Abstract
β-arrestins critically regulate G protein-coupled receptor (GPCR) signaling, not only 'arresting' the G protein signal but also modulating endocytosis and initiating a discrete G protein-independent signal via MAP kinase1–3. Despite enormous recent progress toward understanding biophysical aspects of arrestin function4,5, its cell biology remains relatively poorly understood. Two key tenets underlie the present dogma: (1) β-arrestin accumulates in clathrin-coated structures (CCSs) exclusively in physical complex with its activating GPCR, and (2) MAP kinase activation requires endocytosis of formed GPCR - β-arrestin complexes6–9. We show here, using β1-adrenergic receptors, that β-arrestin-2 (Arrestin 3) accumulates robustly in CCSs after dissociating from its activating GPCR and transduces the MAP kinase signal from CCSs. Moreover, inhibiting subsequent endocytosis of CCSs enhances the clathrin and β-arrestin -dependent MAP kinase signal. These results demonstrate β-arrestin 'activation at a distance', after dissociating from its activating GPCR, and signaling from CCSs. We propose a β-arrestin signaling cycle that is catalytically activated by the GPCR and energetically coupled to the endocytic machinery.
Our motivation to investigate β-arrestin function in living cells stemmed from an intriguing discordance between in vivo and in vitro observations regarding the G protein-independent signaling wave. In vivo, β1-adrenergic receptors (β1ARs) mediate signaling via the ERK1/2 MAP kinase through β-arrestin that is essential for promoting cardiac muscle cell survival under conditions of strong adrenergic drive10. In vitro, however, β1ARs are poorly phosphorylated and weakly engage β-arrestin11–13 and, at the cellular level, β1ARs are internalized poorly, if at all, by clathrin-mediated endocytosis (CME)11,14. This confluence of seemingly contradictory observations presented a puzzle relative to the prevailing current view of β-arrestin function, motivating us to directly testing its underlying tenets.
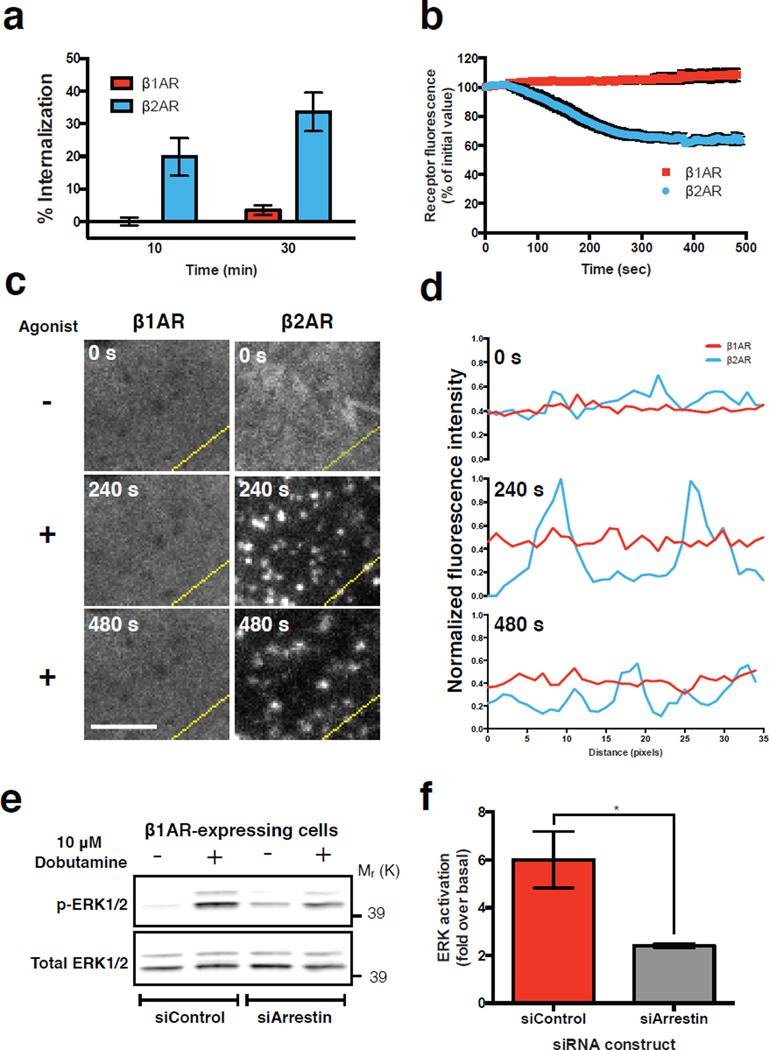
FLAG-tagged β1ARs expressed in human embryonic kidney (HEK 293) cells failed to detectably internalize (Fig. 1a, b, red symbols) or cluster in the plasma membrane (Fig. 1c, d, red lines) after activation by the β-adrenergic agonist isoproterenol. In contrast, β2-adrenergic receptors (β2ARs) internalized and clustered robustly (Fig. 1a-d, blue symbols and lines).
Figure 1. β1ARs do not cluster or internalize but initiate β-arrestin-2 -dependent activation of ERK1/2.
(a) Flow cytometric analysis of FLAG-tagged receptor internalization after exposure of cells to 10 μM isoproterenol for 10 or 30 min (n = 3 independent experiments; 30,000 cells per experiment). (b) Average surface receptor fluorescence after 10 μM isoproterenol treatment in TIR-FM live cell images. (n = 11 cells pooled across 3 independent experiments) (c) Frames of a representative area of cells expressing FLAG-tagged receptors and imaged live with TIR-FM at the indicated time after agonist addition. Scale bar = 5 μm. (d) Fluorescence intensity profiles of FLAG-tagged receptors from lines shown in (c). (e-f) ERK1/2 activation in β1AR-expressing cells. (e) Representative western blot of total and phosphorylated ERK1/2 after 5 minutes of agonist treatment in cells with siRNA treatment as indicated. (f) Quantification of ERK1/2 activation by normalizing phosphorylated ERK1/2 to total ERK1/2 and determining fold over untreated (n = 3 independent experiments, * p = 0.0386 by two-tailed t-test). Error bars correspond to SEM. Raw data of independent repeats are in Supplementary Table 1. Panels c, d, and e show results that are representative from 3 independent experiments. Uncropped western blot scans are shown in Supplementary Fig. 6.
Despite failing to undergo detectable CME, β1ARs clearly activated ERK1/2 (Fig. 1e) and did so to a similar degree as β2ARs when compared in expression-matched cell clones (Supplementary Fig. 1a-c). Further, the β1AR-mediated ERK1/2 response was dependent on both β-arrestin-2 (Fig. 1e, f) and clathrin (Supplementary Fig. 1d). We verified that this effect was indeed mediated by β1ARs by using the β1-selective agonist dobutamine, and verified specificity of both the β-arrestin-2 and clathrin knockdown effects by transgenic rescue using RNAi-resistant constructs (Supplementary Fig. 1d-g).
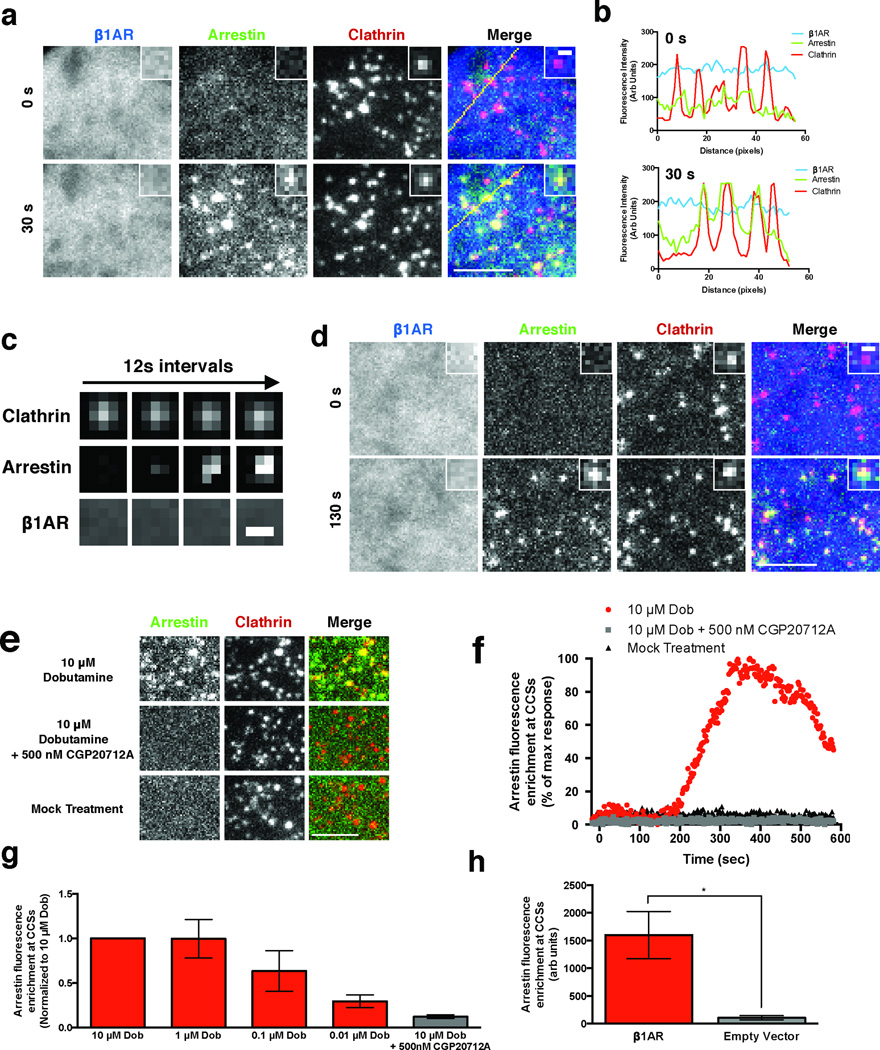
How does a GPCR that is not internalized by CME generate a MAP kinase signal requiring both β-arrestin and clathrin? To address this question, we visualized in living cells the localization of all three proteins using triple-label total internal reflection fluorescence microscopy (TIR-FM). β1ARs remained diffusely distributed in the plasma membrane after application of the β1-selective agonist dobutamine but, unexpectedly, β-arrestin-2 was robustly recruited to spots at the plasma membrane that colocalized with clathrin (Fig. 2a and Supplementary Video 1). Robust recruitment of β-arrestin-2, but not β1ARs, to CCSs was also evident by line scan analysis (Fig. 2b) and by visual inspection of individual examples (Fig. 2c).
Figure 2. β1ARs drive β-arrestin-2 trafficking to CCSs separately from receptors.
(a) Representative images of HEK 293 cells co-expressing FLAG-β1AR (blue), β-arrestin-2-GFP (green), and clathrin light chain-DsRed (red) and imaged live with TIR-FM. Frames captured immediately before (0 s) and 30 seconds after (30 s) bath application of 10 μM dobutamine are shown. Insets show a representative region at higher magnification. Large image scale bar = 5 μm; inset scale bar = 500 nm. (b) Fluorescence intensity profiles from lines shown in (a). (c) Representative time lapse series showing β-arrestin-2 clustering into pre-existing CCSs without β1AR accumulation in CCSs. Images were collected at 0.5 Hz and, for brevity of presentation, one in six frames (12 sec interval) are shown. Scale bar = 500 nm (d) Representative images of COS cells co-expressing FLAG-β1AR (blue), β-arrestin-2-GFP (green), and clathrin light chain-DsRed (red) and imaged live with TIR-FM. Frames captured immediately before (0 s) and 130 seconds after (130 s) bath application of 10 μM dobutamine are shown. Insets show a representative region at higher magnification. Large image scale bar = 5 μm; inset scale bar = 500 nm. (e) Representative images of cells co-expressing FLAG-β1AR, β-arrestin-2-GFP (green), and clathrin light chain-DsRed (red) and imaged live with TIR-FM. Frames shown are after the indicated treatments: 10 μM dobutamine (top), pretreatment with CGP20712A and then 10 μM dobutamine (middle), and mock treatment (bottom). Scale bar = 5 μm. (f) Arrestin enrichment at CCSs for the indicated cells shown in (e). (g) Average maximum arrestin enrichment at CCSs after the indicated treatment normalized to 10 μM dobutamine response (n = 4 cells pooled across 2 independent experiments). (h) Average maximum arrestin enrichment at CCSs in cells expressing either FLAG-β1AR or empty vector after 10 μM dobutamine treatment (n = 11 cells for β1AR pooled across 3 independent experiments or 5 cells for empty vector pooled across 2 independent experiments, * p = 0.036 by two-tailed t-test). Error bars represent SEM. Raw data from independent repeats are in Supplementary Table 1.
β1AR activation drove rapid recruitment of β-arrestin-2 to essentially all CCSs in the plasma membrane, including diffraction-limited spots that include bona fide clathrin-coated pits (CCPs) but also larger structures which correspond to flat clathrin lattices or plaques15. We observed the same behavior in COS cells, another kidney-derived cell line in which diffraction-limited clathrin spots correspond largely to CCPs16,17 (Fig. 2d & Supplementary Video 2). A remarkable property of β-arrestin-2 is that it associates with pre-existing CCSs in response to GPCR activation, in contrast to constitutive endocytic adaptors that must co-assemble at an early stage of CCS formation18,19. β1AR activation clearly drove β-arrestin-2 association with pre-existing CCSs (Supplementary Fig. 2a, b), but we also observed β-arrestin-2 association with clathrin spots as they form, likely representing association at an early stage of CCS assembly (Supplementary Fig. 2c, d). While β-arrestin-2 extensively overlapped with plasma membrane CCSs, there was no detectable recruitment to clathrin associated with Golgi elements or endosomes that dynamically appear in the TIR-FM imaging field (Supplementary Fig. 2e & Supplementary Video 3). Diffraction-limited CCSs were typically seen to abruptly disappear from the evanescent illumination field (Supplementary Fig. 2f, g), consistent with endocytic scission of CCPs and coincident dissociation of β-arrestin-2 as shown previously18,19.
We further characterized β-arrestin-2 associated CCSs using two methods of super-resolution imaging. First, we applied photoactivated localization microscopy (PALM) to image β-arrestin-2 clusters formed after β1AR activation in both HEK 293 (Supplementary Fig. 3a) and COS cells (Supplementary Fig. 3b). β-arrestin-2 localized to structures smaller than 100 nm, consistent with bona fide CCPs as originally defined morphologically, as well as larger structures (Supplementary Fig. 3c & d). A similar size distribution was found by PALM localization of clathrin light chain. Both distributions had a peak at 60 – 90 nanometers but also included smaller and larger structures in lower relative number (Supplementary Fig. 3e). Second, we applied two-color structured illumination microscopy (SIM) to examine β-arrestin-2 localization relative to clathrin in the same living cells. β-arrestin-2 colocalized extensively with clathrin in structures of variable size, including those less than or equal to ~100 nm as well as larger structures, with similar results observed in HEK 293 (Supplementary Fig. 3f, g) and COS cells (Supplementary Fig. 3h, i). Together these observations indicate that β1AR activation drives β-arrestin-2 recruitment to a heterogeneous population of plasma membrane CCSs.
We verified that β-arrestin-2 recruitment to CCSs is indeed mediated specifically by ligand-induced activation of β1ARs both pharmacologically and genetically. Dobutamine-elicited recruitment of β-arrestin-2 to CCSs was concentration-dependent within a range consistent with known β1AR pharmacology20 and blocked by CGP20712A, a highly selective β1AR antagonist (Fig. 2 e-g). Providing genetic proof of specificity, dobutamine failed to elicit detectable recruitment of β-arrestin-2 in cells not expressing β1ARs (Fig. 2h).
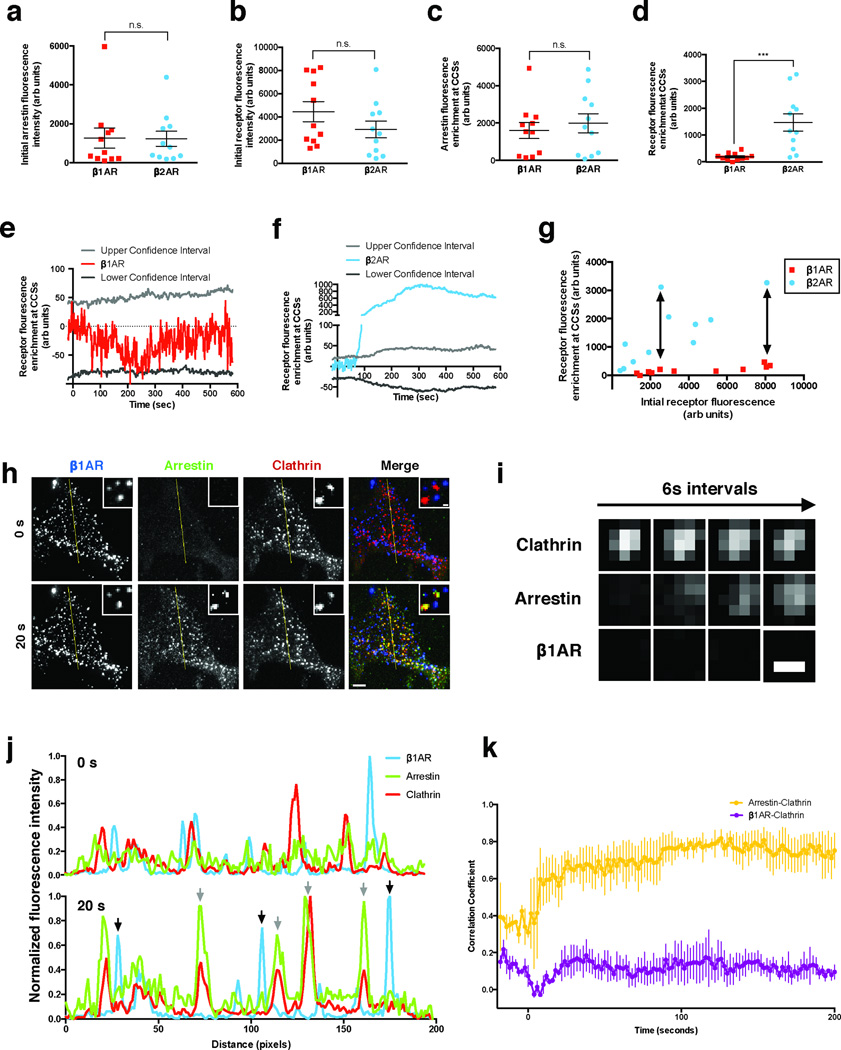
The ability of β1ARs to drive robust localization of β-arrestin-2 to CCSs without detectably concentrating there suggested that the β1AR does not accumulate at CCSs stoichiometrically with activated arrestin. This is contrary to current dogma, which holds that GPCRs accumulate in CCSs together with β-arrestin-2 in a stoichiometric complex. To explicitly test this, we compared the behavior of the β1AR to that of the efficiently clustered β2AR. We reasoned that, if β1ARs indeed accumulate in CCSs sub-stoichiometrically relative to β-arrestin-2, then the intensity of the β1AR fluorescence signal at CCSs after ligand-induced activation should be significantly weaker than the signal of the identically labeled β2ARs, when measured in expression-matched cells and under identical conditions. This was indeed the case. With identical labeling and imaging conditions, and using cell populations carefully matched for β-arrestin-2 and receptor expression (Fig 3a, b), β1ARs and β2ARs were indistinguishable in their ability to drive β-arrestin-2 recruitment to CCSs (Fig 3c) but these GPCRs differed markedly in their ability to accumulate in CCSs (Fig 3d). β1ARs were not significantly enriched at CCSs based on calculation of the 95% confidence interval (Fig 3e). In marked contrast, β2ARs were strongly enriched in a time-dependent manner, exceeding the 95% confidence interval by more than two orders of magnitude within several minutes after receptor activation (Fig 3f). As further verification, and to rule out any possibility that observed differences resulted spuriously from receptor over-expression (such as saturation of a limiting CCS-associated component), we quantified receptor enrichment in CCSs across individual cells expressing receptors over a wide range. The starkly different behavior of β1ARs relative to β2ARs was verified at all levels of relative receptor expression achieved (Supplementary Fig. 3g, see arrows). Thus, while we cannot exclude the possibility that some small number of β1ARs are indeed present in CCSs, it is clear that β1ARs accumulate to a much smaller degree than β2ARs, despite driving comparable β-arrestin-2 recruitment to CCSs. Thus β1ARs could not possibly be present in CCSs in stoichiometric complex with β-arrestin-2.
Figure 3. β1ARs drive β-arrestin-2 trafficking to CCSs even when laterally immobilized.
(a & b) Initial fluorescence intensity of β-arrestin-2 (a) and receptor (b). (c & d) Maximum arrestin (c) or receptor (d) fluorescence enrichment at CCSs in cells expressing β-arrestin-2-GFP, clathrin light chain-DsRed, and either FLAG-β1AR or FLAG-β2AR after treatment with 10 μM dobutamine or 10 μM isoproterenol, respectively. *** p = 0.0009 by two-tailed t-test. (e & f) Average FLAG-β1AR (e) or FLAG-β2AR (f) enrichment at CCSs after treatment with 10 μM dobutamine or 10 μM isoproterenol, respectively. 5th and 95th confidence intervals for significant fluorescence enrichment are shown for each receptor. (g) Maximum receptor enrichment at CCSs as a function of receptor fluorescence when imaged under identical conditions. Arrows indicate expression matched cells that have disparate enrichment at CCSs. (h) Representative TIR-FM images of cells expressing super ecliptic pHluorin (SEP)-β1AR (blue), β-arrestin-2-mApple (green), and clathrin light chain-TagBFP (red) after immobilizing β1ARs with polyclonal antibodies prior to imaging. Frames collected immediately before (0 s) and 20 seconds after (20 s) application of 10 μM dobutamine are shown. Insets show a representative region at higher magnification. Large image scale bar = 5 μm; inset scale bar = 500 nm. (i) Representative time lapse series showing β-arrestin-2 clustering into pre-existing CCSs while immobilized β1ARs do not concentrate in CCSs. Images were collected at 0.5 Hz and, for brevity of presentation, one in three frames (6 sec interval) are shown. Scale bar = 500 nm. (j) Fluorescence intensity profiles from lines shown in (h). Gray arrows indicate arrestin and clathrin overlap. Black arrows indicate immobilized β1ARs without arrestin overlap. (k) Time-dependent correlation coefficient of line scans across cells. Error bars represent SEM. Uncropped original scans of western blots are shown in Supplementary Fig. 6. a-g are representative of 11 cells per condition pooled across 3 independent experiments, k is representative of 3 independent experiments, and raw data from independent repeats are in Supplementary Table 1.
To definitively determine whether β1ARs can drive β-arrestin trafficking to CCSs without themselves moving there, we immobilized receptors in the plasma membrane by surface cross-linking with polyclonal antibodies21 and verified lateral immobilization by fluorescence recovery after photobleaching (FRAP) (Supplementary Fig. 4a, b). Immobilized β1ARs formed clusters that predominantly localized separate from CCSs (Fig. 3h). Nevertheless, immobilized β1ARs still promoted robust recruitment of β-arrestin-2 to CCSs, evident within 20 seconds after application of dobutamine, while β1AR clusters remained separate and immobile (Fig. 3h, Supplementary Video 4 and 5). β1AR-elicited recruitment of β-arrestin-2, but not of β1ARs, to CCSs was visibly apparent in individual examples (Fig. 3i), verified by line scan analysis (Fig. 3j) and further verified across multiple experiments by time-dependent correlation analysis (Fig. 3k). Thus, ligand-activated β1ARs can indeed promote robust trafficking of β-arrestin-2 to CCSs without themselves moving there. The ability of β1ARs to strongly drive this process without entering CCSs, unequivocally establishes that β-arrestin-2 can traffic separately from its upstream activating GPCR - a behavior previously not thought possible.
We next asked if this discrete trafficking behavior of β-arrestin-2 can explain how β1ARs drive β-arrestin-2 and clathrin -dependent ERK1/2 activation without internalizing. The prevailing view is that ligand-activated GPCRs internalize together with β-arrestin in complex, and that internalization of the formed complex is required for full β-arrestin-mediated ERK1/2 activation. Given that β1ARs drive β-arrestin-2 trafficking to CCSs but not to internal membrane structures, we considered the alternate possibility that β-arrestin-2 might activate ERK1/2 from CCSs rather than endosomes. Knockdown of β-arrestin-2 markedly inhibited the magnitude of β1AR-elicited ERK1/2 activation but did not detectably affect its kinetics (Supplementary Fig. 5a, b), a result consistent with either hypothesis.
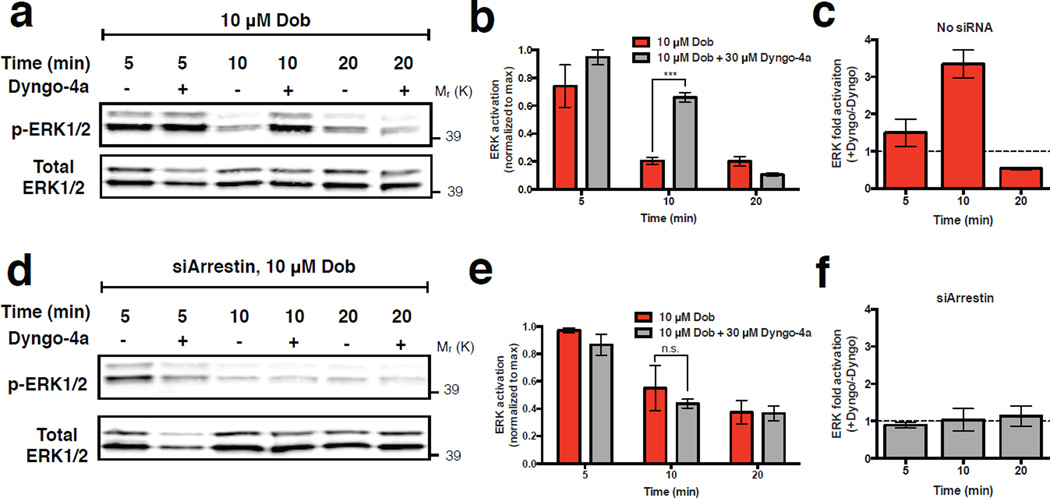
We distinguished these hypotheses by examining the effect of Dyngo-4a, a hydroxylated form of dynasore that acutely blocks all dynamin-dependent endocytic routes including CME22. Acute endocytic blockade did not inhibit β1AR-elicited ERK1/2 activation, contrary to expectation based on the prevailing current view. In fact, endocytic blockade enhanced the β1AR-elicited ERK1/2 response (Fig. 4a-c), and this was specific to the β1AR-elicited signaling because Dyngo-4a did not affect the basal signal detected in the absence of β1AR agonist (Supplementary Fig. 5c). Further, we verified that Dyngo-4a had no detectable effect in cells depleted of β-arrestin-2 (Fig. 4d-f), excluding off-pathway effects of Dyngo-4a in the present interpretation. These observations favor the hypothesis that β-arrestin-2 activated by β1ARs initiates signaling from CCSs rather than endosomes. Dyngo-4a blocks not only CME but also a recently identified clathrin-independent endocytic route (FEME). However, FEME does not require β-arrestin-2 or clathrin23. We found that the β1AR-elicited ERK1/2 signal requires both (as shown in Fig. 1e, f, Supplementary Fig. 1d,e and Supplementary Fig. 5a, b), thus excluding FEME as a potential mechanism of the β1AR-elicited ERK1/2 response.
Figure 4. Surface lifetime of CCSs regulates magnitude and duration of the β-arrestin-2-dependent ERK1/2 signal.
(a) Representative western blot showing phosphorylated ERK1/2 and total ERK1/2 signal in extracts prepared after pre-incubating cells in the absence or presence of 30 μM Dyngo-4a, as indicated, and exposed to 10 μM dobutamine for the indicated time period. (b) Quantification of ERK1/2 activation from western blots in (a). Phosphorylated ERK1/2 band intensity is normalized to ERK1/2 total band intensity and shown as fraction of maximum response observed in each independent experiment. *** p = 0.0001 by two-tailed t-test. (c) Dyngo-4a dependent fold activation of ERK1/2, determined after the indicated time period of dobutamine exposure, in cells expressing β-arrestin-2 at native levels. (d & e) The equivalent experiment of a & b conducted in cells in which β-arrestin-2 was depleted using siRNA. (f) Dyngo-4a dependent fold activation of ERK1/2, determined after the indicated time period of dobutamine exposure, in cells expressing β-arrestin-2 after siRNA-mediated knockdown. ERK1/2 activation in cells treated with Dyngo-4a was normalized to ERK1/2 activation in cells without Dyngo-4a treatment to determine the specific effect of Dyngo-4a. Error bars correspond to SEM. For a-f , n = 4 independent experiments. Raw data from independent repeats are provided in Supplementary Table 1.
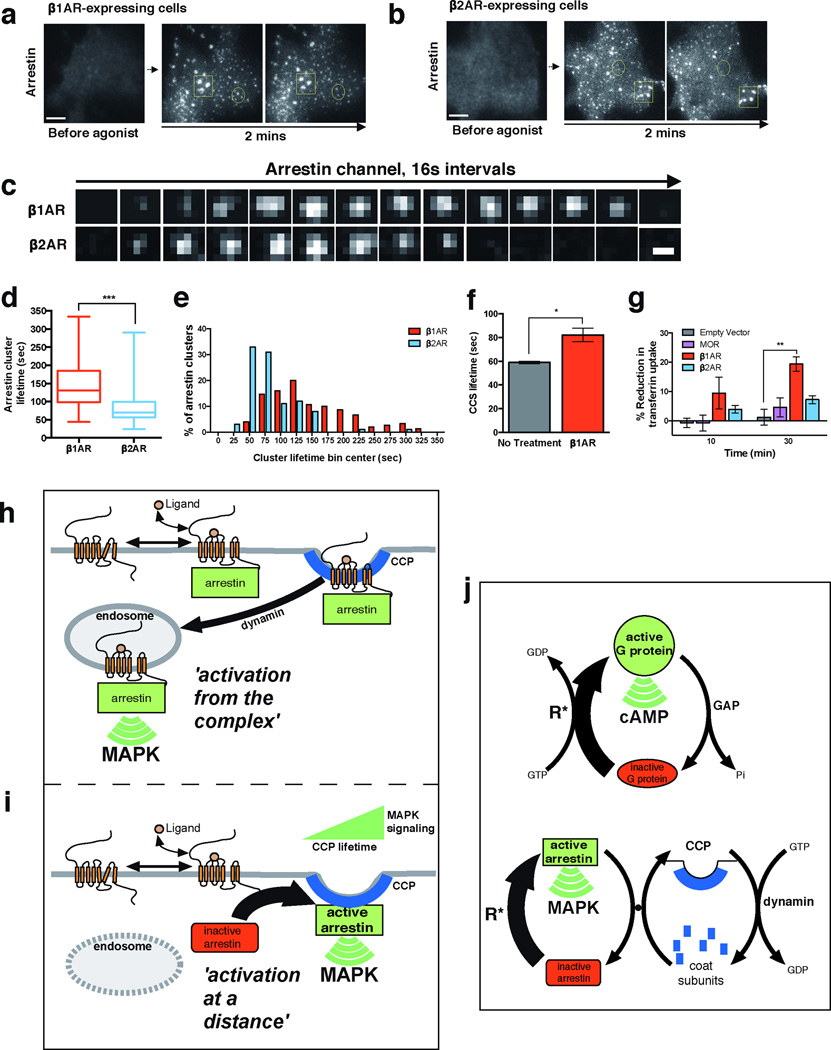
We were intrigued by the observation that Dyngo-4a not only enhanced the maximum degree of β1AR-elicited ERK1/2 activation, but also significantly extended the duration of the ERK1/2 signal (Supplementary Fig. 5d,e). Dyngo-4a, by blocking dynamin activity, effectively stalls CCSs on the plasma membrane22,24,25 and thereby extends the surface lifetime of recruited β-arrestin-2 (Supplementary Fig. 5f). This suggested the possibility that CCSs, in addition to serving as sites of ERK1/2 activation by β-arrestin-2, might also shape the kinetics of β1AR-elicited ERK1/2 signaling according to CCS surface lifetime. To test whether this might be relevant to ERK1/2 signaling elicited by β1ARs under normal conditions, we examined the effect of β1AR activation on the surface lifetime of CCSs in cells not exposed to Dyngo-4a. When diffraction-limited β-arrestin-2 clusters were identified separately from larger structures (Fig. 5a, b; representative examples are shown in circled and boxed regions, respectively), we noticed that their surface lifetime was longer after β1AR activation (Fig. 5c). We verified this observation across multiple specimens and experiments by blinded manual scoring (Fig. 5d, e) and using an automated analysis software (cmeAnalysis) developed and validated previously by others26 (Fig. 5f). Providing a completely independent and inherently unbiased confirmation, β1AR activation also significantly reduced endocytic uptake of labeled transferrin, a constitutive cargo of CME, providing an orthogonal biochemical metric of CME dynamics (Fig. 5g).
Figure 5. β1AR-elicited control of β-arrestin and CME dynamics underlies signaling.
(a & b) Activation of FLAG-β1ARs (a) or FLAG-β2ARs (b) produced similar recruitment of β-arrestin-2-GFP to CCSs. Circled examples indicate diffraction-limited spots representing individual CCSs. Squared examples indicate structures larger than the diffraction limit that were excluded from analysis as they represent groups of CCSs or clathrin plaques. Scale bar = 5 μm. (c) Representative image series of individual arrestin-associated CCSs observed by TIR-FM after β1AR (top row) or β2AR (bottom row) activation. Images were collected at 0.5 Hz and, for brevity of presentation, one in eight frames (16 sec interval) are shown. Scale bar = 500 nm. (d) Average surface lifetime of β-arrestin-2 clusters after indicated receptor activation (whiskers represent min and max, boxplot represents median, and 25th and 75th quartiles; n = 150 CCSs pooled across 3 independent experiments for β1AR and n = 100 CCSs pooled across 2 independent experiments for β2AR, *** p = 0.0001). (e) Frequency distribution analysis of β-arrestin-2 surface lifetimes shown in (d). (f) Automated cmeAnalysis of CCS lifetimes in cells expressing FLAG-β1ARs with and without 10 μM dobutamine treatment (n = 3 independent experiments, * p = 0.0165). (g) Cellular uptake of transferrin after indicated receptor activation (n = 3 independent experiments, ** p = 0.0025). Error bars represent SEM, p values calculated by a two-tailed t-test. Raw data from independent repeats are provided in Supplementary Table 1. (h and i) Schematic summarizing the previous paradigm for β-arrestin function, based on ‘activation from the complex’ (panel h). The different mode of β-arrestin trafficking and signaling, ‘activation at a distance’, which is demonstrated in the present study (panel i). Here, β-arrestin operates after dissociating from its activating GPCR rather than in physical complex with it, and the ERK signal is initiated from CCSs rather than endosomes. (j) Schematic of conventional G protein signaling mechanism based on allosteric coupling to GTP hydrolysis (top diagram) and the proposed β-arrestin signaling mechanism based on intermolecular coupling to the CME cycle (bottom diagram). Activated GPCR form(s) catalyzing the indicated conformational activations are denoted as R*.
Together, the present results reveal a remarkable new aspect of β-arrestin biology. The previous understanding, which we call 'activation from the complex', is based on the tenets that β-arrestin operates in obligate physical complex with an activated GPCR and its signaling activity requires endocytosis of the formed complex (Fig. 5h). We show a different behavior using β1ARs as a relevant model GPCR, which we call 'activation at a distance', where β-arrestin traffics to CCSs separately from its activating GPCR and promotes MAP kinase activation from CCSs before endocytic scission (Fig. 5i). Our results also demonstrate a role of CCS surface dynamics as a physiological control point for β-arrestin signaling, thus revealing an additional relationship between cellular signaling and membrane trafficking that may be relevant more widely in the GPCR family (e.g.,25,27,28).
This newly recognized trafficking and signaling behavior of β-arrestin raises many additional questions, particularly regarding molecular mechanism. Our observations support the hypothesis that β-arrestin undergoes conformational activation through transient contact with an activated GPCR and then dissociates from the GPCR to subsequently bind CCPs while still in a conformationally activated state. We propose that this supports a β-arrestin signaling cycle which is analogous, in overall biochemical logic, to the conventional G protein cycle. In the G protein cycle, the activated GPCR acts as a catalyst for G protein conformational activation, and the cycle is energetically driven by guanine nucleotide hydrolysis through intramolecular coupling to an intrinsic GTPase activity (Fig. 5j, top, catalytic GPCR activity is denoted as R*). We propose that β-arrestin conformational activation is also catalyzed by activated GPCRs, but the energetics of β-arrestin signaling are provided by intermolecular coupling to the endocytic cycle involving dynamic CCP assembly, scission and dissociation. Although different in its biochemical details, this energetic cycle is also fundamentally powered by nucleotide hydrolysis, including that mediated by the dynamin GTPase (Fig. 5j, bottom). Intermolecular coupling between the β-arrestin and endocytic cycles is possible because conformational activation of arrestin releases its C-terminal tail that, in β-arrestins is a binding site for the assembled CCP lattice29–31. Further extending the analogy, the dynamin GTPase operating in the coupled cycle of β-arrestin signaling is positioned similarly to the GTPase activating protein (GAP) activity operating in the conventional G protein cycle. This explains why inhibiting dynamin enhances the β-arrestin-mediated MAP kinase signal, just as inhibiting the GAP enhances the G protein-mediated cAMP signal, and how CCP lifetime controls the magnitude and kinetics of downstream β-arrestin-mediated signaling.
Methods
Cell culture, expression constructs, and transfections
HEK 293 and COS-1 cells (ATCC) were cultured in complete growth Dulbecco’s modified Eagle’s medium (DMEM, Gibco) and supplemented with 10% fetal bovine serum (UCSF Cell Culture Facility). Cell lines cultures were free of mycoplasma contamination.
N-terminally FLAG-tagged versions of the human β1AR and β2AR cloned into pcDNA3 32,33. Super ecliptic pHluorin (SEP)-β1AR was generated by PCR amplification of SEP and in-frame insertion into the FLAG-β1AR construct. β-arrestin-2-GFP was a gift of Dr. Marc Caron and previously described34. β-arrestin-2-mApple was generated by subcloning β-arrestin-2 into pmApple-N1 (Clontech) using HindIII and ApaI. Clathrin-dsRed was a gift of Dr. Wolf Almers and previously described35. Photoactivatable (PA)-mCherry1-clathrin was a gift from Dr. Michael Davidson. Clathrin-TagBFP was generated by PCR amplifying clathrin light chain and subcloning into pTag-BFP-N1 (Evrogen) using EcoRI and ApaI. Clathrin-PA-mCherry1 was a gift from Dr. Michael Davidson. β-arrestin-2-PA-mCherry1 was generated by subcloning β-arrestin-2 into pPA-mCherry1-N1 (gift from Dr. Michael Davidson), using HindIII and ApaI. A non-targetable mutant (NTM) of β-arrestin-2 (β-arrestin-2-GFP-NTM) was generated using site directed mutagenesis (Phusion Site-Directed Mutagenesis Kit, Thermo Scientific) of β-arrestin-2-GFP to introduce synonymous mutations in the nucleotide sequence targeted by ARB2_10 siRNA using primers 5’-CGCCCAGTTCCACGTTCTGTA -3’ and 5’-CTCAGGTCGAGCAGGACGACCAGGTGT -3’. To generate a non-targetable mutant (NTM) of CHC17, clathrin-NTM was generated using a gBlock (IDT) to introduce six synonymous mutations in the nucleotide sequence targeted by the CHC17 siRNA. The clathrin-NTM gBlock was inserted into CHC17 using PmlI and BsteII. All stars negative control (nonsilencing) and silencing siRNAs were obtained from Qiagen. The sense-strand, silencing siRNA sequences are as follows: β-arrestin-2 (ARB2_10), 5′-CGAACAAGAUGACCAGGUATT-3′; clathrin heavy chain, 5’- GCAAUGAGCUGUUUGAAGATT-3’.
Transfections were carried out using Lipofectamine 2000 or RNAi-Max (Life Technologies) for cDNA or siRNA, respectively, according to the manufacturer’s protocol. For DNA expression, cells were transfected 48 hours before experiments. For siRNA knockdown, cells were transfected with siRNA constructs 72 hours before experiments. For siRNA rescue experiments, cells were transfected with indicated siRNA construct 72 hours before experiments and subsequently transfected with a plasmid containing synonymous mutations in the region targeted by siRNA (non-targetable mutants). Experiments were performed 48 hours after DNA rescue construct transfection.
Flow cytometric analysis of receptor endocytosis
The flow cytometric assays to measure endocytosis or receptor surface fluorescence were carried out as previously described36. Surface fluorescence of FLAG-β1AR or FLAG-β2AR expressing HEK 293 cells was used to measure receptor endocytosis. Cells were incubated with 10 μM iso-proterenol for indicated times at 37°C to drive receptor internalization and were subsequently washed with ice-cold PBS, then mechanically lifted and incubated with 1μg/ml Alexa647 (Life Technologies)-conjugated M1 anti-FLAG monoclonal antibody (Sigma F-3040) at 4°C for 45 minutes. Mean fluorescence intensity of 10,000 cells per technical replicate was measured using a FACSCalibur instrument (Becton Dickinson). At least three independent experiments were performed in triplicate for each condition.
Live cell total internal reflection fluorescence microscopy (TIR-FM) imaging
TIR-FM was performed at 37°C using a Nikon Ti-E inverted microscope equipped for through-the-objective TIR-FM and outfitted with a temperature-, humidity-, and CO2-controlled chamber (Okolab). Images were obtained with an Apo TIRF 100X, 1.49 numerical aperture objective (Nikon) with solid-state lasers of 405, 488, 561, and 647 nm (Keysight Technologies). An Andor iXon DU897 EMCCD camera controlled by NIS-Elements 4.1 software was used to acquire image sequences every 2 seconds for 10 minutes. Cells were transfected as indicated according to manufacture protocol 48 hours before imaging and then plated on poly-L-lysine (0.0001%, Sigma) coated 35-mm glass-bottomed culture dishes (MatTek Corporation) 24 hours before imaging. Cells were labeled with M1 monoclonal FLAG antibody (1:1000, Sigma F-3040) conjugated to Alexa 647 dye (Life Technologies) for 10 minutes at 37°C prior to imaging, washed, and imaged live in DMEM without phenol red (UCSF Cell Culture Facility) supplemented with 30 mM HEPES, pH 7.4 (UCSF Cell Culture Facility). Cells were treated with 10 μM dobutamine (Tocris Bioscience) or 10 μM isoproterenol (Sigma) to activate either FLAG-β1ARs or FLAG-β2ARs, respectively, at frame 10 of 301 image sequences. For experiments with the β1AR selective antagonist CGP20712A (Tocris Bioscience), cells were pretreated or 15 minutes with 500 nM CGP20712A, and then treated with 10 μM dobutamine at frame 10 of 301 image sequences. At least two independent experiments were performed for all live cell TIR-FM imaging.
TIR-FM image analysis
Acquired image sequences were saved as stacks of 16 bit TIFF files. All quantitative image analysis was performed on unprocessed images using ImageJ and Fiji software37,38. To quantify change in receptor fluorescence over time in TIR-FM images, fluorescence values were measured in five randomly selected regions of interest (ROIs) in the cell over the entire stack. An area of the coverslip lacking cells was used to estimate background fluorescence. Fluorescence values of the five ROIs were background subtracted, averaged, and normalized to initial fluorescence values before agonist addition. Minimal bleed-through and photobleaching was verified using single-labeled and untreated samples, respectively. Linescan analysis of receptor, β-arrestin, or clathrin fluorescence from the shown line were carried out using the ImageJ plot profile function to measure pixel values from this line. For fluorescence enrichment into CCSs calculations, a mask of CCSs was generated using a thresholded average image of the clathrin channel. Enrichment at CCSs for receptor and arrestin was measured as the difference between the average fluorescence in the mask and average fluorescence outside of the thresholded structures. Confidence intervals of enrichment into CCSs were determined by generating 2000 unique randomized masks, where the position and orientation of the thresholded CCSs is distinct from the original mask or other randomized masks. Enrichment was calculated for every randomization over time, the 2000 values sorted for every time point, and the 5% lowest and highest values used to determine a confidence interval for significant enrichment.
Lifetimes of β-arrestin-2 clusters after β1AR or β2AR activation were measured blindly by calculating the number of frames between cluster appearance and disappearance from the TIR-FM field. Only clusters whose appearance and disappearance could clearly be observed were included in the analysis. cmeAnalysis software package was used to measure the CCP lifetimes.
Live cell photoactivated localization microscopy (PALM) imaging & analysis
PALM was performed at 37°C using a Nikon Ti-E inverted microscope equipped for through-the-objective TIR-FM and outfitted with a temperature-, humidity-, and CO2-controlled chamber (Okolab). Images were obtained with an Apo TIRF 100X, 1.49 numerical aperture objective (Nikon) with solid-state lasers of 405, 488, 561, and 647 nm (Keysight Technologies) as light sources. An Andor iXon DU897 EMCCD camera controlled by NIS-Elements 4.1 software was used to acquire 5000 periods with continuous activation by 405 nm light. Cells were transfected as indicated according to manufacture protocol 48 hours before imaging and then plated on poly-L-lysine (0.0001%, Sigma) coated 35-mm glass-bottomed culture dishes (MatTek Corporation) 24 hours before imaging. Cells were labeled with M1 monoclonal FLAG antibody (1:1000, Sigma F-3040) conjugated to Alexa 647 dye (Life Technologies) for 10 minutes at 37°C, washed one time with imaging media (20 mM HEPES, 135 mM NaCl, 5mM D+ glucose, 5 mM KCl, 0.4 mM MgCl2, 1.8 mM CaCl2, pH 7.4), and imaged live in imaging media. NIS Elements software (Nikon) was used to resolve and analyze the images. Full width at half max calculations were used to measure size of the super resolved structures. PALM imaging experiments were performed at least three independent times.
Live cell structured illumination microscopy (SIM) imaging & analysis
SIM was performed using a Nikon Ti-E inverted microscope equipped with sapphire lasers of 488 and 561 nm (Coherent). Images were obtained using an Apo TIRF 100x 1.49 objective (Nikon) and a 3D EX V-R SIM grating (Nikon). An Andor iXon DU897 EMCCD camera controlled by NIS-Elements 4.12 software was used to acquire images. Cells were transfected as indicated according to manufacture protocol 48 hours before imaging and then plated on poly-L-lysine (0.0001%, Sigma) coated 35-mm glass-bottomed culture dishes (MatTek Corporation) 24 hours before imaging. Cells were imaged live in DMEM without phenol red (UCSF Cell Culture Facility) supplemented with 30 mM HEPES, pH 7.4 (UCSF Cell Culture Facility). NIS-Elements 4.12 software was used to reconstruct images and analyze the images. Full width at half max calculations were used to measure size of observed structures in the reconstructed images. SIM imaging experiments were performed three independent times.
Fluorescence recovery after photobleaching of immobilized receptors
To limit receptor diffusion in the plasma membrane, cells transfected with SEP-β1ARs were treated with a polyclonal rabbit anti-GFP antibody (1:100, Life Technologies A-11122) for 15 minutes at 37°C in order to group multiple receptors together as described previously39. Cells were then washed and imaged live in DMEM without phenol red (UCSF Cell Culture Facility). Fluorescence recovery after photobleaching was performed at 37°C using a Nikon Ti inverted microscope equipped with a temperature-, humidity-, and CO2-controlled chamber (Okolab). Images were obtained with an Apo TIRF 100X, 1.49 numerical aperture objective (Nikon) with 488 and 561 nm solid-state lasers (Keysight Technologies). An Andor Zyla CMOS camera controlled by MicroManager software was used to acquire image sequences. To verify that receptor mobility was slowed after antibody addition, a Rapp Optoelectronic UGA-40 photobleaching system was used to photobleach SEP-β1ARs on an area of the plasma membrane with a 473 laser (Vortran). Cells were imaged every second for 5 minutes to monitor fluorescence recovery after photobleaching. All quantitative image analysis was performed on unprocessed images using ImageJ software. Mean fluorescence intensity was measured in photobleach region, background subtracted using an area outside of the cell, and normalized to initial fluorescence. Imaging experiments were performed three independent times.
Transferrin uptake assay
Cells expressing FLAG-labeled receptors or empty vector plasmid were first incubated in serum-free medium for 90 min at 37°C and subsequently in serum-free media containing 200 μg/mL Alexa647-transferrin (Invitrogen) and 10 μM receptor-specific agonist for the indicated time at 37°C to activate indicated GPCRs and allow for constitutive uptake of Alexa 647-transferrin. After washing with ice-cold PBS, cells were incubated in acid-wash buffer (20 mM sodium-acetate buffer, 1mM CaCl2, 150 mM NaCl, pH 4.8) on ice for 5 minutes to remove surface-bound transferrin. Cells were then mechanically lifted in PBS for 15 minutes at 4°C. Mean fluorescence intensity of 5,000 cells was measured using a FACSCalibur instrument (Becton Dickinson). At least three independent experiments were performed in triplicate for each condition.
ERK1/2 activation assays
To measure activation of ERK1/2, western blot analysis and fixed cell flow cytometry were used. For both methods, cells stably expressing FLAG-β1ARs or FLAG-β2ARs were serum starved for 18 hours prior to assay, incubated with the indicated agonist for the indicated times at 37°C, and then washed on ice with ice-cold PBS. For cells treated with Dyngo-4a, serum-starved cells were pretreated with 30 μM Dyngo-4a for 15 minutes, treated with 10 μM dobutamine for indicated times at 37°C in the presence of 30 μM Dyngo-4a, and then washed on ice with ice-cold PBS. For western blot analysis, cells were directly lysed in sample buffer (NuPAGE LDS Sample Buffer (Life Technologies), 100 mM DTT), sonicated 3 times for 10 seconds, boiled, separated by SDS-PAGE (Life Technologies) and transferred to a nitrocellulose membrane that was blocked with TBS Odyssey blocking buffer (LI-COR) for one hour at room temperature and then incubated overnight at 4°C with a mouse anti-ERK1/2 primary antibody (1:2000, Cell Signaling 4696) and a rabbit anti-phosphorylated-ERK1/2 primary antibody (1:2000, Cell Signaling 4370). Membranes were washed 3 times for 5 minutes in TBS-Tween (0.1% v/v) and incubated with a 680 labeled anti-rabbit secondary antibody (1:5000, LI-COR 926-68073) and a 800 labeled anti-mouse secondary antibody (1:5000, LI-COR 926-32212) for one hour at room temperature. Membranes were washed 3 times for 5 minutes in TBS-Tween (0.1% v/v), imaged using an Odyssey Infrared Imaging System (LI-COR) in the linear range, and quantified by measuring band intensity, background subtracting, and normalizing the phosphorylated ERK1/2 band intensity to the total ERK1/2 band intensity. At least three independent experiments were performed for each condition.
For fixed-cell flow cytometric analysis, cells were fixed on ice for 10 minutes using 4% cytofix solution (BD Cytofix), lifted with a cell scraper, and transferred to a 96 well plate. Cells were permeabilized using cold methanol and incubated overnight at −20°C. Cells were then washed with staining buffer (Ca+/Mg+ PBS, 2% FBS, 2mM EDTA), rehydrated, and incubated for one hour at room temperature with a mouse anti-ERK1/2 primary antibody (1:200, Cell Signaling 4696) and a rabbit anti-phosphorylated ERK1/2 primary antibody (1:200, Cell Signaling 4370) diluted in staining buffer. Cells were then washed 3 times with staining buffer and incubated with a 488 labeled anti-mouse secondary antibody (1:100, Life Technologies A-21202) and a 647 labeled anti-rabbit secondary antibody (1:100, Life Technologies A-21245) for 30 minutes at room temperature. Cells were washed 3 times with staining buffer, separated with a cell strainer, and mean fluorescence intensity of 5,000 cells was measured using a FACSCalibur instrument (Becton Dickinson). Fold activation was determined by normalizing phosphorylated ERK1/2 mean fluorescence to total ERK1/2 mean fluorescence. Mean fluorescence intensity of 5,000 cells was measured using a FACSCalibur instrument (Becton Dickinson). At least three independent experiments were performed in triplicate for each condition.
Statistical Analysis
Quantitative data are expressed as the mean and error bars represent the standard error of the mean (SEM) unless otherwise indicated. Statistical significance between conditions were analyzed using a two-tailed t-test and calculated using Prism 6.0 software (GraphPad Software, Inc). P values were considered to be significant if p < 0.05 when compared with control or no treatment conditions. P values > 0.05 are indicated in the figures as not significant (n.s). Exact values of all significant p values are specified in the appropriate figure legends. All experiments and representative data shown were repeated at least twice and performed with independent samples.
Supplementary Material
Acknowledgements
We thank M. Mettlen, G. Danuser, and S. Schmid for help with the cmeAnalysis package used for measuring CCP lifetime, and J. Benovic, V. Gurevich and B. Shoichet for valuable discussion. We also thank R. Irannejad, G. Peng, N. Michael, K. Ashrafi, O. Weiner, H. Bourne, and other members of the von Zastrow laboratory for useful advice and critical discussion. All live cell imaging experiments were performed in the Nikon Imaging Center at UCSF; we thank K. Thorn and D. Larson for essential advice and assistance. This work was supported by grants from the US National Institutes of Health (DA 010711 and 012864 to M.v.Z.). K.E. is a recipient of a National Science Foundation Graduate Research Fellowship.
Footnotes
Author contributions
K.E. conceived and designed the experiments, performed all experiments, analyzed the data, and wrote the paper. D.J. developed software for analysis of TIR-FM image series. M.v.Z. conceived and designed the experiments, analyzed the data, and wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Shukla AK, Xiao K, Lefkowitz RJ. Emerging paradigms of beta-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem Sci. 2011;36:457–469. doi: 10.1016/j.tibs.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benovic JL, et al. Functional desensitization of the isolated beta-adrenergic receptor by the beta-adrenergic receptor kinase: potential role of an analog of the retinal protein arrestin (48-kDa protein) Proc Natl Acad Sci U S A. 1987;84:8879–8882. doi: 10.1073/pnas.84.24.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchese A, Chen C, Kim YM, Benovic JL. The ins and outs of G protein-coupled receptor trafficking. Trends Biochem Sci. 2003;28:369–376. doi: 10.1016/S0968-0004(03)00134-8. [DOI] [PubMed] [Google Scholar]
- 4.Shukla AK, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang Y, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daaka Y, et al. Essential role for G protein-coupled receptor endocytosis in the activation of mitogen-activated protein kinase. J Biol Chem. 1998;273:685–688. doi: 10.1074/jbc.273.2.685. [DOI] [PubMed] [Google Scholar]
- 7.Tohgo A, et al. The stability of the G protein-coupled receptor-beta-arrestin interaction determines the mechanism and functional consequence of ERK activation. J Biol Chem. 2003;278:6258–6267. doi: 10.1074/jbc.M212231200. [DOI] [PubMed] [Google Scholar]
- 8.Ahn S, Shenoy SK, Wei H, Lefkowitz RJ. Differential kinetic and spatial patterns of beta-arrestin and G protein-mediated ERK activation by the angiotensin II receptor. J Biol Chem. 2004;279:35518–35525. doi: 10.1074/jbc.M405878200. [DOI] [PubMed] [Google Scholar]
- 9.Wei H, Ahn S, Barnes WG, Lefkowitz RJ. Stable interaction between beta-arrestin 2 and angiotensin type 1A receptor is required for beta-arrestin 2-mediated activation of extracellular signal-regulated kinases 1 and 2. J Biol Chem. 2004;279:48255–48261. doi: 10.1074/jbc.M406205200. [DOI] [PubMed] [Google Scholar]
- 10.Patel PA, Tilley DG, Rockman HA. Physiologic and cardiac roles of beta-arrestins. J. Mol. Cell. Cardiol. 2009;46:300–308. doi: 10.1016/j.yjmcc.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T, et al. Distinct regulation of beta 1- and beta 2-adrenergic receptors in Chinese hamster fibroblasts. Molecular Pharmacology. 1992;41:542–548. [PubMed] [Google Scholar]
- 12.Shiina T, Kawasaki A, Nagao T, Kurose H. Interaction with beta-arrestin determines the difference in internalization behavor between beta1- and beta2-adrenergic receptors. J Biol Chem. 2000;275:29082–29090. doi: 10.1074/jbc.M909757199. [DOI] [PubMed] [Google Scholar]
- 13.Liang W, Curran PK, Hoang Q, Moreland RT, Fishman PH. Differences in endosomal targeting of human (beta)1- and (beta)2-adrenergic receptors following clathrin-mediated endocytosis. J Cell Sci. 2004;117:723–734. doi: 10.1242/jcs.00878. [DOI] [PubMed] [Google Scholar]
- 14.Boucrot E, et al. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 2015;517:460–465. doi: 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- 15.Grove J, et al. Flat clathrin lattices: stable features of the plasma membrane. Mol Biol Cell. 2014;25:3581–3594. doi: 10.1091/mbc.E14-06-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zoncu R, et al. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc Natl Acad Sci U S A. 2007;104:3793–3798. doi: 10.1073/pnas.0611733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaidarov I, Santini F, Warren RA, Keen JH. Spatial control of coated-pit dynamics in living cells. Nat Cell Biol. 1999;1:1–7. doi: 10.1038/8971. [DOI] [PubMed] [Google Scholar]
- 18.Santini F, Gaidarov I, Keen JH. G protein-coupled receptor/arrestin3 modulation of the endocytic machinery. J Cell Biol. 2002;156:665–676. doi: 10.1083/jcb.200110132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puthenveedu MA, von Zastrow M. Cargo regulates clathrin-coated pit dynamics. Cell. 2006;127:113–124. doi: 10.1016/j.cell.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 20.Deighton NM, Motomura S, Bals S, Zerkowski HR, Brodde OE. Characterization of the beta adrenoceptor subtype(s) mediating the positive inotropic effects of epinine, dopamine, dobutamine, denopamine and xamoterol in isolated human right atrium. J Pharmacol Exp Ther. 1992;262:532–538. [PubMed] [Google Scholar]
- 21.Mondin M, et al. Neurexin-neuroligin adhesions capture surface-diffusing AMPA receptors through PSD-95 scaffolds. J Neurosci. 2011;31:13500–13515. doi: 10.1523/JNEUROSCI.6439-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCluskey A, et al. Building a better dynasore: the dyngo compounds potently inhibit dynamin and endocytosis. Traffic. 2013;14:1272–1289. doi: 10.1111/tra.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boucrot E, et al. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 2015;517:460–465. doi: 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- 24.Kirchhausen T, Macia E, Pelish HE. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 2008;438:77–93. doi: 10.1016/S0076-6879(07)38006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flores-Otero J, et al. Ligand-specific endocytic dwell times control functional selectivity of the cannabinoid receptor 1. Nat Commun. 2014;5:4589. doi: 10.1038/ncomms5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguet F, Antonescu CN, Mettlen M, Schmid SL, Danuser G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev Cell. 2013;26:279–291. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puthenveedu MA, von Zastrow M. Cargo regulates clathrin-coated pit dynamics. Cell. 2006;127:113–124. doi: 10.1016/j.cell.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 28.Henry AG, et al. Regulation of endocytic clathrin dynamics by cargo ubiquitination. Developmental Cell. 2012;23:519–532. doi: 10.1016/j.devcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurevich VV, Gurevich EV. Extensive shape shifting underlies functional versatility of arrestins. Curr Opin Cell Biol. 2014;27:1–9. doi: 10.1016/j.ceb.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palczewski K, Pulvermuller A, Buczylko J, Hofmann KP. Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. J Biol Chem. 1991;266:18649–18654. [PubMed] [Google Scholar]
- 31.Goodman OB, Jr, et al. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
References
- 32.Cao TT, Deacon HW, Reczek D, Bretscher A, von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- 33.Temkin P, et al. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13:715–721. doi: 10.1038/ncb2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barak LS, Ferguson SS, Zhang J, Caron MG. A beta-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J Biol Chem. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 35.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 36.Yudowski GA, Puthenveedu MA, Henry AG, von Zastrow M. Cargo-mediated regulation of a rapid Rab4-dependent recycling pathway. Mol Biol Cell. 2009;20:2774–2784. doi: 10.1091/mbc.E08-08-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mondin M, et al. Neurexin-neuroligin adhesions capture surface-diffusing AMPA receptors through PSD-95 scaffolds. J Neurosci. 2011;31:13500–13515. doi: 10.1523/JNEUROSCI.6439-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.