Abstract
Integrative analysis of multiple data types to address complex biomedical questions requires the use of multiple software tools in concert and remains an enormous challenge for most of the biomedical research community. Here we introduce GenomeSpace (http://www.genomespace.org), a cloud-based, cooperative community resource. Seeded as a collaboration of six of the most popular genomics analysis tools, GenomeSpace now supports the streamlined interaction of 20 bioinformatics tools and data resources. To facilitate the ability of non-programming users’ to leverage GenomeSpace in integrative analysis, it offers a growing set of ‘recipes’, short workflows involving a few tools and steps to guide investigators through high utility analysis tasks.
Building on the Human Genome Project1 and the advent of high-throughput genomic technologies, the past two decades of biomedical research are yielding a flood of massive and varied biological datasets. As a result, numerous databases and analysis software tools have been developed for researchers to access, visualize, and analyze different data types.
However, integrative analysis of diverse data types through multiple analysis tools remains an enormous challenge for many biologists. There is an ever-growing gap between the need to use various analysis and visualization software tools and the difficulty of getting tools from different sources to work together. Moreover, the wealth of existing and emerging analytical methods makes it difficult – even for experts, but especially for less computationally oriented biologists – to keep up with all of the available tools, and to identify the right recipe to follow, particularly in the absence of an accepted “laboratory manual” for analytic protocols. These difficulties curtail the agility and creativity of researchers and may prevent them from adopting alternative or new methods.
Here, we present GenomeSpace, a cooperative community resource that provides an open-source interoperability platform to enable non-programming scientists to work easily across data types, tools, and analysis methods. GenomeSpace provides a “tool launch pad” into which tools can be seamlessly added, and a “data highway” that handles transfers between tools through format converters, relieving scientists of the burden of identifying and scripting the conversions. The GenomeSpace Recipe Resource is a growing set of high-utility use cases that demonstrate how to leverage multiple tools, and also serve as quick guides to analysis tasks using GenomeSpace and the GenomeSpace tools. The GenomeSpace website, http://www.genomespace.org, serves as a knowledge base, newsstand, and point of online contact and help for the GenomeSpace community of users and tool developers.
Initially seeded by a consortium of biology research labs and development teams of six popular bioinformatics tools (Cytoscape2,3, Galaxy4, GenePattern5, Genomica6, the Integrative Genomics Viewer (IGV)7, and the UCSC Table Browser8), GenomeSpace now connects 20 tools and data resources. Our consortium labs provided real driving biological projects and analytical needs to shape the design and development of the GenomeSpace architecture and software. For example, we recapitulated the steps and results of published analyses9,10 within GenomeSpace (Supplementary Figs. 1-2), dissecting and visualizing the gene regulatory networks in human cancer stem cells (Supplementary Note 1, Supplementary Figs. 2–5). This illustrates how GenomeSpace enables a non-programming biologist to conduct a rich and involved integrative analysis, which previously led to a novel result. The study required diverse data types, analytical steps, methods, tools, and multiple data transfers between the tools. While originally requiring substantial scripting, this work can now be performed using only the tools within, and capabilities of, GenomeSpace.
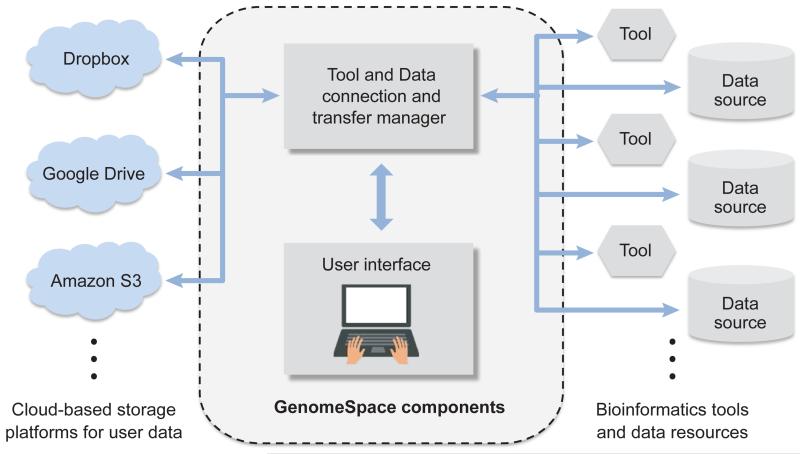
From a user’s perspective (Fig. 1, Supplementary Fig. 6), GenomeSpace has several key features that together facilitate integrative analysis with a low barrier to user entry: (1) The collection of resident tools spanning a broad range of applications (Table 1); (2) Easy dataset management in a variety of cloud storage types, alongside data sharing capabilities. All GenomeSpace account holders receive an allocation of cloud storage, and GenomeSpace also supports connections to other cloud accounts (Dropbox, Google Drive, Amazon S3); (3) The ability to launch tools and to move data and analyses between tools, all facilitated by “behind-the-scenes” file format converters; (4) A lightweight, simple, unifying web interface. In summary, from the web interface a researcher can launch a desired tool and simultaneously feed it input data files, move analysis results into other tools as needed through simple launching operations, and collect additional processed data within their GenomeSpace cloud account, other cloud accounts, or local storage.
Figure 1. The GenomeSpace environment for interoperation of bioinformatics tools.
Table 1. GenomeSpace provides access to a diverse set of bioinformatics tools and resources.
| Tool Name | Organization | Project Website |
|---|---|---|
| Analysis and Visualization Tools | ||
|
| ||
| Cistrome | Dana-Farber Cancer Institute | http://www.cistrome.org |
| Cytoscape 3 * | Cytoscape Consortium | http://www.cytoscape.org |
| Cytoscape 2 * | Cytoscape Consortium | http://www.cytoscape.org |
| Galaxy | Pennsylvania State University; and Johns Hopkins University |
http://www.galaxyproject.org |
| GenePattern | Broad Institute; and UC San Diego | http://www.genepattern.org |
| Genomica | Weizmann Institute of Science | http://genomica.weizmann.ac.il |
| geWorkbench | Columbia University | http://www.geworkbench.org |
| Gitools | University Pompeu Fabra, Barcelona | http://www.gitools.org |
| Integrative Genomics Viewer (IGV) |
Broad Institute; and UC San Diego | http://www.igv.org |
| ISAcreator | University of Oxford | http://www.isa-tools.org |
| Molecular Signatures Database (MSigDB) Online Tools |
Broad Institute; and UC San Diego | http://www.msigdb.org |
|
| ||
| Data Resources | ||
|
| ||
| ArrayExpress | European Bioinformatics Institute | http://www.ebi.ac.uk/arrayexpress |
| InSilicoDB | InSilico Genomics | http://insilicodb.com |
| Synapse | Sage Bionetworks | http://synapse.org |
| UCSC Table Browser | University of California Santa Cruz | http://genome.ucsc.edu |
|
| ||
| Integrated Portals (Data and Analysis) | ||
|
| ||
| Achilles Project | Dana-Farber Cancer Institute; and Broad Institute |
http://broadinstitute.org/achilles |
| Cancer Cell Line Encyclopedia (CCLE) |
Novartis Institutes for BioMedical Research; and Broad Institute |
http://broadinstitute.org/ccle |
| cBioPortal for Cancer Genomics | Memorial Sloan Kettering Cancer Center |
http://www.cbioportal.org |
| Multiple Myeloma Genomics Portal (MMGP) |
Multiple Myeloma Research Consortium; Broad Institute; and Translational Genomics Research Institute (TGen) |
http://broadinstitute.org/mmgp |
| Reactome | Ontario Institute for Cancer Research; European Bioinformatics Institute; and New York University Medical Center |
http://www.reactome.org |
Cytoscape 3 and Cytoscape 2 have different underlying architectures and different user interfaces. Both versions are made available through GenomeSpace to accommodate users who may prefer one to the other.
We developed the GenomeSpace Recipe Resource to aid biomedical researchers in identifying the steps required to perform a genomic analysis – a challenging task even for short analyses. Although pre-constructed pipelines can embody the entire workflow of a study, they may be insufficiently open-ended or flexible for exploratory research. We took an alternative approach by providing a collection or “cookbook” of recipes, i.e., comprehensive descriptions of cross-tool analysis workflows. Recipes are generally short – involving two or three tools – but commoditize important research tasks that investigators can employ in many ways as part of more complex analyses. The notion of our Recipe Resource is modeled after the classical lab guide “Molecular cloning: A laboratory manual”11, which used a similar approach to democratize molecular biology three decades ago.
Each GenomeSpace recipe contains a motivating biological problem, a relevant example dataset, detailed recipe steps, and one possible interpretation of the results, illustrated on the example data. A variety of media accompany the recipe steps including screenshot guides and videos that together walk users through the workflow. The recipes are served on our Recipe web page (http://www.genomespace.org/recipes), the most frequently visited section of the GenomeSpace website after the home page. The current recipes cover diverse genomic analyses as well as basic utilities for using GenomeSpace itself (Supplementary Table 1). Since no single lab can supply the expertise or effort required to create a comprehensive recipe collection, we are adding social media vehicles to make recipe collection a crowd-sourced, collaborative effort through community contributions. We encourage suggestions for new and useful multi-tool recipes and ideas to improve existing recipes.
An illustrative example from the GenomeSpace Recipe Resource is “Find subnetworks of differentially expressed genes and identify associated biological functions”. Briefly, given a gene expression dataset, this recipe identifies network interactions between differentially expressed genes, and annotates the biological functions within subnetworks via the Gene Ontology (GO) (Supplementary Fig. 7). The example dataset provided with this recipe is gene expression data from a study in which granulocyte-macrophage progenitor cells were transformed into leukemia stem cells by introduction of an oncogene, MLL-AF913. Applying the recipe identifies processes that are correlated with transformation from a normal to a leukemic phenotype (Supplementary Fig. 8), such as SMAD1-dependent signaling, a process associated with the regulation of hematopoietic differentiation by TGF-β and BMP14. A second recipe example, “Identify biological functions for genes in copy number variation (CNV) regions”, is described in Supplementary Note 2 (Supplementary Figs. 9–10).
An important GenomeSpace design goal was to facilitate rapid addition of diverse tools contributed by the developer community. This mutually benefits GenomeSpace and independent tool developers by extending the capabilities of GenomeSpace while also giving developers’ tools access to all GenomeSpace-connected tools and data sources, circumventing the need to connect to each one individually. Recent cross-tool interoperability efforts have used one of several approaches: aggregators host a large number of command line tools (Galaxy, GenePattern); plug-in architectures provide a way to extend the functionality of a basic package (Cytoscape, geWorkbench15, MeV16), and messaging systems send data and instructions between tools (MeDICi17, Gaggle18). Our open-source, lightweight, hybrid approach combines aspects of both messaging and aggregating systems. The resulting platform (Supplementary Fig. 11 and Online Methods) provides single sign-on for GenomeSpace tools and data resources; security mechanisms and user-controlled levels of sharing; and a common interface to multiple cloud storage providers. Moreover, this approach supports interoperation among diverse desktop and web-based tools, while minimizing the amount of effort required to connect to the platform (Online Methods).
To further facilitate cross-tool interoperability, GenomeSpace offers a range of file converters for directly converting between pairs of file formats (Supplementary Note 3). Our direct conversion approach confers a number of benefits. Notably, it obviates the development burden of defining and supporting central data models for tools, especially legacy ones, connecting to GenomeSpace. Moreover, since converters are independent and do not rely on a GenomeSpace-specific data model, we can expand the set of supported formats by leveraging converters that are developed outside of GenomeSpace.
In conclusion, GenomeSpace has several key benefits. First, it allows seamless transition between tools. Automatic inter-tool file format converters speed tasks like launching and moving data between tools and obviate the need for custom conversion scripts, an insurmountable barrier for many biologists. Second, the large set of connected tools enriches the interpretation of integrative analyses. Exploring the same data in multiple tools—each designed to highlight distinct features of the data—allows the analysis to be examined in greater depth and diversity than with any single tool. Third, we encourage the inclusion of multiple tools with similar capabilities. Therefore many analysis steps can be performed with alternative tools from the GenomeSpace suite, allowing investigators to test their findings for robustness and reproducibility. It also permits them to use the tool with which they are most familiar. Fourth, recipes play an important role in making integrative analysis accessible. Conceived of as small analysis components, recipes describe short workflows that guide users to perform analysis tasks. Recipes can be assembled into more complex analysis scenarios and can also introduce investigators to new analysis methods and tools. In this way, GenomeSpace and the Recipe Resource can greatly expand the analytic universe accessible to investigators and help to move their research agenda forward.
ONLINE METHODS
GenomeSpace Architecture
From a tool developer’s perspective, GenomeSpace presents a “connection layer” that includes a collection of web services with well defined entry points to the GenomeSpace server that provides the core system functionality (Supplementary Figure 11). The GenomeSpace web user interface also interacts with the server through these entry points. The GenomeSpace server currently runs as an Amazon Machine Instance (AMI) in the Amazon Elastic Compute Cloud (EC2). It consists of three components: (1) An Identity Service manages sign-on credentials, including single sign-on to the GenomeSpace tools, and data access. GenomeSpace leverages the Amazon AWS security mechanisms, which are compliant with the requirements from many standards organizations and government agencies. All data is private by default, but users may share directories or files with other users or groups of users. (2) An Analysis Tools Manager maintains information about tool capabilities and dependencies and coordinates tool launches, including the ability to launch other GenomeSpace tools from within a tool; (3) A Data Manager handles data storage, transfer to/from the cloud (including Amazon S3, Google Drive, and Dropbox), data sharing, and the file format conversions that provide a smooth script-free connection between tools.
Connecting Tools to GenomeSpace
The GenomeSpace connection layer includes a collection of web services with well-defined entry points to the GenomeSpace server that provides the core system functionality. It is available as Java and JavaScript client development kits for tools developed in those languages, or as web services with a RESTful application programming interface (API) for any language. Developers can also take advantage of a number of user interface widgets that are available for common user tasks, including file chooser dialogs and authentication panels. Adding a tool to GenomeSpace, using these resources, typically takes on the order of two programmer days or less, depending on the type of tool. The most recent tool to join the community was cBioPortal (http://www.cbioportal.org) from Memorial Sloan Kettering Cancer Center, and the development team reported that it took an hour to connect this web-based portal as a data source to GenomeSpace. We note that command line tools that do not have their own user interface can join the GenomeSpace community via either of its current aggregator members – GenePattern and Galaxy.
Supplementary Material
ACKNOWLEDGMENTS
We thank other members of the GenomeSpace and GenePattern Teams for their contributions and input: P. Carr, B. Hill-Meyers, S. H. Lee, and T. Tabor (Broad Institute of MIT and Harvard); J. Zhang (Stanford University); and H. Carter and M. Smoot (University of California San Diego). Special thanks to D. Haussler and J. Kent (University of California Santa Cruz) for their involvement in the nascent stages of the GenomeSpace project. We thank J. Bistline for help with the citations and figures, and L. Gaffney for help with the figures. This work has been supported by NIH-NHGRI P01 HG005062 and U41 HG007517, with additional initial support from Amazon Web Services (AWS).
Footnotes
AUTHOR CONTRIBUTIONS
M.R., A.R. and J.P.M. conceived of the GenomeSpace concept. T.L, M.O. and M.R. designed and implemented the GenomeSpace software. K.Q., S.G., F.W., and N.P. implemented the driving biological projects within GenomeSpace with supervision and input from A.R., H.Y.C., and J.P.M. The recipes were implemented by S.G., F.W., and D.B.-R. The GenomeSpace seed tools were added to the system by J.T.R., B.D., T.H., G.B.-A., D.B., G.P.B., B.T.L., R.M.K., A.N., E.S., and T.I, who also consulted on the GenomeSpace architecture. H.T., M.R., A.R., H.Y.C., and J.P.M. supervise the GenomeSpace project. K.Q., S.G., F.W., H.T., M.R., H.C., A.R., and J.P.M. wrote the manuscript. All authors reviewed and approved the final manuscript as submitted.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Lander ES, et al. Nature. 2001;409:860. [Google Scholar]
- 2.Demchak B, et al. F1000Research. 2014;3:151. doi: 10.12688/f1000research.4492.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shannon P, et al. Genome Res. 2003;13:2498. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giardine B, et al. Genome Res. 2005;15:1451. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reich M, et al. Nat Genet. 2006;38:500. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 6.Segal E, Friedman N, Koller D, Regev A. Nat Genet. 2004;36:1090. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 7.Robinson JT, et al. Nat Biotechnol. 2011;29:24. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karolchik D, et al. Nucleic Acids Res. 2004;32:D493. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Porath I, et al. Nat Genet. 2008;40:499. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong DJ, et al. Cell Stem Cell. 2008;2:333. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E, Maniatis T. Vol. 3. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 12.Mostafavi S, et al. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krivtsov AV, et al. Nature. 2006;442:818. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 14.Larsson J, Karlsson S. Oncogene. 2005;24:5676. doi: 10.1038/sj.onc.1208920. [DOI] [PubMed] [Google Scholar]
- 15.Floratos A, et al. Bioinformatics. 2010;26:1779. doi: 10.1093/bioinformatics/btq282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saeed AI, et al. BioTechniques. 2003;34:374. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 17.Gorton I, Wynne A, Almquist J, Chatterton J. Software Architecture; WICSA 2008. Seventh Working IEEE/IFIP Conference on.; 2008.p. 95. [Google Scholar]
- 18.Shannon PT, Reiss DJ, Bonneau R, Baliga NS. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.