Abstract
Natural extracellular matrices (ECMs) are viscoelastic and exhibit stress relaxation. However, hydrogels used as synthetic ECMs for three-dimensional (3D) culture are typically elastic. Here, we report a materials approach to tune the rate of stress relaxation of hydrogels for 3D culture, independently of the hydrogel’s initial elastic modulus, cell-adhesion-ligand density and degradation. We find that cell spreading, proliferation, and osteogenic differentiation of mesenchymal stem cells (MSCs) are all enhanced in cells cultured in gels with faster relaxation. Strikingly, MSCs form a mineralized, collagen-1-rich matrix similar to bone in rapidly relaxing hydrogels with an initial elastic modulus of 17 kPa. We also show that the effects of stress relaxation are mediated by adhesion-ligand binding, actomyosin contractility and mechanical clustering of adhesion ligands. Our findings highlight stress relaxation as a key characteristic of cell-ECM interactions and as an important design parameter of biomaterials for cell culture.
Hydrogels composed of crosslinked networks of polymers, such as poly-ethylene glycol (PEG)1,2, alginate3,4, and hyaluronic acid5,6, that are covalently coupled to integrin binding ligands, such as RGD, are often used for 3D cell culture or as cell-laden biomaterial implants to promote tissue regeneration3,7–11. The use of these hydrogels is often preferred over reconstituted extracellular matrices of collagen, fibrin, or basement membrane due to the independent control over the physical and chemical properties (e.g. matrix elasticity, ligand density, and porosity) possible in these hydrogels4,12–14, as well as their homogeneity at the microscale. However, normal cellular processes such as shape change, migration, and proliferation are inhibited in these hydrogels unless they are designed to degrade over time2,4,6,15. While non-degradable hydrogels can capture some characteristics of physiological ECM, they are typically almost purely elastic. In contrast, reconstituted extracellular matrices, such as collagen, or fibrin2, and various tissues, such as brain16, liver17, adipose tissue18, coagulated bone marrow, initial fracture hematomas, or the soft callus of regenerating bone19, are all viscoelastic and exhibit partial stress relaxation when a constant strain of 15% is applied (Fig. 1a). For comparison, cells typically exert strains of up to 3 – 4% in 2D culture20, and 20 – 30% in 3D culture21 (Supplementary Note 1). Also, peak stresses measured during stress relaxation tests of these tissues ranged from 100 – 1,000 Pa, well within the range of stresses generated by cells in 3D culture21,22 (Supplementary Table 1). A decrease in stress corresponds to a decrease in the relaxation modulus or the resistance to deformation over time. As it has been well established that the mechanical properties of materials regulate adherent cell behavior23–29, the ability of a substrate to either store (purely elastic) or dissipate (viscoelastic) cellular forces could provide a powerful cue to interacting cells. Indeed, recent studies have found an impact of altered substrate viscoelasticity, independent of substrate stiffness, on various cell behaviors using hydrogels as substrates for cell culture30–33. In gels that exhibit stress relaxation, each force or strain a cell applies to the matrix over time is initially resisted with a certain stiffness, defined by the initial elastic modulus, followed by a decrease in resistance over time. For hydrogels formed with weak crosslinks, relaxation arises in part from unbinding of crosslinks and hydrogel flow, so that cellular forces can mechanically remodel the matrix33. Here we investigate the influence of hydrogel viscoelasticity and stress relaxation on cell spreading, proliferation, and MSC differentiation in 3D culture.
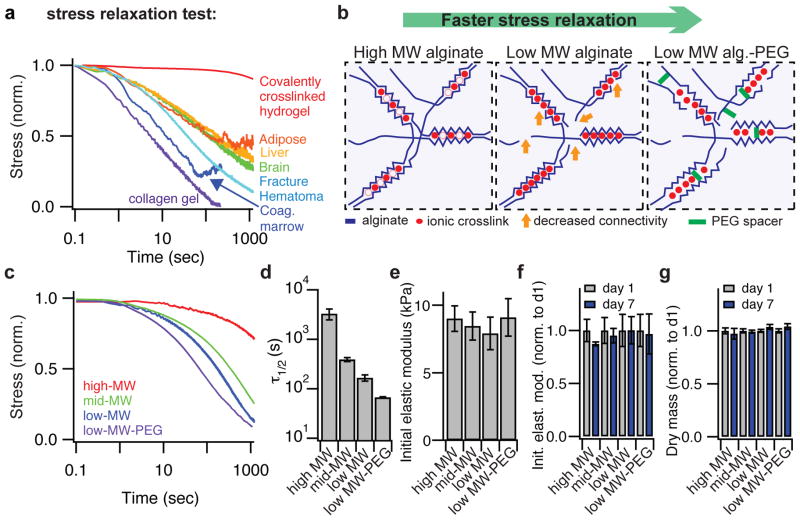
Figure 1. Modulating the nanoscale architecture of alginate hydrogels to modulate stress relaxation properties independent of initial elastic modulus and matrix degradation to capture the viscoelastic behaviors of living tissues.
a, Living tissues are viscoelastic and exhibit stress relaxation. Stress relaxation tests of a crosslinked hydrogel (polyacrylamide), a collagen gel, an initial fracture hematoma (human), and various tissues (rat) at a strain of 15%. Stress is normalized by the initial stress. b, Schematic depicting how lowering the molecular weight (MW) of alginate polymers (blue) crosslinked by calcium (red) decreases entanglement and connectivity (orange arrows) of the network, and coupling of low-MW PEG spacers provides a steric spacing of crosslinking zones in the alginate. Both approaches are predicted to increase the rate of stress relaxation. c, Stress relaxation tests on gels composed of alginates with different molecular weights, or low molecular weight alginate coupled to a PEG spacer (15% compressional strain). d, Quantification of timescale at which the stress is relaxed to half its original value, τ1/2, from stress relaxation tests in c. Timescale of stress relaxation decreases significantly with alteration in architecture (Spearman’s rank correlation coefficient, p < 0.0001). e, Initial modulus measurements of gels in c. Differences between elastic moduli are not significant, and elastic moduli show no statistical trend with altered architecture. f, Initial elastic modulus of alginate hydrogels after 1 day or 7 days in culture, normalized by the value at day 1. g, Measured dry mass of alginate hydrogels after 1 day or 7 days in culture normalized by the value at day 1. All data are shown as mean +/− s.d.
Hydrogels with tunable stress relaxation
First we modulated the nanoscale architecture of hydrogels to develop a set of materials with a wide range of stress relaxation rates, but a similar initial elastic modulus. As hydrogels exhibiting minimal degradation were desired, the polysaccharide alginate was chosen for these studies since mammalian cells do not express specific enzymes that can degrade this polymer34. Alginate presents no intrinsic integrin binding sites for cells and minimal protein absorption, but cell adhesion can be promoted through covalent coupling of the RGD cell adhesion peptide to the alginate chains3. While the stress relaxation properties of hydrogels have been altered previously by changing crosslinking chemistries32,35 or polymer concentration30, we developed an alternative materials approach to control the rate of stress relaxation of hydrogels with a single crosslinker type and the same concentration of alginate. We hypothesized that by using different molecular weight polymers in combination with different crosslinking densities of calcium, which ionically crosslinks alginate, the stress relaxation properties of the resulting hydrogels could be modulated due to the altered connectivity and chain mobility36 in the network (Fig. 1b). Any associated decrease in the initial elastic modulus resulting from decreased polymer molecular weight could be compensated for by increased crosslinking. Further, we hypothesized that covalent coupling of short PEG spacers to the alginate would provide a steric hindrance to crosslinking of alginate chains and enhance stress relaxation in the gel (Fig. 1b). Both approaches would alter the net avidity between individual polymer chains and therefore be expected to control the relaxation behavior. This was confirmed, as it was found that by lowering the molecular weight of the alginate from 280 kDa to 35 kDa, and further by coupling 5 kDa PEG spacers to the 35 kDa alginate, the rate of stress relaxation was enhanced markedly (Fig. 1c). Specifically, the time for the initial stress of the material to be relaxed to half its value during a stress relaxation test (τ1/2) was modulated from ~1 hour to ~1 minute, while holding alginate polymer concentration and the initial gel elastic modulus constant (Fig. 1c–e, Supplementary Table 2). These timescales span a range similar to that measured in various tissues (Fig. 1a). Further, this range of timescales is relevant to cell behaviors as cells are thought to respond to force oscillations over a timescale of ~1 second37, exert traction forces on a timescale of minutes30, and undergo cell spreading on a timescale of minutes to hours38. The stress relaxation behavior of these materials follows that of a 2-element Maxwell-Weichert linear viscoelastic model (Supplementary Note 2, Supplementary Fig. S1). It was previously shown that stress relaxation measured in ionically crosslinked alginate gels represents viscoelasticity of the hydrogel and unbinding of ionic crosslinks followed by matrix flow35. This interpretation was confirmed by a measurement of the frequency dependent rheology of the gels, as an increase in the rate of stress relaxation correlated with a greater rate of decrease in the shear storage modulus with decreasing frequency (Supplementary Fig. S2). Also consistent with this interpretation is the near complete relaxation of the stress, indicating that these matrices can be plastically deformed/mechanically remodeled and that the shorter the τ1/2, the faster the matrices remodel under stress. Homogeneity of the gels at the microscale was confirmed with confocal fluorescence microscopy (Supplementary Fig. S3). Importantly, the mechanical properties of these gels and the dry polymer mass of the gels were both stable over a timescale of at least 7 days under tissue culture conditions (Fig. 1f,g). In combination, these demonstrate that the degradation of these matrices is negligible over this timescale. Taken together, these demonstrate an approach to control matrix stress relaxation, independent of the initial elastic modulus and polymer concentration, and without hydrogel degradation.
Impact of stress relaxation on cell spreading and proliferation
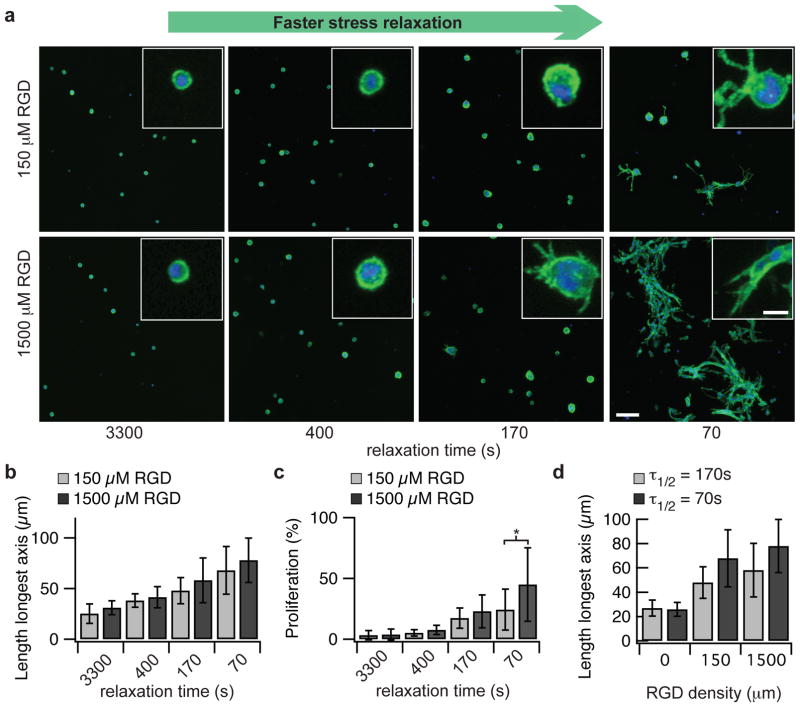
With this set of materials, the effect of the rate of substrate stress relaxation on cell spreading and proliferation in 3D culture was investigated, and striking differences in both were observed. First, 3T3 fibroblasts were encapsulated within RGD coupled alginate hydrogels with varying stress relaxation rates but all with an initial elastic modulus of ~9 kPa (Fig. 2a). Both cell spreading and proliferation were suppressed within materials with long timescales for stress relaxation (τ1/2 ~ 1 hour), and the rounded cell morphologies typical of cells in non-degradable elastic hydrogels were observed under this condition. Strikingly, both spreading and proliferation were found to increase with faster stress relaxation (Fig. 2b–c, Spearman’s rank correlation, p <0.0001 for both). The influence of substrate stress relaxation on cell spreading and proliferation was enhanced when RGD cell adhesion ligand density was increased in gels with faster relaxation, indicating that the effect of stress relaxation is mediated through integrin based adhesions (Fig. 2d, Spearman’s rank correlation, p < 0.0001). This enhancement in spreading and proliferation was attributable to altered stress relaxation alone, as the initial elastic modulus and alginate concentration was constant, and RGD cell adhesion ligand density was also held constant at either 0, 150, or 1500 μM. The observation of striking changes in cell shape in the rapidly relaxing hydrogels suggest mechanical remodeling of the hydrogel by the cells, as these hydrogels are nanoporous and non-degradable so that cell shape change and proliferation must be accommodated by matrix displacement.
Figure 2. Cell spreading and proliferation for fibroblasts encapsulated within gels are enhanced with faster stress relaxation.
a, Representative images of 3T3 cells encapsulated within alginate gels with the indicated τ1/2 for stress relaxation and two RGD concentrations (average initial modulus of 9 kPa). Green color represents actin staining and blue represents nucleus. Images were taken after 7 days in culture. Scale bar is 100 μm for the larger image and 20 μm for the inset. b, Quantification of the longest dimension of the smallest bounding box fully containing individual 3T3 cells for the indicated conditions. ** indicates p<0.01 (student’s t-test). Spreading increases significantly with faster stress relaxation (Spearman’s rank correlation, p <0.0001 for both values of RGD). c, Quantification of proliferating cells. * indicates p<0.05 (student’s t-test). Proliferation was found to increase with faster stress relaxation (Spearman’s rank correlation, p <0.0001 for both values of RGD). d, Quantification of the longest dimension of the smallest bounding box fully containing individual 3T3 cells as a function of RGD density in alginate gels with a relaxation time of 70 or 170 seconds. Spreading increases significantly with increased RGD concentration for both gels (Spearman’s rank correlation, p <0.0001 for both values of τ1/2). Data are shown as mean +/− s.d.
Regulation of MSC differentiation by matrix stress relaxation
Next, we looked at the influence of substrate stress relaxation on the differentiation of a murine mesenchymal stem cell line (D1, MSCs) in 3D culture. A previous study found that D1 MSCs, as well as primary human MSCs, encapsulated in ionically-crosslinked alginate hydrogels undergo predominantly adipogenic differentiation at initial moduli of 1 – 10 kPa, and predominantly osteogenic differentiation at initial moduli of 11 – 30 kPa4. An intuitive expectation might be that cells integrate the elastic modulus over time and feel an effectively lower elastic modulus on a viscoelastic substrate (Supplementary Fig. S4). If this were the case, osteogenic differentiation of MSCs in matrices with an initial elastic modulus of 11 – 30 kPa would be reduced with faster relaxation in the gel. To test whether this is the case, MSCs were encapsulated in alginate hydrogels with various timescales of stress relaxation and initial elastic moduli (Fig. 3a–b, Supplementary Fig. S5). When the initial elastic modulus of the matrix was ~9 kPa, MSCs exhibited primarily adipogenic differentiation, as indicated by staining for neutral lipids, and very low levels of osteogenic differentiation, as indicated by alkaline phosphatase staining and a quantitative assay of alkaline phosphatase activity, for all timescales of stress relaxation probed (Fig. 3a – b). The level of adipogenesis was found to decrease in rapidly relaxing gels, which had a relaxation time of ~1 min. In contrast, at a higher initial elastic modulus of ~17 kPa, no adipogenic differentiation was observed, and osteogenic differentiation was significantly enhanced in gels with faster stress relaxation (Fig. 3a – b, Spearman’s rank correlation, p < 0.0001). This is surprising, as if cells were simply integrating the elastic modulus of the matrix over time, decreased osteogenesis and increased adipogenesis would be expected with faster stress relaxation. Calcium used to crosslink the alginate gels did not influence differentiation (Supplementary Fig. S6), consistent with previous findings4. Faster stress relaxation also promoted a greater degree of cell spreading in MSCs, similar to the result with 3T3s (Supplementary Fig. S7). Even though the initial seeding density of cells was held constant, there appeared to be a higher density of cells in the stiffer gels after 7 days, possibly due to differences in proliferation between the adipogenically and osteogenically differentiated cells (Supplementary Fig. S7). Interestingly, cell morphologies were similar between MSCs in slow relaxing gels (e.g. τ1/2 of 2300 seconds) with an elastic modulus of 9 kPa, in which adipogenesis is observed, and 17 kPa, in which osteogenesis is observed (Supplementary Fig. S7). In addition, cell morphology was similar for MSCs in 17 kPa hydrogels with stress relaxation times of 2300 seconds and 300 seconds, a range over which a striking increase in osteogenesis is observed (Supplementary Fig. S7 and Fig. 3). Thus MSC fate was decoupled from cell shape, consistent with previous findings regarding MSC differentiation in 3D culture4,13.
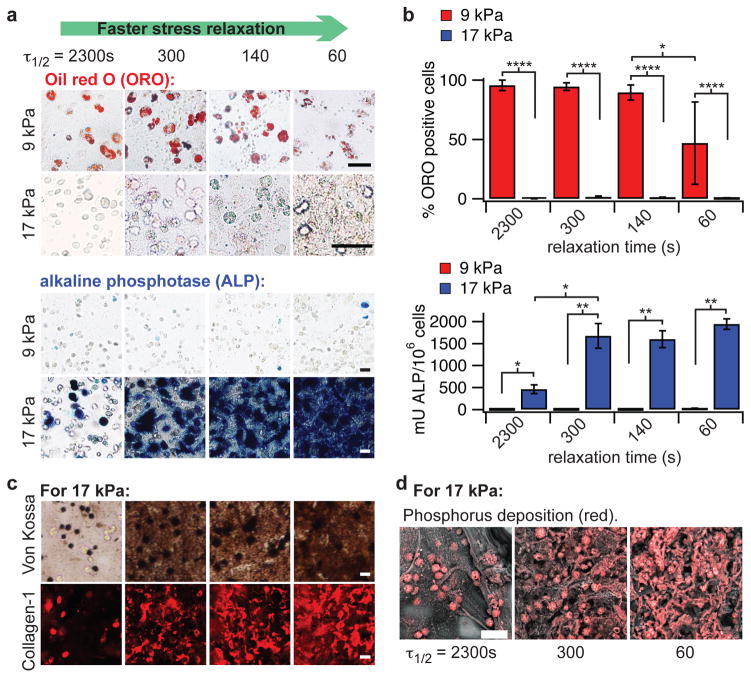
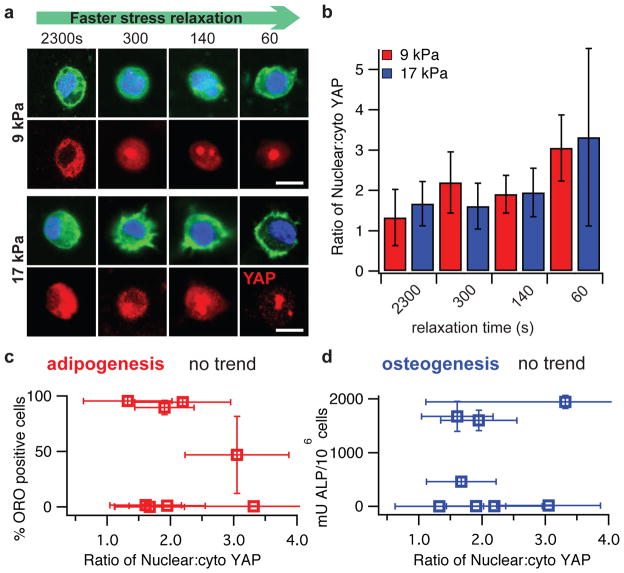
Figure 3. MSCs undergo osteogenic differentiation and form an interconnected mineralized collagen-1 rich matrix only in rapidly relaxing gels.
a, Representative images of cryosections with Oil red O (ORO) staining (red), indicating adipogenic differentiation, and alkaline phosphatase staining (blue), indicating early osteogenic differentiation, for MSC cultured in gels of indicated initial modulus and timescale of stress relaxation for 7 days. RGD density is 1500 μM. Scale bars are 25 μm. b, Quantification of the % of cells staining positive for ORO, and a quantitative assay for alkaline phosphatase activity from lysates of cells in gels from the indicated conditions at 7 days in culture. *, **, and **** indicate p <0.05, 0.01, & 0.0001 respectively (student’s t-test). Bars for % cells staining for ORO in gels with initial modulus of 17 kPa and alkaline phosphatase activity of cells in gels with initial modulus of 9 kPa are barely visible due to the small values relative to the other conditions. Osteogenic differentiation increases significantly with a faster stress relaxation (Spearman’s rank correlation, p < 0.0001). c, Von Kossa (mineralization) and collagen-I stain on cryosections from gels with the indicated conditions after 2 weeks of culture. Scale bars are 25 μm. d, Scanning electron microscope and energy dispersive X-ray spectrometry (SEM-EDS) images of sections of gels with the indicated conditions (all gels at 1500 μM RGD) after 2 weeks of 3D culture of MSCs. Phosphorus elemental maps (P mapped in red) are overlaid on their corresponding backscattered SEM images. Scale bar is 50 μm. All data are shown as mean +/− s.d. For simplicity, τ1/2 values shown in this figure are an average of the τ1/2 values at 9 kPa and 17 kPa. Specific values for each condition are shown in Supplementary Table 2.
In addition to characterizing differentiation of the MSCs, the functional activity of the osteogenically differentiated stem cells was examined. Previous studies have found osteogenic differentiation of MSCs within 3D matrices of slowly relaxing alginate gels4, PEG gels39, degradable hyaluronic acid gels13, or thixotropic PEG silica gels40. However, formation of an interconnected, mineralized, and collagen-1 rich matrix, the three key structural features of bone, by these differentiated cells has not been reported. Here, Von Kossa staining, immunohistochemistry, and energy-dispersive X-ray spectroscopy (EDS) revealed that matrix mineralization and type-1 collagen deposition were both enhanced after 14 days in culture with faster stress relaxation (Fig. 3c–d, Supplementary Fig. S8). Strikingly, MSCs formed an interconnected bone-like matrix in rapidly relaxing gels that exhibit a time constant of stress relaxation on the order of ~1 minute. Intriguingly, this timescale of stress relaxation approaches that of an initial fracture hematoma (Fig. 1a), and is similar to that reported for an early fracture callous19. This result demonstrates that the rapidly relaxing gels not only promote maximal osteogenesis but also enable bone-forming activity in the osteogenically differentiated stem cells.
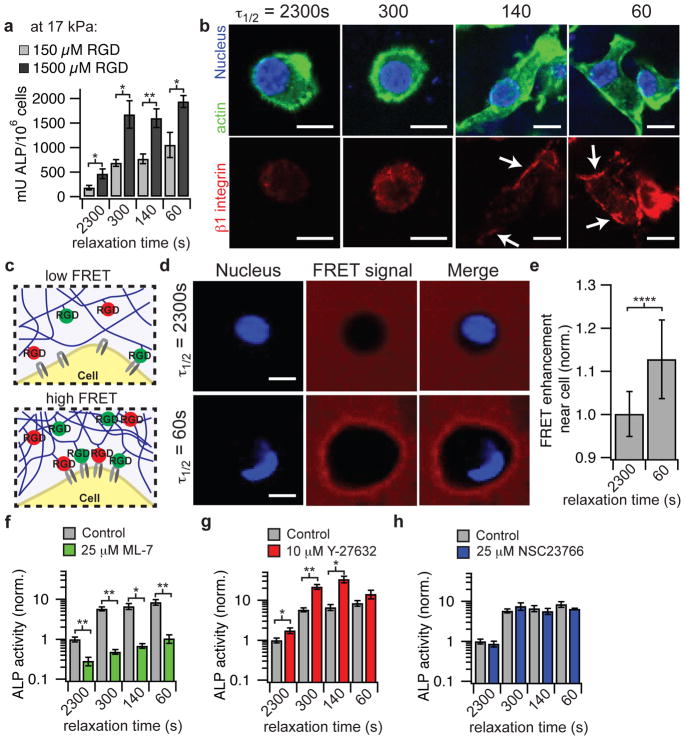
After finding strong effects of the initial elastic modulus and the rate of stress relaxation on MSC differentiation and bone forming activity, we investigated the underlying mechanism by which fast stress relaxation promotes these behaviors. Cells sense mechanical cues in the ECM through binding to ECM ligands, so the impact of RGD density on MSC differentiation was first examined (Fig. 4a, Supplementary Fig. S9). At a lower RGD density of 150 μM, a more graded trend of enhanced osteogenesis with faster stress relaxation is observed in hydrogels with an initial elastic modulus of 17 kPa (Spearman’s rank correlation, p < 0.0001), but the degree of osteogenic differentiation was significantly diminished relative to at a higher RGD density (Fig. 4a). These findings demonstrate that the effect of stress relaxation on osteogenic differentiation is mediated through ECM ligands. Cells bind to ECM ligands through integrin receptors41. An examination of the localization of β1 integrin using an antibody recognizing an epitope of primed β1 integrin revealed increased localization of primed β1 integrin to the periphery of the cell in gels with faster relaxation (Fig. 4b). However, localization of paxillin to the periphery of the cells was not observed, indicating that conventional focal adhesions42 were not being formed in this 3D material system at any level of stress relaxation (Supplementary Fig. S10). As it was previously found that ligand clustering was associated with osteogenic differentiation, the clustering of RGD ligands was next assessed using a Förster Resonance Energy Transfer (FRET) based technique4,43. FRET between fluorescein and carboxytetramethylrhodamine (TAMRA) labeled RGD coupled to the alginate was analyzed in slow and fast relaxing hydrogels with encapsulated cells using confocal microscopy (Fig 4c, d). A higher degree of energy transfer was measured in regions of the hydrogels adjacent to MSCs in gels with fast relaxation relative to MSCs in gels with slower relaxation after eighteen hours of culture (Fig 4e). The FRET signal was typically enhanced in the entire area surrounding the cells in fast relaxing gels, though in some cases a more asymmetric enhancement was observed (Supplementary Fig. S11). As the FRET signal is a highly sensitive function of distance between donor and acceptor fluorophores, this demonstrates nanoscale clustering of RGD ligands and mechanical remodeling of the hydrogel by cells locally in hydrogels with faster relaxation and an initial elastic modulus of 17 kPa. Though a similar trend of increased RGD ligand clustering with faster relaxation was observed for cells in gels with an initial elastic modulus of 9 kPa, the degree of transfer in faster relaxing gels was significantly lower when compared to the stiffer fast relaxing gels (Supplementary Fig. S11).
Figure 4. Osteogenic differentiation of MSCs mediated through ECM ligand density, enhanced RGD ligand clustering, and myosin contractility in stiffer hydrogels.
a, Quantification of ALP activity of MSCs encapsulated in hydrogels with an initial elastic modulus of 17 kPa after 7 days in culture with an RGD density of 150 or 1500 μM. b, Representative immunofluorescence staining for actin (green), nucleus (blue), and β1 integrin (red) in MSCs cultured in the indicated conditions for a week. c, schematic of assay using FRET between RGD-fluorescein and RGD-rhodamine coupled to different alginate chains to monitor mechanical clustering of RGD ligands at the nanoscale by cells locally. d, Representative confocal microscope images of nucleus (DAPI/ blue) and FRET acceptor signal from hydrogel (red) surrounding MSCs cultured in hydrogels with different stress relaxation properties after 18 hours of culture. Blank spot in FRET signal images indicates location of cell. e, Quantification of enhancement of FRET acceptor signal within ~2 – 3 um of cell border relative to the background of the hydrogel. Data are shown as mean +/− s.d. and **** indicates p < 0.0001 (student’s t-test). f, ALP activity of MSCs in the presence of ML-7, a myosin light chain kinase inhibitor. g, ALP activity of MSCs in the presence of a Rho kinase inhibitor, Y-27632. h, ALP activity of MSCs in the presence of a Rac1 inhibitor, NSC 23766. All experiments were done in hydrogels with an initial elastic modulus of 17 kPa and RGD concentration of 1500 μM. All data are shown as mean +/− s.d. *,** indicate p < 0.05, 0.01 respectively (student’s t-test). Scale bars are all 10 μm.
As binding and clustering of integrins activates signaling pathways44,45, we examined the role of signaling pathways in mediating osteogenesis in gels with different levels of stress relaxation. Previous work has found that cells sense the stiffness of ECM substrates through actomyosin contractility46, often mediated through activation of the Rho signaling pathway, and sense the loss modulus of 2D acrylamide substrates through activation of the Rac signaling pathway30. Pharmacological inhibition of myosin, Rho, and Rac1 was performed on MSCs encapsulated within hydrogels with an initial elastic modulus of 17 kPa. Inhibition of myosin light chain kinase with ML-7 diminished osteogenesis, demonstrating that enhanced osteogenesis in substrate with fast stress relaxation involves myosin contractility (Fig. 4f). Interestingly, inhibition of Rho with Y-27632 resulted in enhanced osteogenesis, in three of four rates of stress relaxation (Fig. 4g). Inhibition of Rac1 with NSC 23766 did not have a significant effect on osteogenesis (Fig 4h).
Next, nuclear localization of the YAP transcriptional regulator was examined. The YAP transcriptional regulator is thought to be the key regulatory element controlling the gene expression of cells in response to mechanical or geometric cues47. Nuclear localization of YAP was previously found to direct MSC differentiation into adipogenic or osteogenic lineages47,48 for MSCs cultured on 2D acrylamide substrates in response to altered substrate stiffness. We find nuclear translocation of YAP increases with faster stress relaxation for both values of initial elastic moduli tested (Spearman’s rank correlation, p < 0.0001 for both), indicating that matrix stress relaxation has an impact on transcriptional factor activity. Interestingly, the levels of nuclear YAP spanned the same range for the two different moduli (Fig 5a–b, Supplementary Fig. S12). As adipogenic differentiation is primarily observed in substrates with an elastic modulus of ~9 kPa, and osteogenic differentiation is primarily observed in substrates with an elastic modulus of ~17 kPa, this demonstrates a decoupling of nuclear translocation of YAP from MSC fate (Fig 5c–d). These findings indicate that localization of YAP does not by itself control the differentiation of MSCs in 3D cell culture.
Figure 5. Nuclear localization of YAP is enhanced by faster stress relaxation, but decoupled from MSC fate.
a, Representative immunofluorescence staining for actin (green), nucleus (blue), and YAP (red) in MSCs cultured in the indicated conditions for a week. Scale bar is 10 μm. b, Quantification of the ratio of the concentration of nuclear YAP, to the concentration of YAP in the cytoskeleton. Nuclear YAP increases significantly with faster stress relaxation for both initial elastic moduli (Spearman’s rank correlation, p < 0.0001 for both). c, Quantification of percentage of D1 cells that stain positive for ORO as a function of the relative nuclear YAP. d, Quantification of ALP in differentiated D1 cells as a function of the relative nuclear YAP. No trend is observed in c and d. All data are shown as mean +/− s.d.
Outlook
This work demonstrates an approach to modulating stress relaxation properties in alginate hydrogels, and indicates that substrate stress relaxation has a profound effect on cell biology. Several recent studies have used alternative material approaches to examine the role of altered substrate viscoelasticity on cell biology. These include modulating both the covalent crosslinking density and polymer concentration in acrylamide hydrogels to tune the loss modulus for 2D culture30,31, utilizing different stoichiometry of dynamic covalent crosslinkers with different affinities to modulate stress relaxation in PEG hydrogels32, or use of covalent versus physical crosslinking of alginate hydrogels33. These approaches revealed that an enhanced loss modulus and substrate creep led to increased spreading and osteogenic differentiation of MSCs in 2D culture30,31, and that increased substrate stress relaxation promoted cell spreading and proliferation in 2D culture33 and altered cell morphology in 3D culture32. The approach described in this paper is unique in that only one type of crosslinking is utilized, while maintaining compatibility with 3D culturing of cells and holding alginate concentration constant. The nanoscale architecture of these gels does not capture the fibrillarity of some natural ECM in vivo49. However, this approach does allow modulation of stress relaxation properties over a range similar to that observed in various tissues16 and enables presentation of a homogenous microenvironment to cells, providing a well-controlled system for probing cell-ECM interactions. There is some coupling between the initial elastic modulus and the range of stress relaxation timescales for alginate with a given molecular weight and PEG coupling state, as stiffening the hydrogels from 9 kPa to 17 kPa shifted the range of stress relaxation timescales from 3300s – 70s to 1300s – 40s. Generally, this approach to decouple the initial elastic modulus from relaxation rate, and the associated development of rapidly relaxing gels, may be useful in a variety of material applications.
Mechanistically, we find that the enhancement in cell spreading, proliferation, and osteogenic differentiation of MSCs by faster matrix stress relaxation is mediated through integrin based adhesions, local clustering of RGD ligands, actomyosin contractility, and nuclear localization of YAP (Supplementary Fig. S13). MSC differentiation depended strongly upon the initial elastic modulus in viscoelastic matrices in 3D, with osteogenesis only occurring when the initial elastic modulus was 17 kPa. In contrast, MSC differentiation loses sensitivity to matrix stiffness in covalently crosslinked hydrogels13, which are presumably elastic, highlighting the importance of stress relaxation in cell responding to mechanical cues of ECM. Inhibition of actomyosin contractility completely abrogated osteogenesis, indicating the role of contractile forces in sensing stiffness and stress relaxation, and driving osteogenesis. As forces exerted by cells on the weakly crosslinked alginate hydrogels are relaxed through unbinding of ionic crosslinks and matrix flow, actomyosin contractility coupled to the matrix through binding to RGD ligands mechanically clusters RGD ligands over time in gels with faster relaxation. RGD ligand and integrin clustering in turn is known to activate signaling pathways44,45 and has been associated previously with osteogenic differentiation4, providing a mechanistic link between altered stress relaxation and modulation of biological signaling and long term cell fate. While Rho-mediated contractility has been found to be critical for osteogenesis in 3D culture in degradable hydrogels13, inhibition of Rho did not diminish osteogenesis in the non-degradable alginate hydrogels, and in some cases enhanced it. Further, previous work has found that an increased loss modulus in acrylamide substrates increases Rac activation31, whereas inhibition of Rac did not diminish osteogenesis in alginate hydrogels. These suggest that the role of Rho and Rac in osteogenesis is context dependent.
While many previous studies have highlighted the importance of matrix remodeling through proteolytic degradation on cell function, the altered behavior of cells in rapidly relaxing gels suggests that the ability of cells to mechanically remodel their matrix is also an essential component of cell-ECM interactions (Fig. 6). In particular, it is likely that the greater malleability of fast relaxing hydrogels that enabled increased RGD ligand clustering on short timescales also facilitated the physical aspects of the processes of cell spreading, proliferation, and formation of an interconnected bone-like matrix by osteogenically differentiated MSCs over longer timescales, in the absence of matrix degradation. Similar to the observed trend of diminishing osteogenic differentiation with decreased stress relaxation, a recent study showed that osteogenic differentiation of MSCs was inhibited in non-degradable covalently crosslinked gels, which presumably cannot be remodeled mechanically, though differences in material systems preclude a direct comparison13. Previous work has highlighted the effect of matrix degradation in bone regeneration15 or cartilage formation10 in implantable scaffolds, as well as on cell shape and MSC differentiation in 3D culture in vitro13. Our work suggests the possibility that the effect of degradation may be in part due to enhanced matrix stress relaxation in regions of the matrix that exhibit substantial degradation. Indeed, it was recently found that proteolytic degradation of covalently crosslinked PEG hydrogels by encapsulated MSCs locally converts elastic matrices into viscoelastic fluids50.
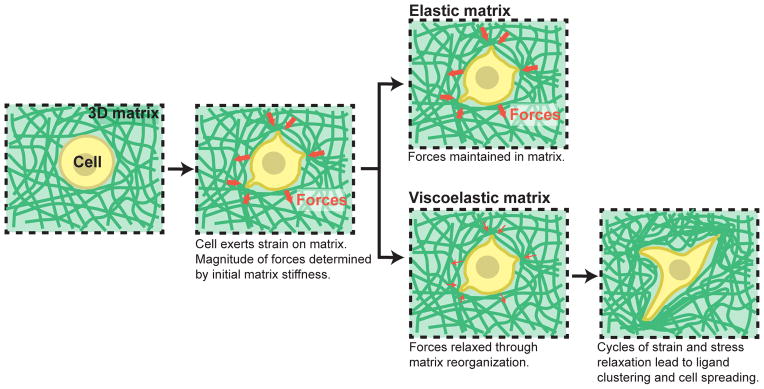
Figure 6. Hypothesis for how initial elastic modulus and stress relaxation properties of matrix regulate cellular behaviors.
A cell in a 3D matrix initially exerts strains on the matrix, resulting in forces/stresses resisting this strain, as determined by the initial elastic modulus of the matrix. In an elastic matrix, these forces are never relaxed so that there is no remodeling of the matrix microenvironment. In a viscoelastic matrix, forces in the matrix can be relaxed over time due to mechanical yielding and remodeling of the matrix. The rate of stress relaxation determines the degree of this mechanical remodeling of the matrix. In fast relaxing matrices, this facilitates adhesion ligand clustering, cell shape change, proliferation, and bone matrix formation by MSCs undergoing osteogenic differentiation.
Broadly, these results highlight the importance of considering matrix stress relaxation as a fundamental signal in understanding the basics of cell-ECM interactions and the underlying biophysics of mechanotransduction, since most physiological extracellular matrices exhibit some degree of stress relaxation. These findings point towards the use of stress relaxation as a design parameter for materials in tissue engineering8, particularly in the context of regulating cell proliferation and promoting bone regeneration.
METHODS
Alginate preparation
Sodium alginate rich in guluronic acid blocks and with a high molecular weight (280 kDa, LF20/40) was purchased from FMC Biopolymer, and was prepared as has been described previously3. Briefly, high molecular weight alginate was irradiated by a 3 or 8 Mrad Cobalt source to produce lower molecular weight alginates. RGD-alginate was prepared by coupling the oligopeptide GGGGRGDSP (Peptides International) to the alginate using carbodiimide chemistry at concentrations such that 2 or 20 RGD peptides were coupled to 1 alginate chain on average for high molecular weight alginate (peptide molar concentrations in low molecular weight alginates were kept the same according to high molecular weight alginate for each degree of substitution, respectively). For FRET experiments, either GGGGRGDASSK(carboxyfluorescein)Y or GGGGRGDASSK(Carboxytetramethylrhodamine)Y were used instead of standard RGD peptide sequence, and were coupled at a concentration of 2 peptides per alginate chain on average for high molecular weight alginate (peptide molar concentrations in low molecular weight alginates were kept the same according to high molecular weight alginate). The coupling efficiency using this procedure was previously characterized using 125I labeled RGD peptides3. These correspond to densities of 150 μM and 1500 μM RGD in a 2% wt/vol alginate gel. Alginate was dialyzed against deionized water for 2–3 days (molecular weight cutoff of 3.5 kDa), treated with activated charcoal, sterile filtered, lyophilized, and then reconstituted in serum free DMEM (Life Technologies).
Polyethylene glycol (PEG)-alginate was prepared by coupling PEG-amine (5 kDa, Laysan Bio) to the low molecular weight alginate (35 kDa) using carbodiimide chemistry with a similar procedure to the RGD coupling3. In brief, 295 mg of PEG-amine was mixed with 50 mL of 10 mg/mL alginate in 0.1 M MES (2-(N-morpholino)ethanesulfonic acid, Sigma-Aldrich) buffer at pH 6.5. Then 242 mg of EDC (N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride, Sigma-Aldrich) and 137 mg of Sulfo-NHS (N-hydroxysulfosuccinimide, Thermo Fisher Scientific) were added into the solution. The reaction was carried out for 20 hours under constant stirring. The product was dialyzed against deionized water for 3 days (molecular weight cutoff of 10 kDa), filtered with activated charcoal, sterile filtered, and lyophilized. The structure of the PEG-alginate was confirmed with nuclear magnetic resonance (NMR) and gel permeation chromatography (GPC). Based on the change of molecular weight of alginate before and after PEG coupling (from 35 kDa to 45 kDa), an average of 2 PEG molecules were coupled to 1 alginate chain. This number was confirmed by 1H NMR spectroscopy (Supplementary Fig. S14).
Alginate characterization
Molecular weights of alginates and PEG-alginate were analyzed with a Malvern Viscotek 270max GPC equipped with a GPCmax solvent and sample delivery module, an Eldex Ch-150 temperature-controlled column holder, a VE 3580 refractive index (RI) detector, viscotek 270 Dual Detector featuring intrinsic viscosity (IV-DP) and right angle light scattering (RALS), and OmniSec software. Samples were dissolved in 0.1 M NaNO3 buffer solution at a concentration of 5 mg/mL, and 200 μL of sample was injected. Polymers separated through a set of two TSK-gel columns (G4000PWXL and G3000PWXL) were analyzed with the triple detector system. Malvern PEO and pullulan standards were used in molecular weight calculation, and weight average molecular weights (Mw) were used.
High-resolution 1H NMR spectra were obtained in deuterium oxide (D2O) using a Varian Unity-400 (400 MHz) NMR spectrometer (Varian). 1H NMR was used to characterize PEG coupling of alginate and degree of functionalization of PEG-alginate.
Mechanical characterization
Rheology measurements were made with an AR-G2 stress controlled rheometer (TA Instruments). Alginate gels were deposited directly onto the surface plate of the rheometer immediately after mixing with the crosslinker. A 20 mm plate was immediately brought down, forming a 20 mm disk of gel with an average thickness of ~1.8 mm. The mechanical properties were then measured over time until the storage modulus reached an equilibrium value. The storage modulus at 0.5% strain and at 1 Hz was recorded periodically for 45 minutes. Then, a strain sweep was performed to confirm this value was within the linear elastic regime, followed by a frequency sweep. No prestress was applied to the gels for these measurements.
The initial elastic moduli and stress relaxation properties of alginate gels were measured from compression tests of the gel disks (15 mm in diameter, 2 mm thick, equilibrated in DMEM for 24 hr) using a previously published method4,35. The gel disks were compressed to 15% strain with a deformation rate of 1 mm/min using an Instron 3342 single column apparatus. Within 15% compression, the stress vs. strain relations of the gels are almost linear, and the slope of the stress-strain curves (first 5–10% of strain) gives the initial elastic modulus. Subsequently, the strain was held constant, while the load was recorded as a function of time. Compression and stress relaxation measurements of polyacrylamide hydrogels and biological tissues were performed using the same procedure. Polyacrylamide hydrogels were formed following previously established protocols28. In brief, 0.2 g of acrylamide and 0.02 g of bis-acrylamide were dissolved in 2 mL of water. Then 60 μL of 137 mg/mL ammonium persulfate and 60 μL of 70 mg/mL tetramethylethylenediamine (TEMED) were added into above mixture. The solution was mixed and allowed to gel for 6 hours. The hydrogel was then equilibrated in PBS for 24 hours before mechanical testing. We note that some stress relaxation of the covalently crosslinked hydrogels is observed at longer timescales, but this was previously found to arise from water leaving the hydrogel under bulk compression35. Sprague Dawley Rats (male, 7 weeks of age, Charles River Lab) were euthanized in compliance with National Institutes of Health and institutional guidelines. Brain, liver, and adipose were collected immediately after euthanization and tested with Instron 3342 single column apparatus. Bone marrow from multiple rat femurs and tibias were collected fresh after euthanization and allowed to coagulate for 1 hr before compression testing. A fracture hematoma from human patient was retrieved from the bone fracture site at the moment of bone stabilization surgery. The surgery took place 7 days after occurrence of the fracture, when the surrounding soft tissue trauma around the fracture gap was sufficiently stabilized. The complete hematoma was collected and processed for mechanical testing within 1h after surgery. The same procedure of compression and relaxation measurements was performed as with the rat samples but on a Bose TestBench LM1 system using a 250g load cell. Care was taken to not test samples that contained bone chips. Fracture hematoma collection was approved by the Institutional Review Board of the Charité University Hospital Berlin, where the collection and testing were performed, and the participant gave written informed consent. Stress relaxation tests that were noisy were smoothed with a Savitzky-Golay filter in Igor Pro (Wavemetrics) with a 4s window.
Characterization of gel degradation
For characterization of gel degradation, hydrogels were formed and incubated in culture media at 37 °C for 1 or 7 days. Then, the hydrogels were removed from the incubation medium, frozen, and then lyophilized. The dry mass of the hydrogels was then measured following lyophilization.
Characterization of gel homogeneity
Gels were formed with fluorescein-RGD coupled alginates for each rate of stress relaxation with an initial elastic modulus of 9 kPa. Gels were incubated in cell culture media in the incubator for one day. Multiple fluorescence images were taken of at least 3 gels for each of the conditions with a 20X objective using a laser scanning confocal microscope (Zeiss, LSM 710). ImageJ was used to collect histograms of pixel intensity from each image.
Cell culture
3T3 fibroblasts (ATCC) were cultured in standard Dulbecco’s Modified Eagles Medium (DMEM, Invitrogen) with 10% Fetal Bovine Serum (Invitrogen) and 1% penicillin/streptomycin (Invitrogen). D1 cells, clonally derived mouse bone marrow stromal mesenchymal stem cells, were originally obtained from Balb/c mice51. The D1s were maintained at sub-confluency in DMEM containing 10% Fetal Bovine Serum and 1% Pen/Strep. For differentiation experiments, the culture medium was supplemented with 50 μg/mL L-ascorbic acid (Sigma), 10 mM β-glycerophosphate (Sigma), and 0.1 μM dexamethasone (Sigma)51. The medium was changed every 3–4 days.
Encapsulation of cells within hydrogels
Cells in flasks were trypsinized using 0.05% trypsin/EDTA (Invitrogen), washed once in serum free DMEM, and resuspended in serum free media at 10X the final concentration. The concentration of the cells was determined using a Coulter counter (Beckman Coulter). Cells were then mixed well with alginate, also reconstituted in serum free DMEM using luerlock syringes (Cole-Parmer) and a female-female luerlock coupler (Value-plastics). The cell-alginate solution was then rapidly mixed with DMEM containing the appropriate concentration of calcium sulfate, and then deposited between two glass plates spaced 1 mm apart. The solutions were allowed to gel for 45 minutes, and then disks of hydrogel were punched out and transferred to well plates where they were immersed in media.
Immunohistochemistry
For immunohistochemical staining, media was first removed from the gels. The gels were then fixed with 4% paraformaldehyde in serum free DMEM at 37°C for 30 – 45 minutes. Gels were then washed 3 times in PBS containing calcium (cPBS), and incubated overnight in 30% sucrose in cPBS at 4°C. The gels were then placed in a mix of 50% of a 30% sucrose in cPBS solution, and 50% OCT (Tissue-Tek) for several hours. Then the media was removed, the gels were embedded in OCT and frozen. The frozen gels were sectioned with a cryostat (Leica CM1950) to a thickness of 30 – 100 μm, and stained following standard immunohistochemistry protocols. The following antibodies/reagents were used for immunohistochemistry: Rabbit-anti-mouse Collagen I polyclonal antibody (Abcam, cat. # 34710), YAP antibody (Cell signaling, cat. # 4912), Paxillin antibody (Abcam, cat. # 32084), β1 integrin antibody (BD Biosciences, cat. # 550531), Prolong Gold antifade reagent with DAPI (Invitrogen), AF-488 Phalloidin to stain actin (Invitrogen), Goat anti-Rabbit IgG AF 647 (Invitrogen). The Click-IT EdU cell proliferation assay (Invitrogen) was used to identify proliferating cells.
Image analysis
For measurements of YAP nuclear localization in 3D, images of DAPI/phalloidin/YAP antibody stained cells were taken with an NA=1.40 63X PlanApo oil immersion objective with a laser scanning confocal microscope (Zeiss, LSM710). Images were thresholded on each color channel to determine the nuclear area and cell/cytoskeleton area outside of the nucleus. The YAP nuclear localization ratio was then determined as the summed intensity of the YAP signal within the nucleus normalized by the nuclear area divided by the summed intensity of the YAP signal outside of the nucleus normalized by the non-nuclear cytoskeleton area. For quantification of the percentage of MSCs with nuclear YAP shown in Supplementary Fig. S12, the number of cells exhibiting nuclear YAP in cryosection stains was manually counted and then this number was divided by the total number of cells and multiplied by 100 to get a percentage.
Spreading of 3T3 cells was quantified using Imaris software (Bitplane). Z-stack images of DAPI/phalloidin stained cells were taken with a laser scanning confocal microscope. The stacks were analyzed using Imaris with embedded cell body algorithm. The DAPI channel was used for nuclei detection and the phalloidin channel was used for cell body detection. Statistics of the cells were generated by the algorithm, and the longest dimension of the object-oriented bounding box of each cell was determined as an indication of cell spreading.
Analysis of MSC differentiation
Oil Red O staining was performed on 100 μm frozen sections to probe for neutral lipids. Slides were equilibrated in 60% isopropanol and stained in 1.8 mg/ml Oil Red O in 60% isopropanol for 30 minutes. Frozen sections were probed for alkaline phosphatase by Fast Blue staining. The slides were equilibrated in alkaline buffer (100 mM Tris-HCl, 100 mM NaCl, 0.1% Tween-20, 50 mM MgCl2, pH 8.2) for 15 minutes and stained in 500 μg/ml naphthol AS-MX phosphate (Sigma) and 500 μg/ml Fast Blue BB Salt Hemi (ZnCl) salt (Sigma) in alkaline buffer for 60 minutes. The sections were then washed in alkaline buffer and neutralized in PBS. Von Kossa staining was performed on 30 μm frozen section to probe for mineralized matrix. Samples were equilibrated in distilled water and exposed to 3% silver nitrate solution under UV light for 1 min. After several dips in distilled water, a 2.5% sodium thiosulphate solution in 50 mM HEPES with 25 mM CaCl2 was added for 2 minutes, followed by washes in distilled water.
To quantify ALP enzyme activity, cells were retrieved from the gels after 7 days of culture. Cells were collected by incubation in trypsin for 10 minutes followed by a PBS wash, and soaking gels in 50mM EDTA in PBS for 10 minutes at room temperature. The cells were washed, counted using a Z2 Coulter Counter and lysed for 30 min in lysis buffer (50 mM Tris-HCl, 0.1% Triton X-100) at 4°C. 10 μL of each lysate was added to 100 μL 4-Methylbelliferyl phosphate (4-MUP) substrate (Sigma) and incubated for 25 minutes at 37°C. Bovine ALP (Sigma) was used to create a standard curve. After incubation, fluorescence was read by a fluorescent plate reader (BioTek) and measured ALP activity was normalized to cell counts.
Color micrographs of Oil Red O, Fast Blue, and von Kossa staining were acquired using a Nikon E800 upright microscope and an Olympus DP-70 color camera.
Structural and compositional analyses of the alginate gels were performed with a Tescan Vega environmental scanning electron microscope (SEM) equipped with a Bruker XFlash 5030 energy dispersive X-ray spectrometer (EDS). Frozen sections of alginate gels with a thickness of 100 μm were attached to a silicon wafer with conductive carbon tape. The gel sections were washed in DI water for 4 times, 5 minutes each time to remove any soluble Ca2+ and phosphate, and dried under vacuum overnight before SEM-EDS. Elemental mapping and compositional analysis of phosphorus for each sample was performed under identical conditions at an accelerator voltage of 20 keV and 12 Pa of pressure.
FRET measurements of RGD clustering
FRET-based analysis of ligand clustering was carried out through imaging energy transfer between fluorescein-labeled RGD donors and tetramethylrhodamine-labeled RGD acceptors coupled to different alginate chains using confocal microscopy, following a previously established technique4,43. High-MW and low-MW-PEG alginate were coupled to RGD-fluorescein (RGD-FITC) and RGD-tetramethylrhodamine (RGD-TAMRA) separately as described above. MSCs were encapsulated in hydrogels that had a final concentration of 40 μM RGD-FITC and 40 uM RGD-TAMRA, an initial elastic modulus of 17 kPa and 9kPa, and different levels of stress relaxation as described above. The gels were then incubated in phenol red free complete culture media for 18 hours following encapsulation, and then treated with DAPI to stain the nuclei of cells for 10 minutes. Images of the FRET signal were taken with a laser scanning confocal microscope (Zeiss, LSM710) using a 20X objective (NA = 0.8), by exciting the fluorescein donor (488 nm) and collecting emissions from the TAMRA acceptor (580 – 720 nm). A confocal 3D stack of images of DAPI and FRET signal were taken for each cell. The z-slice corresponding to the center of the cell body, or where the cell body took up the maximum area, was selected for image analysis. FRET enhancement for each cell was calculated as the average FRET signal within ~2 – 3 μm of the cell border divided by the background FRET signal in the hydrogel, which was determined as the average FRET signal ~10 – 20 μm away from each cell. At least 30 cells were analyzed for each condition. Images were analyzed using ImageJ.
Inhibition studies
For pharmacological inhibition studies, the inhibitors were added to the culture media following encapsulation. The concentrations used for each inhibitor were: 25 μM for ML-7 (Tocris Bioscience); 10 μM for Y-27632 (Tocris Bioscience); and 25 μM for NSC 23766 (Tocris Bioscience). These concentrations matched those used in similar studies13,31. Analysis of alkaline phosphatase activity was performed as described above after 7 days of culture.
Supplementary Material
Acknowledgments
The authors acknowledge the help of Sandeep Koshy, Manav Mehta, Catia Verbeke, Xuanhe Zhao (now at MIT), and other members of the Mooney lab. The authors also thank the Weitz lab for use of rheometer, Oktay Uzun for help with GPC, and Simon Reinke and Dag Wulsten for support in bone fracture hematoma testing. This work was supported by an NIH Grant to DJM (R01 DE013033), NIH F32 grant to OC (CA153802), an Einstein Visiting Fellowship for DJM, funding of the Einstein Foundation Berlin through the Charité – Universitätsmedizin Berlin, Berlin-Brandenburg School for Regenerative Therapies GSC 203, ZonMW-VICI grant 918.11.635 (The Netherlands) for DK, and Harvard MRSEC for DJM (DMR-1420570). This work was performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network (NNIN).
Footnotes
AUTHOR CONTRIBUTIONS
OC, LG, DK, MD, NH, and DM designed the experiments. OC and LG conducted most of the experiments. DK helped with experiments involving the MSCs, SB helped with alginate characterization, JW helped with EDS experiments and analysis, and HPL assisted with mechanical characterization. EL and GD carried out fracture hematoma measurement. OC and LG analyzed the data. OC, LG, DK, and DM wrote the manuscript.
References
- 1.Burdick JA, Anseth KS. Photoencapsulation of osteoblasts in injectable RGD-modified PEG hydrogels for bone tissue engineering. Biomaterials. 2002;23:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 2.Raeber GP, Lutolf MP, Hubbell JA. Molecularly engineered PEG hydrogels: a novel model system for proteolytically mediated cell migration. Biophys J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 4.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park YD, Tirelli N, Hubbell JA. Photopolymerized hyaluronic acid-based hydrogels and interpenetrating networks. Biomaterials. 2003;24:893–900. doi: 10.1016/s0142-9612(02)00420-9. [DOI] [PubMed] [Google Scholar]
- 6.Burdick JA, Chung C, Jia X, Randolph MA, Langer R. Controlled degradation and mechanical behavior of photopolymerized hyaluronic acid networks. Biomacromolecules. 2005;6:386–391. doi: 10.1021/bm049508a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 8.Langer R, Tirrell DA. Designing materials for biology and medicine. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 9.Healy KE, Rezania A, Stile RA. Designing biomaterials to direct biological responses. Ann N Y Acad Sci. 1999;875:24–35. doi: 10.1111/j.1749-6632.1999.tb08491.x. [DOI] [PubMed] [Google Scholar]
- 10.Metters AT, Anseth KS, Bowman CN. Fundamental studies of biodegradable hydrogels as cartilage replacement materials. Biomed Sci Instrum. 1999;35:33–38. [PubMed] [Google Scholar]
- 11.Nguyen KT, West JL. Photopolymerizable hydrogels for tissue engineering applications. Biomaterials. 2002;23:4307–4314. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 12.Peyton SR, Raub CB, Keschrumrus VP, Putnam AJ. The use of poly(ethylene glycol) hydrogels to investigate the impact of ECM chemistry and mechanics on smooth muscle cells. Biomaterials. 2006;27:4881–4893. doi: 10.1016/j.biomaterials.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Khetan S, et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater. 2013 doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen JH, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alsberg E, et al. Regulating bone formation via controlled scaffold degradation. J Dent Res. 2003;82:903–908. doi: 10.1177/154405910308201111. [DOI] [PubMed] [Google Scholar]
- 16.Levental I, Georges PC, Janmey PA. Soft biological materials and their impact on cell function. Soft Matter. 2007;3:299–306. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Bilston L. On the viscoelastic character of liver tissue: experiments and modelling of the linear behaviour. Biorheology. 2000;37:191–201. [PubMed] [Google Scholar]
- 18.Geerligs M, Peters GWM, Ackermans PAJ, Oomens CWJ, Baaijens FPT. Linear viscoelastic behavior of subcutaneous adipose tissue. Biorheology. 2008;45:677–688. [PubMed] [Google Scholar]
- 19.McDonald SJ, et al. Early fracture callus displays smooth muscle-like viscoelastic properties ex vivo: implications for fracture healing. J Orthop Res Off Publ Orthop Res Soc. 2009;27:1508–1513. doi: 10.1002/jor.20923. [DOI] [PubMed] [Google Scholar]
- 20.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 21.Legant WR, et al. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Meth. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legant WR, et al. Microfabricated tissue gauges to measure and manipulate forces from 3D microtissues. Proc Natl Acad Sci U S A. 2009;106:10097–10102. doi: 10.1073/pnas.0900174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelham RJ, Jr, Wang Yl. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA. Neurite branching on deformable substrates. Neuroreport. 2002;13:2411–2415. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engler A, et al. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong HJ, et al. Non-viral gene delivery regulated by stiffness of cell adhesion substrates. Nat Mater. 2005;4:460–464. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 27.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Engler A, Sen S, Sweeney H, Discher D. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhuri O, et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014 doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 30.Cameron AR, Frith JE, Cooper-White JJ. The influence of substrate creep on mesenchymal stem cell behaviour and phenotype. Biomaterials. 2011;32:5979–5993. doi: 10.1016/j.biomaterials.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Cameron AR, Frith JE, Gomez GA, Yap AS, Cooper-White JJ. The effect of time-dependent deformation of viscoelastic hydrogels on myogenic induction and Rac1 activity in mesenchymal stem cells. Biomaterials. 2014;35:1857–1868. doi: 10.1016/j.biomaterials.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 32.McKinnon DD, Domaille DW, Cha JN, Anseth KS. Biophysically Defined and Cytocompatible Covalently Adaptable Networks as Viscoelastic 3D Cell Culture Systems. Adv Mater. 2013 doi: 10.1002/adma.201303680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaudhuri O, et al. Substrate stress relaxation regulates cell spreading. Nat Commun. 2015;6:6364. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee KY, Mooney DJ. Hydrogels for tissue engineering. Chem Rev. 2001;101:1869–1879. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 35.Zhao X, Huebsch N, Mooney DJ, Suo Z. Stress-relaxation behavior in gels with ionic and covalent crosslinks. J Appl Phys. 2010;107:63509. doi: 10.1063/1.3343265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graessley WW. Synthesis and Degradation Rheology and Extrusion. Vol. 47. Springer-Verlag; pp. 67–117. [Google Scholar]
- 37.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 38.Mooney DJ, Langer R, Ingber DE. Cytoskeletal filament assembly and the control of cell spreading and function by extracellular matrix. J Cell Sci. 1995;108( Pt 6):2311–2320. doi: 10.1242/jcs.108.6.2311. [DOI] [PubMed] [Google Scholar]
- 39.Parekh SH, et al. Modulus-driven differentiation of marrow stromal cells in 3D scaffolds that is independent of myosin-based cytoskeletal tension. Biomaterials. 2011;32:2256–2264. doi: 10.1016/j.biomaterials.2010.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pek YS, Wan ACA, Ying JY. The effect of matrix stiffness on mesenchymal stem cell differentiation in a 3D thixotropic gel. Biomaterials. 2010;31:385–391. doi: 10.1016/j.biomaterials.2009.09.057. [DOI] [PubMed] [Google Scholar]
- 41.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc Natl Acad Sci U S A. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnold M, et al. Activation of Integrin Function by Nanopatterned Adhesive Interfaces. ChemPhysChem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 45.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113( Pt 10):1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 46.Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 48.Swift J, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultz KM, Kyburz KA, Anseth KS. Measuring dynamic cell-material interactions and remodeling during 3D human mesenchymal stem cell migration in hydrogels. Proc Natl Acad Sci U S A. 2015;112:E3757–3764. doi: 10.1073/pnas.1511304112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diduch DR, Coe MR, Joyner C, Owen ME, Balian G. Two cell lines from bone marrow that differ in terms of collagen synthesis, osteogenic characteristics, and matrix mineralization. J Bone Joint Surg Am. 1993;75:92–105. doi: 10.2106/00004623-199301000-00012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.