Abstract
Pathogens attack host cells by deploying toxins that perturb core host processes. Recent findings from the nematode C. elegans and other metazoans indicate that surveillance or ‘effector-triggered’ pathways monitor functioning of these core processes and mount protective responses when they are perturbed. Despite a growing number of examples of surveillance immunity, the signaling components remain poorly defined. Here we show that CEBP-2, the C. elegans ortholog of mammalian CCAAT-enhancer binding protein gamma, is a key player in surveillance immunity. We show that CEBP-2 acts together with the bZIP transcription factor ZIP-2 in the protective response to translational block by P. aeruginosa Exotoxin A, as well as to perturbations of other processes. CEBP-2 serves to limit pathogen burden, promote survival upon P. aeruginosa infection, and also promote survival upon Exotoxin A exposure. These findings may have broad implications for the mechanisms by which animals sense pathogenic attack and mount protective responses.
Introduction
The innate immune system serves to defend hosts against pathogen infection, without the need for prior exposure to these pathogens (Kumar et al., 2011). A key component of the innate immune system is the detection of molecules characteristic of pathogens, so-called Pathogen-Associated Molecular Patterns or PAMPs. Hosts use pattern recognition receptors that are tuned to detect these PAMPs and trigger defense, often through upregulation of immune response gene expression. However, PAMPs are usually molecules found in broad classes of microbes and do not necessarily represent the presence of a pathogenic microbe. For example, lipopolysaccharide is a PAMP found in Gram-negative bacterial species, both pathogenic and non-pathogenic alike. Thus, PAMPs may be more accurately defined as Microbe-Associated Molecular Patterns, or MAMPs. MAMPs provide hosts information about the presence of microbes, but not necessarily whether those microbes are pathogenic (Ausubel, 2005; Sanabria et al., 2010).
A growing theme in animal immunity is that hosts specifically detect pathogen attack with surveillance or ‘effector-triggered’ immune pathways, which detect the effects of pathogen-delivered toxins and virulence factors, rather than recognizing the molecular structure of the factors themselves (Cohen and Troemel, 2015; Rajamuthiah and Mylonakis, 2014; Spoel and Dong, 2012; Stuart et al., 2013). For example, many bacterial toxins inhibit host mRNA translation elongation (Beddoe et al., 2010; Lee et al., 2013; Lemaitre and Girardin, 2013; Lemichez and Barbieri, 2013; Mohr and Sonenberg, 2012), and these toxins are quite prevalent in the environment, with up to 29% of soil samples in one study harboring DNA for translation-blocking Shiga toxin (Casas et al., 2006). Translation-blocking toxins are made by diverse bacterial pathogens including P. aeruginosa (Iglewski et al., 1977) Corynebacterium diphtheriae (Pappenheimer, 1977), Vibrio cholera (Jorgensen et al., 2008), Legionella pneumophila (Belyi et al., 2006) Shigella spp, and Shiga toxin-producing E. coli (Pacheco and Sperandio, 2012). Because these toxins are diverse in structure, it is arguably an efficient defense strategy for hosts to detect the common block in translation elongation caused by these toxins to trigger defense.
Recent findings indicate that C. elegans uses surveillance pathways for defense against toxins delivered by the bacterial pathogen P. aeruginosa that block not only mRNA translation, but also mitochondria, the proteasome and histones (Dunbar et al., 2012; Liu et al., 2014; McEwan et al., 2012; Melo and Ruvkun, 2012; Pellegrino et al., 2014). P. aeruginosa causes a lethal intestinal infection in its nematode host, and in the early response to infection C. elegans upregulates mRNA expression of many defense genes, including candidate anti-microbial peptides, detoxifying enzymes and efflux pumps (Shapira et al., 2006; Troemel et al., 2006). We identified the bZIP transcription factor ZIP-2 as a key mediator of this infection-induced gene expression, and showed that it promotes a defense response (Estes et al., 2010). The transcriptional response to P. aeruginosa infection appears to be predominantly a response to pathogenicity, triggered in part by the translation-blocking Exotoxin A (ToxA) (Dunbar et al., 2012; Estes et al., 2010; McEwan et al., 2012). In previous studies we showed that C. elegans intestinal cells appear to endocytose ToxA, which blocks mRNA translation specifically in the intestine, and this block is sensed by the host to upregulate defense gene expression. Surprisingly, this translational block appears to trigger an increase in protein levels of ZIP-2, apparently through regulation in cis by an upstream open reading frame (Dunbar et al., 2012). Thus, ZIP-2 appears to function in effector-triggered immunity in C. elegans to respond to the translational block caused by P. aeruginosa-delivered ToxA. However, ZIP-2 does not have an obvious mammalian ortholog, which made it unclear which transcription factor might be involved in this type of immunity in mammals.
Here we show that ZIP-2 acts together with another bZIP transcription factor called CEBP-2 in C. elegans surveillance immunity. Intriguingly, CEBP-2 is the C. elegans ortholog of mammalian CCAAT-enhancer binding protein gamma (C/EBP-gamma), which plays a role in the acute response to infection and inflammation in mammals, together with other C/EBP transcription factors (Gao et al., 2002; Parkin et al., 2002; Tsukada et al., 2011), although its role in effector-triggered defense has not been shown. We show that CEBP-2 is required to upregulate a transcriptional response to ToxA in C. elegans, and promote defense against insult by this toxin as well as against pathogen infection. We also show that CEBP-2 is required for upregulation of defense gene expression in response to RNAi against genes that function in the mitochondria and transcription-related processes, and that perturbation of these processes increases levels of ZIP-2 protein, similar to perturbation of translation (Dunbar et al., 2012). Thus, ZIP-2/CEBP-2 appears to be a key transcription factor in C. elegans surveillance immunity that promotes defense against pathogenic microbes.
Results and Discussion
CEBP-2 is required for induction of ZIP-2-dependent genes in response to P. aeruginosa infection
Previously, we demonstrated that the bZIP protein ZIP-2 mediates induction of candidate defense genes and promotes survival upon P. aeruginosa infection (Estes et al., 2010). As bZIP proteins canonically act as dimers, we were interested in identifying a heterodimeric partner that could act together with ZIP-2 in C. elegans host defense. A comprehensive study of bZIP transcription factor protein-protein interactions in vitro identified C. elegans C48E7.11 as the highest affinity binding partner for ZIP-2 (Reinke et al., 2013). C48E7.11 is the top BLAST hit in the C. elegans genome for human CCAAT/enhancer binding protein gamma (C/EBPγ) transcription factor (NP_001797). It shares 37% amino acid identity with the human protein and has a similar domain structure, with most of the protein being composed of a bZIP domain. Therefore, we renamed C48E7.11 CEBP-2, for CCAAT/enhancer binding protein 2.
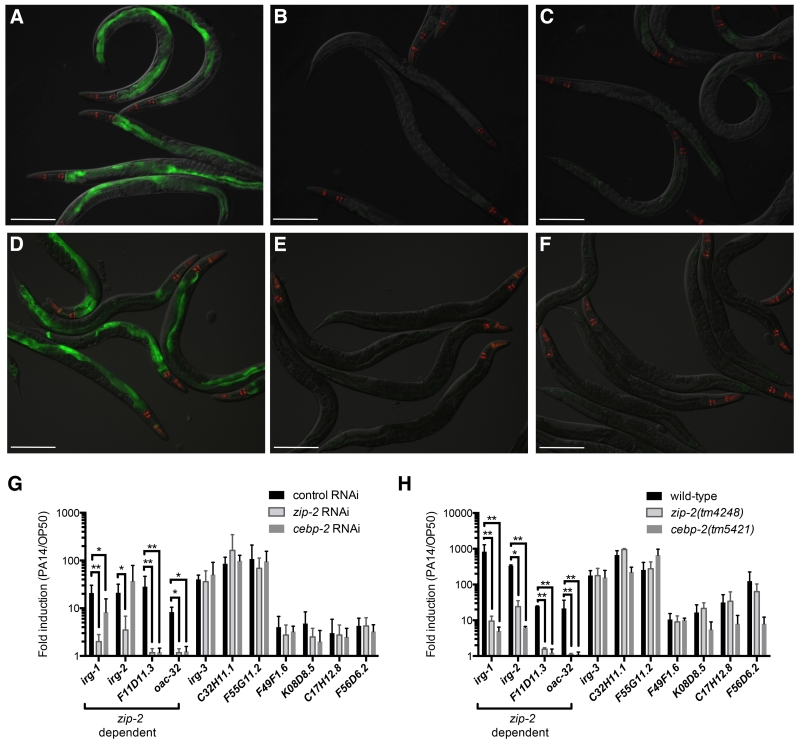
We investigated whether CEBP-2 mediates a protective response to P. aeruginosa strain PA14 infection of C. elegans, which would support the hypothesis that CEBP-2 acts together with ZIP-2 as a heterodimeric transcription factor in mediating defense against infection in vivo. First, we examined whether cebp-2 regulates expression of target genes known to be induced by zip-2 upon infection. In particular, we examined P. aeruginosa-induced expression of a GFP reporter for infection response gene-1 (irg-1p::GFP) in cebp-2-deficient animals, using either cebp-2 RNAi-treated animals or cebp-2(tm5421) mutant animals, and found greatly reduced induction of GFP compared to control, similar to that seen in worms lacking zip-2 function (Figure 1A-F). This result indicates that cebp-2, like zip-2, regulates irg-1 induction in response to P. aeruginosa infection.
Figure 1. cebp-2 is required for infection response gene induction upon P. aeruginosa PA14 infection.
(A-C) irg-1p::GFP animals treated with either (A) L4440 RNAi control, (B) zip-2 RNAi, or (C) cebp-2 RNAi and infected with PA14. (D-F) irg-1p::GFP expression in (D) wild-type, (E) zip-2(tm4248), or (F) cebp-2(tm5421) animals infected with PA14. In (A)-(F), green is irg-1p::GFP, red is myo-2::mCherry expression in the pharynx as a marker for presence of the transgene. Images are overlays of green, red and Nomarski channels and were taken with the same camera exposure for all. Scale bar, 200 μm. (G) qRT-PCR comparison of PA14-induced gene expression in control RNAi (L4440), zip-2 RNAi, and cebp-2 RNAi treated animals. (H) qRT-PCR comparison of PA14-induced gene expression in wild-type, zip-2(tm4248), and cebp-2(tm5421) animals. For (G) and (H), results shown are the average of two independent biological replicates, error bars are SD. ** p < 0.01, * p < 0.05 with a two-tailed t test.
We next confirmed that cebp-2 controls pathogen induction of endogenous irg-1 mRNA expression by using qRT-PCR to measure RNA levels in cebp-2-deficient animals. We found that cebp-2 RNAi-treated animals had decreased induction of irg-1 in response to P. aeruginosa, as compared to control, as well as decreased induction of two other zip-2-dependent genes, F11D11.3 and oac-32 (Figure 1G). However, induction of another zip-2-dependent gene, infection response gene 2, irg-2, was not decreased in cebp-2 RNAi-treated animals (Figure 1G). cebp-2(tm5421) mutant animals had a stronger phenotype, with greatly reduced mRNA induction of irg-1, irg-2 F11D11.3 and oac-32 (Figure 1H). We also tested a panel of infection response genes whose induction in response to P. aeruginosa infection does not require zip-2 and found that most of these genes were induced normally in cebp-2-deficient animals (Figure 1G-H). The fact that cebp-2 is required for induction of the infection response genes that also require zip-2 for their induction supports the model that CEBP-2 and ZIP-2 work together as a transcription factor to mediate a transcriptional response to P. aeruginosa infection in C. elegans.
CEBP-2 and ZIP-2 promote resistance against P. aeruginosa infection
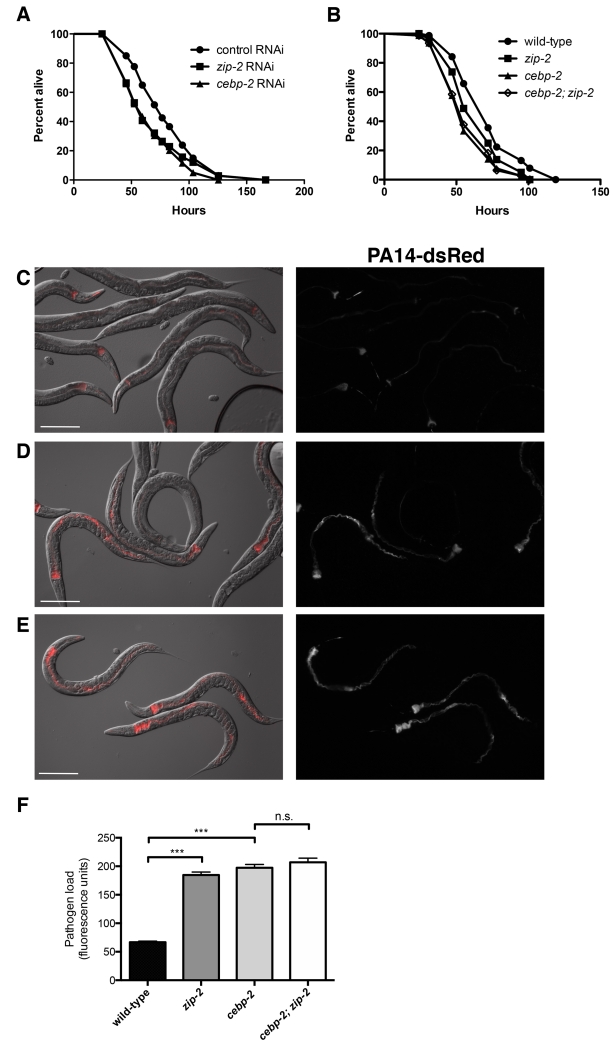
Our previous studies indicated that zip-2-mediated gene expression promotes a defense response, as zip-2-defective animals have modestly decreased survival upon infection with P. aeruginosa (Estes et al., 2010). To determine if cebp-2 is also important for defense against killing by P. aeruginosa, we tested the survival of cebp-2-defective animals upon infection with P. aeruginosa. Indeed, we found that cebp-2 RNAi-treated animals, like zip-2 RNAi-treated animals, had a modest but significant decrease in survival upon infection (Figure 2A), indicating that cebp-2, like zip-2, promotes host defense. In addition, we found that cebp-2 and zip-2 mutants had a modest decrease in survival upon PA14 infection (Figure 2B). cebp-2 mutants had slightly decreased survival compared to zip-2 mutants, perhaps due to the decreased overall health of these animals (see results below). If however, cebp-2 and zip-2 were working together to regulate gene expression that promotes survival upon PA14 infection, then a cebp-2;zip-2 mutant should not have a further decrease in survival compared to the cebp-2 single mutant alone. Consistent with this model, we found that cebp-2;zip-2 mutants did not have a greater decrease in survival compared to the single cebp-2 mutant (Figure 2B).
Figure 2. cebp-2 and zip-2 control pathogen burden and promote survival upon P. aeruginosa infection.
(A) Survival of RNAi control (L4440), zip-2 RNAi, and cebp-2 RNAi treated animals on P. aeruginosa PA14. zip-2 RNAi and cebp-2 RNAi treated animals were more susceptible to killing by PA14 than control (p < 0.0001 for each). (B) Survival of wild-type, zip-2(tm4248), cebp-2(tm5421), and cebp-2(tm5421);zip-2(tm4248) animals on PA14. zip-2(tm4248), cebp-2(tm5421), and cebp-2(tm5421);zip-2(tm4248) animals were more susceptible to killing by PA14 than wild-type (p < 0.001 for each); there was not a significant difference between cebp-2(tm5421), and cebp-2(tm5421);zip-2(tm4248), p=0.7. For (A) and (B), graph shows a representative assay of three independent replicates. (C-E) Images of (C) wild-type, (D) zip-2(tm4248) and (E) cebp-2(tm5421) animals after 16 hours of exposure to dsRed-expressing PA14. In each panel, the left image shows an overlay of Nomarski with red fluorescence and the right image shows red fluorescence alone. Scale bar, 200 μm. (F) Quantification of dsRed fluorescence levels in the intestine of wild-type, zip-2(tm4248), cebp-2(tm5421), and cebp-2(tm5421); zip-2(tm4248) animals after 16 hours of infection with dsRed-expressing PA14. Fluorescence was measured with a COPAS Biosort machine. Results shown are a representative assay of two independent replicates, with at least 500 animals measured for each sample. Error bars are SEM. ***, p < 0.001 with a two-tailed t test; n.s., not significant.
We next investigated whether zip-2 and cebp-2 promote increased survival upon infection with P. aeruginosa by restricting pathogen accumulation in the intestine, or by improving tolerance of the pathogen (Medzhitov et al., 2012). To distinguish between these possibilities we measured fluorescence levels of P. aeruginosa PA14-dsRed (Djonovic et al., 2013) in animals deficient for either zip-2 or cebp-2 at 16 hours post-infection and found that zip-2 and cebp-2-deficient animals accumulated significantly more intestinal PA14-dsRed than control animals (Figure 2C-F, S1A-D). This result indicates that both zip-2 and cebp-2 likely contribute to defense against killing by P. aeruginosa in part by controlling pathogen burden in the intestine. In addition, we found that the cebp-2;zip-2 double mutant had a similar increase in PA14-dsRed levels in the intestine as the single mutants (Figure 2F). Together these results indicate that zip-2 and cebp-2 act to promote resistance to the pathogen P. aeruginosa, perhaps functioning together as a heterodimeric transcription factor to induce genes that limit pathogen accumulation in the intestine and promote survival upon infection.
CEBP-2 and ZIP-2 are both expressed in intestinal nuclei during P. aeruginosa infection
Previous studies indicated that much of the P. aeruginosa-mediated induction of infection response genes such as irg-1 was due to pathogen-induced perturbation of core processes, including inhibition of mRNA translation (Dunbar et al., 2012; McEwan et al., 2012). Indeed, a key trigger of irg-1 induction appears to be the P. aeruginosa translational inhibitor Exotoxin A (ToxA), because heterologous expression of ToxA in non-pathogenic E. coli is sufficient to induce irg-1 mRNA expression in C. elegans, in a zip-2-dependent manner (McEwan et al., 2012). The induction of irg-1 mRNA upon infection is likely mediated by an increase in ZIP-2 protein levels, which could then serve to increase irg-1 transcription. Indeed, ZIP-2 protein levels increase upon P. aeruginosa infection and also with pharmacological inhibition of translation by the elongation inhibitor cycloheximide (Dunbar et al., 2012). Consistent with this model, we show here that ZIP-2 protein levels increase upon exposure to ToxA. Animals carrying a ZIP-2::GFP transgene had virtually no GFP expression when feeding on E. coli carrying the empty expression vector, but had strong GFP expression with nuclear localization in intestinal cells when feeding on E. coli expressing ToxA (Figure S2A-B,E).
We next investigated whether cebp-2 was required for the increased ZIP-2 protein levels seen after exposure to ToxA, because one possible explanation for the similar defects seen in zip-2 and cebp-2-deficient animals in response to P. aeruginosa infection is that cebp-2 is required for ZIP-2 protein expression. However, we found that cebp-2 was not required for ZIP-2 protein expression in response to ToxA, as cebp-2 RNAi-treated animals had robust induction of ZIP-2::GFP in intestinal nuclei after feeding on E. coli expressing ToxA (Figure S2C-E). This result indicates that the similar phenotypes of zip-2 and cebp-2-deficient animals are not due to regulation of ZIP-2 expression by CEBP-2.
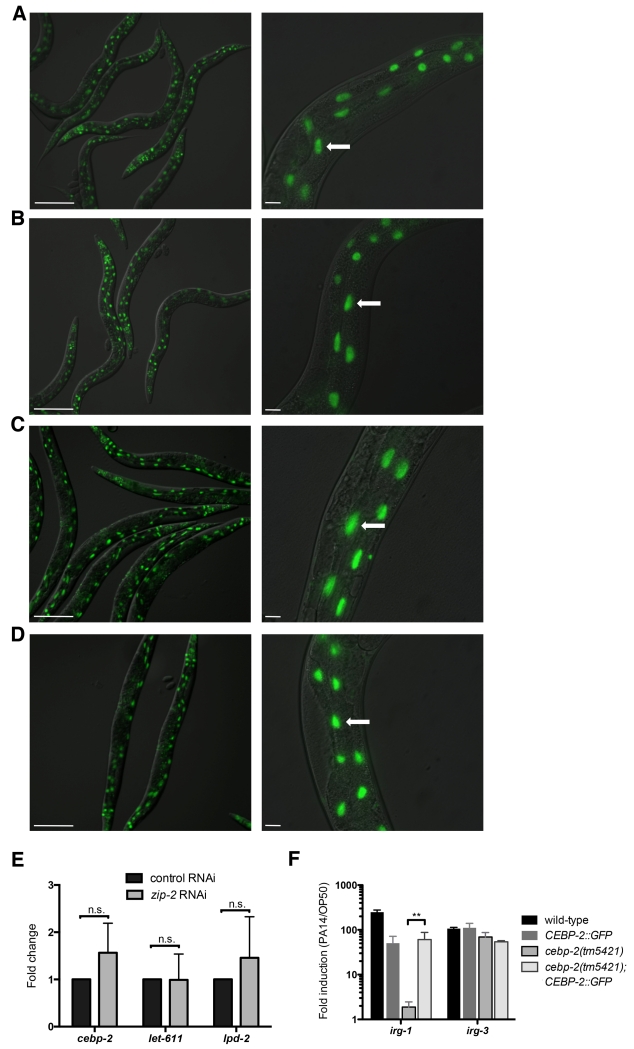
If CEBP-2 and ZIP-2 function together in the response to P. aeruginosa infection, these proteins should be expressed at the same time and in the same location. To test this model, we generated a transgene that contains 1.1 kb of genomic DNA upstream of the predicted cebp-2 ATG start site followed by the cebp-2 genomic coding region with GFP fused to the C terminus. We found that animals carrying this CEBP-2::GFP transgene express GFP broadly in somatic tissues including the intestine, with strong nuclear localization (Figure 3A). We did not see any change in CEBP-2::GFP expression or localization in animals infected with P. aeruginosa (Figure 3B), indicating that CEBP-2 is constitutively expressed, unlike ZIP-2. In addition, we did not find that zip-2 was required for CEBP-2 expression, as CEBP-2::GFP expression did not change after zip-2 RNAi treatment (Figure 3C-D). Furthermore, there was not a change in cepb-2 mRNA expression (or other genes in the cebp-2 operon) after zip-2 RNAi (Figure 3E), further supporting the conclusion that zip-2 is not required for cebp-2 expression.
Figure 3. CEBP-2::GFP is expressed in intestinal nuclei independently of infection and independently of ZIP-2.
(A) CEBP-2::GFP transgenic animals express GFP throughout the body with strong nuclear localization. (B) CEBP-2::GFP transgenic animals show nuclear GFP in the intestine when infected with P. aeruginosa. (C-D) CEBP-2::GFP transgenic animals have GFP in intestinal nuclei after treatment with (C) control RNAi or (D) zip-2 RNAi. (A-D) Images are overlays of green and Nomarski channels, with left panels imaged at 10× (scale bar, 200 μm) and right panels imaged at 40× (scale bar, 20 μm). White arrows indicate intestinal nuclei. (E) qRT-PCR comparison of gene expression in control RNAi (L4440) and zip-2 RNAi treated animals, shown as the fold change relative to L4440. (F) qRT-PCR measurement of gene expression shows that the CEBP-2::GFP transgene rescues the irg-1 induction defect of cebp-2(tm5421) mutants. For (E) and (F), results shown are the average of two independent biological replicates, error bars are SD. ** p < 0.01 with a two-tailed t test; n.s., not significant.
To confirm that the CEBP-2::GFP expression construct was functional, and thus likely to reflect endogenous expression of CEBP-2 protein, we analyzed whether it could rescue the cebp-2 mutant phenotype. Indeed, we found that this CEBP-2::GFP construct could rescue the defects in gene induction in response to PA14 in the cebp-2(tm5421) mutant (Figure 3F). This result also confirms that the cepb-2 gene induction phenotype in the cebp-2(tm5421) mutant strain is not due to a background mutation, and rather due to a mutation in the cebp-2 gene itself.
Taken together these results indicate that ZIP-2 and CEBP-2 do not function to regulate expression or localization of each other, and are both present in intestinal nuclei during infection with P. aeruginosa when there is robust gene induction of irg-1 and other infection response genes. These results are consistent with the model that ZIP-2 and CEBP-2 function together as a heterodimeric transcription factor to induce genes in the context of pathogen infection.
cebp-2 mutants have a decrease in body size and reproductive output
Although zip-2 and cebp-2 mutants appear to have nearly identical phenotypes in terms of their response to pathogen infection, they do differ in terms of overall health and vigor. In particular, cebp-2(tm5421) mutants had reduced body size compared to wild-type animals during normal well-fed conditions (Figure S3A), a defect that was rescued by the CEBP-2::GFP transgene. In contrast, zip-2(tm4248) mutants had no decrease in body size compared to wild-type animals. We also found that cebp-2(tm5421) mutants had a significantly reduced brood size compared to wild-type animals, while zip-2(tm4248) mutants had no reduction in brood size (Figure S3B). These differences in overall health between cebp-2 and zip-2 mutants may be due to CEBP-2 acting in a dimer with a different bZIP transcription factor to regulate growth and reproduction. Notably, mammalian C/EBP-gamma does not appear able to regulate transcription on its own, but rather partners with several different C/EBP factors to regulate distinct outputs (Tsukada et al., 2011). Indeed, CEBP-2 has been shown in vitro to interact with several other binding partners (Reinke et al., 2013), and recently was shown to have a role in fat metabolism as well (Xu et al., 2015), which may explain its effects on body size and reproduction.
CEBP-2 mediates a transcriptional response to inhibition of mRNA translation and inhibition of other core processes, which increase ZIP-2 protein expression
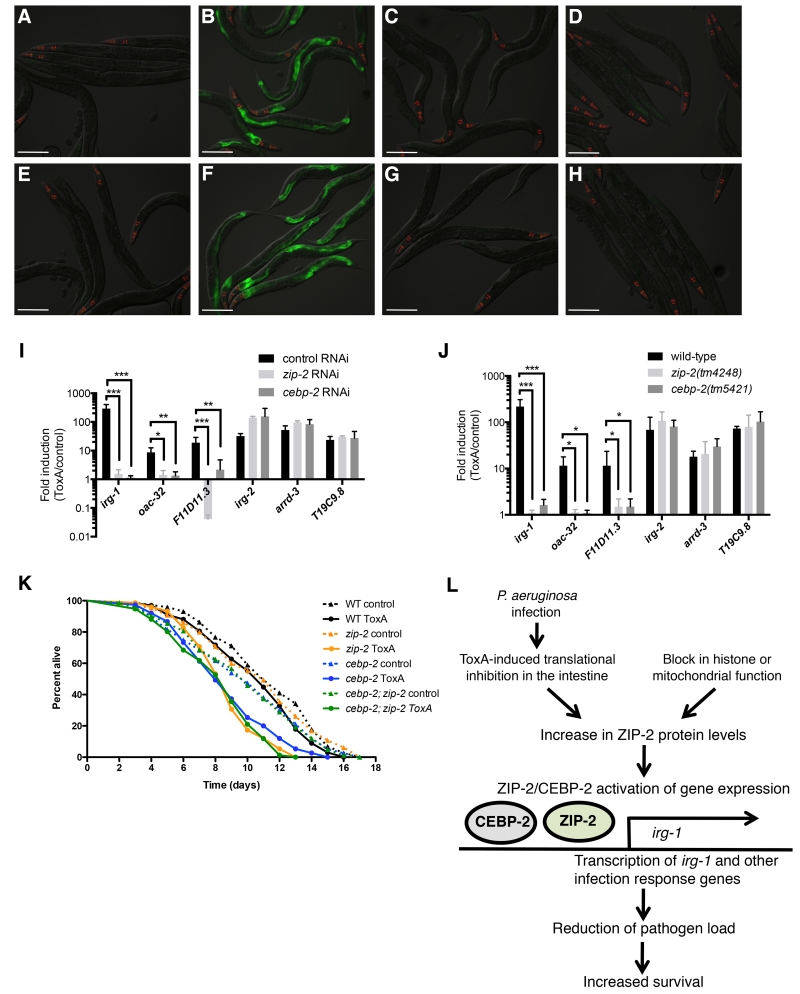
As mentioned above, previous studies indicate that ToxA-mediated translational inhibition appears to be responsible for a subset of the P. aeruginosa infection-induced transcriptional response in C. elegans. This gene induction is partially dependent on zip-2, and the zip-2 signaling pathway was shown to protect C. elegans from killing by ToxA. We therefore investigated whether cebp-2 is similarly required for ToxA-mediated gene induction and for survival after exposure to ToxA. We first examined whether cebp-2 is required for ToxA-induced expression of the irg-1p::GFP reporter. In both cebp-2 RNAi-treated animals and cebp-2(tm5421) mutant animals we found greatly reduced induction of irg-1p::GFP after exposure to ToxA as compared to control, similar to that seen in worms lacking zip-2 function (Figure 4A-H). This result indicates that cebp-2, like zip-2, regulates irg-1 induction after ToxA treatment. We next tested whether cebp-2 regulates endogenous irg-1 induction as well as induction of two other zip-2-dependent gene, oac-32 and F11D11.3, in response to ToxA exposure. We used qRT-PCR to measure RNA levels in cebp-2-deficient animals and found that cebp-2, like zip-2, is required for induction of both irg-1, oac-32 and F11D11.3 after ToxA-mediated translational inhibition (Figure 4I, J). We also tested an additional three genes whose induction in response to ToxA treatment does not require zip-2 (McEwan et al., 2012) and found that these genes were induced normally in cebp-2-deficient animals (Figure 4I, J).
Figure 4. cebp-2 is required for induction of gene expression and survival upon translational inhibition by ToxA.
(A-B) RNAi control (L4440) treated irg-1p::GFP animals after exposure to E. coli expressing either (A) the empty vector control or (B) ToxA. (C) zip-2 RNAi and (D) cebp-2 RNAi treated irg-1p::GFP animals after exposure to E. coli expressing ToxA. (E-F) Wild-type irg-1p::GFP animals after exposure to E. coli expressing either (E) the empty vector control or (F) ToxA. (G-H) irg-1p::GFP expression in (G) zip-2(tm4248) and (H) cebp-2(tm5421) animals after exposure to E. coli expressing ToxA. (A-H) Images are overlays of green, red, and Nomarski channels and were taken with the same camera exposure for all. Green is irg-1p::GFP, red is myo-2::mCherry expression in the pharynx as a marker for presence of the transgene. Scale bar, 200 μm. (I) qRT-PCR comparison of ToxA-induced gene expression in control RNAi (L4440), zip-2 RNAi, and cebp-2 RNAi treated animals. (J) qRT-PCR comparison of ToxA-induced gene expression in wild-type, zip-2(tm4248), and cebp-2(tm5421) animals. For (I) and (J), results shown are the average of two independent biological replicates, error bars are SD. *** p < 0.001, ** p < 0.01, * p < 0.05 with a two-tailed t test. n.s., not significant. (K) Survival of wild-type N2, zip-2(tm4248), cebp-2(tm5421), and cebp-2(tm5421); zip-2(tm4248) animals on E. coli expressing either ToxA or an empty expression vector starting at the L4 stage. Graph shows a representative assay of three independent replicates. N2 worms had no difference in life span when fed E. coli expressing either ToxA or the vector control (p = 0.7), while zip-2(tm4248), cebp-2(tm5421), and cebp-2(tm5421); zip-2(tm4248) animals had significantly shorter life spans on E. coli expressing ToxA as compared to the vector control (p < 0.001 for each). (L) Model for ZIP-2/CEBP-2 activation of gene expression after P. aeruginosa infection.
Surveillance pathways in C. elegans monitor not only mRNA translation, but also core processes mediated by mitochondria, the proteasome and transcriptional machinery (Bakowski et al., 2014; Dunbar et al., 2012; Liu et al., 2014; Melo and Ruvkun, 2012). Our previous screen found RNAi clones against not only translation factors, but also mitochondrial pathways and histones can induce irg-1p::GFP in a ZIP-2-dependent manner (Dunbar et al., 2012). Thus, we investigated whether cebp-2 was required for surveillance of these processes. Indeed, we found that RNAi against the histone H2A his-57 and the mitochondrial enzyme dihydrolipoamide dehydrogenase dlat-1 no longer induced irg-1p::GFP in cebp-2 mutants (Figure S4A-F). Thus, cebp-2 appears to be important for gene induction upon perturbation of multiple core processes.
Previously, we had found that either genetic or chemical inhibition of mRNA translation caused an increase in ZIP-2 protein levels (Dunbar et al., 2012), explaining how translational inhibition could lead to an induction of ZIP-2-dependent gene expression. Here we extend those analyses to blockade of other core processes, such as mitochondrial function and histone function. In particular we found that RNAi against the histone H2A his-57 and the mitochondrial enzyme dihydrolipoamide dehydrogenase dlat-1 caused an increase in ZIP-2::GFP protein expression (Figure S4G-L). Thus, perturbation of several core processes appears to increase ZIP-2 protein expression, where it could act together with the constitutively expressed CEBP-2 to promote a transcriptional response to xenobiotic insults.
CEBP-2 mounts a protective response against ToxA-mediated killing
Previous studies found that wild-type animals mount a defense response against ToxA, as ToxA treatment does not compromise survival unless immune pathways are defective (McEwan et al., 2012). zip-2 mutants have a substantially shorter life span when fed E. coli expressing ToxA as compared to control. To determine whether cebp-2 is important for defense against killing by ToxA, we exposed cebp-2(tm5421) mutant animals to E. coli expressing either a vector control or ToxA. We found that cebp-2 mutants, like zip-2 mutants, have greatly decreased survival upon treatment with ToxA, with relatively normal survival on the vector control (Fig. 4K). Thus, cebp-2 is required for the defense response against the pathogen-derived toxin ToxA. Furthermore, we found that the cebp-2;zip-2 double mutant had a similar decrease in survival upon ToxA exposure as the cebp-2 and zip-2 single mutants (Fig. 4K). Together these results support the model that cebp-2 is acting together with zip-2 to mount a protective response against ToxA-mediated killing (Fig. 4L).
Concluding remarks: CEBP-2 and ZIP-2 act together in surveillance immunity in C. elegans
A growing theme in animal innate immunity is that hosts are able to discriminate pathogens from other microbes through the use of surveillance pathways that monitor disruption of host processes commonly targeted by pathogens. This ‘effector-triggered’ immunity is critical for epithelial cells that encounter a wide variety of microbial species. In addition to the responses to the bacterial pathogen P. aeruginosa described here and in other publications, recent findings suggest that defense against natural eukaryotic pathogens in C. elegans can also be triggered by perturbing core processes (Bakowski et al., 2014). Our discovery that CEBP-2 and ZIP-2 act as a potential heterodimeric transcription factor in surveillance immunity against P. aeruginosa in C. elegans sheds light on this process and also provides a mammalian connection to be explored. CEBP-2 is the ortholog of C/EBP-gamma in mammals, which is a bZIP transcription factor that heterodimerizes with several other bZIP transcription factors to regulate upregulation of cytokines such as IL-6 and IL-8 in response to classic PAMPs like LPS (Gao et al., 2002), although it has not yet been shown to play a role in effector-triggered immunity. Interestingly, interindividual variation in C/EBP-γ transcript expression levels has been implicated as a risk factor for altered severity of lung disease in cystic fibrosis (Gu et al., 2009) – a genetic disease in which chronic P. aeruginosa pneumonia is a pathological hallmark. Future studies could investigate the role that C/EBP-gamma and its binding partners play in surveillance immunity in mammals in order to better understand how animals discriminate pathogens from other microbes to fight off infection.
Experimental Procedures
RNAi experiments
RNAi experiments were performed as described (Estes et al., 2010; Kamath et al., 2003). Overnight cultures of RNAi feeding clones were seeded onto RNAi plates and incubated at 25°C for 1 day. Synchronized L1 stage animals were fed RNAi for two or three days at 20°C. All experiments with feeding RNAi used an unc-22 positive control RNAi clone, which resulted in twitching animals in all experiments. See Supplemental Experimental Procedures for further details.
Pathogen infection experiments
Pathogen infection experiments were performed as described (Troemel et al., 2006). Briefly, overnight cultures of P. aeruginosa strain PA14 were seeded onto SK plates, then incubated at 37°C for 24 h, followed by 25°C for 24 h. Animals at the L4 stage were washed onto plates and were harvested 4 h later for qRT-PCR experiments, or viewed 16-20 h later for GFP experiments.
ToxA assays
ToxA and vector control assay plates were prepared as described (McEwan et al., 2012). Overnight cultures of E. coli were diluted 1:20, grown for 2 h at 37°C, induced with 0.84 M IPTG, and grown another hour at 37°C. Concentrated bacteria (10×) were seeded on NGM plates containing 5 mM IPTG and 1mM carbenicillin and used immediately. Animals at the L4 stage were washed onto assay plates and were harvested 24 h later for qRT-PCR experiments, or viewed 18-24 h later for GFP experiments.
ToxA and P. aeruginosa survival assays
ToxA survival assays and P. aeruginosa slow-killing experiments were performed as described, with the addition of FUDR (100 μg/ml) to inhibit progeny formation (McEwan et al., 2012; Troemel et al., 2006). Thirty to fifty L4 stage animals were transferred to assay plates prepared as described above and incubated at 25°C, using three plates per strain in each experiment. Survival was monitored over time until all animals had died.
Gene expression analysis
RNA extraction, reverse transcription and qRT-PCR were performed as described (Troemel et al., 2006). qRT-PCR primer sequences are available upon request. For all qRT-PCR experiments, each biological replicate was measured in duplicate and normalized to the control gene nhr-23, which did not change expression upon infection or exposure to ToxA.
Supplementary Material
Acknowledgements
Some C. elegans strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank the National BioResource Project for the cebp-2(tm5421) deletion strain. Supported by NIH T32 GM07240 training grant to TLD; NIH R01AI087528, R01GM114139, the Searle Scholars Program, Packard Foundation, and Burroughs Wellcome Fund fellowships to ERT; NIH R01AG040061 to CMH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
KCR, TLD, AMN, CMH and ERT designed the experiments. KCR, TLD, and AMN performed the experiments. KCR and ERT wrote the manuscript. CMH and ERT secured funding.
Supplemental Information contains four figures, and Supplemental Experimental Procedures.
References
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nature immunology. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TA, Lopez-Moyado IF, Rifkin SA, Cuomo CA, Troemel ER. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS pathogens. 2014;10:e1004200. doi: 10.1371/journal.ppat.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddoe T, Paton AW, Le Nours J, Rossjohn J, Paton JC. Structure, biological functions and applications of the AB5 toxins. Trends Biochem Sci. 2010;35:411–418. doi: 10.1016/j.tibs.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi Y, Niggeweg R, Opitz B, Vogelsgesang M, Hippenstiel S, Wilm M, Aktories K. Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16953–16958. doi: 10.1073/pnas.0601562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas V, Miyake J, Balsley H, Roark J, Telles S, Leeds S, Zurita I, Breitbart M, Bartlett D, Azam F, et al. Widespread occurrence of phage-encoded exotoxin genes in terrestrial and aquatic environments in Southern California. FEMS Microbiol Lett. 2006;261:141–149. doi: 10.1111/j.1574-6968.2006.00345.x. [DOI] [PubMed] [Google Scholar]
- Cohen LB, Troemel ER. Microbial pathogenesis and host defense in the nematode C. elegans. Current opinion in microbiology. 2015;23:94–101. doi: 10.1016/j.mib.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djonovic S, Urbach JM, Drenkard E, Bush J, Feinbaum R, Ausubel JL, Traficante D, Risech M, Kocks C, Fischbach MA, et al. Trehalose biosynthesis promotes Pseudomonas aeruginosa pathogenicity in plants. PLoS pathogens. 2013;9:e1003217. doi: 10.1371/journal.ppat.1003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar TL, Yan Z, Balla KM, Smelkinson MG, Troemel ER. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell host & microbe. 2012;11:375–386. doi: 10.1016/j.chom.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2153–2158. doi: 10.1073/pnas.0914643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Parkin S, Johnson PF, Schwartz RC. C/EBP gamma has a stimulatory role on the IL-6 and IL-8 promoters. The Journal of biological chemistry. 2002;277:38827–38837. doi: 10.1074/jbc.M206224200. [DOI] [PubMed] [Google Scholar]
- Gu Y, Harley IT, Henderson LB, Aronow BJ, Vietor I, Huber LA, Harley JB, Kilpatrick JR, Langefeld CD, Williams AH, et al. Identification of IFRD1 as a modifier gene for cystic fibrosis lung disease. Nature. 2009;458:1039–1042. doi: 10.1038/nature07811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglewski BH, Liu PV, Kabat D. Mechanism of action of Pseudomonas aeruginosa exotoxin Aiadenosine diphosphate-ribosylation of mammalian elongation factor 2 in vitro and in vivo. Infection and immunity. 1977;15:138–144. doi: 10.1128/iai.15.1.138-144.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R, Purdy AE, Fieldhouse RJ, Kimber MS, Bartlett DH, Merrill AR. Cholix toxin, a novel ADP-ribosylating factor from Vibrio cholerae. The Journal of biological chemistry. 2008;283:10671–10678. doi: 10.1074/jbc.M710008200. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- Lee MS, Kim MH, Tesh VL. Shiga toxins expressed by human pathogenic bacteria induce immune responses in host cells. Journal of microbiology. 2013;51:724–730. doi: 10.1007/s12275-013-3429-6. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Girardin SE. Translation inhibition and metabolic stress pathways in the host response to bacterial pathogens. Nat Rev Microbiol. 2013;11:365–369. doi: 10.1038/nrmicro3029. [DOI] [PubMed] [Google Scholar]
- Lemichez E, Barbieri JT. General aspects and recent advances on bacterial protein toxins. Cold Spring Harbor perspectives in medicine. 2013;3:a013573. doi: 10.1101/cshperspect.a013573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DL, Kirienko NV, Ausubel FM. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell host & microbe. 2012;11:364–374. doi: 10.1016/j.chom.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science (New York, NY. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 2012;149:452–466. doi: 10.1016/j.cell.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr I, Sonenberg N. Host translation at the nexus of infection and immunity. Cell host & microbe. 2012;12:470–483. doi: 10.1016/j.chom.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Sperandio V. Shiga toxin in enterohemorrhagic E.coli: regulation and novel anti-virulence strategies. Front Cell Infect Microbiol. 2012;2:81. doi: 10.3389/fcimb.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappenheimer AM., Jr. Diphtheria toxin. Annual review of biochemistry. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- Parkin SE, Baer M, Copeland TD, Schwartz RC, Johnson PF. Regulation of CCAAT/enhancer-binding protein (C/EBP) activator proteins by heterodimerization with C/EBPgamma (Ig/EBP) The Journal of biological chemistry. 2002;277:23563–23572. doi: 10.1074/jbc.M202184200. [DOI] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 2014;516:414–417. doi: 10.1038/nature13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamuthiah R, Mylonakis E. Effector triggered immunity: Activation of innate immunity in metazoans by bacterial effectors. Virulence. 2014;5 doi: 10.4161/viru.29091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke AW, Baek J, Ashenberg O, Keating AE. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science (New York, NY. 2013;340:730–734. doi: 10.1126/science.1233465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria NM, Huang JC, Dubery IA. Self/nonself perception in plants in innate immunity and defense. Self Nonself. 2010;1:40–54. doi: 10.4161/self.1.1.10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan MW. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nature reviews Immunology. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- Stuart LM, Paquette N, Boyer L. Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nature reviews. 2013;13:199–206. doi: 10.1038/nri3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS genetics. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54:6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Xu XY, Hu JP, Wu MM, Wang LS, Fang NY. CCAAT/enhancer-binding protein CEBP-2 controls fat consumption and fatty acid desaturation in Caenorhabditis elegans. Biochem Biophys Res Commun. 2015 doi: 10.1016/j.bbrc.2015.10.106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.