SUMMARY
Suppression of innate immune responses during filoviral infection contributes to disease severity. Ebola (EBOV) and Marburg (MARV) viruses each encode a VP35 protein that suppresses RIG-I-like receptor signaling and interferon-α/β (IFN-α/β) production by several mechanisms, including direct binding to double-stranded RNA (dsRNA). Here, we demonstrate that in cell culture MARV infection results in a greater upregulation of IFN responses as compared to EBOV infection. This correlates with differences in the efficiencies by which EBOV and MARV VP35s antagonize RIG-I signaling. Furthermore, structural and biochemical studies suggest that differential recognition of RNA elements by the respective VP35 C-terminal IFN inhibitory domain (IID) rather than affinity for RNA by the respective VP35s is critical for this observation. Our results reveal functional differences in EBOV versus MARV VP35 RNA binding result in unexpected differences in the host response to deadly viral pathogens.
Keywords: filovirus, immune evasion, pattern associated molecular pattern (PAMP), RIG-I like receptor, type I interferon, VP35
INTRODUCTION
Zaire ebolavirus (EBOV) and Marburg marburgvirus (MARV) are members of the Filoviridae family of negative sense ssRNA viruses and cause highly lethal hemorrhagic fever in humans (Bray and Murphy, 2007). The virulence of filoviruses is due in part to the potent inhibition of the innate immune system (Basler and Amarasinghe, 2009; Messaoudi and Basler, 2015). Although both EBOV and MARV inhibit the production of interferon (IFN)-α/β and the ability of cells to respond to IFNs, the mechanisms of inhibition differ. For example, the EBOV VP24 protein inhibits IFN-induced Jak-STAT signaling by blocking karyopherin alpha mediated nuclear accumulation of tyrosine phosphorylated STAT1 whereas MARV VP40 prevents STAT protein tyrosine phosphorylation (Mateo et al., 2010; Reid et al., 2006; Reid et al., 2007; Valmas and Basler, 2011; Valmas et al., 2010; Xu et al., 2014).
EBOV VP35 (eVP35) and MARV VP35 (mVP35) also block IFN production by binding dsRNAs through the C-terminal IFN inhibitory domain (IID) and prevent retinoic-acid inducible gene-I (RIG-I)-like receptor (RLR) activity (Albarino et al., 2015; Cardenas et al., 2006; Hartman et al., 2006; Leung et al., 2010a; Prins et al., 2010; Ramanan et al., 2012; Yen et al., 2014). Mutation of VP35 residues critical for dsRNA binding results in increased IFN-α/β responses, reduced viral replication, and attenuation of EBOV in animal models, demonstrating the importance of VP35 as a virulence determinant (Hartman et al., 2008b; Prins et al., 2010). Despite functional and structural similarities, comparison of the crystal structures of eVP35 and mVP35 IIDs in complex with dsRNA suggests differences in how eVP35 and mVP35 interact with dsRNA. Specifically, eVP35 interacts with the phosphodiester backbone and caps the ends of dsRNA (Kimberlin et al., 2010; Leung et al., 2010), preventing PAMP recognition by RIG-I. However, evidence for end-capping interactions by mVP35 is lacking and mVP35 appears to interact with the dsRNA backbone only (Ramanan et al., 2012). The biological consequences of these differences are unclear.
Here, we compared antiviral responses to EBOV and MARV infections in THP-1 cells and investigated the mechanistic basis for the suppression of IFN-α/β responses by EBOV and MARV VP35s. Our data reveal that MARV infections trigger a greater IFN response than does EBOV, which correlates with a stronger inhibition of RLR signaling by eVP35 compared to mVP35. This functional difference can be mapped to VP35 IID and its capacity to block PAMP recognition by RLRs. Our data, for the first time, implicate the mode of interaction of viral VP35 with immunostimulatory RNA as a determinant of early host IFN response to filovirus infection. These observations also demonstrate that complete suppression of IFN-α/β responses is not a prerequisite for MARV to cause severe disease.
RESULTS
MARV and EBOV show differential expression of IFN-regulated genes
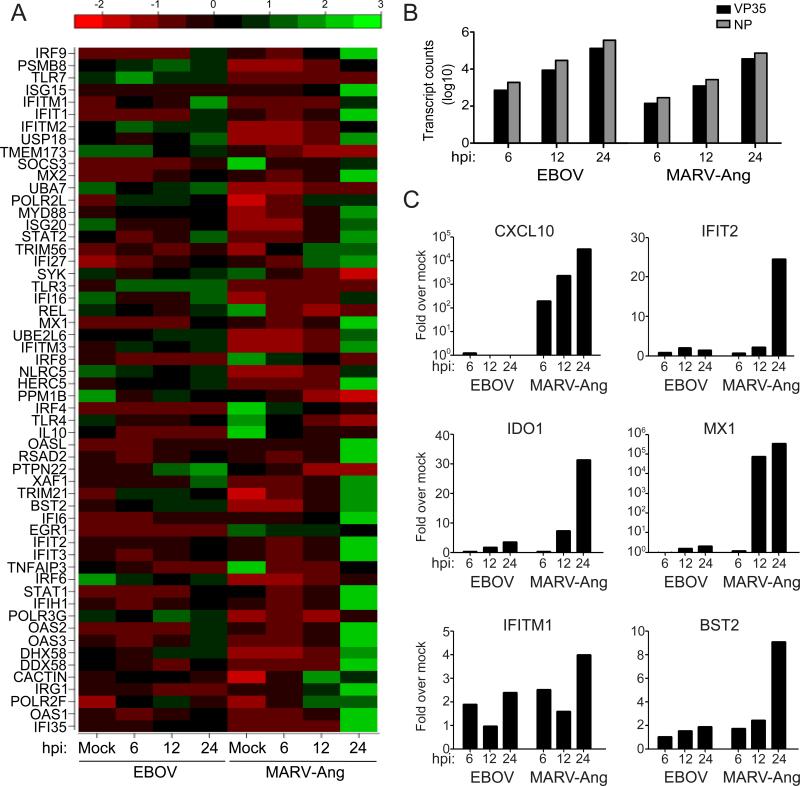
RNA sequencing of mRNAs from THP-1 cells infected with EBOV or the MARV Angola strain (MARV-Ang) at a multiplicity of infection (MOI) of 3 demonstrated a substantial upregulation of IFN-stimulated genes (ISGs) by 24 hours post-infection (hpi) by MARV-Ang but not EBOV (Figure 1A). MARV NP and VP35 mRNA levels were modestly lower than EBOV VP35 mRNA levels at the three time points assayed, suggesting slightly slower replication for MARV (Figure 1B). The enhanced induction of ISGs in MARV-infected versus EBOV-infected cells was confirmed by performing quantitative RT-PCR (qRT-PCR) for six representative ISG mRNAs (Figure 1C). To determine whether the induction of an IFN response was specific to the MARVAng strain, the expression of the same six ISGs was assessed following new infections of THP-1 cells with MARV-Ang, MARV Musoke (MARV-Mus) or EBOV at an MOI of 1 (Figure S1). MARV-Ang and EBOV NP mRNA levels were comparable, but MARV-Mus NP mRNA levels were lower at 6 hpi (Figure S1A). By 24 hpi however, NP mRNA levels were comparable among all three infections. qRT-PCR also confirmed the lack of ISG induction in EBOV-infected cells in contrast to the upregulation of ISGs by MARV-Ang. MARV-Mus also exhibited upregulation of several ISGs, although the levels of induction were not as uniform or robust as for MARV-Ang, including at 24 hpi when viral replication levels were comparable (Figure S1B). These data suggest that MARV infection activates a greater IFN response than does EBOV, with some differences observed between MARV strains.
Figure 1.
EBOV and MARV show differential expression of IFN-regulated genes. (A-C) THP-1 cells were either mock-infected or infected with EBOV or MARV-Ang at an MOI of 3. (A) RNA from infected THP-1 cells was subjected to expression analysis by mRNA deep sequencing (mRNA-seq). The heatmap displays the expression profile of IFN response genes at 6, 12, and 24 hpi. The expression levels have been normalized to account for varying library sizes. These have been scaled and centered, with green indicating larger number of transcripts and red fewer transcripts. (B) Filoviral VP35 and NP mRNA expression levels represented in terms of transcript counts. (C) Expression levels of 6 ISGs from EBOV and MARV-Ang infected cells were analyzed by qRT-PCR at 6, 12 and 24 hpi. Values were normalized to RPS11. The y-axis scale for CXCL10 and MX1 is log10. The y-axis scale for IFIT2, IDO1, IFITM1 and BST2 is linear.
MARV VP35 and EBOV VP35 inhibit virus induced IFN-β production with different efficiencies
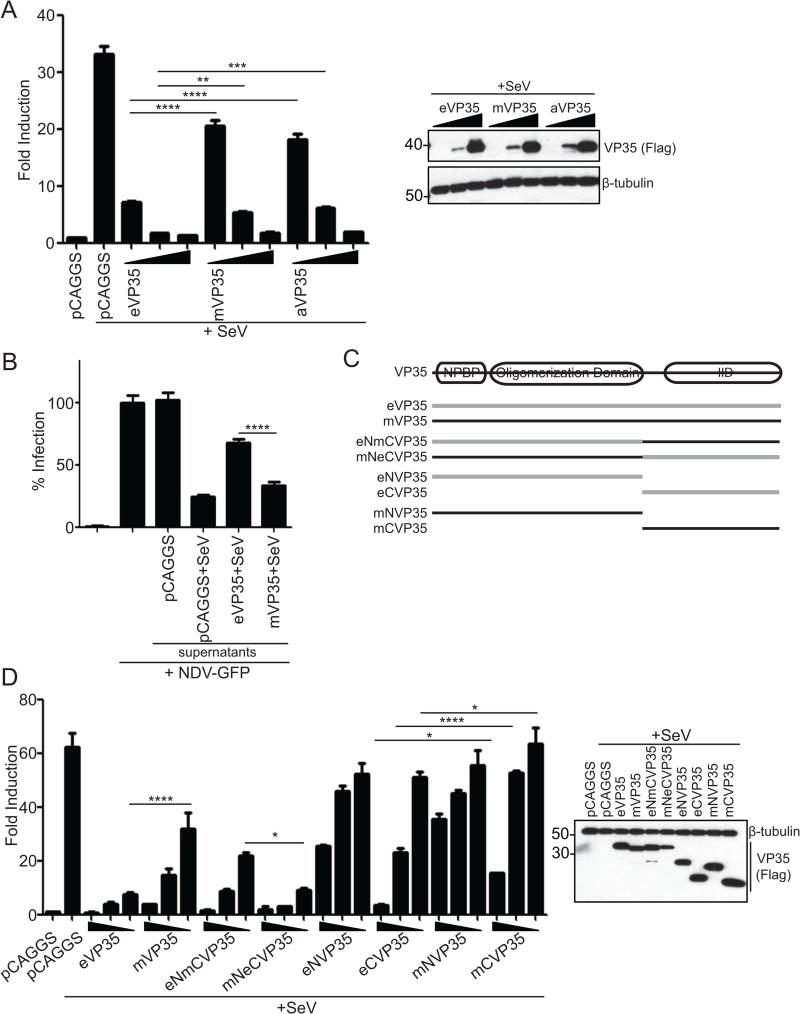
To determine whether the differences in IFN gene expression might be attributable to VP35 RIG-I inhibitory activity, Flag-tagged EBOV VP35 (eVP35), MARV-Mus (mVP35) or MARV-Ang (aVP35) were evaluated for inhibition of IFN-β promoter activity induced by Sendai virus (SeV) infection, a documented RIG-I activator (Kato et al., 2006). Although all VP35 proteins tested inhibited IFN-β promoter activity, eVP35 was a more potent inhibitor compared to mVP35 or aVP35 at comparable protein expression levels (Figure 2A). As both mVP35 and aVP35 inhibit SeV induced IFN-β reporter activity to statistically indistinguishable degrees, mVP35 was used in subsequent studies.
Figure 2.
eVP35 and mVP35 inhibit virus induced IFN-β production with different efficiencies. (A) An IFN-β luciferase reporter assay in the presence of empty vector (pCAGGS) or increasing concentrations of eVP35, mVP35 or aVP35 (2.5, 25, and 250ng). 24 h post transfection cells were infected with SeV and IFN-β activity was assessed by measuring luciferase activity 18 hours later. VP35 expression was assessed by western blot for Flag (Right). (B) An IFN bioassay in the presence of empty plasmid (pCAGGS), eVP35, or mVP35. Transfected cells were infected with SeV to activate RIG-I signaling. Supernatants were harvested 18 hpi and UV inactivated. Serial 5-fold dilutions of the supernatant were used to overlay Vero cells. 24 h later, Vero cells were infected with NDV-GFP (MOI=1) and the percent infected cells was determined 18 hours later by monitoring GFP. The GFP signal obtained for Vero cells untreated by supernatants and infected with NDV-GFP was set at 100%; all other conditions were normalized to this control. Shown are results from 1:625 dilution of supernatants. (C) Schematic of chimeric and truncated VP35 constructs. Filovirus VP35 proteins contain an N-terminal nucleoprotein binding peptide (NPBP) followed by an oligomerization domain and a C-terminal dsRNA-binding domain termed the interferon inhibitory domain (IID). Chimeric VP35s were designed by swapping the C-terminus of EBOV (gray) and MARV (black). (D) Same assay protocol as (A) but transfected with wildtype eVP35, wildtype mVP35, chimeric VP35, or N- or C-terminal VP35s. Protein expression for the controls and the samples receiving the highest concentration of VP35 constructs was assessed by western blot for Flag (Right). In (A), (B) and (D), values represent the mean and SEM of triplicate samples, and statistical significance was assessed by a one-way ANOVA comparing columns as indicated: *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
We used an IFN bioassay to measure the release of endogenous IFN-α/β in cells expressing either eVP35 or mVP35, where replication of an IFN-sensitive recombinant Newcastle disease virus (NDV) that expresses GFP serves as a readout for antiviral activity. Supernatant from empty vector-transfected SeV-infected cells inhibited NDV-GFP infection, indicating the presence of IFN. Supernatants from mVP35 expressing cells also showed high levels of antiviral activity with a corresponding decrease in NDV-GFP infection (Figure 2B), consistent with the elevated expression levels of ISGs observed in cells infected with MARV (Figure 1). In contrast, supernatant from eVP35 expressing cells displayed reduced antiviral activity, consistent with better suppression of IFN-α/β production by eVP35.
To determine the efficiency by which eVP35 and mVP35 inhibit different steps in the RLR signaling pathway, eVP35 and mVP35 were co-expressed with activators of the IFN-α/β response, including a constitutively active form of RIG-I (RIG-IN), TBK1, or IKKε. Inhibition of IFN induction by all these signaling proteins was comparable between eVP35 and mVP35 (Figure S2A-C). However, when full-length RIG-I was expressed in RIG-I knockout cells infected with SeV, eVP35 was the more potent inhibitor than mVP35 (Figure S2D), suggesting differential inhibition of the RIG-I pathway at a point upstream of activated RIG-I.
To map the determinant of the different inhibitory efficiencies, we generated truncated and chimeric VP35 proteins (Figure 2C). As previously shown, wildtype eVP35 was more efficient at inhibiting SeV induced IFN-β reporter activity than wildtype mVP35 (Figure 2D). The N-terminus alone of eVP35 or mVP35 was only modestly inhibitory. The C-terminal IID of eVP35 displayed more robust inhibitory activity compared to mVP35 IID (Figure 2D). The eNmCVP35 construct, which contains the eVP35 N-terminus and mVP35 IID, lost inhibitory activity compared to wildtype eVP35 and more closely resembled wildtype mVP35 (Figure 2D). Conversely, mNeCVP35, which contains the mVP35 N-terminus and eVP35 IID, behaved similarly to wildtype eVP35. These results suggest that the IID is the source of functional differences between eVP35 and mVP35.
eVP35 IID and mVP35 IID have different affinities and binding modes for dsRNA
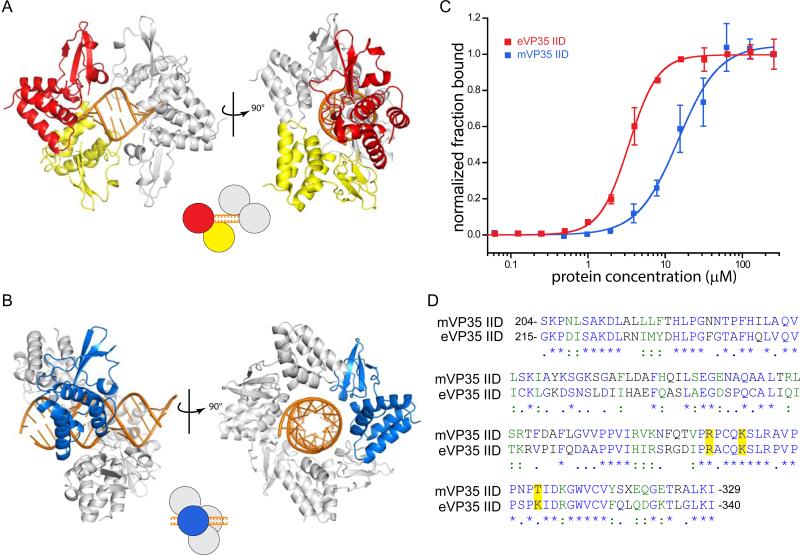
The crystal structures of eVP35 IID and mVP35 IID in complex with dsRNA and corresponding biochemical analyses revealed that eVP35 IID bound to both the backbone and blunt ends of dsRNA (Figure 3A) (Leung et al., 2010b), whereas mVP35 IID bound only to the dsRNA backbone (Figure 3B) (Ramanan et al., 2012; Bale et al., 2012). To understand the significance of the different modes of dsRNA interaction, we measured the affinities of eVP35 IID and mVP35 IID for 25 bp dsRNA using a dot blot assay and found that eVP35 IID has a 4-fold higher affinity for dsRNA than mVP35 IID (KD,eIID = 3.40 ± 0.070 μM vs. KD,mIID = 14.8 ± 1.1 μM) (Figure 3C).
Figure 3.
eVP35 IID and mVP35 IID have different interaction modes and binding affinities for dsRNA. (A) The two types of interactions are highlighted in the cartoon representation of the asymmetric unit of PDB 3L25. eVP35 IID recognizes both the blunt ends (red molecule) and double stranded backbone of dsRNA (yellow molecule). Symmetry related eVP35 IID molecules are colored gray. (B) mVP35 IID interacts with the backbone of dsRNA. Shown are the mVP35 IID molecules in the crystal structure (PDB: 4GHL). One mVP35 IID molecule is colored blue and the symmetry mates are shown in gray. (C) Dot blot assay shows that eVP35 IID (gray) has a higher affinity for 25 bp than mVP35 IID (black). 0 to ~250 μM of IID (with 2-fold dilutions) were incubated with 5 nM of P32-25 bp dsRNA. Fraction bound of dsRNA was normalized and plotted against protein concentration (μM). (D) Sequences of mVP35 IID and eVP35 IID are aligned using ClustalW. Fully conserved residues are colored blue and labeled with an asterisk (*); strongly conserved green and with a colon (:); and weakly conserved navy and with a period (.). Residues, R305, K309, and K319, are highlighted.
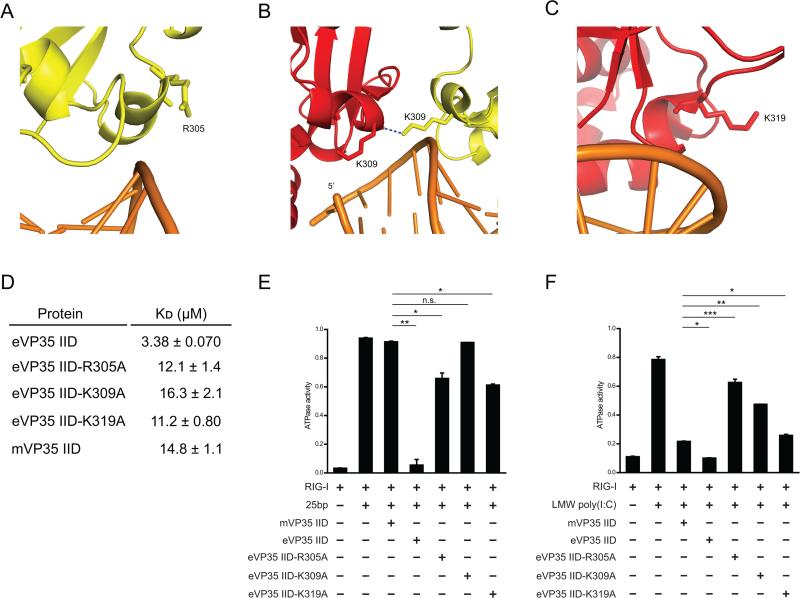
To test if the differences in binding affinities for dsRNA are the main contributors to relative RIGI inhibition, we generated mutant eVP35 IIDs with reduced dsRNA affinity to serve as functional mimics of mVP35 IID. eVP35 residues R305, K309, and K319 were mutated because of their proximity to basic residues that are directly involved in dsRNA interactions (R312, R322, K339, and F239). While the side chains of R305, K309, and K319 do not form direct interactions with dsRNA, they are involved in van der Waals contacts or water-mediated contacts with dsRNA and therefore contribute to electrostatic interactions that are important for dsRNA binding (Figure 4A-C). Dot blot assays demonstrated that mutation of residues R305, K309, and K319 results in a decrease in affinity for 25 bp dsRNA, yielding KD values comparable to that of mVP35 IID (Figure 4D).
Figure 4.
Differential inhibition of RLR activity by eVP35 IID and mVP35 IID. (A-C) Three residues in the central basic patch of eVP35 IID were selected for mutational analysis. Color scheme is the same as in Figure 3. Residues (A) R305, (B) K309, and (C) K319 are shown in stick representation. Hydrogen bonding is shown as blue dotted lines. (D) Dissociation constants (KD) are calculated from the dot blot assay data for VP35 IID (wildtype and mutants) and 25 bp dsRNA. (E-F) ATPase assays were used to test inhibitory effects of eVP35 IID wildtype, eVP35 IID mutants, and mVP35 IID. RIG-I (1 μM) was activated by (E) 10 ng of 25 bp dsRNA or (F) 50 ng of LMW poly(I:C) in the presence and absence of IID at 25 μM. Statistical significance is as indicated: n.s. not significant, *p<0.05, **p<0.01 and ***p<0.001.
To test whether eVP35 and mVP35 IID inhibition of RIG-I can be attributed to interaction with a RNA element, namely the dsRNA backbone, the dsRNA blunt ends, or both, we measured inhibition of RIG-I ATPase activity by VP35 IID in vitro. We used short 5’-triphosphate dsRNA (25 bp), longer dsRNA, or poly(I:C) as RIG-I activators. By varying the lengths of dsRNA, we varied the ratio of blunt ends to double strandedness, which enabled us to assess the role of each RNA element. When RIG-I is activated with 25 bp dsRNA, eVP35 IID is a potent inhibitor of RIG-I activity whereas mVP35 IID displayed >1000-fold lower level of inhibition (Figure 4E, bars 2-4). This observation was consistent over a 50-fold concentration range (0.0005 mM to 0.025 mM) for mVP35 IID, while only 0.025 mM eVP35 IID was required to achieve near-complete inhibition of RIG-I ATPase activity (Figure S3A-B). We also tested full-length VP35, which is tetrameric due to the presence of an N-terminal oligomerization domain (Table S1). While oligomerization enhanced the overall inhibition observed for eVP35 IID and mVP35 IID, eVP35 was the better inhibitor of RIG-I activity (Figure S3A, S3B). Therefore, when blunt ends are the more prevalent PAMP than double-strandedness, eVP35 is a better inhibitor.
We next tested the eVP35 IID R305A, K309A, and K319A mutants and observed that all mutants were impaired in their ability to inhibit RIG-I compared to wildtype eVP35 IID (Figure 4E), suggesting that the decreased affinity for dsRNA corresponds to diminished inhibition of RIG-I ATPase activity. However, eVP35 IID R305A and K319A inhibit RIG-I activation by short dsRNA more efficiently than eVP35 IID K309A (Figure 4E, bars 2 and 5-7). eVP35 IID K309A and mVP35 IID have affinities for dsRNA comparable to the other eVP35 IID mutants, yet these two proteins exhibit more severe defects as compared to the R305A and K319A mutants. This suggests that VP35-mediated inhibition of RIG-I does not strictly depend upon the IID affinity for dsRNA. Instead, the capacity of wildtype eVP35 and the R305A and K319A mutants to mask the 5’-triphosphate on the blunt ends of dsRNA further contributes to effective suppression of RIG-I.
We next used LMW poly(I:C) to activate RIG-I in the ATPase assay. Unlike 25 bp dsRNA, LMW poly(I:C) (0.2-1 kbp) contains longer stretches of double-stranded RNA and therefore, double-strandedness is likely the predominant PAMP. Our results show that both eVP35 IID and mVP35 IID inhibit RIG-I ATPase activity with similar efficiency when stimulated with poly(I:C) (Figure 4F, lanes 2-4). Therefore, when double-strandedness predominates, the difference in inhibitory activity between eVP35 and mVP35 decreases, suggesting that the VP35 binding mode is a determinant of RLR inhibition.
eVP35 IID R305A, K309A and K319A mutants are less effective inhibitors of RIG-I activation by poly(I:C) than wildtype eVP35 and mVP35 IIDs. This can be attributed to the fact that key residues within the central basic patch (CBP) are important for both dsRNA binding and protein-protein interactions between protomers of VP35 IID that can stabilize the dsRNA bound conformation of the VP35 IID when bound to dsRNA (Bale et al., 2013; Leung et al., 2010a). Therefore, the effect of mutating CBP residues reflects not only the impact of the binding modes (i.e. potential loss of blunt end binding) but also the impact of binding affinities between VP35 IID and dsRNA. Together, these data demonstrate that eVP35 IID and mVP35 IID use multiple modes of interaction with RNA PAMPs to prevent RLR activation.
To further assess the ability of the VP35s to inhibit dsRNAs with different ratios of double-strandedness to blunt ends in a cell-based system, we used SeV defective interfering (DI) RNAs that were previously described as specific activators of RIG-I (Patel et al., 2013). These DI RNAs have the same sized loop region with differing lengths of complementary dsRNA stem regions (25, 46, and 94 bp). Consistent with previous studies, we observe a corresponding increase in IFN-β promoter activity as the length of the double-stranded region increases (Figure S3C) (Patel et al., 2013). Furthermore, eVP35 displays modest differences in inhibition of IFN-β reporter activity compared to mVP35 at all lengths of SeV DI dsRNA tested (Figure S3C), further supporting our in vitro observations.
eVP35 IID and mVP35 IID inhibit RIG-I activation by PACT with similar efficiency
Because previous studies demonstrated that PACT activation of RIG-I is inhibited by eVP35 (Luthra et al., 2013), we also performed the ATPase assay in the presence of PACT (Figure S4). mVP35 IID was able to inhibit RIG-I activation by PACT with the same efficiency as eVP35 IID (Figure S4A). Furthermore, there was no significant difference in the ability of eVP35 or mVP35 to inhibit PACT-mediated enhancement of RIG-I induced IFNβ reporter assay when PACT was the only stimulus (Figure S4B). However, in the presence of SeV infection, eVP35 inhibited IFNβ reporter activity more efficiently than mVP35 (Figure S4C). These observations suggest that inhibition of PACT does not account for the differences in IFN suppression activity between eVP35 and mVP35 and point to differences related to suppression by an RNA stimulus.
DISCUSSION
Our data reveals a surprising difference in how EBOV and MARV VP35 proteins modulate IFN responses. Prior studies have demonstrated that the VP35 proteins of EBOV and MARV suppress IFN-α/β production induced via the RLR pathway (Bale et al., 2012; Bale et al., 2013; Cardenas et al., 2006; Leung et al., 2010a; Prins et al., 2009; Ramanan et al., 2012) and mutation of residues in VP35 IIDs results in attenuation of recombinant EBOV or MARV in IFN-producing cells (Albarino et al., 2015; Hartman et al., 2006; Prins et al., 2010). For EBOV, the VP35 IFN-antagonist function was also required for robust growth and virulence in rodent models (Hartman et al., 2008a; Prins et al., 2010). Therefore, it was surprising that in macrophage-like THP-1 cells either of two MARV strains induced an IFN response that is absent in EBOV infected cells. Importantly, the MARV-Ang isolate used in our studies was from the Angola outbreak in 2005, which had an 88 percent case fatality rate and is highly lethal in non-human primates (Geisbert et al., 2007). Therefore, the increased IFN responses observed for MARV do not appear to attenuate the virus. These observations suggest that an undefined mechanism(s) may allow MARV to tolerate a higher level of IFN response than EBOV.
Direct comparison of the dsRNA-binding properties of eVP35 and mVP35 provides an explanation as to why EBOV and MARV induce different degrees of IFN response. We, and others, have shown that filoviral VP35 proteins function to inhibit IFN responses via the IID through direct interaction with dsRNA, thereby competing with the cytosolic PAMP receptors RIG-I and MDA5 (Bale et al., 2012; Bale et al., 2013; Kimberlin et al., 2010; Leung et al., 2010a; Ramanan et al., 2012). However, it was unclear from previous studies if the lack of dsRNA blunt end binding by mVP35 IID resulted in a functionally significant outcome. Our data here show that select eVP35 IID mutants (R305A and K319A) retain some ability to bind dsRNA blunt ends and limit IFN production resulting from blunt end mediated RLR activation, suggesting that the binding mode by which filoviral VP35 IID interacts with dsRNA plays a more significant role in the cellular IFN response to filovirus infections than previously appreciated. Whether the inhibitory activities of either MARV VP40 or EBOV VP24, which antagonize IFN-induced Jak-STAT signaling, also contribute to the differential IFN response requires further study.
Our findings have implications for viral infection, diagnosis and therapy. EBOV and MARV each induce severe hemorrhagic fever with high case fatality rates and similar pathogenesis (Martines et al., 2015). The viruses also share many similarities in their replication cycles (Sanchez et al., 2007). Despite these similarities, our present data and other studies demonstrate important differences in the molecular biology of these pathogens. Accumulating data point to differential interactions with host signaling pathways including pathways connected to host IFN responses (Edwards and Basler, 2015; Edwards et al., 2014; Page et al., 2014; Reid et al., 2006; Shabman et al., 2014; Valmas et al., 2010; Xu et al., 2014). The results presented here identify additional differences with regard to suppression of an IFN response that should be taken into account when developing antifiloviral countermeasures.
METHODS
Constructs
Expression plasmids in the pCAGGs backbone include Flag-tagged eVP35, mVP35 IKKε, TBK-1 and HA-tagged RIG-I have been described previously (Cardenas et al., 2006; Ramanan et al., 2012). aVP35 was amplified from RNA isolated from MARV-Ang infected THP-1 cells and cloned into pCAGGS with an N-terminal Flag tag. Flag-tagged truncations of eVP35 or mVP35 included eNVP35 (residues 1-198), eCVP35 (residues 199-340), mNVP35 (residues 1-187) and mCVP35 (residues 188-329). eNmCVP35 contains residues 1-198 from eVP35 and 188-329 from mVP35. mNeCVP35 contains residues 1-187 from mVP35 and 199-340 from eVP35.
Generation of proteins and RNA
VP35 wildtype and mutant proteins were expressed and purified as previously described (Leung et al., 2010a; Ramanan et al., 2012). Mutations were introduced into the sequence through overlap PCR and confirmed by DNA sequencing. 25 bp dsRNA and LMW poly(I:C) were purchased from IDT Technologies (Coralville, IA) and Invivogen (San Diego, CA), respectively, and desalted prior to use. The sequence of 25 bp dsRNA used in the ATPase and filter binding assays is rArArArCrUrGrArArArGrGrGrArGrArArGrUrGrArArArGrUrG for the sense strand and rCrArCrUrUrUrCrArCrUrUrCrUrCrCrCrUrUrUrCrArGrUrUrU for the antisense strand.
RNA Binding Assays
25 bp dsRNA was labeled by 32P-ATP and purified by 10% urea gel. 5 nM RNA was incubated with VP35 IID constructs at various concentrations for 15 min, loaded onto a dot blot apparatus (Bio-Rad), and passed through nitrocellulose (NC) and nylon (NY) membranes. Membranes were scanned using a Typhoon 9410 Variable Model Imager (GE Healthcare) and the amount of dsRNA bound to NC and NY membranes was quantified from the radioactivity detected. The fraction of RNA bound to IID proteins was calculated using the equation shown below:
Data were fit to the Hill equation and dissociation constants (KD) were calculated using Origin software.
ATPase Assays
MBP tagged RIG-I (1 μM) was incubated with 32P-ATP (1 μCi), and dsRNA or MBP tagged PACT (0.7 μM) was used as the agonist in the absence or presence of VP35 IID proteins for 30 min. One microliter of the reactions was loaded onto a PEI (polyethyleneimine)-cellulose TLC plate and developed in 3M KH2PHO4 buffer. The ATPase activity of RIG-I was quantified by the relative amount of 32P inorganic phosphate (Pi) present using the following equation:
Cells and viruses
HEK293T cells were maintained in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum and cultured at 37°C and 5% CO2. SeV and NDV were grown in 10-day-old embryonated chicken eggs for two days at 37 °C.
Western blots and antibodies
Lysates were run on 10% acrylamide SDS-PAGE gels and transferred to PVDF membranes. Membranes were probed with monoclonal mouse anti-Flag M2 (Sigma Aldrich) and anti-β-tubulin antibodies (Sigma Aldrich) prior to development with Western Lightning ECL (Perkin-Elmer).
Filovirus Infections
Infections were performed under BSL-4 conditions at the Galveston National Laboratory. THP-1 cells were differentiated overnight with 100 nM PMA and infected with MARV-Ang (MOI = 3 or 1), MARV-Mus (MOI = 1), or EBOV (MOI = 3). Viral total RNA was extracted with TRIzol at the indicated time points for analysis by deep sequencing or qRT-PCR. For deep sequencing, mRNA was purified with Oligo(dT) magnetic beads (Invitrogen). cDNA libraries were generated (NEBNext; New England Biolabs) and sequenced on the Illumina HiSeq 2500 platform as described (Shabman et al., 2014). The sequenced reads were aligned to human genome hg19 using TopHat2 (Kim et al., 2013) and gene-wise transcript numbers were counted and normalized using DESeq2 (Love et al., 2014). To visualize the transcription changes in IFN-α/β and ISG mRNA levels, we used these gene-wise transcript counts, centered and scaled them based on the mean and variance of all the counts from all time points. The heatmap was generated using levelplot function in R. To quantify the viral transcripts, the reads were aligned to respective viral genomes using Bowtie2 (Langmead and Salzberg, 2012) and gene-wise transcript numbers calculated using intersectBed function from BEDTools package (Quinlan and Hall, 2010). For qRT-PCR, cDNA was generated with Oligo(dT) primers, and relative expression for each gene of interest was determined by normalizing to RPS11.
IFN Bioassay
An NDV-GFP bioassay was used to quantify secreted IFN-α/β levels according to previously described methods (Shaw et al., 2004). Briefly, supernatants from HEK293T cells transfected with empty plasmid, eVP35 or mVP35 and infected with SeV were UV irradiated for 10 minutes on ice to inactivate the SeV. Vero cells grown in 96-well format were overlaid with serial 5-fold dilutions of the supernatants for 24 h, after which they were infected with NDV-GFP as indicated at an MOI of 3. 24 hpi cells were fixed with 4% paraformeldehyde. GFP fluorescence was measured using the Acumen Explorer HTS plate reader. GFP fluorescence from Vero cells untreated by supernatant and infected with NDV-GFP was set at 100% infection and all other conditions were normalized to this. Error bars represent the mean and SEM of triplicate samples. Statistical significance was assessed by a one-way ANOVA and Student's T-test.
IFN-β Luciferase Reporter Assays
SeV-induced reporter assays: HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen) with the IFN-β firefly luciferase reporter plasmid, a constitutively expressed Renilla luciferase reporter plasmid (pRLTK, Promega), and the indicated expressions plasmids. 24 h post transfection, cells were mock infected or infected with SeV to induce reporter activation. 18 hpi luciferase activity was determined. RIG-I and kinase overexpression induced reporter assays: HEK293T cells were transfected as before with the addition of RIG-IN, IKKε or TBK-1 as indicated. 18 hpi, luciferase activity was determined. Reporter assays in RIG-I KO cells: RIG-I KO HEK293T cells were transfected as before with the addition of RIG-I as indicated. 24 h post transfection cells were infected with SeV or transfected with LMW poly(I:C) (250ng) (Invivogen) for 18 hours after which luciferase activity was determined. SeV DI RNA induced reporter assay: SeV DI RNAs were generated as previously described (Patel et al., 2013). HEK293T cells were transfected as before. 24 h post transfection cells were transfected with SeV DI RNAs (35fmol) to induce reporter activity for 24 h after which luciferase activity was determined. For all reporter assays, a dual luciferase assay (Promega) was performed and firefly luciferase values were normalized to Renilla luciferase values. Error bars represent the mean and SEM of triplicate samples. Statistical significance was assessed with a one-way ANOVA followed by an independent samples t-test with the Bonferroni correction.
Supplementary Material
Acknowledgments
This work is supported in part by NIH grants (R01AI107056 to D.W.L.; R01AI059536 to C.F.B.; U19AI109945 (Basler-PI) to C.F.B., G.K.A., I.M. and T.W.G. and U19AI109664 (Basler-PI) to C.F.B. and G.K.A.; and by Department of the Defense, Defense Threat Reduction Agency grant HDTRA1-12-1-0051 (to C.F.B., G.K.A. and T.W.G.). The content of the information does not necessarily reflect the position or the policy of the federal government, and no official endorsement should be inferred. We thank Christine Schwall-Pecci for help with manuscript editing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, M.R.E., G.L., R.S.S., D.W.L., T.W.G., G.K.A. and C.F.B.; Investigation, M.R.E., G.L., C.E.M., P.L. and R.S.S.; Formal Analysis, S.S.; Resources, B.Y.; Writing – Original Draft, M.R.E., G.L., D.W.L., G.K.A. and C.F.B.; Writing – Review & Editing, M.R.E., G.L., D.W.L., G.K.A. and C.F.B.; Funding Acquisition, D.W.L., I.M., T.W.G., G.K.A. and C.F.B. Supervision, D.W.L., I.M., T.W.G., G.K.A. and C.F.B.
References
- Albarino CG, Wiggleton Guerrero L, Spengler JR, Uebelhoer LS, Chakrabarti AK, Nichol ST, Towner JS. Recombinant Marburg viruses containing mutations in the IID region of VP35 prevent inhibition of Host immune responses. Virology. 2015;476:85–91. doi: 10.1016/j.virol.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Julien JP, Bornholdt ZA, Kimberlin CR, Halfmann P, Zandonatti MA, Kunert J, Kroon GJ, Kawaoka Y, MacRae IJ, et al. Marburg virus VP35 can both fully coat the backbone and cap the ends of dsRNA for interferon antagonism. PLoS pathogens. 2012;8:e1002916. doi: 10.1371/journal.ppat.1002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Julien JP, Bornholdt ZA, Krois AS, Wilson IA, Saphire EO. Ebolavirus VP35 coats the backbone of double-stranded RNA for interferon antagonism. Journal of virology. 2013;87:10385–10388. doi: 10.1128/JVI.01452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Amarasinghe GK. Evasion of interferon responses by Ebola and Marburg viruses. J Interferon Cytokine Res. 2009;29:511–520. doi: 10.1089/jir.2009.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Murphy FA. Filovirus research: knowledge expands to meet a growing threat. J Infect Dis 196 Suppl. 2007;2:S438–443. doi: 10.1086/520552. [DOI] [PubMed] [Google Scholar]
- Cardenas WB, Loo YM, Gale M, Jr., Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Basler CF. Marburg Virus VP24 Protein Relieves Suppression of the NF-kappaB Pathway Through Interaction With Kelch-like ECH-Associated Protein 1. The Journal of infectious diseases. 2015 doi: 10.1093/infdis/jiv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Johnson B, Mire CE, Xu W, Shabman RS, Speller LN, Leung DW, Geisbert TW, Amarasinghe GK, Basler CF. The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep. 2014;6:1017–1025. doi: 10.1016/j.celrep.2014.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Daddario-DiCaprio KM, Geisbert JB, Young HA, Formenty P, Fritz EA, Larsen T, Hensley LE. Marburg virus Angola infection of rhesus macaques: pathogenesis and treatment with recombinant nematode anticoagulant protein c2. The Journal of infectious diseases. 2007;196(Suppl 2):S372–381. doi: 10.1086/520608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Bird BH, Towner JS, Antoniadou ZA, Zaki SR, Nichol ST. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of ebola virus. Journal of virology. 2008a;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Bird BH, Towner JS, Antoniadou ZA, Zaki SR, Nichol ST. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of ebola virus. J Virol. 2008b;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Dover JE, Towner JS, Nichol ST. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. Journal of virology. 2006;80:6430–6440. doi: 10.1128/JVI.00044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin CR, Bornholdt ZA, Li S, Woods VL, Jr., MacRae IJ, Saphire EO. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:314–319. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Prins KC, Basler CF, Amarasinghe GK. Ebolavirus VP35 is a multifunctional virulence factor. Virulence. 2010a;1:526–531. doi: 10.4161/viru.1.6.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol. 2010b;17:165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, et al. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe. 2013;14:74–84. doi: 10.1016/j.chom.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol. 2015;235:153–174. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J Virol. 2010;84:1169–1175. doi: 10.1128/JVI.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi I, Basler CF. Immunological features underlying viral hemorrhagic fevers. Curr Opin Immunol. 2015;36:38–46. doi: 10.1016/j.coi.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A, Volchkova VA, Reid SP, Mateo M, Bagnaud-Baule A, Nemirov K, Shurtleff AC, Lawrence P, Reynard O, Ottmann M, et al. Marburgvirus hijacks nrf2-dependent pathway by targeting nrf2-negative regulator keap1. Cell Rep. 2014;6:1026–1036. doi: 10.1016/j.celrep.2014.02.027. [DOI] [PubMed] [Google Scholar]
- Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KC, Cardenas WB, Basler CF. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, Ramanan P, Cardenas WB, Amarasinghe GK, Volchkov VE, et al. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, Otwinowski Z, Liu G, Huh J, Basler CF, et al. Structural basis for Marburg virus VP35-mediated immune evasion mechanisms. Proc Natl Acad Sci U S A. 2012;109:20661–20666. doi: 10.1073/pnas.1213559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, Volchkov VE, Nichol ST, Basler CF. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J Virol. 2007;81:13469–13477. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg and Ebola Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 1410–1448. [Google Scholar]
- Shabman RS, Jabado OJ, Mire CE, Stockwell TB, Edwards M, Mahajan M, Geisbert TW, Basler CF. Deep sequencing identifies noncanonical editing of Ebola and Marburg virus RNAs in infected cells. MBio. 2014;5:e02011. doi: 10.1128/mBio.02011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw ML, Garcia-Sastre A, Palese P, Basler CF. Nipah Virus V and W Proteins Have a Common STAT1-Binding Domain yet Inhibit STAT1 Activation from the Cytoplasmic and Nuclear Compartments, Respectively. J Virol. 2004;78:5633–5641. doi: 10.1128/JVI.78.11.5633-5641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmas C, Basler CF. Marburg virus VP40 antagonizes interferon signaling in a species-specific manner. J Virol. 2011;85:4309–4317. doi: 10.1128/JVI.02575-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmas C, Grosch MN, Schumann M, Olejnik J, Martinez O, Best SM, Krahling V, Basler CF, Muhlberger E. Marburg virus evades interferon responses by a mechanism distinct from ebola virus. PLoS Pathog. 2010;6:e1000721. doi: 10.1371/journal.ppat.1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edwards MR, Borek DM, Feagins AR, Mittal A, Alinger JB, Berry KN, Yen B, Hamilton J, Brett TJ, et al. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe. 2014;16:187–200. doi: 10.1016/j.chom.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen B, Mulder LC, Martinez O, Basler CF. Molecular basis for ebolavirus VP35 suppression of human dendritic cell maturation. J Virol. 2014;88:12500–12510. doi: 10.1128/JVI.02163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.