor-ligand interactions, such as those mediated by a surface adhesion molecule intimin or a bacterial cell wall component LPS. Importantly, host-derived signals, such as host cytoskeletal rearrangements, lead to pedestal formation, a mode of attachment known to be induced by LEE gene expression. However, pedestal formation in itself is not required for attachment induced LEE activation. Furthermore, GrlA-dependent LEE induction is enhanced by levels of fluid shear stress similar to peristaltic forces between the intestinal brush border surface (5 dynes/cm2) and the microvilli (2–3 dynes/cm2). Although further studies are needed to clarify the detailed mechanisms through which the bacterial envelope components are involved in the sensing of mechanical cues, these findings suggest that enteric pathogens sense and respond to mechanical cues as they strive to adapt to their environment (49).
Viscosity
The colonic mucus barrier is composed of two distinct layers and has a total thickness of 800 µm (50). The top layer of mucus is thicker and less viscous than the bottom layer, providing the habitat for a large number of commensal bacteria. In contrast, the bottom layer is thinner and denser; its role is to prevent commensal bacteria from having access to host tissue (51). Certain pathogens have evolved to sense mucus viscosity and control their virulence gene expression accordingly. As described earlier in this review, the virulence factors of V. cholerae (CT and TCP) are down-regulated when bile concentration is high (intestinal lumen). However, it has been shown that once the pathogen reaches a high-viscosity environment (bottom layer), it is able to detect the change in viscosity, presumably by a signaling process that involves the sensing of flagellar rotation rates. In turn, the expression of CT and TCP is induced, presumably due to reduced bile concentration. Although the detailed mechanism remains elusive, it is appears that motility and changes in membrane sodium flux, the driving force of the flagellar motor, affect virulence gene expression in V. cholerae (52).
Quorum sensing
Quorum sensing (QS) is a sensing mechanism that allows bacteria to monitor their surrounding environment for the presence of other bacteria. In QS, bacteria release small signaling molecules called autoinducers (AIs). As the bacterial population increases, AIs accumulate in the micro milieu and bacteria monitor this information to track changes in bacterial density in the defined environmental space. Although it is now known that a large portion of bacterial behavior can be influenced by QS, QS is also used by pathogens to modulate virulence factor expression used in pathogenesis (53). In enteric infection, QS is thought to enable the pathogen to minimize host immune responses by delaying the production of virulence factors with high antigenicity until sufficient bacteria accumulate and are ready to overwhelm host defense. By utilizing AIs, pathogens can control their behavior to reflect population density and act as a group. This strategy provides several advantages used in pathogenesis. In the case of Staphylococcus aureus, the agr system is central to virulence gene regulation and intracellular survival in host cells (54, 55). At low population densities, the concentration of the pathogen-derived autoinducing peptide (AIP) is not high enough in the extracellular environment to induce subsequent signal activation. However, once the population density is high enough and AIP concentration has reached a threshold level, AIP activates the transmembrane receptor histidine kinase AgrC that triggers autophosphorylation and phosphotransfer to AgrA. This event, in turn, promotes the transcription of several virulence genes associated with toxins, endosomal escape, and intracellular survival/replication. The agr system is also known to be evolutionarily conserved across the phylum Firmicutes, including C. difficile, Listeria monocytogenes and E. faecalis (56).
Regulation of virulence: a strategy for pathogens to maximize their fitness
As described above, pathogens use various types of virulence regulation strategies to sense the local milieu. However, the expression of virulence genes is energetically costly for the organism. It is known that virulence-competent pathogens grow less than avirulent mutants, suggesting that uncontrolled virulence can be a threat to successful colonization. In this context, recent studies pointed out the importance of bistable virulence regulation that leads to phenotypic heterogeneity of enteric pathogens as means to maximize their in vivo fitness. As described above, S. typhimurium utilizes the type III secretion system 1 (T1) to induce gut inflammation, attenuating colonization resistance by commensal microbes and shaping a better niche for its growth. Meanwhile, the T1 mutant strain (T1m), which harbors an irreversible gene mutation and cannot express T1 virulence proteins, emerges from the pool of pathogens (57). Since virulence factor expression is costly, T1m mutant bacteria can grow faster than wild-type S. typhimurium, which expresses virulence factors (T1+), eventually superseding the virulent populations (57). To prevent the rise of virulence deficient mutant strains, infectious pathogens employ bistable virulence regulation. A phenotypically avirulent subpopulation (T1−) arises from the infectious population through the down-regulation of virulence factors (57). The T1− subpopulation can grow faster due to a lack of virulence, as is observed with the T1m strain, thus preventing the growth of undesired avirulent mutants (57). Thus, although the phenotypically avirulent subpopulation per se does not contribute to the induction of inflammation, its rise plays a key role in stable, long lasting S. typhimurium infection. Virulence expression heterogeneity has also been reported in pathogenic E. coli-related strains. As described above, EHEC uses the intestinal fucose gradient to regulate expression of the LEE genes (47). The LEE virulence genes are down-regulated in the lumen (high fucose), allowing the bacterium to focus its energy on replication rather than virulence expression. In the epithelial pathogen-specific niche (low fucose), where not many commensal competitors reside, EHEC is free to launch its virulence mechanisms to elicit inflammation without regard to the competition. Similarly, the rodent specific, EHEC/EPEC-related bacterium C. rodentium expresses the LEE virulence factors in a fashion similar to EHEC (58). Thus, although the detailed mechanisms remain elusive, this apparent plasticity of virulence factor expression is thought to allow the pathogens to hedge the risk of their extinction in the gut and provides a growth advantage that can be used to compete with the commensals.
Host immunity and the microbiota cooperatively control heterogeneous pathogen populations
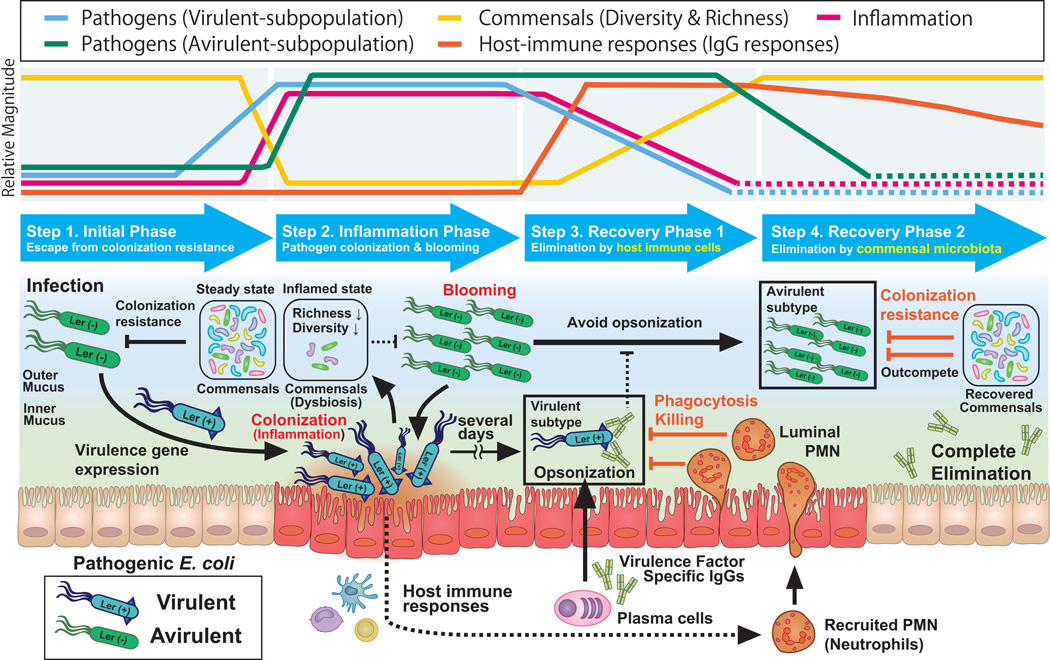
Although colonization resistance mediated by the commensal microbiota is a potent first line defense mechanism against enteric infection, pathogens, through virulence factor utilization, are able to breach this key defense mechanism. However, successful pathogen colonization is not permanent, except in the case of fatal infections. For example, in most non-lethal food-borne infections, including Salmonella or pathogenic E. coli, the pathogen will eventually be eliminated from the gut. In other words, pathogen virulence can overcome commensal microbiota-mediated colonization resistance only in the early phase of infection; the commensal microbiota regains the upper hand in the late phase of infection, outcompeting and eliminating the pathogen. In this context, the remaining question is what tips the balance of power between commensal and pathogenic bacteria during the course of infection. Since virulence is a key strategy employed by pathogens to counter colonization resistance by the commensal microbiota, it is conceivable that virulence is suppressed in the late phase of infection. If the virulence genes are down-regulated, the commensals can outcompete the pathogens. Indeed, C. rodentium expresses the LEE virulence genes in the early phase of infection, but down-regulates them in the late phase of infection (9) (58). In accord with the dynamics of virulence gene expression, the balance of power between C. rodentium and the commensal microbiota shifts from pathogen dominant to commensal dominant during the course of infection. This is because regulation of pathogen virulence also plays a key role in host defense. Host immunity is tolerant of harmless microorganisms, such as the commensal microbiota. The host immune system has developed effective strategies to selectively target pathogens by recognizing the virulence factors they express as danger signals. Recently, it became evident that host adaptive immunity, particularly its antibodies, regulates virulence gene expression in the gut (9) (58). The antibodies induced during infection selectively bind to virulence factors, thus promoting pathogen clearance (58). Importantly, host immunity-mediated protection against virulent pathogens is not sufficient for a complete elimination of pathogens from the gut, because the avirulent subpopulations also arise in the gut during infection. Since it is difficult for the host to discriminate between avirulent pathogens and commensal bacteria, host immunity seems to be anergic to avirulent pathogens. However, since the avirulent subpopulation can acquire a virulent phenotype under certain conditions, it is incumbent upon the host to eliminate the pathogen completely. To this end, the commensal microbiota cooperatively works with host immunity and effects the elimination of avirulent pathogens from the gut (9) (58). Here we illustrate the dynamic interplay among the pathogen, host immunity, and the microbiota during the course of infection using the C. rodentium infection model (Figure 1).
-
Step 1: Pathogens counter colonization resistance mediated by the commensal microbiota through the expression virulence factor, resulting in an increase in the virulent subpopulation (Initial phase, 0–2 days after infection).
After entering the gut, the phenotypically high-virulent (expressing LEE virulence genes) and the low- or avirulent (not expressing LEE virulence genes) subpopulations of C. rodentium arise (58). Although it remains unknown what environmental cues trigger expression of the LEE genes, the LEE-encoded virulence factors enable the pathogen to localize in proximity to the intestinal epithelial surface, allowing it to avoid competition with the commensals for common nutrients (9).
-
Step 2: Inflammation induced by the virulent subpopulation causes dysbiosis, resulting in the bloom of the avirulent subpopulation (Inflammation phase, 2–8 days after infection).
The virulent subpopulation of C. rodentium, localized in the epithelial niche, initiates an extensive remodeling of the host cellular cytoskeleton by forming attaching and effacing (AE) lesions, resulting in the formation of pedestals. Additionally, it induces epithelial damage through the use of the T3SS, a molecular syringe the pathogen employs to inject effector proteins into host cells, resulting in intestinal inflammation (59). Inflammation in the gut induces disruption of the microbiota. As a consequence, the microbiota-mediated colonization resistance is impaired and the conditions in the intestinal lumen favor the formation of bacterial blooms, presumably the fast-growing avirulent subpopulation of C. rodentium.
-
Step 3: Selective killing of the virulent subpopulation of C. rodentium by host immunity results in the diminishment of inflammation and restoration of the commensal microbiota (Recovery phase 1, Day 8–11 post infection).
In the late phase of infection, host immune responses against pathogens have been developed. Particularly, the immunoglobulin (Ig) response is known to be a key adaptive immune mechanism that controls pathogen clearance (60, 61). Notably, the IgGs specific to the LEE encoded virulence factors are primarily induced, specifically targeting the virulent subpopulation of C. rodentium (58). The opsonization of bacteria by IgG most likely does not directly kill the pathogen, but the IgG-flagged bacteria are more efficiently recognized by host innate immune cells, such as neutrophils. In this recovery phase of infection, neutrophils transmigrate from the lamina propria to the gut lumen where they selectively capture and kill the IgG-flagged bacteria. On the other hand, the avirulent subpopulation of C. rodentium and the commensal microbiota are not targeted by host innate immune cells. As a consequence of this selective elimination of the virulent subpopulation, C. rodentium no longer causes intestinal inflammation because the remaining bacteria are phenotypically avirulent. Hence, intestinal inflammation starts to resolve and the perturbed commensal microbiota community concomitantly restores. Consistently, if the host’s ability to produce virulence factor specific IgGs is compromised or the host is neutrophil-deficient, the phenotypically virulent C. rodentium subpopulation stays intact in the gut, thereby continuing to cause epithelial damage, bacterial penetration of the lamina propria, and eventually host lethality (58).
-
Step 4: Elimination of the remaining avirulent subpopulation and a full restoration of commensal microbiota-mediated colonization resistance (Recovery phase 2, Day 12–21 post infection).
Although the phenotypically avirulent subpopulation of C. rodentium is not harmful to the host, it still needs to be eradicated because the down-regulation of virulence factors is plastic and the bacteria can revert to its virulent phenotype. However, host immunity (e.g. IgG and neutrophils) cannot target the avirulent C. rodentium due to the failure of recognition. In this context, the host uses its gut microbiota as a secret weapon. By now the inflammation-induced perturbation of microbial communities is has been resolved due to the absence of intestinal inflammation. The restored and fully functional commensal microbiota is able to outcompete the remaining avirulent C. rodentium bacteria through nutritional competition (99).
Figure 1. Host immunity and the microbiota cooperatively control the elimination of enteric pathogens from the gut.
Enteric pathogens are eliminated by a cooperative mechanism that involves host immunity and the gut resident microbiota as follows: Step 1: Pathogens evade commensal-mediated colonization resistance through virulence gene expression, resulting in a rise of the virulent subpopulation. The virulent subpopulation resides in proximity to the intestinal epithelium. Step 2: The virulent subpopulation induces inflammation. Intestinal inflammation leads to the bloom of the avirulent subpopulation in the gut lumen due to disruption of colonization resistance. Step 3: Virulence factor-specific IgGs are formed and "flag" the virulent subpopulation. The IgG-opsonized virulent subpopulation is targeted and eliminated by intraluminal neutrophils. Elimination of virulent pathogens results in a recovery from inflammation. Microbiota-mediated colonization resistance begins to be restored. Step 4: The remaining avirulent subpopulation is outcompeted by the commensal microbiota; colonization resistance has been fully restored.
Conclusions
Pathogens have developed a wide variety of virulence factors to enhance their own survival in the host. These virulence factors are spatiotemporally regulated in response to the local milleu. The host has also evolved and developed strategies to combat the invading pathogens through a dynamic interplay between the commensal microbiota and its own immune system. However, many questions still remain unaddressed. How does the host discriminate between virulent pathogens and commensals, promoting the specific IgG response only against virulent pathogens? Are there any other mechanisms, aside from opsonization, that regulate pathogens through IgG-binding? Furthermore, it is still unclear to what extent the compromised interplay among pathogen virulence, host immunity and the microbiota contributes to non-infectious GI diseases. For example, in patients with inflammatory bowel disease (IBD), the selective elimination of IgG-bound, potentially pathogenic bacteria seems to be impaired, as it has been reported that the levels of IgG-bound bacteria are increased in the stool of IBD patients (62). However, it is also possible that IBD patients may not appropriately induce IgG against potentially pathogenic bacteria. An impaired IgG response may lead to improper control of pathogenic bacteria; an accumulation of pathogens or abnormal IgG induction against beneficial bacteria may influence the balance of beneficial and pathogenic microorganisms in the gut. Taken together, a more complete understanding of these questions may lead to the development of novel preventative and therapeutic strategies for the control of enteric pathogens as well as non-infectious GI disorders, including IBD.
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad (to H. N.-K. and S. K.), the Crohn’s and Colitis Foundation of America, the Young Investigator Grant from the Global Probiotics Council, and the Michigan Gastrointestinal Research Center grant DK034933 (to N. K.).
Footnotes
Disclosure: The authors declared no conflict of interest.
References
- 1.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. Epub 2010/03/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. Epub 2002/06/11. [DOI] [PubMed] [Google Scholar]
- 3.Atarashi K, Tanoue T, Shima T, et al. Science. 6015. Vol. 331. New York, NY: 2011. Induction of colonic regulatory T cells by indigenous Clostridium species; pp. 337–341. Epub 2011/01/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. Epub 2009/10/17. [DOI] [PubMed] [Google Scholar]
- 5.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. Epub 2009/10/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamada N, Chen GY, Inohara N, et al. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685–690. doi: 10.1038/ni.2608. Epub 2013/06/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. doi: 10.1038/nri3430. Epub 2013/04/27. [DOI] [PubMed] [Google Scholar]
- 8.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13(11):790–801. doi: 10.1038/nri3535. Epub 2013/10/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamada N, Kim YG, Sham HP, et al. Science. 6086. Vol. 336. New York, NY: 2012. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota; pp. 1325–1329. Epub 2012/05/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10(4):336–347. doi: 10.1016/j.chom.2011.10.002. Epub 2011/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.May T, Mackie RI, Fahey GC, Jr, et al. Effect of fiber source on short-chain fatty acid production and on the growth and toxin production by Clostridium difficile. Scand J Gastroenterol. 1994;29(10):916–922. doi: 10.3109/00365529409094863. Epub 1994/10/01. [DOI] [PubMed] [Google Scholar]
- 12.Van Immerseel F, De Buck J, Pasmans F, et al. Invasion of Salmonella enteritidis in avian intestinal epithelial cells in vitro is influenced by short-chain fatty acids. Int J Food Microbiol. 2003;85(3):237–248. doi: 10.1016/s0168-1605(02)00542-1. Epub 2003/07/25. [DOI] [PubMed] [Google Scholar]
- 13.Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2(2):119–129. doi: 10.1016/j.chom.2007.06.010. Epub 2007/11/17. [DOI] [PubMed] [Google Scholar]
- 14.Sekirov I, Tam NM, Jogova M, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76(10):4726–4736. doi: 10.1128/IAI.00319-08. Epub 2008/08/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertin Y, Girardeau JP, Chaucheyras-Durand F, et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011;13(2):365–377. doi: 10.1111/j.1462-2920.2010.02334.x. Epub 2010/09/21. [DOI] [PubMed] [Google Scholar]
- 16.Perna NT, Plunkett G, 3rd, Burland V, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409(6819):529–533. doi: 10.1038/35054089. Epub 2001/02/24. [DOI] [PubMed] [Google Scholar]
- 17.Winter SE, Thiennimitr P, Winter MG, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467(7314):426–429. doi: 10.1038/nature09415. Epub 2010/09/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreyra JA, Wu KJ, Hryckowian AJ, et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16(6):770–777. doi: 10.1016/j.chom.2014.11.003. Epub 2014/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturm A, Heinemann M, Arnoldini M, et al. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 2011;7(7):e1002143. doi: 10.1371/journal.ppat.1002143. Epub 2011/08/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolling G, Wu M, Guerrant RL. Enteric pathogens through life stages. Front Cell Infect Microbiol. 2012;2:114. doi: 10.3389/fcimb.2012.00114. Epub 2012/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellani F, Ghidini V, Tafi MC, et al. Fate of pathogenic bacteria in microcosms mimicking human body sites. Microb Ecol. 2013;66(1):224–231. doi: 10.1007/s00248-013-0239-7. Epub 2013/05/10. [DOI] [PubMed] [Google Scholar]
- 22.Shin S, Castanie-Cornet MP, Foster JW, et al. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol Microbiol. 2001;41(5):1133–1150. doi: 10.1046/j.1365-2958.2001.02570.x. Epub 2001/09/14. [DOI] [PubMed] [Google Scholar]
- 23.Abe H, Tatsuno I, Tobe T, et al. Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2002;70(7):3500–3509. doi: 10.1128/IAI.70.7.3500-3509.2002. Epub 2002/06/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caparon MG, Geist RT, Perez-Casal J, et al. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J Bacteriol. 1992;174(17):5693–5701. doi: 10.1128/jb.174.17.5693-5701.1992. Epub 1992/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tauschek M, Yang J, Hocking D, et al. Transcriptional analysis of the grlRA virulence operon from Citrobacter rodentium. J Bacteriol. 2010;192(14):3722–3734. doi: 10.1128/JB.01540-09. Epub 2010/05/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Tauschek M, Hart E, et al. Virulence regulation in Citrobacter rodentium: the art of timing. Microb Biotechnol. 2010;3(3):259–268. doi: 10.1111/j.1751-7915.2009.00114.x. Epub 2011/01/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan A, Yang J, Tauschek M, et al. Autogenous transcriptional regulation of the regA gene, encoding an AraC-Like, essential virulence regulator in Citrobacter rodentium. J Bacteriol. 2011;193(7):1777–1782. doi: 10.1128/JB.01224-10. Epub 2011/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cummings LA, Wilkerson WD, Bergsbaken T, et al. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol. 2006;61(3):795–809. doi: 10.1111/j.1365-2958.2006.05271.x. Epub 2006/06/29. [DOI] [PubMed] [Google Scholar]
- 29.Cummings LA, Barrett SL, Wilkerson WD, et al. FliC-specific CD4+ T cell responses are restricted by bacterial regulation of antigen expression. Journal of immunology. 2005;174(12):7929–7938. doi: 10.4049/jimmunol.174.12.7929. Epub 2005/06/10. [DOI] [PubMed] [Google Scholar]
- 30.Winter SE, Winter MG, Godinez I, et al. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog. 2010;6(8):e1001060. doi: 10.1371/journal.ppat.1001060. Epub 2010/09/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida T, Cai S, Inouye M. Interaction of EnvZ, a sensory histidine kinase, with phosphorylated OmpR, the cognate response regulator. Mol Microbiol. 2002;46(5):1283–1294. doi: 10.1046/j.1365-2958.2002.03240.x. Epub 2002/11/28. [DOI] [PubMed] [Google Scholar]
- 32.Winter SE, Winter MG, Thiennimitr P, et al. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol Microbiol. 2009;74(1):175–193. doi: 10.1111/j.1365-2958.2009.06859.x. Epub 2009/08/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fink RC, Evans MR, Porwollik S, et al. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s) J Bacteriol. 2007;189(6):2262–2273. doi: 10.1128/JB.00726-06. Epub 2007/01/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marteyn B, West NP, Browning DF, et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465(7296):355–358. doi: 10.1038/nature08970. Epub 2010/05/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunn JS. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2(8):907–913. doi: 10.1016/s1286-4579(00)00392-0. Epub 2000/08/30. [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee A, Dutta PK, Chowdhury R. Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect Immun. 2007;75(4):1946–1953. doi: 10.1128/IAI.01435-06. Epub 2007/01/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowden MJ, Skorupski K, Pellegrini M, et al. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci U S A. 2010;107(7):2860–2865. doi: 10.1073/pnas.0915021107. Epub 2010/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta S, Chowdhury R. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect Immun. 1997;65(3):1131–1134. doi: 10.1128/iai.65.3.1131-1134.1997. Epub 1997/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81(3):1031–1064. doi: 10.1152/physrev.2001.81.3.1031. Epub 2001/06/28. [DOI] [PubMed] [Google Scholar]
- 40.Binder HJ. Role of colonic short-chain fatty acid transport in diarrhea. Annu Rev Physiol. 2010;72:297–313. doi: 10.1146/annurev-physiol-021909-135817. Epub 2010/02/13. [DOI] [PubMed] [Google Scholar]
- 41.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62(1):67–72. doi: 10.1079/PNS2002207. Epub 2003/05/13. [DOI] [PubMed] [Google Scholar]
- 42.Lawhon SD, Maurer R, Suyemoto M, et al. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol. 2002;46(5):1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. Epub 2002/11/28. [DOI] [PubMed] [Google Scholar]
- 43.Robbe C, Capon C, Coddeville B, et al. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. The Biochemical journal. 2004;384(Pt 2):307–316. doi: 10.1042/BJ20040605. Epub 2004/09/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liquori GE, Mastrodonato M, Mentino D, et al. In situ characterization of O-linked glycans of Muc2 in mouse colon. Acta Histochem. 2012;114(7):723–732. doi: 10.1016/j.acthis.2011.12.009. Epub 2012/01/21. [DOI] [PubMed] [Google Scholar]
- 45.Jaswal VM, Babbar HS, Mahmood A. Changes in sialic acid and fucose contents of enterocytes across the crypt-villus axis in developing rat intestine. Biochem Med Metab Biol. 1988;39(1):105–110. doi: 10.1016/0885-4505(88)90064-3. Epub 1988/02/01. [DOI] [PubMed] [Google Scholar]
- 46.Chow WL, Lee YK. Free fucose is a danger signal to human intestinal epithelial cells. Br J Nutr. 2008;99(3):449–454. doi: 10.1017/S0007114507812062. Epub 2007/08/19. [DOI] [PubMed] [Google Scholar]
- 47.Pacheco AR, Curtis MM, Ritchie JM, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492(7427):113–117. doi: 10.1038/nature11623. Epub 2012/11/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atarashi K, Tanoue T, Ando M, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015 doi: 10.1016/j.cell.2015.08.058. Epub 2015/09/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alsharif G, Ahmad S, Islam MS, et al. Host attachment and fluid shear are integrated into a mechanical signal regulating virulence in Escherichia coli O157:H7. Proc Natl Acad Sci U S A. 2015;112(17):5503–5508. doi: 10.1073/pnas.1422986112. Epub 2015/04/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atuma C, Strugala V, Allen A, et al. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280(5):G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. Epub 2001/04/09. [DOI] [PubMed] [Google Scholar]
- 51.Corazziari ES. Intestinal mucus barrier in normal and inflamed colon. J Pediatr Gastroenterol Nutr. 2009;48(Suppl 2):S54–S55. doi: 10.1097/MPG.0b013e3181a117ea. Epub 2009/03/28. [DOI] [PubMed] [Google Scholar]
- 52.Hase CC, Mekalanos JJ. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A. 1999;96(6):3183–3187. doi: 10.1073/pnas.96.6.3183. Epub 1999/03/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sifri CD. Healthcare epidemiology: quorum sensing: bacteria talk sense. Clin Infect Dis. 2008;47(8):1070–1076. doi: 10.1086/592072. Epub 2008/09/11. [DOI] [PubMed] [Google Scholar]
- 54.Chan WC, Coyle BJ, Williams P. Virulence regulation and quorum sensing in staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J Med Chem. 2004;47(19):4633–4641. doi: 10.1021/jm0400754. Epub 2004/09/03. [DOI] [PubMed] [Google Scholar]
- 55.Cheung AL, Bayer AS, Zhang G, et al. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40(1):1–9. doi: 10.1016/S0928-8244(03)00309-2. Epub 2004/01/22. [DOI] [PubMed] [Google Scholar]
- 56.Wuster A, Babu MM. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J Bacteriol. 2008;190(2):743–746. doi: 10.1128/JB.01135-07. Epub 2007/10/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diard M, Garcia V, Maier L, et al. Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature. 2013;494(7437):353–356. doi: 10.1038/nature11913. Epub 2013/02/22. [DOI] [PubMed] [Google Scholar]
- 58.Kamada N, Sakamoto K, Seo SU, et al. Humoral Immunity in the Gut Selectively Targets Phenotypically Virulent Attaching-and-Effacing Bacteria for Intraluminal Elimination. Cell Host Microbe. 2015;17(5):617–627. doi: 10.1016/j.chom.2015.04.001. Epub 2015/05/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collins JW, Keeney KM, Crepin VF, et al. Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol. 2014;12(9):612–623. doi: 10.1038/nrmicro3315. Epub 2014/08/05. [DOI] [PubMed] [Google Scholar]
- 60.Bry L, Brenner MB. Critical role of T cell-dependent serum antibody, but not the gut-associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. Journal of immunology. 2004;172(1):433–441. doi: 10.4049/jimmunol.172.1.433. Epub 2003/12/23. [DOI] [PubMed] [Google Scholar]
- 61.Maaser C, Housley MP, Iimura M, et al. Clearance of Citrobacter rodentium requires B cells but not secretory immunoglobulin A (IgA) or IgM antibodies. Infect Immun. 2004;72(6):3315–3324. doi: 10.1128/IAI.72.6.3315-3324.2004. Epub 2004/05/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158(5):1000–1010. doi: 10.1016/j.cell.2014.08.006. Epub 2014/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]