Abstract
To determine whether the expression of histone modification enzymes is regulated in physiological and pathological conditions, we took an experimental database mining approach pioneered in our labs to determine a panoramic expression profile of 164 enzymes in 19 human and 17 murine tissues. We have made the following significant findings: 1) Histone enzymes are differentially expressed in cardiovascular, immune and other tissues; 2) Our new pyramid model showed that heart and T cells are among a few tissues in which histone acetylation/deacetylation, histone methylation/demethylation are in the highest varieties; and 3) Histone enzymes are more downregulated than upregulated in metabolic diseases and Treg polarization/differentiation, but not in tumors. These results have demonstrated a new working model of “sand out and gold stays,” where more downregulation than upregulation of histone enzymes in metabolic diseases makes a few upregulated enzymes the potential novel therapeutic targets in metabolic diseases and Treg activity.
Keywords: Histone modification enzymes, metabolic diseases, regulatory T cell, epigenetic regulation, gene expression and inflammation
Introduction
Hyperlipidemia, an independent metabolic risk factor for cardiovascular diseases (CVDs), including myocardial infarction and stroke, is defined as pathologically elevated plasma concentrations of cholesterol and other lipids [1]. Reports from our lab and others support the idea that hyperlipidemia, along with other metabolic/environmental risk factors, promotes atherosclerosis via several cellular mechanisms, including induction of endothelial cell activation and injury [2–5], increase of inflammatory monocyte differentiation and recruitment [6–9], decrease of the population of regulatory T cells (Tregs) [10–14], and impairment of the vascular repair ability of bone marrow-derived progenitor cells [15,16,5]. However, the molecular mechanisms underlying gene expression regulation in these pathological processes remain incompletely understood.
Atherogenesis, the process of atherosclerotic development, has been found to be promoted by certain genetic factors. These include monogenic heredity, such as 600 mutations identified in the low-density lipoprotein receptor gene, and polygenic heredity as shown in recent studies [17,18]. In addition, metabolic/environmental risk factors, such as hyperlipidemia, hyperglycemia, hyperhomocysteinemia, hypertension, smoking, autoimmunity and chronic kidney disease have been identified to play critical roles in accelerating atherogenesis [19]. Atherosclerosis affects vascular cells, such as endothelial and smooth muscle cells, and immune cells. As a result, one question that naturally arises is, how do these cells respond to the aforementioned risk factors? At the cellular level, signaling cascades initiated at the pathogen- /danger metabolic signal-associated molecular pattern receptors (PAMP-Rs/DAMP-Rs) [20] on the cell surface relay messages, via effectors, that reach the nucleus. In the nucleus, the targeted transcription factors, which include three categories of inflammatory transcription factors [21,22], interplay with epigenetic mechanisms, including histone modification machineries, in order to decide whether to open or shut down a particular gene expression signature [23]. Histones undergo several types of post-translational modifications [24,25] in response to metabolic/environmental change-initiated signaling pathways, which are associated with the pathogenesis of cardiovascular diseases [26–28].
In addition, Tregs play a critical role in suppressing various immune responses and inflammation as well as atherosclerosis [29]. In response to metabolic/environmental stimuli, Tregs undergo “transcriptional remodeling” where they differentiate into tissue-specific Tregs and present subset/lineage plasticity [30]. Recently, numerous reports have been focused on the epigenetic regulation of Treg differentiation and polarization. In our recently invited review, we pointed out that epigenetic enzymes are novel therapeutic targets for Treg activity [31–33].
So far, seven types of histone modification enzymes have been identified, including histone acetyltransferases, histone deacetylases, histone methyltransferases, histone demethylases, histone serine kinases, histone ubiquitination enzymes and histone SUMOylation enzymes [34,25]. Several studies have shown that mutations in epigenetic enzymes/modifiers have been associated with global chromatin remodeling in cancers and carcinogenesis [35]. In fact, Romidepsin, a histone deacetylase inhibitor, has been approved for the treatment of cutaneous T-cell lymphoma and peripheral T-cell lymphomas [36,37]. On the other hand, emerging evidence has indicated that epigenetic regulators critically confer future cell memories in the regulation of vascular, immune and other tissue-specific gene expression relating to diabetic complications and within atherosclerotic lesions [27,38]. For example, HDAC3 (histone deacetylase 3) plays a critical role in endothelial function, while HDAC7 (histone deacetylase 7) plays a critical role in vascular smooth muscle cell function [39]. In spite of recent significant progress in this front, several important questions need to be addressed: first, in physiological conditions, whether all the histone modification enzymes are differentially expressed among tissues. If not, whether some tissues exhibit higher epigenetic varieties of histone modification enzymes than other tissues; second, whether the expression of certain histone modification enzymes is modulated, either up-regulated or down-regulated, in atherogenic and metabolic disease-related pathological conditions; and third, whether the variation scales of histone modification enzymes are increased or decreased in polarization and differentiation of Tregs. Thus, we hypothesized that the expression of histone modification enzymes, as one of many important regulatory events, is under regulation of tissue differentiation signals, cell polarization signals and pathogenic signals. To test this hypothesis, we took “panoramic frames” to profile the tissue expression patterns of 164 identified histone modification enzymes. Our results demonstrated that histone modification enzymes are differentially expressed among tissues in physiological conditions. We also found that the expression of certain histone modification enzymes is more downregulated than upregulated in atherogenic conditions, metabolic diseases and in the ontogenesis of Tregs but not in tumors, another important pathological condition we considered. These findings have suggested that a few upregulated histone modification enzymes can be novel therapeutic targets for inhibiting metabolic, cardiovascular diseases and strengthening immune suppression during chronic metabolic diseases.
Methods
Tissue expression profiles of genes encoding categories of histone modification enzymes
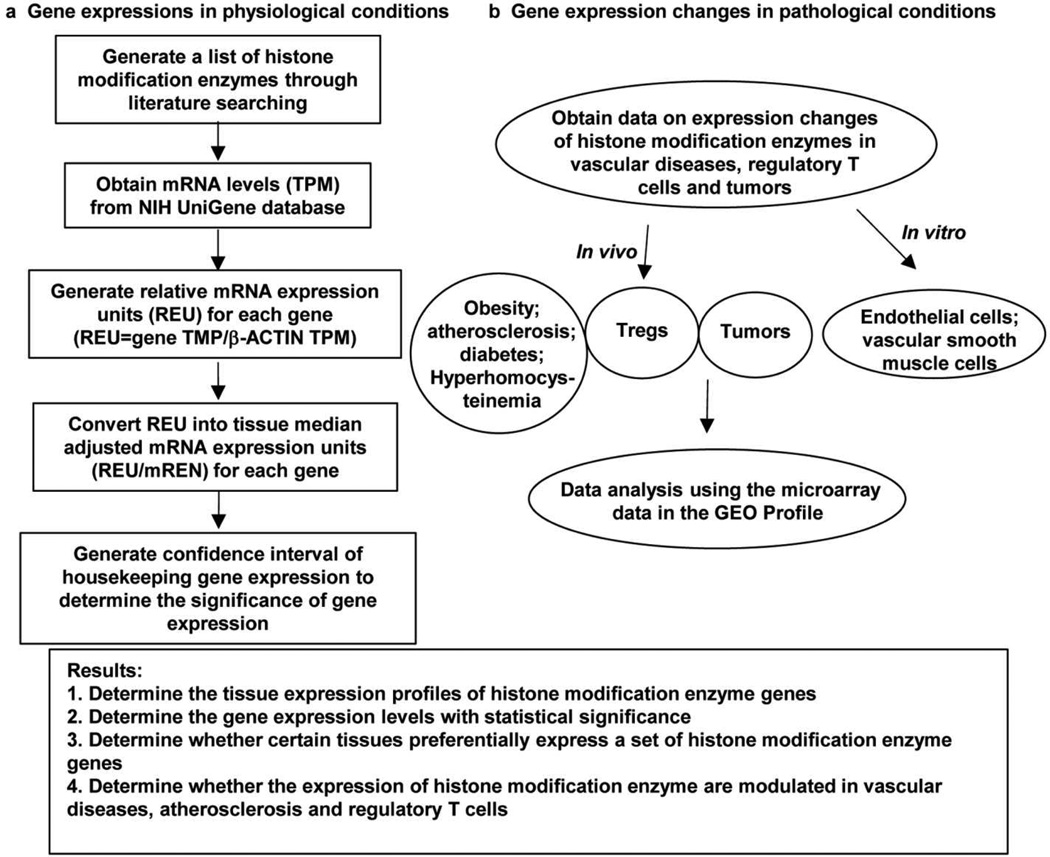
An experimental data mining strategy (Fig. 1), as we reported [40,20], was used to analyze the expression profiles of mRNA transcripts of 164 genes in cardiovascular and other tissues in humans and mice by mining experimentally verified human and mouse mRNA expression levels in the expressed sequence tag (EST) databases of the National Institutes of Health (NIH)/National Center of Biotechnology Information (NCBI) UniGene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene). Transcripts per million of genes of interest were normalized with those of house-keeping gene, β-actin, in each given tissue to calculate the arbitrary units of gene expression. The confidence interval of the expression variation of house-keeping genes was generated by calculating the mean plus two times the standard deviation of the arbitrary units of three randomly selected house-keeping genes (GAPDH, ALDOA and RPL19) normalized by β-actin in the given tissues (Fig. 2). If the increased expression level of a given gene in the tissues was larger than the upper limit of the confidence interval of the house-keeping gene expression variations, the gene in the tissue was considered as having statistically significant high expression level. Any given gene transcript lower than one per million was represented as no expression.
Fig.1. Novel strategies of experimental database mining and data organizing.
Tissue mRNA levels (TMP, Transcripts per million) were retrieved from NIH-NCBI-UniGene database. Generated relative mRNA expression units (REU) and tissue median-adjusted mRNA expression units were evaluated. Confidence interval of housekeeping gene expression was determined. In addition, gene expression levels in disease conditions were collected form GEO Profiles (GEO, Gene Expression Omnibus database in NIH-NCBI databases).
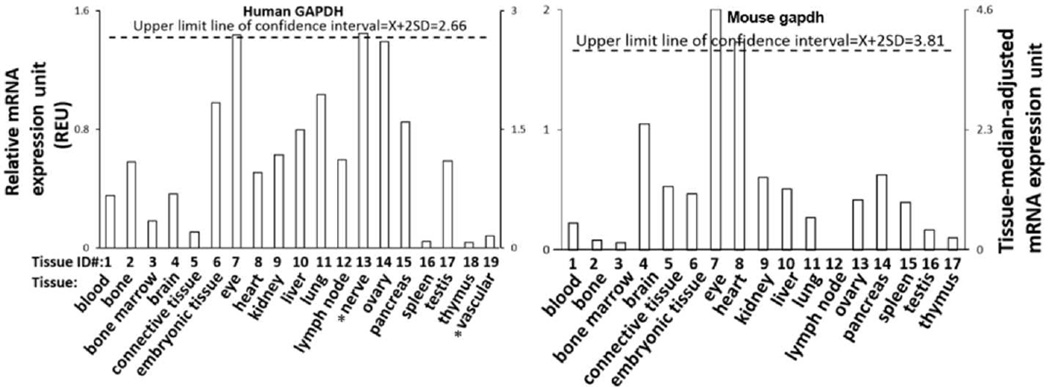
Fig.2. The histone modification enzyme genes are differentially expressed in human and mouse tissues.
A) Data Presentation Format (The data presented in X, Y axis and tissue order are applied to all the genes examined. As an example, the gene expression profiles of human house-keeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), aldolase A, fructose-bisphosphate (ALDOA), ribosomal protein L19 (RPL19) in the nineteen tissues, including blood, bone, bone marrow, brain, connective tissue, embryonic tissue, eye, heart, kidney, liver, lung, lymph node, nerve, ovary, pancreas, spleen, testis, thymus and vascular tissue are presented, with the tissue names and position numbers shown on the X-axis. The gene expression data were normalized by β-actin (Hs. 520640) expression data from the same tissue, which are presented on the left Y-axis. The expression ratios among tissues were generated by normalizing the arbitrary units of the gene in the tissues with the median level of the arbitrary units of the gene in all the tissues which are presented on the right Y-axis. In order to define confidence intervals for statistically higher expression levels of given genes, we calculated the confidence intervals of tissue expression using the formula, X+ 2SD = 2.66 (where X is the mean and SD is the standard deviation) for three house-keeping genes, i.e., glyceraldehyde-3-phosphate dehydrogenase (GAPDH Hs.544577), aldolase A, fructose-bisphosphate (ALDOA Hs.513490), and ribosomal protein L19 (RPL19 Hs.381061). The expression of a given gene in tissues that it was larger than 2.66-fold, was defined as high expression level with statistical significance (the right Y-axis). To discriminate between the genes from two species, human histone modification enzyme family members are designated with capital letters and those belonging to mouse are designated with lower case letters. *The data in the underlined tissues were only available for human tissues but not mouse tissues.
Expression profiles of histone modification enzymes in disease models and cell activity
Gene expression profiles were collected from twelve microarray datasets in the NIH GEO profiles. For metabolic disease studies, we examined the enzymes in the following conditions: 1) adipose tissue and liver in high fat diet-induced obese mouse model versus normal diet controls; 2) aortic arch segment in the atherogenic mouse model, apolipoprotein E gene knock-out (ApoE−/−) mice, versus that of wild-type controls; 3) pancreatic islets and white fat of leptin receptor mutant db/db type II diabetes mice versus those of control mice; 4) oxidized low-density lipoprotein (oxLDL)-stimulated mouse endothelial cells versus control endothelial cells; and 5) high concentration homocysteine (Hcy)-treated vascular smooth muscle cells (VSMCs) versus low concentration homocysteine (Hcy)-treated VSMCs. For Treg polarization/differentiation studies, we examined the expression changes of the enzymes in Tregs versus effector T cells in mice, in vivo as well as in vitro, with T cell antigen receptor ligation using anti-CD3 antibody and T cell co-stimulation with anti-CD28 antibody and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) ligation. In addition, we studied two representative tumor datasets (human cutaneous T cell lymphoma and human breast adenocarcinoma) as non-metabolic disease controls. Specific samples were chosen as disease groups and parallel controls, where the number of samples was normally more than three except for the pooled samples. By matching the gene symbols from the microarray data, we selected the enzymes with statistically significant changes (p<0.05) in various disease models. The enzymes that showed expression change of more than 1 fold were defined as upregulated ones while the enzymes with expression change of less than 1 fold were defined as downregulated ones.
Results
Histone methylation and acetylation are dominant modifications since 128 out of 164 enzymes modify only 24 lysine and arginine residues – a striking functional redundancy
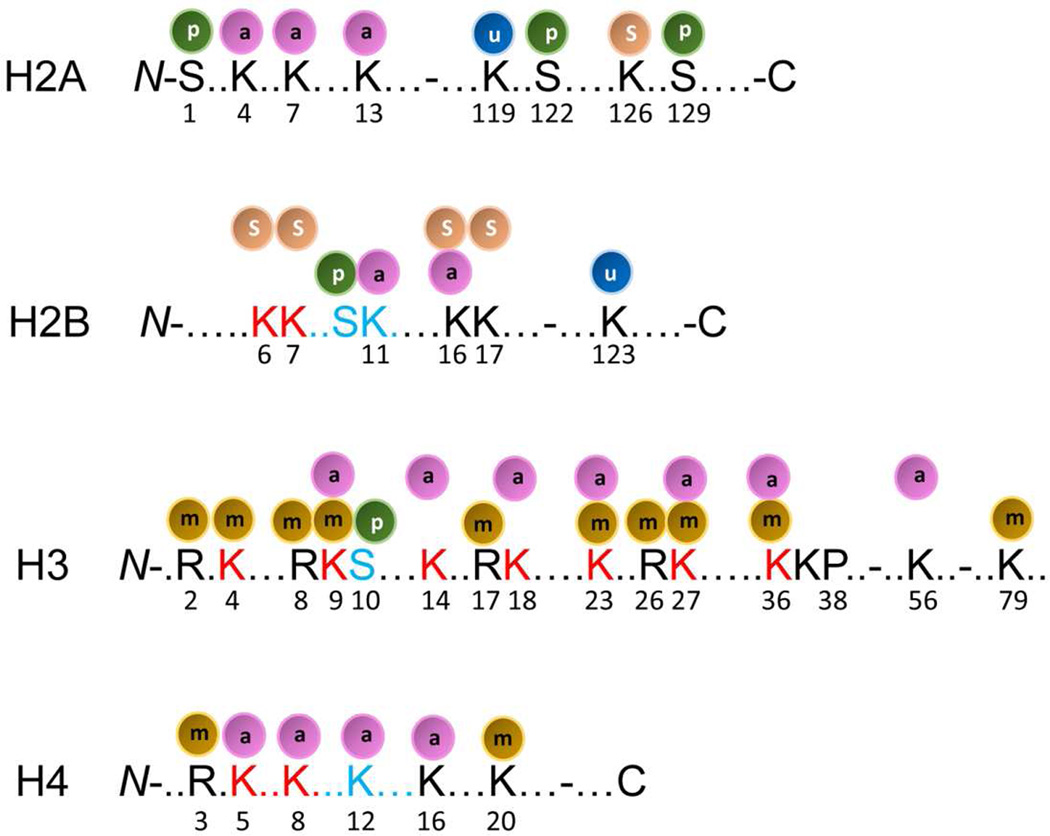
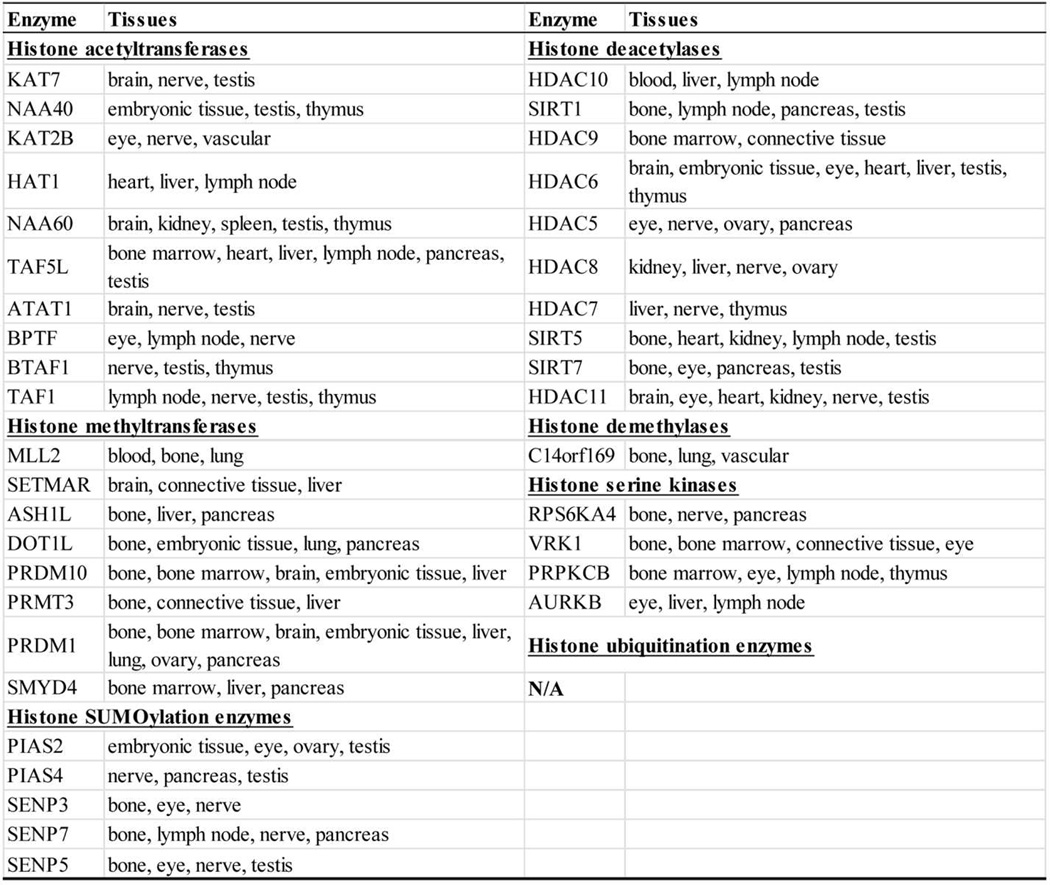
In order to examine the tissue expression profile of histone modification enzymes, as shown in Supplementary Tab. 1, we collected 31 histone acetyltransferases, 18 histone deacetylases, 55 histone methyltransferases, 24 histone demethylases, 15 histone serine kinases, 8 histone ubiquitination enzymes and 13 histone SUMOylation enzymes, which have been previously reported [34]. In order to have a comprehensive understanding of the functional models of those histone modification enzymes, we wanted to examine current findings on various histone modifications [41]. As shown in Fig. 3, among four histones (H2A, H2B, H3 and H4), methylation has been observed on 12 positively-charged amino acid residues, including 7 lysine (K) residues and 5 arginine (R) residues of N-terminal histone tails of H3 and H4. These are identified as H3R2, H3K4, H3R8, H3K9, H3R17, H3K23, H3R26, H3K27, H3K36, H3K79, H4R3, and H4K20 [41]. Common sites of methylation associated with gene activation include H3K4 and H3K79. Common sites for gene inactivation include H3K9 and H3K27. In addition, among four histones (histone 2A, histone 2B, histone 3 and histone 4), acetylation has been observed on 16 lysine (K) residues (but not arginine residues) of N-terminal histones, including H2AK4, H2AK7, H2AK13, H2BK11, H2BK16, H3K9, H3K14, H3K18, H3K23, H3K27, H3K36, H3K56, H4K5, H4K8, H4K12 and H4K16 [41]. Of note, four lysine residues of histone 3 (H3K9, H3K23, H3K27 and H3K36) can be either methylated or acetylated, suggesting that methylation and acetylation on the same amino acid residue are mutually exclusive. One lysine residue, H2BK16, can be either acetylated or SUMOylated. Moreover, histone phosphorylation has been identified in five serine residues of three histones; H2AS1, H2A122S, H2A129S, H2B10S, and H3S10. Furthermore, histone ubiquitination has been identified in only two histones, H2A and H2B, at two lysine residues, H2AK119 and H2BK123. Finally, similar to histone ubiquitination, histone SUMOylation has been observed in only two histones at five lysine residues; H2AK126, H2BK6, H2BK7, H2BK16, and H2BK17. Since histone ubiquitination and SUMOylation are only observed in H2A and H2B but not in H3 and H4, while histone methylation is only observed in H3 and H4 but not in H2A and H2B, it seems to be the case that histone ubiquitination, SUMOylation, and methylation are mutually exclusive due to potential physical inaccessibility of enzymes to histones or mutually exclusive events for two types of enzymes. The collection of enzymes in Supplementary Tab. 1 showed that as many as 128 enzymes modulate histone methylation and acetylation, which are much more than the 24 modifiable histone lysine and arginine residues. This observation suggests the following points: first, histone methylation and acetylation are a highly fine-tuned process in presumably context-dependent manners (tissue-specific, cell-specific, differentiation stage-specific, physiological status-specific, and pathological status-specific, etc.). For example, five to seven lysine residues, including H3K4, H3K9, H3K27, H3K36 and H4K20, can be mono- (me1), di- (me2) or tri-methylated (me3) [42]. The binding of specific proteins, which recognize methylated lysine positions, can have different biological functions [43]; second, such striking functional redundancy exists among those enzymes having single nucleotide polymorphisms in order to overcome the potential risk of loss of function due to gene mutations. In addition, those enzymes may also have evolved in order to overcome the potential functional variations due to differences of tissue- or cell-specific histone enzyme expression patterns. For example, five histone methyltransferases (Kmt2a, Kmt2b, Kmt2c, Kmt2d, Kmt2e) can all methylate H3K4 [43]; and third, histone methylation and acetylation, with 128 out of 164 enzymes (78%) acting on 24 residues (average 5.33 enzymes/residue), are much more varied in histone modification enzymes than other types of histone modifications, including serine phosphorylation, ubiquitination and SUMOylation, with 36 enzymes acting on 12 residues (average 3 enzymes/residue). Based on the numbers of enzymes involved and histone residues modified, these results suggest that histone methylation and acetylation are dominant modifications.
Fig.3. Post-translational modifications identified in histones.
Modified from a recent review entitled, “Transcription-associated histone modifications and cryptic transcription” by M. Smolle & J.L. Workman (PMID: 22982198).
Histone modification enzymes are differentially expressed in cardiovascular, immune systems and other tissues in humans and mice
To dissect the mechanisms underlying the functional redundancy of histone modification enzymes, we hypothesized that in order to keep gene expression and other histone functions and structure in check, various tissues express certain levels and certain types of histone modification enzymes under physiological conditions. To examine this hypothesis, the expression of seven categories of enzyme genes in 19 human tissues and 17 mouse tissues were examined (fewer mouse tissues were examined due to the fact that gene expression data for two of the mouse counterparts, i.e., nerve and vascular tissues, were not available in the NIH UniGene database) (Fig. 2). Due to the space limit, we summarized the gene expression data in human tissues in Supplementary Tab. 2A. The results showed that histone methyltransferases were highly expressed in various tissues in comparison to other histone modification enzymes in human tissues, except spleen. In addition, human bones, eyes, lymph nodes, nerves, testes and thymus are among the tissues with the highest numbers of high histone methyltransferase and histone demethylase expression, suggesting that the gene expression, differentiation and function of these tissues are largely physiologically regulated by histone methylation. Moreover, eyes, lymph nodes, nerves and testes are among the tissues with the highest numbers of high histone acetyltransferase and histone deacetylase expression, suggesting that the gene expression, differentiation and function of these tissues are also largely physiologically regulated by histone acetylation. Of note, human eyes have high expression of three histone acetyltransferases but have high expression of six histone deacetylases, suggesting that histone deacetylation in human eyes has a more fine-tuned function than histone acetylation for the gene expression, differentiation and function of human eyes. Furthermore, nearly half of the tissues examined, including blood, bone, bone marrow, connective tissue, embryonic tissue, heart, kidney, liver, lung, spleen and vascular tissue, did not highly express histone ubiquitination enzymes; and several tissues, including blood, heart, lung, spleen and vascular tissue, do not highly express histone SUMOylation enzymes. These suggest that the gene expression, differentiation and function of these tissues are physiologically regulated by histone ubiquitination and SUMOylation the least. Of note, human vascular tissue does not have a high variety of histone modification enzymes in physiological conditions, since only four out of 164 enzymes examined, i.e., a histone acetyltransferase (KAT2B), histone methyltransferase (SUV39H2), histone demethylase (C14orf169), and histone serine kinase (MAPK8), are highly expressed.
Similarly, due to the space limit, we summarized the gene expression data in mouse tissues in Supplementary Tab. 2B. These results showed that eyes, lymph nodes, ovaries and pancreas are among the tissues with the highest numbers of high histone methyltransferase and histone demethylase expression, suggesting that the gene expression, differentiation and function of these tissues are most physiologically regulated by histone methylation. In contrast to human tissue, mouse bones, bone marrow, liver and thymus did not have high expression of histone acetyltransferases, suggesting that histone acetyltransferases may have evolved to be adapted for more sophisticated gene regulatory functions in humans. In addition, brain, eyes, lymph nodes and ovaries are among the tissues with the highest numbers of high histone acetyltransferase and histone deacetylase expression, suggesting that the gene expression, differentiation and function of these tissues are more physiologically regulated by histone acetylation than by other modifications. Similar to those in human, histone ubiquitination enzymes and histone SUMOylation enzymes were not highly expressed in half of the tissues in mouse, suggesting that these enzymes are not highly involved in the physiological functions of mouse tissues.
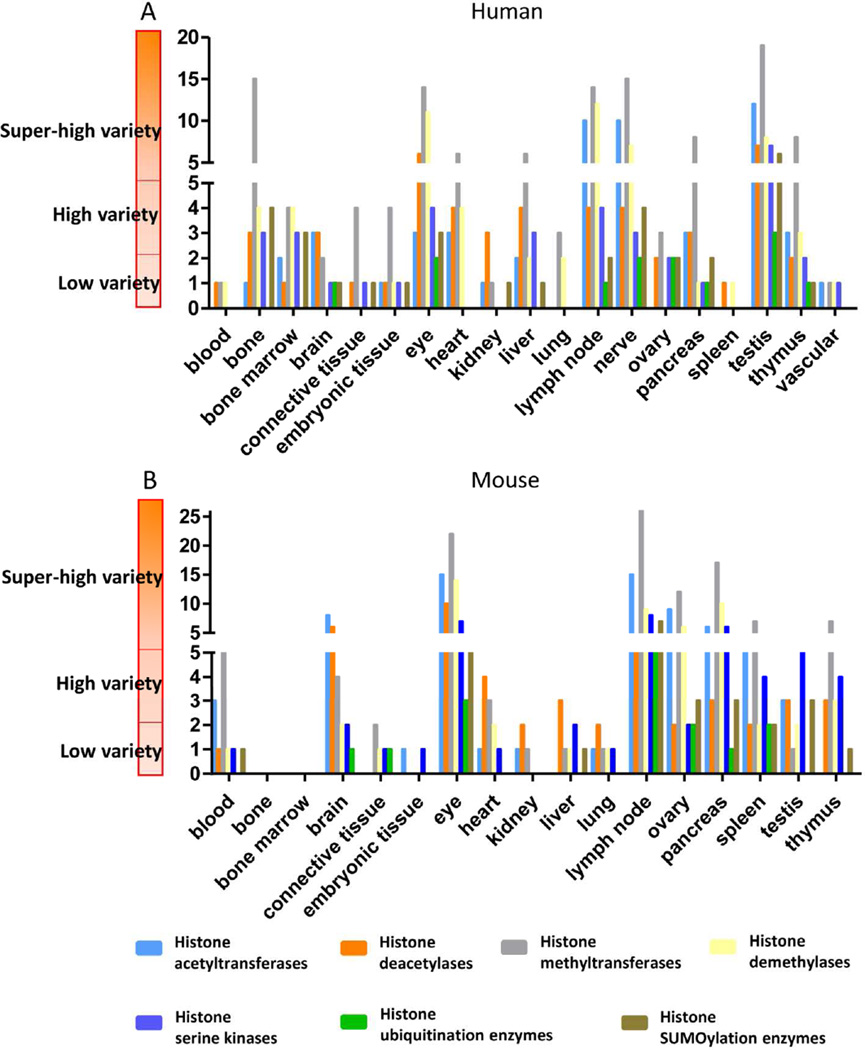
Our new pyramid model showed that heart and T cell compartments are among a few tissues in which the varieties of histone acetylation/deacetylation, histone methylation/demethylation are high
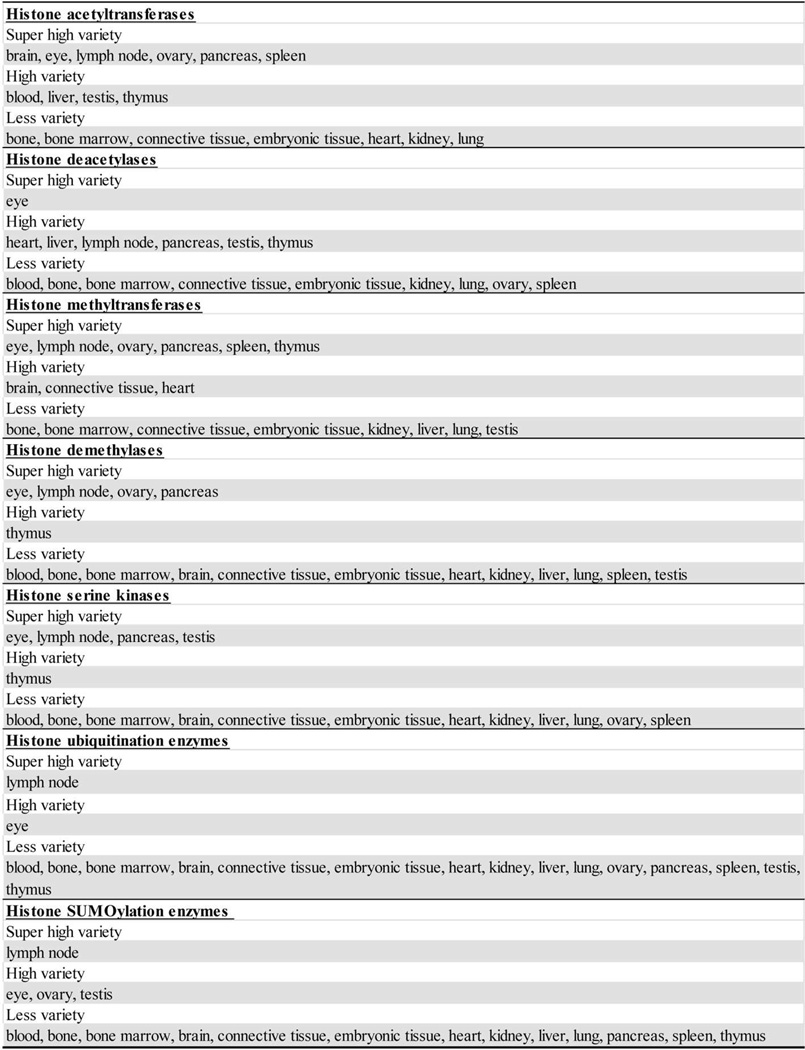
Based on the tissue expression pattern of key components of histone modification enzymes, we classified the tissues as one of three statuses based on their expression levels of each category of enzymes in human (Fig. 4A) and mouse (Fig. 4B). Thus, tissues that highly express less than two enzymes were categorized as the “low variety” status. Tissues that highly express more than two but less than five enzymes were classified as the “high variety” status. Tissues that highly express five or more enzymes were placed as the “super high variety” status. In human tissues, in terms of expression levels of histone acetyltransferases/deacetylases and histone methyltransferases/demethylases, lymph nodes have super high variety status in three categories and high variety status in histone deacetylases, while heart has high variety status in three categories and super high variety status in histone methyltransferases. In sharp contrast, vascular tissue has low variety status in all categories. In mouse, T cell compartments, i.e., lymph nodes and thymus, have super high variety status in three categories and high variety status in histone deacetylases, while heart has high variety status in histone deacetylases and histone methyltransferases. Of note, the lack of availability of mouse vascular data is a limitation. Taken together, this classification allows us to propose novel histone modification pyramids for human and mouse tissues (Fig. 5). The results suggest that the super high variety and high variety tissues in human and mouse, including testis, eye, lymph nodes, heart, thymus and pancreas, may use histone acetylation/deacetylation, histone methylation/demethylation pathways the most to regulate gene expression in response to developmental, physiological and environmental stimuli. In addition, those tissues that are low variety in histone modification enzymes may use less functional redundancy and less fine-tuned strategy in response to the above-mentioned stimuli.
Fig.4. Heart and lymph nodes are among the tissues in which the varieties of histone acetylation/deacetylation, histone methylation/demethylation are high.
Human(A) and mouse(B) tissues were classified into three groups. Tissues defined as “low variety,” “high variety,” and “super high variety” contained high expression level enzymes of each category that were ≤2, 5>X>2, and ≥5, respectively.
Fig.5. Our newly proposed “histone modification pyramids” model.
This model indicates that the histone modification statuses in heart, lymph nodes and thymus of human and mouse are highly regulated by histone acetyltransferases and methyltransferases.
A minority of human histone modification enzymes are highly expressed in multiple tissues
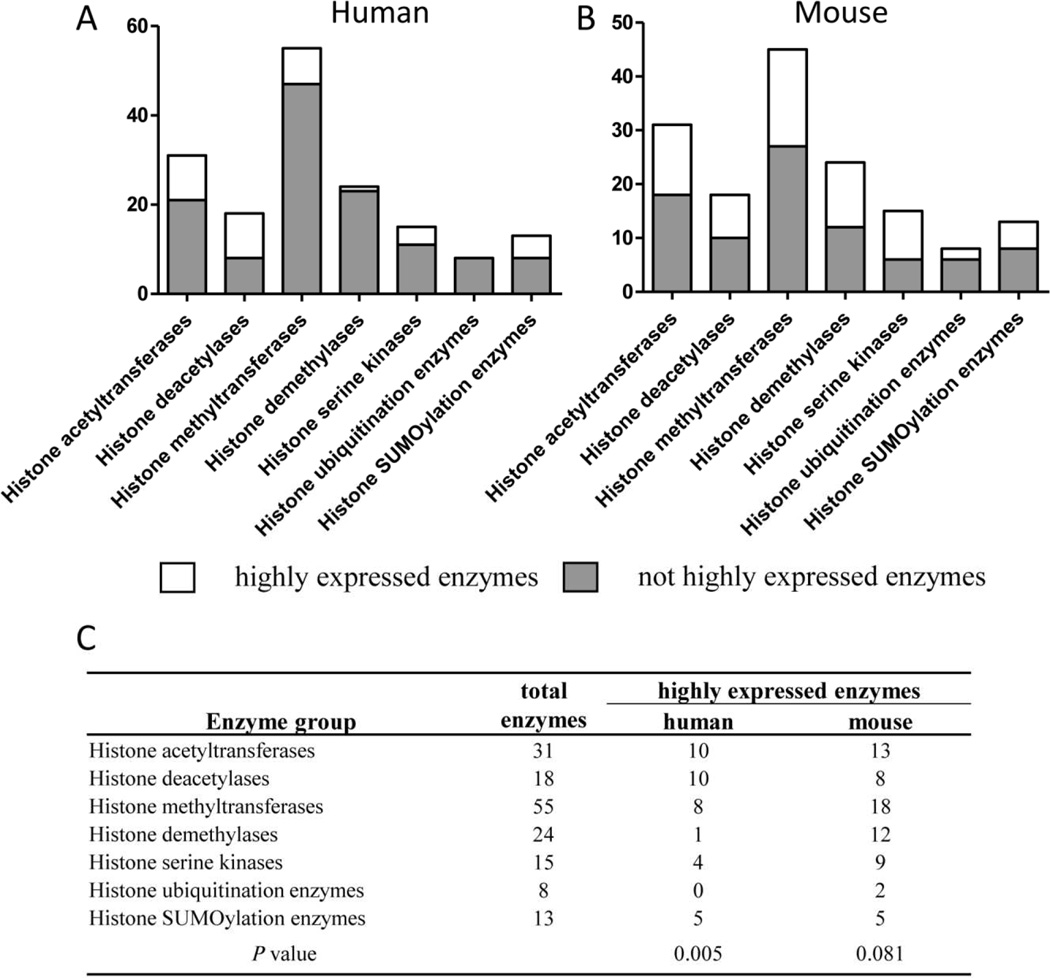
We further hypothesized that some histone modification enzymes are more tissue-specific than others. To examine this issue, we listed the histone modification enzymes that are highly expressed in three or more tissues in Fig. 6 and Supplementary Tab. 3A and 3B. The results showed that among the human enzymes (Fig. 6A), 10 out of 31 histone acetyltransferases, 10 out of 18 histone deacetylases, 8 out of 55 histone methyltransferases, 1 out of 24 histone demethylases, 4 out of 15 histone serine kinases, 0 out of 8 histone ubiquitination enzymes, and 5 out of 13 histone SUMOylation enzymes were highly expressed in three or more tissues. In addition, the results showed that among the mouse enzymes (Fig. 6B), 13 out of 31 histone acetyltransferases, 8 out of 18 histone deacetylases, 18 out of 55 histone methyltransferases, 12 out of 24 histone demethylases, 9 out of 15 histone serine kinases, 2 out of 8 histone ubiquitination enzymes, and 5 out of 13 histone SUMOylation enzymes were highly expressed in three or more tissues. The statistical analysis showed that histone modification enzymes with high expression levels in multiple tissues (>3) are the statistical minority in human (p<0.01), but are not statistically significant in mice (p=0.081) (Fig. 6C). Furthermore, while human histone demethylases and histone ubiquitination enzymes are the most tissue-specific, the mouse counterparts are not. These results suggest that the majority of histone modification enzymes play an important specific physiological role in tissues and that the minority of histone modification enzymes fulfill tissue-shared common physiological tasks in epigenetic regulation of gene expression in fine-tuned, less-specific, and less directional manners.
Fig.6. The minority of histone enzymes are highly expressed in multiple human tissues.
(A) The number of histone modification enzymes with high expression in more than 3 tissues in human. (B) The number of histone modification enzymes with high expression in more than 3 tissues in mouse. (C)Statistic analysis using t-test.
Histone modification enzymes are more downregulated than upregulated in chronic inflammatory metabolic diseases
Our findings indicate that histone modification enzymes are differentially expressed in human and mouse tissues in response to tissue differentiation signals, and that the majority of the enzymes are highly expressed in less than three tissues in physiological conditions, which further strengthen the argument that the expression levels of the majority of histone modification enzymes may be changed in response to other stimuli, such as pathological stimuli. We hypothesized that the expression of histone modification enzymes are subject to regulation in systemic metabolic diseases, including cardiovascular diseases, diabetes, adipose tissue dysfunction and hyperhomocysteinemia [44–48]. To test the hypothesis, we examined the 164 enzymes’ gene expression levels in the microarray datasets in NIH-GEO database originally designated for studies in metabolic disorders but not histone modification studies. The most consistent and striking results showed that metabolic diseases and pathological stimuli upregulated a few histone modification enzymes, which were significantly fewer than the downregulated enzymes. The results suggest that first, downregulated enzymes are functional in those tissues and cells in physiological conditions; second, upregulated enzymes play less significant roles in physiological conditions; and third, the pathological conditions require specific, strong and directional histone modifications than physiological settings.
Metabolic diseases induce more focused upregulations of histone enzymes in diseased tissues
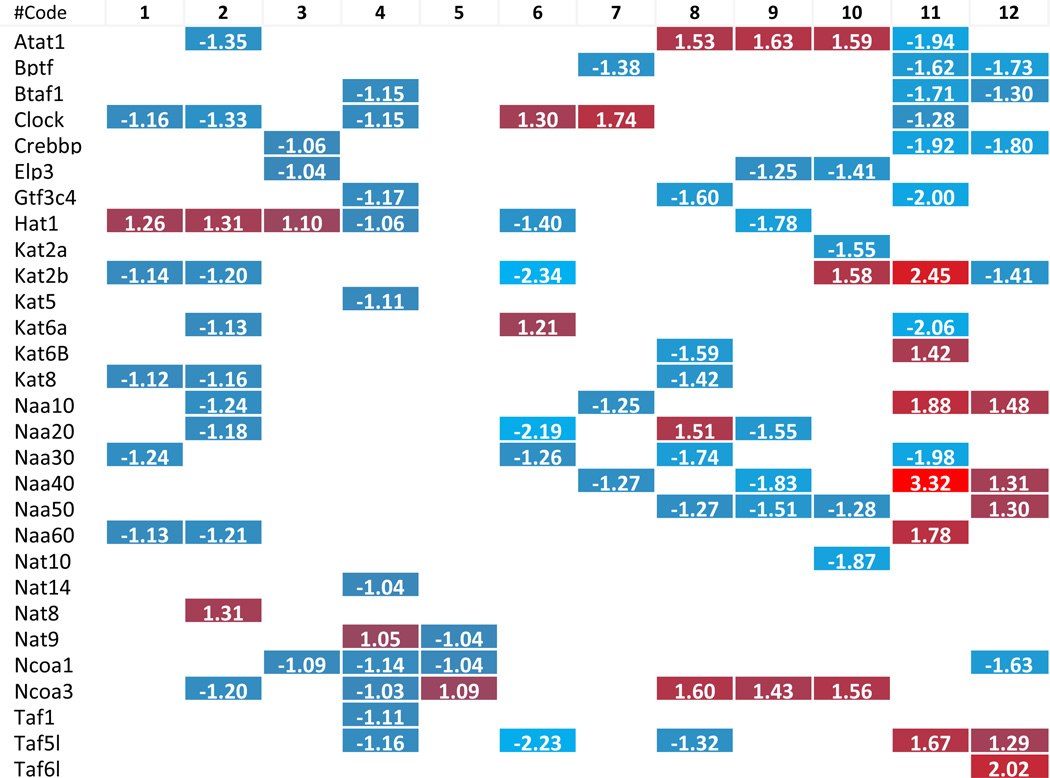
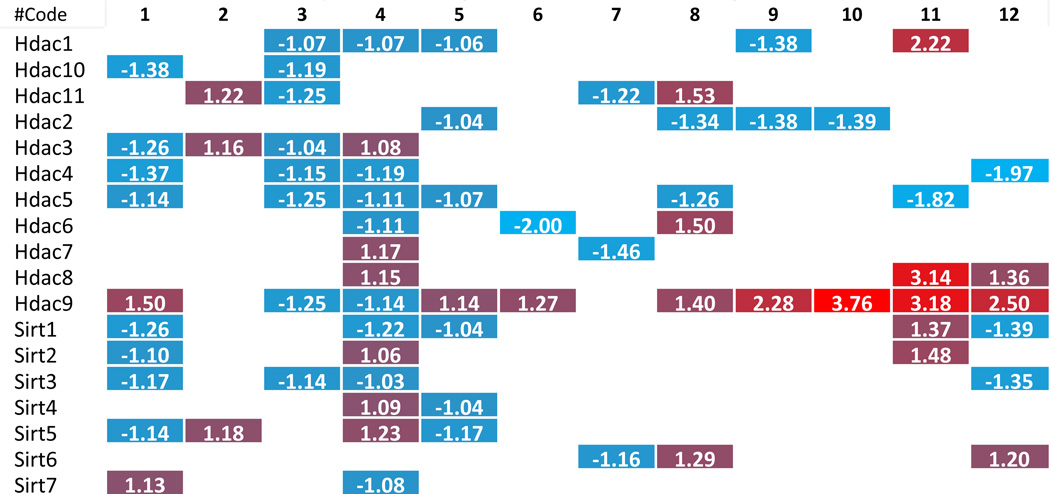
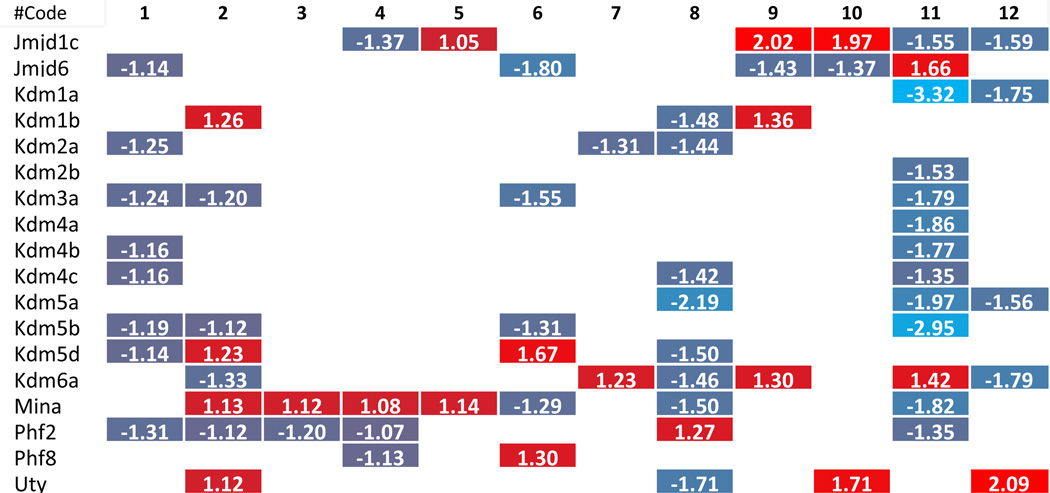
To dissect the potential mechanism underlying the phenotype where more histone modification enzymes experience downregulation than upregulation in metabolic diseases, we carefully analyzed the large amount of data that has been included in Tab. 2–8. Due to the space limit, the analyzed results have been summarized in Supplementary Tab. 4, which show that i) the minority (i.e., 6 out of 31 histone acetyltransferases) were upregulated in three different metabolic diseases; two pathologically stimulated aortic endothelial cells and vascular smooth muscle cells; ii) 2 out of 6 upregulated histone acetyltransferases were the enzymes with the highest expression levels in less than three tissues (Supplementary Tab. 3A and 3B); iii) the upregulated enzymes induced by one pathological stimulation were not the same as those induced by another pathological stimulation, suggesting the histone acetyltransferases were upregulated in response to specific pathological stimuli; iv) histone acetyltransferase (Hat1) was the only histone acetyltransferase induced in three tissues with hyperlipidemic environment, including adipose tissue, liver of diet-induced obesity mice and ApoE−/− mouse aorta; v) histone deacetylase (Hdac9) and histone demethylase (Kdm5d) were the only two histone modification enzymes upregulated in both oxidized low-density lipoprotein (oxLDL)-stimulated aortic endothelial cells and metabolic diseases; and vi) histone acetyltransferase (Clock), histone methyltransferases (Setd7 and Suv420h1), and histone demethylase (Kdm6a) were the only four enzymes upregulated in homocysteine-stimulated VSMCs. Taken together, these results suggest that pathological stimulations induce more focused upregulation of enzymes in diseased tissues. Furthermore, these enzymes were often not shared among different diseased tissues/cells. These results emphasize the regulatory specificities of pathological effects on histone modification enzyme expression and presumably the functions as well.
Table 2.
Histone acetyltransferase change in related pathological status.
The numbers in the cells are representing the fold change with significance. The color scales are showing the upregulation (more red means more increasing) or downregulation (more blue means more decreasing) levels. The numbers highlight in
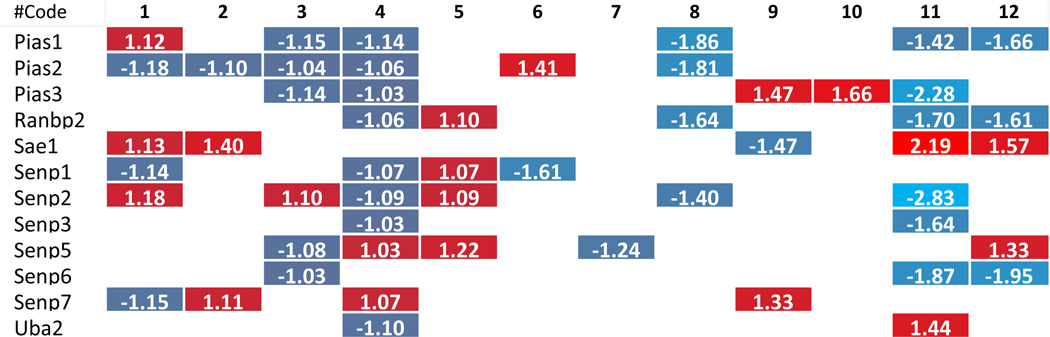
Table 8.
Histone SUMOylation enzyme change in related pathological status.
Polarizing/differentiating effects in Treg generation also induce less upregulations than downregulations of histone modification enzymes
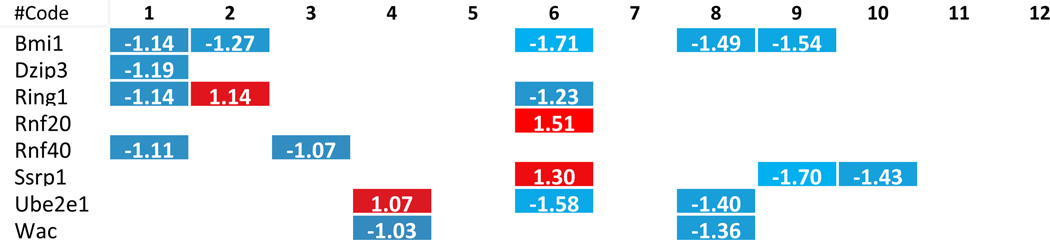
We hypothesized that histone modification enzymes play important roles in polarizing/differentiating Tregs from naïve CD4 T helper cells (Th0), forming inducible Tregs and tissue-specific Tregs. To test this hypothesis, we examined the expression changes of 164 histone modification enzymes in Tregs versus effector T cells in mice, in vivo as well as in vitro. The results in Tab. 2–8 and Supplementary Tab. 4 showed that first, histone modification enzymes are less upregulated than downregulated, and only 4 out of 31 histone acetyltransferases, 4 out of 18 histone deacetylases, 8 out of 55 histone methyltransferases, 5 out of 24 histone demethylases, 2 out of 15 histone serine kinases, and 2 out of 13 histone SUMOylation enzymes are upregulated; second, some histone acetyltransferases (Atat1, Naa20), histone deacetylases (Hdac6), histone methyltransferases (Kmt2a, Setd2), histone demethylases (Phf2) and histone SUMOylation enzymes (Pias3) are not overlapped with the enzymes upregulated in metabolic diseases and tumors; and third, no histone ubiquitination enzyme is upregulated in Tregs. These results suggest that i) similar to the results in chronic metabolic diseases, polarizing/differentiating effects of Tregs also induce less upregulations than downregulations of histone modification enzymes, indicating that epigenetic regulations for disease development and Treg differentiation/polarization are more specific and directional but less fine-tuned than those in physiological condition; ii) increase of a few, specific enzymes may play vital roles in Treg polarization/differentiation.
Most histone acetyltransferases upregulated in tumors are not shared with those in chronic metabolic diseases or in Treg polarization/differentiation
Recent progress resulting from the use of inhibitors of epigenetic regulators in clinical cancer treatments or in preclinical trials has significantly improved our understanding of the epigenetic mechanisms in tumorigenesis and tumor progression [49]. We hypothesized that, during tumorigenesis, specific signaling modulates the expression changes of histone modification enzymes in order to fulfill gene expression regulation requirements. To examine this hypothesis, we selected two representative tumor datasets and also summarized the data into Tab. 2–8 and Supplementary Tab. 4. The data showed that 1) upregulated histone modification enzymes, compared with downregulated enzymes, in tumors were much more abundant than those in chronic metabolic diseases; 2) upregulated histone deacetylases and serine kinases were dramatically more abundant than downregulated in patients with cutaneous T cell lymphoma, suggesting that they are potential therapeutic targets; 3) histone ubiquitination enzymes were insensitive in tumorigenesis, suggesting that they play more physiological functions; and 4) 7 out of 8 histone acetyltransferases, 2 out of 6 histone deacetylases, 4 out of 13 histone methyltransferases, 1 out of 3 histone demethylases, 2 out of 9 histone serine kinases, and 1 out of 3 histone SUMOylation enzymes upregulated in tumorigenesis were neither shared with those upregulated in Treg development nor with those upregulated in chronic metabolic diseases. These results suggested that: first, the upregulated histone modification enzymes may play a critical role in properties unique to tumors compared with metabolic diseases or with Treg polarization/differentiation, such as the promotion of tumorigenesis; and second, most categories of histone modification enzymes are active in tumorigenesis, except histone ubiquitination enzymes.
Most upregulated enzymes in chronic metabolic disease show smaller increase scale than those in Treg polarization or tumorigenesis
To further examine the features of the expression changes of histone modification enzymes in pathophysiology conditions, we analyzed the fold changes scale of enzyme expression levels listed in Tab. 2–8, which show that 1) except for a few enzymes, including histone acetyltransferases (Clock) from VSMCs after Hcy stimulation, histone deacetylases (Hdac9), histone serine kinases (Aurkb) from hyperlipidemic mouse adipose tissues, histone methyltransferases (Setdb1), histone demethylases (Kdm5d), and histone ubiquitination enzymes (Rnf20) from mouse endothelial cells after oxLDL stimulation, most enzymes induced in metabolic disease tissue show less than 1.5 fold increase; 2) most upregulated enzymes in Treg polarization/differentiation show more than 1.5-fold increase in expression, especially histone acetyltransferases (Atat1), histone deacetylases (Hdac9), histone methyltransferases (Prdm1) and histone demethylases (Jmjd1c); 3) Two representative human cancers induce higher fold changes in histone modification enzyme expression levels, especially histone acetyltransferases, histone deacetylases, histone methyltransferases and histone serine kinases. Taken together, these results suggest that low scale expression changes of histone modification enzymes induced by metabolic diseases may result from the features of low scale, chronic multicellular pathologies in comparison to single cell type differentiation in Treg polarization or dramatic genome-unstable tumorigenesis.
Discussion
A previous study identified that histone modifications that are stable among different human cell types are evolutionarily conserved between mice and humans in the same cell type. Histone modifications that are stable among cell types are also likely to be conserved between other species [50]. These modified regions showed distinct genetic and epigenetic properties, such as clustered transcription factor binding sites (TFBSs), high GC content, and CCCTC-binding factor (CTCF) binding at flanking sides. In mammalian genomes, a common mechanism maintains histone modifications against both genetic and environmental (cellular) changes [50], which emphasizes the common aspect of histone modification enzymes. However, previous reports have not addressed the following important questions: first, which are the tissues that have more histone modification enzyme expression than other tissues at physiological conditions? Second, which histone modification enzymes have tissue-specific expression patterns? Third, which are the enzymes that are upregulated in response to disease/pathological stimulations? To fill in these important knowledge gaps, we took an experimental database mining approach that was pioneered and developed in our lab throughout the years [51,52,20,53] and determined a panoramic profile on the expression levels of 19 tissue/cell histone modification machinery using 164 enzymes [34]. We have made the following significant findings: 1) histone methylation and acetylation are dominant modifications since 128 enzymes modify only 24 lysine and arginine residues – a striking functional redundancy; 2) histone modification enzymes are differentially expressed in cardiovascular and immune systems as well as in other tissues in humans and mice; 3) Our new pyramid model showed that heart, liver and T cell compartments are among a few tissues, in which the varieties of histone acetylation/deacetylation, histone methylation/demethylation are the highest among tissues examined; 4) a minority of human histone modification enzymes are highly expressed in multiple tissues; 5) histone modification enzymes are more downregulated than upregulated in chronic metabolic diseases; 6) metabolic diseases induce more focused upregulation of histone enzymes in diseased tissues; 7) polarizing/differentiating effects in Treg generation also induce less upregulation than downregulation of histone modification enzymes; 8) most histone acetyltransferases upregulated in tumors are neither shared in chronic metabolic diseases nor in Treg polarization; and 9) most upregulated enzymes in chronic metabolic disease show smaller increase scales than those in Treg polarization or tumorigenesis. These results have clearly demonstrated a principle that more histone modification enzyme downregulation than upregulation makes a few upregulated enzymes the novel therapeutic targets in chronic metabolic diseases and Treg activity.
By analyzing cDNA cloning and DNA sequencing data from tissue cDNA libraries, we were able to study expression profiles of histone modification enzymes in various tissues. Since this data is collected from cDNA cloning and DNA sequencing experiments rather than theoretical data derived from computer modeling, the data require no further experimental verification. Since EST databases have been established based on precise DNA sequencing data, the data obtained by EST database mining are more precise in providing the tissue expression profiles of genes than traditional hybridization- and primer annealing-based approaches like Northern blots and RT-PCRs [20].
Of note, since the UniGene database does not have much non-tumor cell line-related gene expression data, we mined the NIH-GEO Datasets for analyzing the enzyme expression changes in disease settings.
Previous reports have confirmed the role of histone modification in various pathologies. For example, HDAC1, HDAC2, HDAC3, HDAC4, HDAC5 and HDAC9 play critical roles in pathological gene expression in the heart; and histone methylation and demethylation regulate the expression of 13 pathological genes in the heart [54]. Also, histone modifications are also very important in mediating alcohol-induced gene expression changes in liver [55]. Although a transcriptional network and epigenetic modification network have started to be characterized in T cell development [56], an important issue of the role histone acetylation, deacetylation, methylation and demethylation play in physiological conditions in heart and liver remains poorly characterized. As previously mentioned, in order to clearly summarize our findings, we proposed a novel pyramid model to highlight the histone acetylation/methylation activities in those tissues. This pyramid model is significant as it improves our understanding of the tissue differences of histone modification enzyme machinery. This model is also significant for understanding the potential pharmacological side effects of new drugs targeting histone acetylation/methylation in those tissues.
The reason for paying more attention to histone methylation is that histone methylation is among the most stable histone modifications [57,58], and have been implicated in the inheritance of epigenetic traits, so-called epigenetic memory [59]. Similar findings were obtained in the pathological setting. In alcohol-induced epigenetic changes, histone phosphorylation lasts 12–24 hours. By comparison, alcohol-induced histone acetylation lasts longer than 24 hours, and alcohol-induced histone methylation lasts even longer [55]. As far as probable regulatory mechanisms of this process are concerned, we and others have shown that cellular methyl-generating reaction for S-adenosine methionine (SAM) in the one-carbon cycle and its inhibitor, S-adenosine homocysteine (SAH), is under extensive regulation during an independent cardiovascular disease risk factor, hyperhomocysteinemia [47,48], and other metabolic cardiovascular diseases [60,48]. The metabolite, acetyl-CoA, is not only an intermediate in energy metabolism generated in mitochondria but is also an acetyl group donor after it is transported out of the mitochondria and re-generated in cytosol, for protein and histone acetylation. This process is under regulation by acetyl-CoA availability and also energy metabolic status of cells [61,62]. The findings provide insight into how metabolism in mitochondria regulates histone methylation, acetylation, and ultimately innate and adaptive immune responses and inflammation [63].
In addition to the histone modification enzymes acting as “writers” and “erasers” that we discussed here, a number of histone modification reader proteins have been characterized [24]. However, here we focused our efforts on catalytic functions of histone modification machinery rather than histone modification recognition systems such as the reader proteins.
Of note, our findings in tissue regulation and disease regulation of histone modification enzyme expression do not lower the significance of other regulatory processes on these sets of enzymes, including the regulation of enzymatic activities. Progress in the field has found that in addition to gene mutation-caused functional loss of epigenetic genes, the expression changes of histone modification enzymes also result in serious pathologies, including systemic lupus erythematosus, asthma, Huntingdon’s disease and cancer [64]. Recent progress in understanding the regulation of histone modification enzymes by the ubiquitin-proteasome system [65] supports our finding that the expression of histone modification enzymes is important. Therefore, it is justified for us to argue that upregulated histone modification enzymes are therapeutic targets, ultimately relieving the dysregulation of gene expression and activities in response to environmental/pathological stimuli or cell development and differentiation [66,67] (Tab. 9).
Table 9.
Upregulated histone modification enzymes are therapeutic targets to relieve the dysregulation of gene activity and expression.
| Enzymes | Modification residues |
Proposed function |
|---|---|---|
| Histone acetyltransferases | ||
| Hat1* | H2AK4, H2AK5, H4K5,H4k12 | transcriptional repression |
| Ncoa3 | - | stimulates transcriptional activities; pluripotency maintenance |
| Kat2b | H3K9, H3K14 | promote transcriptional activation |
| Kat6b | - | both positive and negative regulation of transcription |
| Taf5L | - | coactivator of transcription initiation |
| Histone deacetylases | ||
| Hdac9 | H3K9, H3K14, H3K18 | transcriptional repressor |
| Sirt2 | H3K56, H4K16 | - |
| Sirt6 | H3K9, H3K56 | - |
| Histone methyltransferases | ||
| Setd7 | H3K4 | transcriptional activation |
| Prdm1 | H3K9 | transcription repression |
| Prdm5 | - | transcription repression |
| Whsc1 | H3K4, H3K27, H3K36, H4K20 | suppresses IL5 transcription |
| Carm1 | H3R2me1, H3R17me1, H3R17me2, H3R26me1 | activates transcription |
| Suv39h1 | H3K9 | displays features of a long-range repressor capable of acting over several kilobases to silence basal promoters |
| Histone demethylases | ||
| Mina | H3K9me3 | leads to an increase in ribosomal RNA expression; binds to the IL4 promoter where it represses IL4 expression |
| Kdm1b | H3K4 | transcriptional corepressor |
| Jmjd6 | H3R2me2a, H3R2me2s, H4R3me | regulates promoter-proximal pause release and permits activation |
| Histone serine kinases | ||
| Aurkb | H3S10ph H3S28ph | phosphates p53/TP53 to negatively regulate transcriptional activity; inhibits RNF2/RING1B-mediated ubiquitination of histone H2A; arrests B- and T-lymphocytes |
| Rps6ka3 | H3S10 | controls cell growth and differentiation |
| Histone ubiquitylation enzymes | ||
| Ring1 | H2AK119 | transcriptional repression |
| Rnf20 | H2BK123, H2BK120 | transcriptional activation; prerequisite for histone H3K4 and H3K79 methylation |
| Histone SUMOylation enzymes | ||
| Pias1 | - | inhibits STAT1-mediated gene activation and DNA binding activity |
With currently available information, we summarized part of the upregulated enzymes mined in our system.
Based on the new findings presented in this report, we propose a new working model, “Sand out and gold stays” (Fig. 7): first, in physiological conditions, many metabolic disease-downregulated histone modification enzymes regulate histone modifications in a “non-directional, relatively-weak, fine-tuned” manner in order to maintain homeostasis in tissues; second, in response to non-controllable environmental/metabolic risk factors, histone modification machinery experiences a pathological remodeling in significantly downregulating the majority of histone modification enzymes and upregulating only a few histone modification enzymes. This allows the cells in question to deliver a strong “directional and specific” regulation in order to fulfill gene regulation for the pathogenesis of metabolic diseases. To improve the explanation of our interesting findings and this new model, we used a day-to-day example of gold mining process. Since the upregulated enzymes can be potential therapeutic targets and are potentially more therapeutically valuable than the downregulated enzymes, we refer to the few upregulated enzymes as “gold” and to the mostly downregulated enzymes as “sand”; third, the strong “directional and specific” regulation on histone modification machinery is very similar to what happens in Treg differentiation/polarization; and fourth, tumorigenesis emphasizes more de-differentiation and cellular proliferation. Thus, tumorigenesis-specific pathological remodeling of histone modification machinery is different from strong “directional and specific” regulation patterns. In summary, newly identified upregulated histone modification enzymes in chronic metabolic diseases, Treg polarization and tumorigenesis are novel therapeutic targets for inhibiting metabolic cardiovascular diseases, strengthening immune suppression during chronic metabolic diseases and suppressing tumorigenesis.
Fig.7. A new working model presents “sand out and gold stays,” indicating that more downregulation than upregulation of histone enzymes in metabolic diseases makes a few upregulated enzymes the novel therapeutic targets in metabolic diseases and Treg activity.
In this figure, “a” in white represents histone acetyltransferases, “a” in black represents histone deacetylases, “m” in white represents histone methyltransferases, “m” in black represents histone demethylases, “p” represents histone serine kinases, “u” represents histone ubiquitination enzymes, “s” represents histone SUMOylation enzymes.
Clinical Relevance of the Study
An important issue of how the expression of histone modification enzymes is globally regulated in physiological and pathological conditions remains unknown. We demonstrated that in response to non-controllable metabolic risk factors and in the process of Treg polarization/differentiation, histone modification machinery experiences a pathological remodeling in significantly downregulating the majority of histone modification enzymes and upregulating only a few histone modification enzymes. Our new finding would lead to the future development of using upregulated histone enzymes as biomarkers for new assays for disease diagnosis as well as novel therapies for inhibiting metabolic cardiovascular diseases, and for strengthening immune suppression during chronic metabolic diseases.
Supplementary Material
Table 1.
Histone modification enzymes are more down-regulated than up-regulated in metabolic diseases and Treg polarization but not in cancer.
| Pathologic status |
Metabolic diseases | Treg generation | Cancer | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse hyperlipidemia |
ApoE knock out |
Mouse diabetes (db/db) |
oxLDL stimulation |
Hcy stimulation |
Normal condition |
CD3 and CD28 stimulation |
CTLA -4 stimulation |
Cutaneus T cell lymphoma |
Adenocarcinoma of breast |
|||
| Tissue /cell |
adipose | liver | aortic arch |
pancreas | white fat |
mouse endothe lial cells |
VSMC | mouse lymph node |
mouse Treg | human skin biopsy |
breast epithelium |
|
| GSE No. | 54189 | 18443 | 31953 | 39264 | 9490 | 11775 | 42276 | 59307 | 61304 | |||
| #Code | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
Table 3.
Histone deacetylase change in related pathological status.
Table 4.
Histone methyltransferase change in related pathological status.
Table 5.
Histone demethylase change in related pathological status.
Table 6.
Histone serine kinase change in related pathological status.
Table 7.
Histone ubiquitination enzyme change in related pathological status.
Acknowledgments
We express our heartfelt thanks to Drs. Xinyuan Li, Yafeng Li, and Xin Wang in our laboratories at Temple University Lewis Katz School of Medicine for the excellent technical assistance.
Funding
This work was partially supported by the National Institutes of Health Grants to XFY and HW.
Abbreviations
- Treg
Regulatory T cell
- CVDs
Cardiovascular diseases
- PAMP-Rs
Pathogen metabolic signal-associated molecular pattern receptors
- DAMP-Rs
Danger metabolic signal-associated molecular pattern receptors
- EST
Expressed sequence tag
- ApoE−/−
Apolipoprotein E gene knock-out
- oxLDL
Oxidized low-density lipoprotein
- Hcy
Homocysteine
- VSMC
Vascular smooth muscle cell
- CTLA-4
Cytotoxic T-lymphocyte-associated protein 4
Footnotes
Disclosures
The authors declare that they have no competing interests.
Compliance with Ethical Standards
Human subjects/informed consent statement
No human studies were carried out by the authors for this article.
Animal studies.
None
Author contributions
Y. Shao, V. Chernaya, and XF. Yang designed, performed research, and analyzed the data; Y. Shao, XF. Yang wrote the manuscript; V. Chernaya, C. Johnson, WY. Yang, X. Sha, Y. Zhang, X. Qin, E.T. Choi, and H. Wang performed the critical readings.
References
- 1.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. [Review] Nature. 2011;473(7347):317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 2.Yin Y, Li X, Sha X, Xi H, Li YF, Shao Y, et al. Early Hyperlipidemia Promotes Endothelial Activation via a Caspase-1-Sirtuin 1 Pathway. Arterioscler Thromb Vasc Biol. 2015;35(4):804–816. doi: 10.1161/ATVBAHA.115.305282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang X, Yang F, Tan H, Liao D, Bryan RM, Jr, Randhawa JK, et al. Hyperhomocystinemia impairs endothelial function and eNOS activity via PKC activation. Arterioscler Thromb Vasc Biol. 2005;25(12):2515–2521. doi: 10.1161/01.ATV.0000189559.87328.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao Y, Cheng Z, Li X, Chernaya V, Wang H, Yang XF. Immunosuppressive/anti-inflammatory cytokines directly and indirectly inhibit endothelial dysfunction- a novel mechanism for maintaining vascular function. J Hematol Oncol. 2014;7(1):80. doi: 10.1186/s13045-014-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li YF, Ren LN, Guo G, Cannella LA, Chernaya V, Samuel S, et al. Endothelial progenitor cells in ischemic stroke: an exploration from hypothesis to therapy. J Hematol Oncol. 2015;8(1):33. doi: 10.1186/s13045-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang D, Jiang X, Fang P, Yan Y, Song J, Gupta S, et al. Hyperhomocysteinemia promotes inflammatory monocyte generation and accelerates atherosclerosis in transgenic cystathionine beta-synthase-deficient mice. [Research Support, N.I.H., Extramural] Circulation. 2009;120(19):1893–1902. doi: 10.1161/CIRCULATIONAHA.109.866889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Combadiere C, Potteaux S, Rodero M, Simon T, Pezard A, Esposito B, et al. Combined inhibition of CCL2, CX3CR1, and CCR5 abrogates Ly6C(hi) and Ly6C(lo) monocytosis and almost abolishes atherosclerosis in hypercholesterolemic mice. Circulation. 2008;117(13):1649–1657. doi: 10.1161/CIRCULATIONAHA.107.745091. [DOI] [PubMed] [Google Scholar]
- 8.Zhang D, Fang P, Jiang X, Nelson J, Moore JK, Kruger WD, et al. Severe hyperhomocysteinemia promotes bone marrow-derived and resident inflammatory monocyte differentiation and atherosclerosis in LDLr/CBS-deficient mice. [Research Support, N.I.H., Extramural] Circ Res. 2012;111(1):37–49. doi: 10.1161/CIRCRESAHA.112.269472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang P, Zhang D, Cheng Z, Yan C, Jiang X, Kruger WD, et al. Hyperhomocysteinemia potentiates hyperglycemia-induced inflammatory monocyte differentiation and atherosclerosis. [Research Support, N.I.H., Extramural] Diabetes. 2014;63(12):4275–4290. doi: 10.2337/db14-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong Z, Yan Y, Song J, Fang P, Yin Y, Yang Y, et al. Expression of TCTP antisense in CD25(high) regulatory T cells aggravates cuff-injured vascular inflammation. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't] Atherosclerosis. 2009;203(2):401–408. doi: 10.1016/j.atherosclerosis.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ait-Oufella H, Salomon BL, Potteaux S, Robertson AK, Gourdy P, Zoll J, et al. Natural regulatory T cells control the development of atherosclerosis in mice. [Research Support, Non-U.S. Gov't] Nat Med. 2006;12(2):178–180. doi: 10.1038/nm1343. [DOI] [PubMed] [Google Scholar]
- 12.Yan Y, Xiong Z, Zhang S, Song J, Huang Y, Thornton AM, Wang H, Yang X-F. CD25high T cells with a prolonged survival inhibit development of diabetes. International Journal of Immunopathology and Pharmacology. 2008;21(4):767–780. doi: 10.1177/039463200802100401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Z, Song J, Yan Y, Huang Y, Cowan A, Wang H, et al. Higher expression of Bax in regulatory T cells increases vascular inflammation. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't] Front Biosci. 2008;13:7143–7155. doi: 10.2741/3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klingenberg R, Gerdes N, Badeau RM, Gistera A, Strodthoff D, Ketelhuth DF, et al. Depletion of FOXP3+ regulatory T cells promotes hypercholesterolemia and atherosclerosis. [Research Support, Non-U.S. Gov't] J Clin Invest. 2013;123(3):1323–1334. doi: 10.1172/JCI63891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y-F, Huang Xiao, Li Xinyuan, Gong Ren, Huang Cong-xin, Yin Ying, Nelson Jun, Gao Erhe, Zhang Hongyu, Hoffman Nicholas E, Madesh Muniswamy, Tilley Douglas E, Choi Eric T, Jiang Xiaohua, Wang Hong, Yang Xiao-Feng. Caspase-1 mediates hyperlipidemia-weakened progenitor cell vessel repair. Frontiers in Bioscience (Landmark Edition) 2015;20:1–1. doi: 10.2741/4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson J, Wu Y, Jiang X, Berretta R, Houser S, Choi E, et al. Hyperhomocysteinemia suppresses bone marrow CD34+/VEGF receptor 2+ cells and inhibits progenitor cell mobilization and homing to injured vasculature-a role of beta1-integrin in progenitor cell migration and adhesion. Faseb J. 2015 doi: 10.1096/fj.14-267989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Rollins J, Paigen B, Wang X. Genetic and genomic insights into the molecular basis of atherosclerosis. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Review] Cell Metab. 2007;6(3):164–179. doi: 10.1016/j.cmet.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovacic S, Bakran M. Genetic susceptibility to atherosclerosis. Stroke Res Treat. 2012;2012:362941. doi: 10.1155/2012/362941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang XF, Yin Y, Wang H. VASCULAR INFLAMMATION AND ATHEROGENESIS ARE ACTIVATED VIA RECEPTORS FOR PAMPs AND SUPPRESSED BY REGULATORY T CELLS. Drug Discov Today Ther Strateg. 2008;5(2):125–142. doi: 10.1016/j.ddstr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Y, Yan Y, Jiang X, Mai J, Chen NC, Wang H, et al. Inflammasomes are differentially expressed in cardiovascular and other tissues. Int J Immunopathol Pharmacol. 2009;22(2):311–322. doi: 10.1177/039463200902200208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. [Research Support, Non-U.S. Gov't Review] Nat Rev Immunol. 2009;9(10):692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 22.Tarakhovsky A. Logic of the inflammation-associated transcriptional response. [Review] Adv Immunol. 2013;119:107–133. doi: 10.1016/B978-0-12-407707-2.00004-7. [DOI] [PubMed] [Google Scholar]
- 23.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Review] Nat Rev Mol Cell Biol. 2013;14(4):211–224. [Google Scholar]
- 24.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. [Research Support, Non-U.S. Gov't Review] Nat Rev Drug Discov. 2014;13(5):337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 25.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. [Review] Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Miao X, Liu Y, Li F, Liu Q, Sun J, et al. Dysregulation of histone acetyltransferases and deacetylases in cardiovascular diseases. [Research Support, Non-U.S. Gov't Review] Oxid Med Cell Longev. 2014;2014:641979. doi: 10.1155/2014/641979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wierda RJ, Geutskens SB, Jukema JW, Quax PH, van den Elsen PJ. Epigenetics in atherosclerosis and inflammation. [Research Support, Non-U.S. Gov't Review] J Cell Mol Med. 2010;14(6A):1225–1240. doi: 10.1111/j.1582-4934.2010.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turunen MP, Aavik E, Yla-Herttuala S. Epigenetics and atherosclerosis. [Research Support, Non-U.S. Gov't Review] Biochim Biophys Acta. 2009;1790(9):886–891. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Pastrana JL, Sha X, Virtue A, Mai J, Cueto R, Lee IA, et al. Regulatory T cells and Atherosclerosis. J Clin Exp Cardiolog. 2012;2012(Suppl 12):2. doi: 10.4172/2155-9880.S12-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripathi SK, Lahesmaa R. Transcriptional and epigenetic regulation of T-helper lineage specification. [Review] Immunol Rev. 2014;261(1):62–83. doi: 10.1111/imr.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang WY, Shao Ying, Lopez-Pastrana Jahaira, Mai Jietang, Wang Hong, Yang Xiao-feng. Pathological conditions re-shape physiological Tregs into pathological Tregs. Burns & Trauma. 2015;3:1–11. doi: 10.1186/s41038-015-0001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Pastrana J, Shao Y, Chernaya V, Wang H, Yang XF. Epigenetic enzymes are the therapeutic targets for CD4(+)CD25(+/high)Foxp3(+) regulatory T cells. [Research Support, N.I.H., Extramural Review] Transl Res. 2015;165(1):221–240. doi: 10.1016/j.trsl.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allan RS, Nutt SL. Deciphering the epigenetic code of T lymphocytes. [Research Support, Non-U.S. Gov't Review] Immunol Rev. 2014;261(1):50–61. doi: 10.1111/imr.12207. [DOI] [PubMed] [Google Scholar]
- 34.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. [Research Support, Non-U.S. Gov't Review] Nat Rev Drug Discov. 2012;11(5):384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 35.Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. [Research Support, Non-U.S. Gov't Review] Nat Rev Genet. 2013;14(11):765–780. doi: 10.1038/nrg3554. [DOI] [PubMed] [Google Scholar]
- 36.Whittaker SJ, Demierre MF, Kim EJ, Rook AH, Lerner A, Duvic M, et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. [Clinical Trial, Phase II Multicenter Study Research Support, Non-U.S. Gov't] J Clin Oncol. 2010;28(29):4485–4491. doi: 10.1200/JCO.2010.28.9066. [DOI] [PubMed] [Google Scholar]
- 37.Piekarz RL, Frye R, Prince HM, Kirschbaum MH, Zain J, Allen SL, et al. Phase 2 trial of romidepsin in patients with peripheral T-cell lymphoma. [Clinical Trial, Phase II Multicenter Study Research Support, N.I.H., Intramural Research Support, Non-U.S. Gov't] Blood. 2011;117(22):5827–5834. doi: 10.1182/blood-2010-10-312603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper ME, El-Osta A. Epigenetics: mechanisms and implications for diabetic complications. [Research Support, Non-U.S. Gov't Review] Circ Res. 2010;107(12):1403–1413. doi: 10.1161/CIRCRESAHA.110.223552. [DOI] [PubMed] [Google Scholar]
- 39.Zhou B, Margariti A, Zeng L, Xu Q. Role of histone deacetylases in vascular cell homeostasis and arteriosclerosis. [Research Support, Non-U.S. Gov't Review] Cardiovasc Res. 2011;90(3):413–420. doi: 10.1093/cvr/cvr003. [DOI] [PubMed] [Google Scholar]
- 40.Chen NC, Yang F, Capecci LM, Gu Z, Schafer AI, Durante W, et al. Regulation of homocysteine metabolism and methylation in human and mouse tissues. Faseb J. 2010;24(8):2804–2817. doi: 10.1096/fj.09-143651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smolle M, Workman JL. Transcription-associated histone modifications and cryptic transcription. [Review] Biochim Biophys Acta. 2013;1829(1):84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maes T, Carceller E, Salas J, Ortega A, Buesa C. Advances in the development of histone lysine demethylase inhibitors. [Review] Curr Opin Pharmacol. 2015;23:52–60. doi: 10.1016/j.coph.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Dambacher S, Hahn M, Schotta G. Epigenetic regulation of development by histone lysine methylation. [Research Support, Non-U.S. Gov't Review] Heredity (Edinb) 2010;105(1):24–37. doi: 10.1038/hdy.2010.49. [DOI] [PubMed] [Google Scholar]
- 44.Chatterjee TK, Basford JE, Knoll E, Tong WS, Blanco V, Blomkalns AL, et al. HDAC9 knockout mice are protected from adipose tissue dysfunction and systemic metabolic disease during high-fat feeding. [Research Support, N.I.H., Extramural] Diabetes. 2014;63(1):176–187. doi: 10.2337/db13-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christensen DP, Dahllof M, Lundh M, Rasmussen DN, Nielsen MD, Billestrup N, et al. Histone deacetylase (HDAC) inhibition as a novel treatment for diabetes mellitus. [Research Support, Non-U.S. Gov't Review] Mol Med. 2011;17(5–6):378–390. doi: 10.2119/molmed.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markus HS, Makela KM, Bevan S, Raitoharju E, Oksala N, Bis JC, et al. Evidence HDAC9 genetic variant associated with ischemic stroke increases risk via promoting carotid atherosclerosis. [Research Support, Non-U.S. Gov't] Stroke. 2013;44(5):1220–1225. doi: 10.1161/STROKEAHA.111.000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jamaluddin MD, Chen I, Yang F, Jiang X, Jan M, Liu X, et al. Homocysteine inhibits endothelial cell growth via DNA hypomethylation of the cyclin Agene. Blood. 2007;110(10):3648–3655. doi: 10.1182/blood-2007-06-096701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jamaluddin MS, Yang X, Wang H. Hyperhomocysteinemia, DNA methylation and vascular disease. Clin Chem Lab Med. 2007;45(12):1660–1666. doi: 10.1515/CCLM.2007.350. [DOI] [PubMed] [Google Scholar]
- 49.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. [Research Support, N.I.H., Extramural Review] Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woo YH, Li WH. Evolutionary conservation of histone modifications in mammals. [Research Support, N.I.H., Extramural] Mol Biol Evol. 2012;29(7):1757–1767. doi: 10.1093/molbev/mss022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng Z, Elmes M, Kirkup SE, Abayasekara DR, Wathes DC. Alteration of prostaglandin production and agonist responsiveness by n-6 polyunsaturated fatty acids in endometrial cells from late-gestation ewes. [Comparative Study Research Support, Non-U.S. Gov't] J Endocrinol. 2004;182(2):249–256. doi: 10.1677/joe.0.1820249. [DOI] [PubMed] [Google Scholar]
- 52.Yang XF, Mirkovic D, Zhang S, Zhang QE, Yan Y, Xiong Z, et al. Processing sites are different in the generation of HLA-A2.1-restricted, T cell reactive tumor antigen epitopes and viral epitopes. Int J Immunopathol Pharmacol. 2006;19(4):853–870. doi: 10.1177/039463200601900415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Mai J, Virtue A, Yin Y, Gong R, Sha X, et al. IL-35 is a novel responsive anti-inflammatory cytokine--a new system of categorizing anti-inflammatory cytokines. [Research Support, N.I.H., Extramural] PLoS One. 2012;7(3):e33628. doi: 10.1371/journal.pone.0033628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathiyalagan P, Keating ST, Du XJ, El-Osta A. Chromatin modifications remodel cardiac gene expression. [Research Support, Non-U.S. Gov't Review] Cardiovasc Res. 2014;103(1):7–16. doi: 10.1093/cvr/cvu122. [DOI] [PubMed] [Google Scholar]
- 55.Shukla SD, Lim RW. Epigenetic effects of ethanol on the liver and gastrointestinal system. [Review] Alcohol Res. 2013;35(1):47–55. doi: 10.35946/arcr.v35.1.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh H. Transcriptional and epigenetic networks orchestrating immune cell development and function. [Introductory] Immunol Rev. 2014;261(1):5–8. doi: 10.1111/imr.12210. [DOI] [PubMed] [Google Scholar]
- 57.Rintisch C, Heinig M, Bauerfeind A, Schafer S, Mieth C, Patone G, et al. Natural variation of histone modification and its impact on gene expression in the rat genome. [Research Support, Non-U.S. Gov't] Genome Res. 2014;24(6):942–953. doi: 10.1101/gr.169029.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. [Research Support, Non-U.S. Gov't] J Biol Chem. 2010;285(5):3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Review] Nat Rev Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNeil CJ, Beattie JH, Gordon MJ, Pirie LP, Duthie SJ. Differential effects of nutritional folic acid deficiency and moderate hyperhomocysteinemia on aortic plaque formation and genome-wide DNA methylation in vascular tissue from ApoE−/− mice. Clin Epigenetics. 2011;2(2):361–368. doi: 10.1007/s13148-011-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi L, Tu BP. Protein acetylation as a means to regulate protein function in tune with metabolic state. [Research Support, N.I.H., Extramural Review] Biochem Soc Trans. 2014;42(4):1037–1042. doi: 10.1042/BST20140135. [DOI] [PubMed] [Google Scholar]
- 62.Carrer A, Wellen KE. Metabolism and epigenetics: a link cancer cells exploit. [Review] Curr Opin Biotechnol. 2014;34:23–29. doi: 10.1016/j.copbio.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. [Research Support, N.I.H., Extramural Review] Immunity. 2015;42(3):406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. [Research Support, N.I.H., Extramural Review] Nat Biotechnol. 2010;28(10):1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zou C, Mallampalli RK. Regulation of histone modifying enzymes by the ubiquitin-proteasome system. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, Non-P.H.S. Review] Biochim Biophys Acta. 2014;1843(4):694–702. doi: 10.1016/j.bbamcr.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones PA, Baylin SB. The epigenomics of cancer. [Research Support, N.I.H., Extramural Review] Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. [Review] Cell. 2008;132(4):567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.