SUMMARY
Ribosome-footprint profiling provides genome-wide snapshots of translation, but technical challenges can confound its analysis. Here, we use improved methods to obtain ribosome-footprint profiles and mRNA abundances that more faithfully reflect gene expression in Saccharomyces cerevisiae. Our results support proposals that both the beginning of coding regions and condos matching rare tRNAs are more slowly translated. They also indicate that emergent polypeptides with as few as three basic residues within a 10-residue window tend to slow translation. With the improved mRNA measurements, the variation attributable to translational control in exponentially growing yeast was less than previously reported, and most of this variation could be predicted with a simple model that considered mRNA abundance, upstream open reading frames, cap-proximal structure and nucleotide composition, and lengths of the coding and 5’-untranslated regions. Collectively, our results provide a framework for executing and interpreting ribosome-profiling studies and reveal key features of translational control in yeast.
INTRODUCTION
Although most cellular mRNAs use the same translation machinery, the dynamics of translation can vary between mRNAs and within mRNAs, often with functional consequences. For example, strong secondary structure within the 5' untranslated region (UTR) of an mRNA can impede the scanning ribosome, thereby reducing the rate of protein synthesis (Kozak, 1986; Andersson and Kurland, 1990; Bulmer, 1991; Kudla et al., 2009; Tuller et al., 2010; Plotkin and Kudla, 2011; Tuller et al., 2011; Ding et al., 2012; Bentele et al., 2013). The accessibility of the 5' cap (Godefroy-Colburn et al., 1985; Richter and Sonenberg, 2005) and the presence of small ORFs within 5' UTRs referred to as upstream ORFs (uORFs) (Kozak, 1986; Ingolia et al., 2009; Brar et al., 2012; Zur and Tuller, 2013) can also modulate the rate of translation initiation (Sonenberg and Hinnebusch, 2009). Likewise, codon choice, mRNA structure, and the identity of the nascent polypeptide can influence elongation rates (Varenne et al., 1984; Brandman et al., 2012). In addition, differences in elongation rates can influence co-translational protein folding, localization of the mRNA or protein, and in extreme cases the rate of protein production (Kimchi-Sarfaty et al., 2007; Xu et al., 2013; Zhou et al., 2013). Finally, stop-codon read-through can introduce alternative C-terminal regions that affect protein stability, localization, or activity (Dunn et al., 2013). Despite known examples of regulation at each of these stages of translation, translation is largely controlled at initiation, which is rate limiting for most mRNAs (Andersson and Kurland, 1990; Bulmer, 1991; Chu and von der Haar, 2012; Shah et al., 2013).
Variation in protein abundances observed in yeast cells largely reflects variation in mRNA abundances, indicating that much of gene regulation occurs at the level of mRNA synthesis and decay (Greenbaum et al., 2003; Csardi et al., 2015). However, differences in translation rates also contribute. Studies using microarrays for global polysome profiling indicate that ribosome densities for different mRNAs vary over a 100-fold range (from 0.03 to 3.3 ribosomes per 100 nucleotides), indicating extensive translation control in Saccharomyces cerevisiae (Arava et al., 2003). More recently, the use of ribosome-footprint profiling has enabled transcriptome-wide analyses of translation using high-throughput sequencing, which again suggested a nearly 100-fold range of translational efficiencies (TEs) in log-phase yeast (Ingolia et al., 2009).
The ribosome-profiling method has itself undergone refinements over the last few years. Here, we build upon these advances and present improved ribosome-profiling and mRNA-seq datasets for log-phase yeast. Comparisons to many previous datasets reveal protocol-specific biases that can influence interpretation of ribosome-profiling experiments. With these insights, we then address several classical questions and on-going debates in protein translation, such as the influence of tRNA abundances and nascent-peptide sequence on elongation rates. Our improved datasets also constrict the differences in TEs observed in log-phase yeast, such that the gene-to-gene variability that does remain can be largely predicted using a simple statistical model that considers only six features of the mRNAs.
RESULTS
Less perturbed ribosome footprints reveal the dynamics of elongation
Protocols for analyzing polysome profiles or capturing ribosome footprints (referred to as ribosome-protected fragments, or RPFs) typically involve treating cells with the elongation inhibitor cycloheximide (CHX) to arrest the ribosomes prior to harvest (Ingolia et al., 2009; Gerashchenko et al., 2012; Zinshteyn and Gilbert, 2013; Artieri and Fraser, 2014; McManus et al., 2014). An advantage of CHX pre-treatment is that it prevents the run-off of ribosomes that can otherwise occur during harvesting (Ingolia et al., 2009). However, this treatment can also have some undesirable effects. Because CHX does not inhibit translation initiation or termination, pre-treatment of cultures leads to ribosome accumulation at start codons and depletion at stop codons (Ingolia et al., 2011; Guydosh and Green, 2014; Pelechano et al., 2015). In addition, because CHX binding to the 80S ribosome is both non-instantaneous and reversible, the kinetics of CHX binding and dissociation presumably allow newly initiated ribosomes to translocate beyond the start codon. Another possible effect of CHX treatment is that ribosomes might preferentially arrest at specific codons that do not necessarily correspond to codons that are more abundantly occupied by ribosomes in untreated cells. Although effects of CHX pre-treatment have minimal consequence for analyses performed at the gene level, i.e., comparisons of the same gene in different conditions, or comparisons between different genes after discarding reads in the 5' regions of ORFs, CHX pre-treatment may have severe consequences for analyses that require single-codon resolution.
The potential effects of CHX pre-treatment near the start codon have been discussed since the introduction of ribosome profiling, where an alternative protocol with flash-freezing and no CHX pre-treatment is also presented (Ingolia et al., 2009). Indeed, many recent ribosome-profiling experiments avoid CHX pre-treatment (Gardin et al., 2014; Gerashchenko and Gladyshev, 2014; Guydosh and Green, 2014; Jan et al., 2014; Lareau et al., 2014; Pop et al., 2014; Williams et al., 2014; Nedialkova and Leidel, 2015). However, consensus on the ideal protocol has not yet been reached, in part because the influence of alternative protocols on the interpretation of translation dynamics has not been systematically analyzed.
Here, we implemented a filtration and flash-freezing protocol to rapidly harvest yeast cultures. Importantly, this protocol minimized the time the cells experience starvation, which leads to rapid ribosome run-off (Ingolia et al., 2009; Gardin et al., 2014; Guydosh and Green, 2014). The protocol did include CHX in the lysis buffer to inhibit elongation that might occur during RNase digestion, although we doubt this precaution was necessary.
The original ribosome-profiling protocol also used cDNA circularization (Ingolia et al., 2009), while some subsequent protocols instead ligate to a second RNA adapter prior to cDNA synthesis (Guo et al., 2010). Both approaches can introduce sequence-specific biases at the 5' ends of reads, which are not expected to influence results of analyses performed at the level of whole mRNAs but might influence results of codon-resolution analyses. Borrowing from methods developed for small-RNA sequencing (Jayaprakash et al., 2011; Sorefan et al., 2012), we minimized these biases by ligating a library of adapter molecules that included all possible sequences at the eight nucleotides nearest to the ligation junction. Using this ligation protocol with a rapidly harvested, flash-frozen sample, we generated 74.3 million RPFs for log-phase yeast.
The 5' ramp of ribosomes
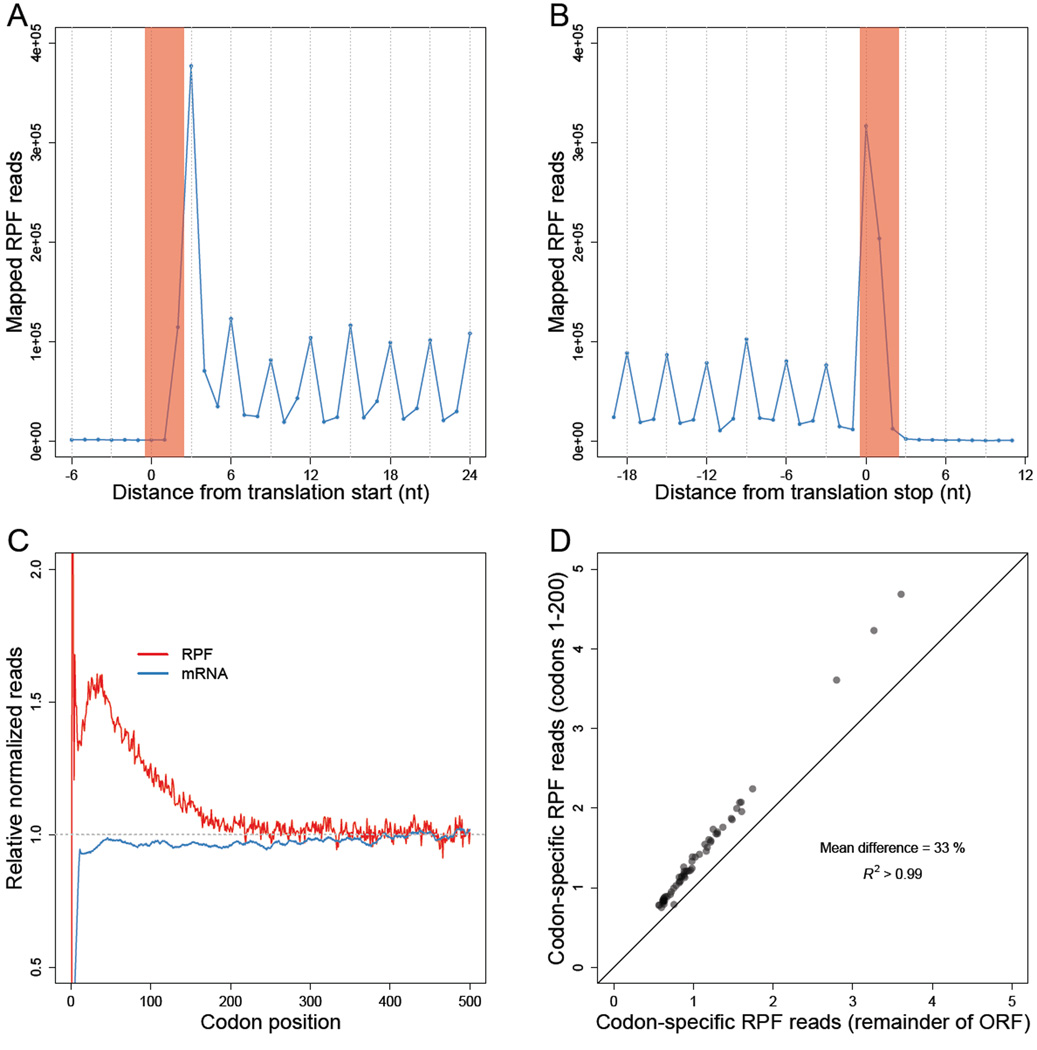
Using the 5' ends of RPFs, we inferred the codon at the A site of each footprint (Ingolia et al., 2009). Analysis of all mapped reads revealed the expected three-nucleotide periodicity along the ORFs, as well as ribosome accumulation at the start and stop codons (Figure 1A–B).
Figure 1. Less perturbed RPFs reveal a codon-independent 5' ramp.
(A–B) Metagene analyses of RPFs. Coding sequences were aligned by their start (A) or stop (B) codons (red shading). Plotted are the numbers of 28–30-nt RPF reads with the inferred ribosomal A site mapping to the indicated position along the ORF.
(C) Metagene analyses of RPFs and RNA-seq reads (mRNA). ORFs with at least 128 total mapped reads between ribosome-profiling (red) and RNA-seq (blue) samples were individually normalized by the mean reads within the ORF, and then averaged with equal weight for each codon position across all ORFs (e’j Eqn S10 and h’j Eqn S14).
(D) Comparison of codon-specific RPFs as a function of the 5' ramp. For each of the codons, densities of RPFs with ribosomal A sites mapping to that codon were calculated using either only the ramp region of each ORF (codons 1–200) or the remainder of each ORF (v5k Eqn S16 and v3k Eqn S17, respectively). The diagonal line indicates the result expected for no difference between the two regions.
See also Figure S1.
To examine the global landscape of 80S ribosomes, we averaged the position-specific RPF densities of individual genes into a composite metagene, in which each gene was first normalized for its overall density of RPFs (i.e., RPKM of RPFs) and then weighted equally in the average (Eqn S10). A small excess of ribosome density was observed in the first ~200 codons compared to the remainder of the ORF (Figure 1C). The trend towards decreasing ribosome density with codon position was also evident on a gene-by-gene basis: 82% of genes exhibited declining raw RPF reads along their entire gene-length, based on linear-regression of RPF reads with codon position (binomial test, p < 10−15), with the 5'-to-3' decrease in ribosome densities for a gene of average length (~500 codons) averaging ~43%.
Much larger 5' ramps are observed in other studies (Ingolia et al., 2009; Gerashchenko et al., 2012; Zinshteyn and Gilbert, 2013; Artieri and Fraser, 2014; Guydosh and Green, 2014; McManus et al., 2014), which is attributed to their use of CHX pre-treatment (Ingolia et al., 2009; Gerashchenko and Gladyshev, 2014) (Figure S1). However, CHX pre-treatment cannot explain the more modest ramp observed in our dataset, since our protocol did not involve such treatment.
The 5' ramp of ribosomes has previously been attributed to slower elongation due to preferential use of codons corresponding to low-abundance cognate tRNAs at the 5’ ends of genes (Tuller et al., 2010). To determine the contribution of codon usage, we tested whether differences in RPF densities between the 5' and 3' ends of genes depended on codon choice. Surprisingly, for each of the 61 sense codons, the average density of RPFs was 33% greater on average when the codon fell within the first 200 codons of an ORF (Figure 1D and S1), which showed that differential codon usage alone cannot explain the 5' ramp. Consistent with these experimental results, simulation of protein translation in a yeast cell, using a whole-cell stochastic model of yeast translation (Shah et al., 2013), indicated that codon ordering could account for at most a 20% ramp (Figure S1). Thus, codon ordering might explain some of the ~60% ramp observed in our dataset, but the majority of this ramp is likely caused by other mechanisms (see Discussion).
Codon-specific elongation dwell times are inversely correlated with tRNA abundances
The 61 sense codons varied in their average RPF densities by more than 6 fold (Figure 1D), indicating that different codons are decoded at different rates. Molecular biologists have long assumed that such differences in elongation rates are caused by corresponding differences in the cellular abundances of cognate tRNAs (Andersson and Kurland, 1990; Bulmer, 1991). Several early experiments provide empirical support for this view (Varenne et al., 1984; Sorensen and Pedersen, 1991), but early analyses of ribosome-profiling results do not find any relationship between ribosome density and cognate tRNA abundance expected from this model (Ingolia et al., 2011; Li et al., 2012; Qian et al., 2012; Charneski and Hurst, 2013; Zinshteyn and Gilbert, 2013). However, the datasets analyzed in these studies were all from experiments that used CHX pre-treatment.
At least three considerations help explain why CHX pre-treatment would disrupt the correlation between tRNA abundances and measured ribosome densities at the A site. The first is that CHX, once bound to a ribosome, allows for an additional round of elongation before halting ribosomes (Schneider-Poetsch et al., 2010; Gardin et al., 2014; Lareau et al., 2014), which alone would remove any correlation at the A-site. Second, CHX binding is reversible, and at concentrations typically used in ribosome-profiling protocols, additional rounds of elongation might occur between CHX-binding events. Third, CHX prevents translocation of the ribosome by binding to the E-site, with space for a deacylated tRNA (Schneider-Poetsch et al., 2010), and thus CHX binding affinity presumably varies with features of the E site and the tRNA in it. Thus, in the presence of CHX pre-treatment, the ribosome density at a site is likely more a function of the on and off rates of CHX binding than a function of differential isoaccepting tRNA availability. Indeed, recent analyses of profiling results obtained without CHX pre-treatment have observed modest correlations between tRNA abundances and ribosome-densities at the A-site (Gardin et al., 2014; Lareau et al., 2014).
When examining earlier ribosome-profiling datasets, we found that whenever CHX pre-treatment was employed, the relationship between ribosome occupancy and tRNA abundance was both insignificant (p > 0.05) and in the opposite direction than expected (Figure S2C–E). Moreover, the concordance between these CHX pre-treatment datasets indicated a systematic bias (Figure S2), suggesting that an orthogonal set of mRNA sequence biases influence CHX binding. In contrast, for every dataset without CHX pre-treatment, we found that ribosome densities were inversely correlated with tRNA abundances (Figure S2C–E).
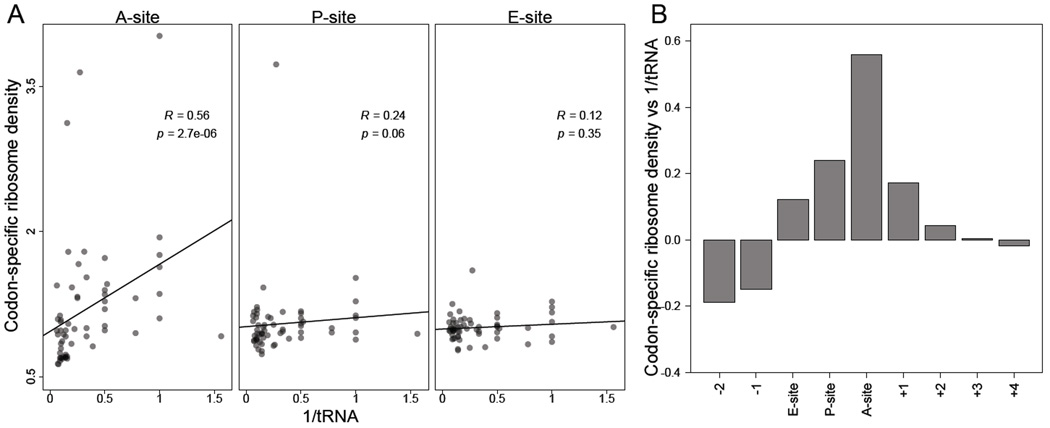
In our dataset, we found that codon-specific excess ribosome densities (vk, Eqn S19) were strongly anti-correlated with cognate tRNA abundances, as estimated by copy numbers of tRNA genes and wobble parameters (Figure 2A–B). This strong anti-correlation was also observed with direct estimates of tRNA abundances obtained from our RNA-seq measurements (Figure S2A, Table S1). As expected, the correlation was specific to the codon within the A site, with residual correlations at the P and E sites, which were potentially caused by some 5' heterogeneity of RPFs.
Figure 2. Codons corresponding to lower-abundance tRNAs are decoded more slowly.
(A) Correlation between codon-specific excess ribosome densities and cognate tRNA abundances. Codons within RPFs were assigned to the A-, P-, and E-site positions based on the distance from the 5' ends of fragments, and codonspecific excess ribosome densities were calculated (vk, Eqn S19). Cognate tRNA abundances for each codon were estimated using the genomic copy numbers of iso-accepting tRNAs and wobble parameters (Table S2). Spearman R values are shown, with their significance (p values).
(B) The correlations of codon–tRNA abundance at different positions relative to the A site. Analysis was as in (A) using varying offsets from the A-site position within RPFs (x axis) to calculate Spearman correlations (y axis).
See also Figures S2–3 and Tables S1–2.
Taken together, these results strongly support the idea that differential cognate tRNA abundances influence differential elongation times of codons in the absence of CHX. Without CHX pre-treatment we also observed widespread pausing after polybasic tracts (Figure S3) but not at P-site proline codons (Figure S2), which has been the subject of some debate (Supplemental Text).
Slower elongation at regions encoding inter-domain linkers
The modulation of elongation rates by either tRNA abundances (Figure 2A) or polybasic stretches (Figure S3) might influence the kinetics of co-translational folding. Indeed, slower elongation rates within inter-domain linkers relative to the adjacent domains is reported to coordinate co-translational folding of nascent polypeptides (Thanaraj and Argos, 1996; Kimchi-Sarfaty et al., 2007; Pechmann and Frydman, 2013). However, systematic experimental evidence for such differences in elongation rates has been lacking.
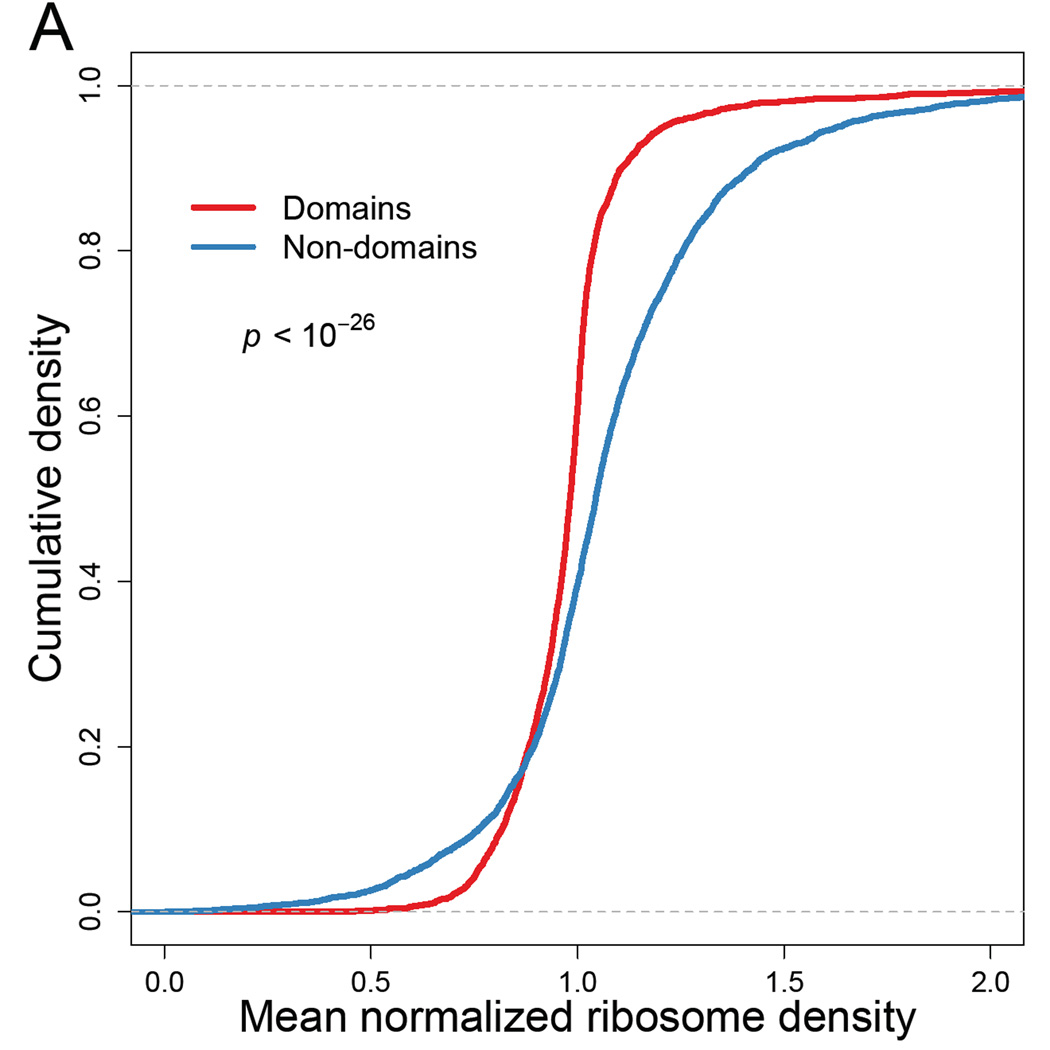
To examine whether our ribosome-profiling data reveals such differences, we first used InterProScan classifications (Jones et al., 2014) based on the Superfamily database (Wilson et al., 2009) to partition coding sequences into domain and linker regions. We then calculated the mean normalized RPF densities (zij, Eqn S7) for codons within the domain- and linker-encoding regions and found significantly lower densities in regions of genes that fell within domains compared those that fell outside of domains (Figure 3A, mean difference 0.094, paired t-test, p < 10−26). To eliminate any influence of the 5' ramp, we repeated the analysis excluding the first 200 codons. Although the size of the effect diminished (mean diff = 0.029), the difference in mean ribosome densities remained significant (p = 0.0002), indicating that the 5' ramp was not solely responsible for lower ribosome densities within domains (Figure S4A).
Figure 3. Elongation dynamics correlate domain architecture.
(A) Cumulative distributions of normalized ribosome densities within and outside of protein-folding domains. Mean normalized RPF densities (zij, Eqn S7) for codons within the domain-encoding and non-domain-encoding regions were individually calculated for each ORF. Domain assignments were based on InterProScan classifications (Jones et al., 2014) obtained from the Superfamily database (Wilson et al., 2009). Statistical significance was evaluated using paired t-test (p < 10−26).
See also Figure S4.
The trend towards relatively lower ribosome densities in domain regions held even when restricted to each individual amino acid, with the exceptions of cysteine residues and the single-codon-encoded methionine and tryptophan residues (Figure S4). Thus, differences in amino-acid content between domains and linkers could not account for the observed differences in bound ribosome densities. Moreover, for 54 out of 61 sense codons, we found significantly higher ribosome densities in domains compared to linkers (one-sided t-test, p < 0.05). For 26 out of 61 codons, we found significantly higher ribosome densities in domains even after excluding the first 200 codons (one-sides t-test, p < 0.05). This result implied that differences in synonymous codon usage between domain and linker regions cannot alone account for the differences in ribosome densities. One possible mechanism for differential ribosome occupancy, independent of codon usage, is differential recruitment of chaperones and their associated effects on co-translational folding (Ingolia, 2014).
Similar results for densities in domain and linker regions were obtained when using InterProScan classifications (Bateman et al., 2002) instead of the Superfamily database (Figure S4B). Finally, consistent with other computational analyses (Pechmann and Frydman, 2013), differences in elongation rate were found at the level of protein secondary structures as well: Regions corresponding to helices and sheets exhibited significantly lower RPF densities than regions corresponding to loops (Figure S4C). Taken together, these results provided systematic empirical support for the claim that co-translational folding requirements influence elongation rates. Nonetheless, the magnitude of this signal was very small, suggesting that slower inter-domain elongation either has very little impact or impacts very few genes.
Estimates of protein-synthesis rates
Our results thus far indicated that the ribosome density at a given codon position is influenced by the abundance of cognate tRNAs and whether the codon is immediately downstream of a polybasic stretch, falls within a protein domain, or lies in the 5’ region of the ORF. The non-uniform ribosome densities along individual ORFs imply that the overall RPF density on each gene (i.e., RPKM of RPFs) does not directly reflect the rate of protein synthesis (Li et al., 2014). For example, the RPF densities of genes enriched in more slowly elongated codons would tend to overestimate their protein-synthesis rates; and the same would be true for shorter ORFs. To more accurately quantify the protein-synthesis rates of individual genes from RPF densities, we used empirically derived correction factors to account for the position- and codon-specific effects we observed (fj, Eqn S23). With these correction factors, the ~74.3 million sequenced RPFs enabled reliable estimates of protein-synthesis rates for 4839 genes (Eqn S28).
Accurate measurement of yeast mRNA abundances
In addition to improving measurements of ribosome densities, we sought to improve measurements of mRNA abundances, which is also critical for accurately quantifying translational control. Prior experiments have typically measured yeast mRNA abundances by performing RNA-seq on poly(A)-selected RNA (Ingolia et al., 2009; Gerashchenko et al., 2012; Zinshteyn and Gilbert, 2013; Artieri and Fraser, 2014; Guydosh and Green, 2014; McManus et al., 2014). However, poly(A) selection might bias mRNA-abundance measurements. For example, mRNAs that lack a poly(A) tail of sufficient length to stably hybridize to oligo(dT) might not be as efficiently recovered. Although S. cerevisiae is not known to contain translated mRNAs that altogether lack a poly(A) tail, the lengths of poly(A) tails found on S. cerevisiae mRNAs are relatively short, with a median length of 27 nt (Subtelny et al., 2014). Another source of potential bias in poly(A)-selection is partial recovery of mRNAs endonucleolytically cleaved during RNA isolation or poly(A)-selection. The 5' fragments resulting from mRNA cleavage are not recovered by poly(A) selection, which causes a 3' bias in the resulting RNA-seq data (Nagalakshmi et al., 2008). Indeed, analyses of published RNA-seq datasets from ribosome-profiling studies revealed a severe 3' bias in poly(A)-selected RNA-seq reads, ranging from 19–130% excess reads (Eqn S15) (Figure S5). Because longer mRNAs have a higher probability of being cleaved, the abundances of longer mRNAs might be systematically underestimated by poly(A) selection (Table S3).
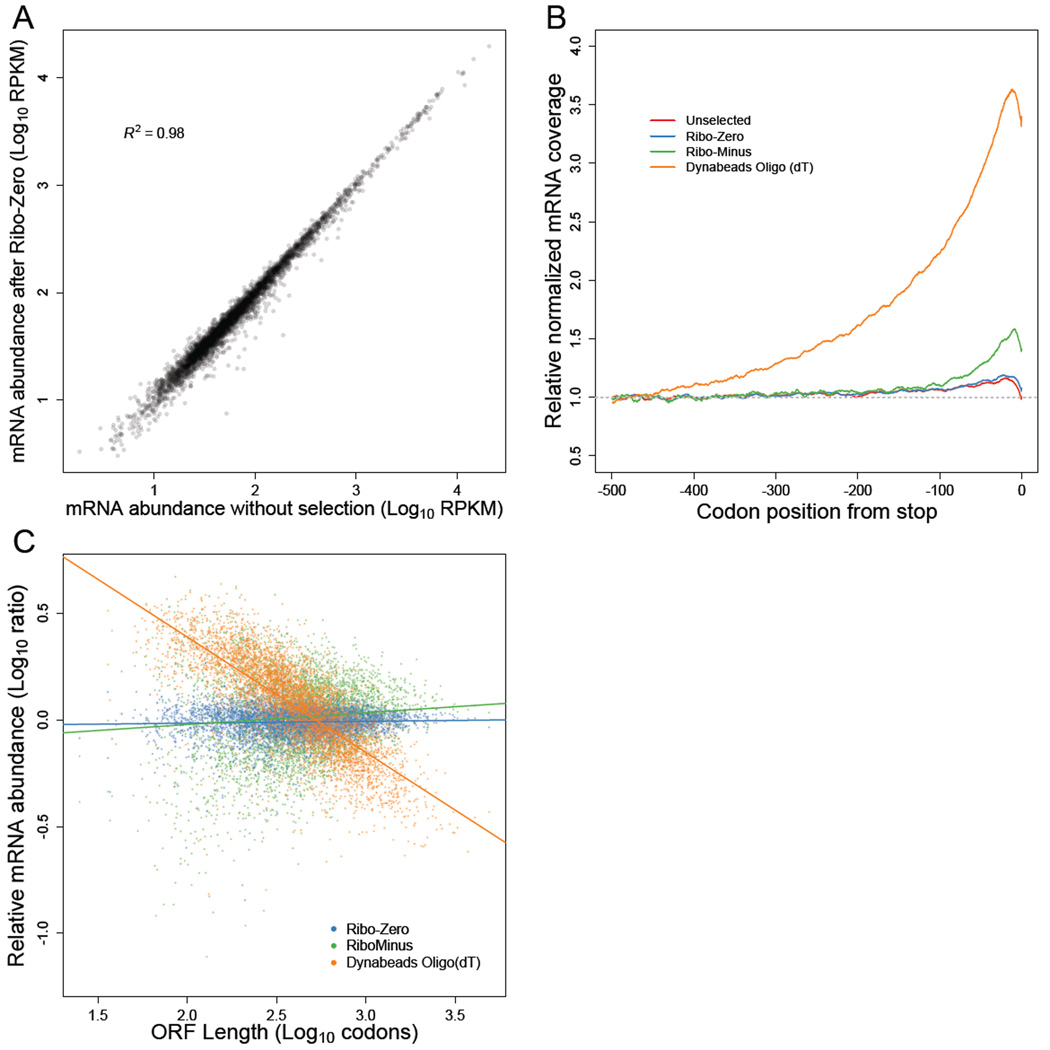
An alternative to poly(A) selection is ribosomal RNA (rRNA) depletion, which enriches mRNAs by removing rRNA using subtractive hybridization. A concern with subtractive hybridization is the potential depletion of mRNAs that either cross-hybridize to the oligonucleotides used to remove rRNA sequences or adhere to the solid matrix to which the oligonucleotides are attached. To investigate the extent to which unintended mRNA depletion occurs when using reagents sold for yeast RNA-seq library preparations, we subjected the same total RNA to each of three procedures: Dynabeads oligo(dT)25 (Life Technologies), RiboMinus Yeast Transcriptome Isolation Kit (Life Technologies), or Ribo-Zero Yeast Magnetic Gold Kit (Epicentre). As a reference, we also generated an RNA-seq library from the total RNA that was not enriched for mRNA and thus contained primarily rRNA (90.2% of 199.7 million genome-mapping reads). We also note that we started with RNA extracted from the lysate that was used for ribosome-footprint profiling, as opposed to RNA extracted from whole cells as done in the original ribosome-profiling study (Ingolia et al., 2009). When comparing the 4540 mRNAs for which we obtained at least 64 reads in our total RNA library, only the Ribo-Zero-treated sample faithfully recapitulated the mRNA abundances observed in total RNA (R2 = 0.98, Figure 4A, Figure S5). The poly(A)-selected and RiboMinus-treated samples each had significantly lower correlations with total RNA (R2 = 0.85 and R2 = 0.87, respectively), indicating a skewed representation of the transcriptome. Compared to RNA-seq data from published ribosome-profiling studies, our Ribo-Zero-treated sample also exhibited the highest correlations with microarray-based estimates of mRNA abundances (Table S3).
Figure 4. mRNA enrichment methods can bias mRNA abundance measurements.
(A) mRNA abundances measured by RNA-seq of Ribo-Zero-treated RNA compared to those measured by RNA-seq of total unselected RNA. Pearson R2 is indicated.
(B) Metagene analysis of RNA-seq read density in total unselected or mRNA-enriched RNA samples. Coding sequences were aligned by their stop codons, and RNA-seq reads were individually normalized by the mean reads within the ORF and then averaged with equal weight for each codon position across all ORFs (h’’j Eqn S15).
(C) mRNA abundances for mRNA-enriched samples relative to total unselected RNA, as a function of ORF length.
See also Figures S5–7 and Tables S3–4.
As anticipated, the poly(A)-selected sample contained a strong 3' bias (Figure 4B), which caused a systematic underestimation of the abundances of longer genes (Figure 4C). After accounting for this strong bias in the poly(A)-selected sample, we did not detect a relationship between poly(A)-tail length and poly(A)-selection efficiency, suggesting that tail-length differences did not significantly contribute to the biases of poly(A)-selected RNA-seq data. For the RiboMinus-treated sample, cross-hybridization to the depletion probes might have skewed the mRNA abundances, which might have been largely avoided in the Ribo-Zero protocol because of its more stringent hybridization conditions. The RiboMinus-treated sample also had substantial rRNA contamination (44.5% of reads, originating primarily from the 5S rRNA).
Interestingly, the total-RNA and the Ribo-Zero datasets both contained a small 3' bias (Figure 4B), with median 3'/5' excess reads of 22% and 28%, respectively (Table S4). This bias was consistent with reports that yeast mRNAs are primarily degraded in the 5'-to-3' direction (Hu et al., 2009; Pelechano et al., 2015). The decay intermediates of this vectorial degradation process would contribute more reads toward the 3' ends of mRNAs, giving rise to the observed bias, especially when considering that our RNA samples were enriched for cytoplasmic RNA, which would diminish the countervailing vectorial mRNA synthesis process occurring in the nucleus. Nonetheless, the 3' biases in the total-RNA and Ribo-Zero datasets were smaller than those in poly(A)-selected samples, for which median 3'/5' excess mRNA reads ranged from 42% to 275% (Table S4). Because Ribo-Zero treatment enabled deep coverage of the yeast transcriptome without substantially biasing mRNA abundances, we used mRNA abundances estimated from Ribo-Zero-treated RNA for all subsequent analyses.
A narrow range of initiation efficiencies in log-phase yeast
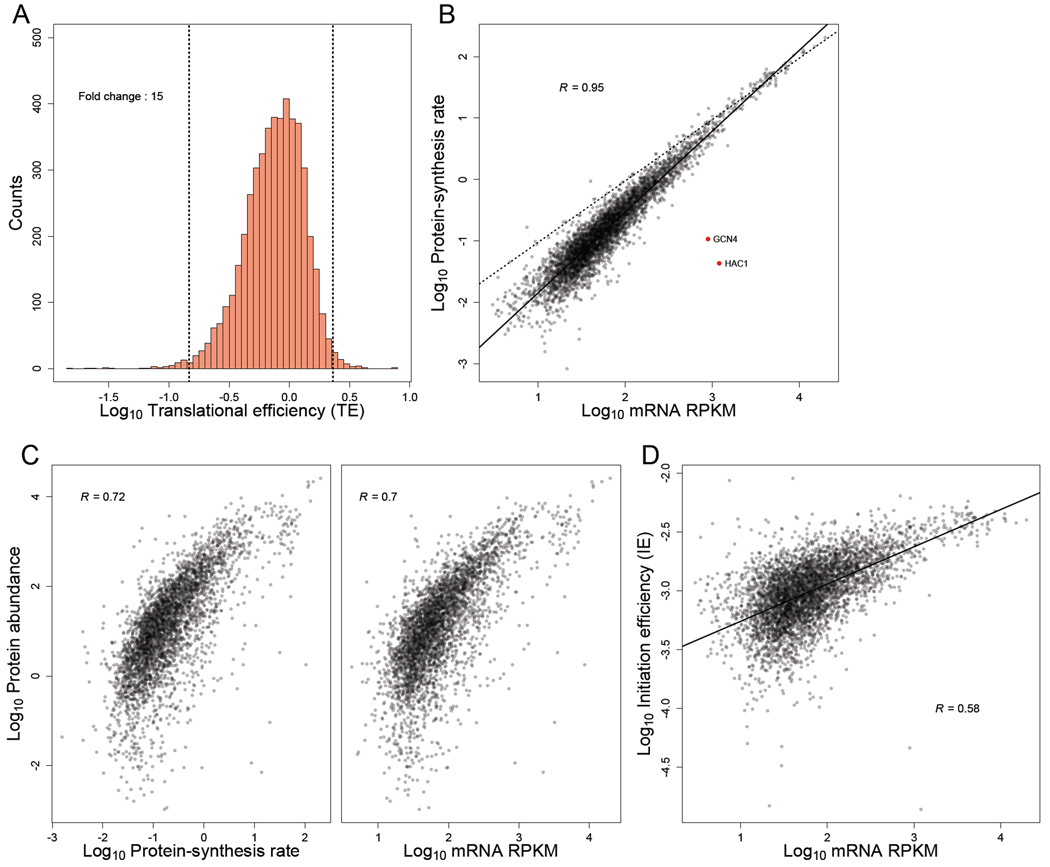
Because protein synthesis is typically limited by the rate of translation initiation (Andersson and Kurland, 1990; Bulmer, 1991; Shah et al., 2013), we defined the initiation efficiency (IE) of each gene as its protein-synthesis rate divided by its mRNA abundance (Eqn S27). Thus, the IE measure quantified the efficiency of protein production per mRNA molecule of a gene, in a typical cell. To facilitate comparisons with published datasets, we also calculated the translational efficiency (TE) of each gene, defined as its RPF density normalized by its mRNA abundance (Ingolia et al., 2009). Because TE is calculated based on the RPF density rather than the protein-synthesis rate, TE does not account for differential rates of elongation associated with the 5' ramp or codon identity. Nonetheless, IE and TE were highly correlated (R = 0.951, Figure S6A).
A wide range of IEs (or TEs) among genes would indicate that protein production is under strong translational control, whereas a narrow range would indicate that protein production is typically governed by mRNA abundances, and hence protein-synthesis rate is primarily controlled by mRNA transcription and decay. The first ribosome-profiling study suggested a large amount of translational control in yeast, with the range of TEs reported to span roughly 100 fold (Ingolia et al., 2009). Indeed, we found that the 1–99 percentile range of TEs in those data spanned 73 fold (Figure S6C). In contrast, the range of TEs observed in our data was narrower, with the 1–99 percentile spanning only a 15-fold range (Figure 5A). Although the range of IEs was marginally wider than that of TEs (1–99 percentile spanning 21 fold, Figure S6B), it was still substantially smaller than the range of TEs initially reported (Ingolia et al., 2009). The relatively narrow range of IEs in our data was also reflected by the high correlation between mRNA abundance and protein-synthesis rate (R = 0.948; Figure 5B), supporting the conclusion that protein-synthesis rates are largely dictated by mRNA abundances (Csárdi et al. 2015). Interestingly, the slope of the regression between mRNA and protein-synthesis rates was >1 on the log-scale, indicating that translation regulation mostly amplifies the effect of differential mRNA abundances rather than buffering it (Csardi et al., 2015). Further indicating that mRNA abundance (when accurately measured) is a strong predictor of total protein production, mass-spectrometry-based measurements of steady-state protein abundance (de Godoy et al., 2008) correlated as well with mRNA abundances as they did with protein-synthesis rates (Figure 5C).
Figure 5. TEs and IEs span a narrow range in log-phase yeast cells.
(A) Distribution of TE measurements, with vertical dashed lines marking the 1st and 99th percentiles, and the fold-change separating these percentiles indicated. All ORFs with at least 128 total reads between the ribosome-profiling and RNA-seq datasets were included (except YCR024C-B, which was excluded because it is likely the 3' UTR of PMP1 rather than an independently transcribed gene).
(B) Relationship between estimated protein-synthesis rate and mRNA abundance for genes shown in (A). GCN4 and HAC1 (red points) were the only abundant mRNAs with exceptionally low protein-synthesis rates. The best linear least-squares fit to the data is shown (solid line), with the Pearson R. For reference, a one-to-one relationship between protein-synthesis rate and mRNA abundance is also shown (dashed line).
(C) Relationship between with experimentally measured protein abundance (de Godoy et al., 2008) and either (left) or mRNA abundance (right). The 3,845 genes from (A) for which protein-abundance measurements were available were included in these analyses. Pearson correlations are shown (R).
(D) Relationship between mRNA abundance and IE for genes shown in (A). The best linear least-squares fit to the data is shown, with the Pearson R.
See also Figures S8–9 and Table S5.
When we examined the range of TEs in other published datasets, we also found more narrow ranges (as low as 11 fold from 1–99 percentiles) than that of Ingolia et al. (2009) (Figure S6C). However, the TEs in published datasets—which are all generated using poly(A)-selected mRNA—were not particularly well correlated with each other (Table S5). These discrepancies in TEs were largely due to differences in measured mRNA abundances, whereas the RPF abundances correlated almost perfectly (Table S5). Collectively, these results indicate that the amount of translational control in log-phase yeast has been overestimated due to inaccuracies in TE measurements, largely caused by challenges in accurately measuring mRNA levels.
We also noticed that the shape of the TE distribution from our data, which was asymmetric, differed from that of the Ingolia data, which is highly symmetric. In particular, in our data there were relatively few genes in the right tail of the distribution (Figure 5A, note the location of the mode closer to the 99th than the 1st percentile). This observation implied that mRNAs from very few genes contain elements that impart an exceptionally high initiation efficiency and are thereby “translationally privileged”. Rather, most mRNAs either initiate close to a maximum possible rate (likely set by the availability of free ribosomes or initiation factors) or contain features that modestly reduce the initiation rate.
To the extent that differences in IE were observed, the genes with lower IE tended to be expressed at lower mRNA levels, with IE increasing roughly linearly with mRNA expression levels (Figure 5D). These results were consistent with the notion that abundant mRNAs have undergone evolutionary selection to be efficiently translated (Sharp and Li, 1987; Andersson and Kurland, 1990; Plotkin and Kudla, 2011; Shah and Gilchrist, 2011). Interestingly, in the plots comparing protein-synthesis rate or IE with mRNA level, the points for 11 of the 12 highest expressed mRNAs fell below the regression lines (Figure 5B and Figure 5D, dashed lines), suggesting that the efficiency for the highest expressed mRNAs might have saturated.
Two notable outliers appeared in the comparison of mRNA abundances and synthesis rates (Figure 5B, red dots). These two, which corresponded to relatively abundant mRNAs with exceptionally low synthesis rates, were HAC1 and GCN4. These are the two most well-known examples of translational control in log-phase yeast and are both involved in rapid stress responses, during which translational repression is relieved (Ruegsegger et al., 2001)(Mueller and Hinnebusch, 1986)(Dever et al., 1992). The observation that HAC1 and GCN4 were the only abundant mRNAs that were strongly regulated at the translational level further emphasized that translational control only modestly influences the protein production of most yeast genes. Nevertheless, the tuning of synthesis rates via translational control can help maintain the proportional synthesis of the subunits of multiprotein complexes (Figure S6D–G, Supplemental Text).
Determinants of initiation efficiencies in yeast
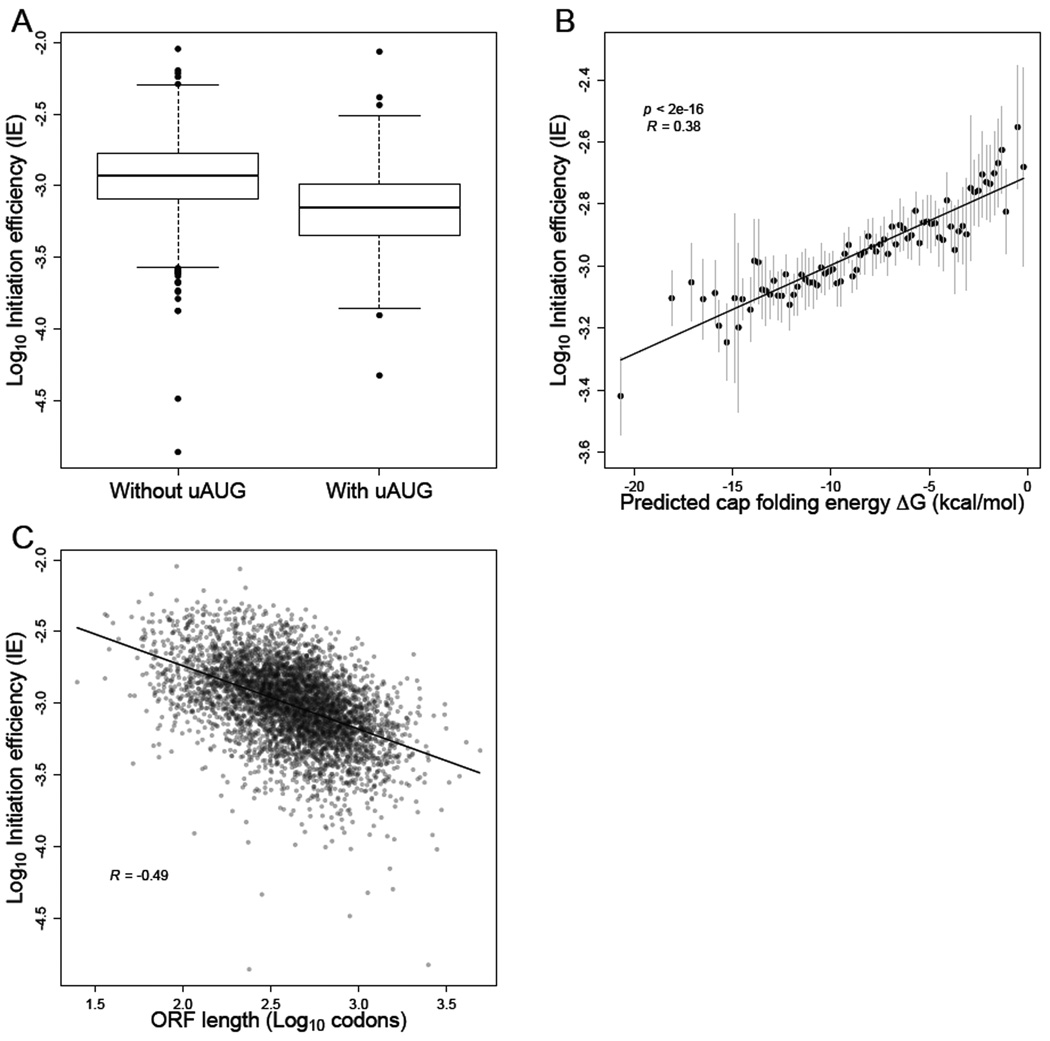
Next, we sought to identify sequence-based features that explain the variation in IE values that remained among genes after improving the RPF and mRNA measurements. First we considered uORFS, which can inhibit translation by serving as decoys to prevent initiation at the start codons of bona fide ORFs (Zur and Tuller, 2013), as occurs for GCN4 (Mueller and Hinnebusch, 1986)(Dever et al., 1992), one of two genes with the greatest translational repression (Figure 5B). Using high-resolution 5' UTR annotations (Arribere and Gilbert, 2013), we identified upstream AUGs (uAUGs) in 303 out of the 2549 genes that had reproducibly uniform transcription-start sites. Those genes containing uAUGs had significantly lower IEs than genes without uAUGs, even after controlling for 5' UTR lengths (Figure 6A, t-test p < 10−16). These results confirmed that a general feature of uORFs is to decrease the translation of downstream ORFs, and that the presence of uAUGs can explain some of the variance in IEs (Arribere and Gilbert, 2013; Zur and Tuller, 2013).
Figure 6. mRNA sequence, structure, and length correlate with IE.
(A) Reduced IE values for genes with at least one upstream AUG (i.e., an AUG codon located within the annotated 5' UTR). The plots indicated the median (line), quartile (box) and 1st and 99th percentiles (whiskers) of the distributions.
(B) Inverse relationship between IE and the folding energy of predicted RNA secondary structure near the cap (Cap-folding energy). RNAfold was used to estimate folding energies for the first 70 nt of the mRNA. Gray bars indicate 1 SD of IE values for genes binned by predicted folding energy. The best linear least-squares fit to the data is shown (solid line), with the Pearson R.
(C) Inverse relationship between IE and ORF length. The best linear least-squares fit to the data is shown (solid line), with the Pearson R.
See also Figure S7.
Another feature that has been linked to differences in synthesis rates is mRNA secondary structure. Structure located near the 5' cap might interfere with binding of the eIF4F cap-binding complex, while structure within the 5' UTR could disrupt the scanning 40S ribosome. An open structure around the start codon might also be important for facilitating joining of the 60S subunit. Previous genome-wide structure analyses revealed a weak but significant inverse correlation between start-codon-proximal structure and TE (Kertesz et al., 2010), but the accessibility of the 5' UTR more generally was not reported, and the TE values used in those studies were affected by RNA-seq biases. For each mRNA with a single reproducible 5' end (Arribere and Gilbert, 2013), we predicted the accessibility of the 5' cap by calculating the predicted folding energy of the sequence spanning increasing distances from the cap. For all distances examined, we observed a significant correlation between predicted cap accessibility and IE (t-test, p < 10−6 for each window; Figure 6B, Figure S7). This correlation rapidly increased with window length, approaching a maximum at 70–90 nt (Pearson correlation, R ~ 0.37 for windows 70–90 nt) and then steadily declined for larger windows (Figure S7), consistent with local folding of the 5' end determining cap accessibility. Together, these results confirmed that mRNAs with less-structured 5' UTRs tend to be initiated more efficiently (Godefroy-Colburn et al., 1985; Shah et al., 2013), which is consistent with eIF4F binding, 40S recruitment, or scanning as influential regulatory steps during eukaryotic initiation. Notably, the correlations that we observed between predicted mRNA structure and translation were the largest that have been reported between these features in eukaryotes, which emphasized the utility of our accurate IE measurements and of predicting structure near the cap as opposed to more downstream regions.
Gene length has also been reported to correlate with translational efficiency. Although global polysome-profiling studies indicate strong anti-correlation between ORF length and ribosome density (Arava et al., 2003), analysis of published ribosome-footprint-profiling data revealed essentially no correlation (or even a positive correlation in some cases) between length and TE (Figure S7). In contrast, we observed a striking negative correlation in our IE (and correspondingly in our TE) data (Figure 6C, Figure S7). Our IE measure already corrected for the elevated ribosome densities in the first 200 codons, and the negative correlation between ORF length and TE persisted even after removing the first 250 codons of each ORF, which further confirmed that this correlation was not caused by the 5’ ramp (Figure S7). The discrepancy between our data and earlier ribosome-profiling datasets was likely due to the RNA-seq 3'-bias caused by poly(A) selection (Figure 4B, Figure S5). Indeed, an anti-correlation between ORF length and TE was observed in most other datasets when we controlled for the 3' bias by estimating mRNA abundances based on mapped RNA-seq reads from only the 3' ends of genes (Figure S7). Together, these results showed that the original report of shorter mRNAs having relatively higher initiation efficiencies (Arava et al., 2003) is correct, even after accounting for the CHX-enhanced 5' ramp that confounded that analysis.
A statistical model that predicts initiation efficiencies
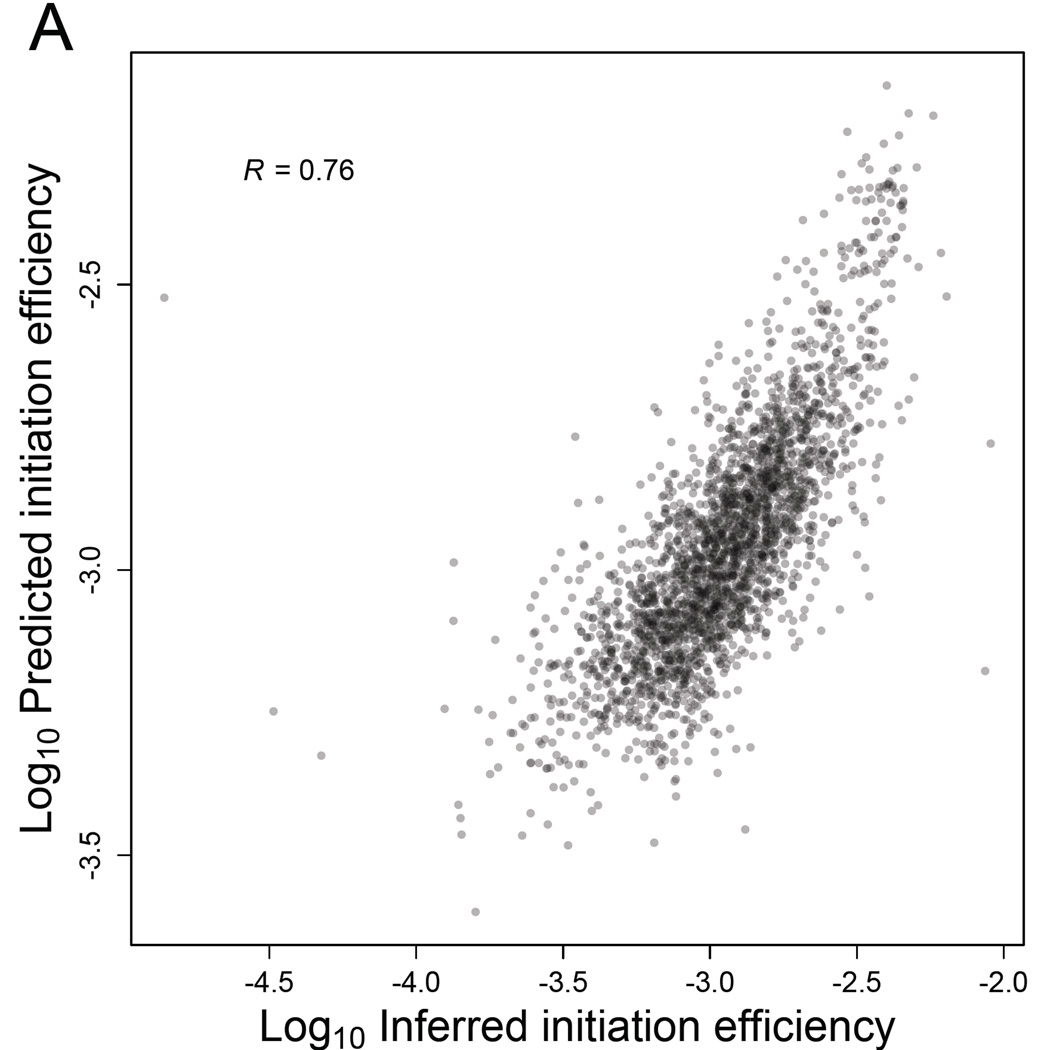
Based on these results, we used multiple linear regression to build a model that considered number of uAUGs, predicted cap-proximal RNA-folding energy (and also GC content of the 5' UTR as another metric for structure), and lengths of the ORF and the 5' UTR to explain the variance in IE observed among genes. We also included an mRNA-abundance term in the model because IE is greater for more abundant mRNAs (Figure 5D). To identify the most informative features, we used Akaike's Information Criteria (AIC) for model selection and both step-up and step-down model-selection procedures (using the stepAIC function in the MASS package in R). The multiple regression model that best explained the variation in IE included all six variables, even after penalizing for model complexity (Figure 7A, Table S6). The dominant explanatory variable was mRNA abundance, which alone accounted for ~40% of the variance in IE. Collectively, a model containing all six variables explained ~58% of the variance in IE. A model that excluded mRNA abundance, and therefore depended on only sequence-based features, still explained ~39% of the variance in IE. These results of our statistical modeling should help motivate mechanistic studies of how each of these mRNA features impacts translation.
Figure 7. Sequence-based features of mRNAs largely explain yeast IEs.
(A) Correspondence between predicted IEs and IEs inferred directly from the RPF and RNA-seq data. Initiation efficiencies were predicted using a multiple-regression model, based on mRNA abundance and sequence-based features of the 2549 genes with empirically determined 5'-UTRs. Shown is the Pearson R.
See also Table S6.
DISCUSSION
We have shown that improved measurements of both mRNA abundances and RPFs can provide insights into the regulation and dynamics of eukaryotic translation. The RPFs that we isolated and sequenced are indicative of a dynamic and heterogeneous elongation process, with ribosomes transiting along mRNA molecules at variable rates depending on the distance from the start codon, codon identity, and nascent polypeptide sequence.
What might explain the 5' ramp of ribosomes observed even in the absence of CHX pre-treatment (Figure 1C)? Codon usage accounted for about a third of it, but even the same codons were differentially occupied by ribosomes depending upon whether they occurred in the 5' or 3' ends of genes (Figure 1D), indicating that additional mechanisms must be involved. Although we cannot rule out ribosome drop-off as a contributing factor, we favor the idea that elongation is slower during the early phase of translation. Perhaps an initiation factor remains engaged with the 80S ribosome during early elongation, and the bound factor maintains the ribosome in a slower state until it stochastically dissociates from the ribosome within the first 200 codons. The eIF3 complex is a promising candidate for such a factor, as it binds the solvent-exposed face of the 40S ribosome (Siridechadilok et al., 2005) and can therefore bind to 80S ribosomes as well (Beznoskova et al., 2013). Maintaining eIF3 on early elongating ribosomes might also facilitate re-initiation after translation of short uORFs (Szamecz et al., 2008; Zur and Tuller, 2013).
A practical finding of our studies is that the choice of mRNA enrichment method can have a significant impact on yeast mRNA-abundance measurements. rRNA depletion using the Ribo-Zero kit was the only method that enriched for mRNAs without introducing substantial and systematic biases (Figure 4A, Figure S5). One caveat of rRNA depletion is that nascent pre-mRNAs that lack a poly(A) tail may also be recovered, which can inflate mRNA abundance measurements with respect to the pool of translatable mRNA molecules. This effect may be more pronounced in metazoans that contain long introns and correspondingly long transcription times. The extent to which poly(A)-selection biases affect metazoan mRNA abundance data and thereby influence TE measurements remains to be determined.
The initial report that TE spans a roughly 100-fold range across mRNAs in budding yeast spurred intensive investigation of the underlying TE determinants, with varying degree of success (Kertesz et al., 2010; Tuller et al., 2011; Charneski and Hurst, 2013; Zur and Tuller, 2013; Bentele et al., 2013; Rouskin et al., 2014). Our results showed that this apparently wide range of TEs is partly explained by inaccurate mRNA-abundance measurements. After identifying and minimizing this source of inaccuracy, we observed a narrower range of TEs and IEs (Figure 5A, Table S3), suggesting a more limited degree of translational control. The TE range that we observed in yeast resembled the range observed in mouse embryonic stem cells (Ingolia et al., 2011), suggesting that limited translational control is a general principle of gene regulation in rapidly dividing eukaryotic cells.
Using our IE measurements, we were able to generate a statistical model that explained a majority of the IE variance (Figure 7A, Table S6). Based on this model, secondary structure within the 5' UTR, most especially cap-proximal structure, appears to be an important determinant of IE. These results are in agreement with early mechanistic studies demonstrating that cap accessibility correlates with initiation efficiency (Godefroy-Colburn et al., 1985) and that stable 5'-UTR secondary structures block the scanning ribosome (Kozak, 1986). One caveat of our structure analyses is that we used in silico prediction of mRNA structure, which does not always accurately capture the in vivo structure of mRNA (Rouskin et al., 2014). Further indicating the inadequacy of in silico predictions was the benefit of also including 5'-UTR GC content as a feature in our model. Likewise, the inclusion of mRNA abundance might have helped compensate for the inadequacy of in silico structure predictions, as highly expressed genes have less predicted structure in 5' UTRs than do lowly expressed genes (Gu et al., 2010), and presumably these differences would be even greater when looking at actual 5'-UTR structure. Therefore, mRNA structure presumably explains even more variation in IE than our analyses suggested.
We also found that longer ORFs tended to be more poorly translated in log-phase yeast, even after accounting for the 5' ramp (Figure 6C). Given that initiation occurs at the 5' ends of mRNAs, how might initiation rates be sensitive to ORF lengths? One possibility is that shorter mRNAs, which include ribosomal proteins and other housekeeping genes (Hurowitz and Brown, 2003), might be under selection for faster initiation rates by virtue of their high expression. However, our stepwise regression showed that ORF length was informative even after accounting for mRNA abundance. Another possibility is that the 5'-UTR-bound initiation machinery can sense and be affected by ORF length via the closed-loop structure. In eukaryotes, translating mRNAs are thought to adopt a pseudo-circularized structure in which the 5'- and 3'-ends are in close proximity, enhancing translation and mRNA stability (Christensen et al., 1987). Previous biochemical analysis of the closed loop in yeast extracts revealed that only short mRNAs adopt a stable closed-loop structure in vitro (Amrani et al., 2008), presumably due to the relatively short distance between the mRNA termini. If the same principle applies in vivo, then inefficient closed-loop formation of long mRNAs could explain their relatively low IEs.
EXPERIMENTAL PROCEDURES
Yeast culture, harvesting, and lysate preparation
S. cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was grown at 30°C in 500 ml YPD to OD600 0.5. Cells were harvested by filtration using a Kontes Ultra-Ware Microfiltration Assembly with a Supor 450 Membrane Disc Filter that had been pre-wet with YPD. As the last liquid flowed through, the filtration apparatus was rapidly disassembled, cells were gently scraped off of the filter using a cell lifter, and the scraper was immediately submerged in a 50-ml conical tube filled with liquid nitrogen. Once the liquid nitrogen had boiled off, the pellet was stored in the conical tube at −80°C until lysis. To lyse cells under cryogenic conditions, the cell pellet was transferred into a pre-chilled mortar that was surrounded and filled with liquid nitrogen. The pellet was ground to a fine powder with a pre-chilled pestle, transferred into a 50-ml conical tube filled with liquid nitrogen, and after boiling off the liquid stored at −80°C. Crude lysate was prepared by briefly thawing the cell powder on ice for 1 minute and then resuspending in 4 ml Polysome Lysis Buffer [10 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM dithiothreitol, 100 µg/mL cycloheximide, 500 U/mL RNasin Plus RNase Inhibitor (Promega), cOmplete EDTA-free Protease Inhibitor Cocktail (Roche)]. The lysate was centrifuged at 1300g for 10 minutes, and the supernatant was flash frozen in single-use aliquots.
RNA-seq
Total RNA was extracted from an aliquot of frozen yeast lysate using TRI Reagent (Ambion) according to the manufacturer’s protocol. Aliquots of the same sample were subjected to either no enrichment (the total RNA sample), poly(A) selection using 30 µg total RNA and 100 µl Dynabeads oligo(dT)25 (Life Technologies) according to the manufacturer’s instructions, rRNA depletion using 4 µg total RNA and the RiboMinus Yeast Transcriptome Isolation Kit (Life Technologies) according to the manufacturer’s instructions, rRNA depletion using 10 µg total RNA and the Ribo-Zero Gold Yeast rRNA Removal Kit (Illumina) according to the manufacturer’s instructions. RNA samples were then diluted to 90 µl with water and precipitated with 10 µl 3 M NaCl, 30 µg GlycoBlue (Life Technologies), and 250 µl ethanol. RNA-seq was performed as described (Subtelny et al., 2014), using 5 cycles of PCR.
Ribosome profiling
RPFs were isolated from an aliquot of frozen yeast lysate and sequenced on the Illumina HiSeq platform, as described (Subtelny et al., 2014). Detailed protocols for RNA-seq and ribosome profiling are available at http://bartellab.wi.mit.edu/protocols.html. RNase I treatment was performed using 0.2 U/µL lysate. Subtractive hybridization to remove contaminating rRNA fragments was performed using a mixture of three biotinylated oligonucleotides (Integrated DNA Technologies): 5'-GATCGGTCGATTGTGCACCTC/3Bio/; 5'-CGCTTCATTGAATAAGTAAAG/3Bio/; 5'-GACGCCTTATTCGTATCCATC/3Bio/
Analyses
Equations and detailed procedures for analyses are provided in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank J. Weissman and L. Lareau for helpful discussions, and the Whitehead Genome Technology Core for sequencing. This work was supported by the UCSF Program for Breakthrough Biomedical Research (funded in part by the Sandler Foundation, D.W.) and by NIH grants DP5OD017895 (D.W.) and GM061835 (D.P.B.), the Burroughs Wellcome Fund (J.B.P.), the David and Lucile Packard Foundation (J.B.P.), US Department of the Interior Grant D12AP00025 (J.B.P.), and US Army Research Office Grant W911NF-12-1-0552 (J.B.P.). D.P.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE75897.
AUTHOR CONTRIBUTIONS
D.E.W., P.S., S.W.E., J.B.P., and D.P.B. designed the study. D.E.W., P.S., J.B.P., and D.P.B. wrote the manuscript, with help from the other authors. D.E.W. and S.W.E. prepared RNA-seq and ribosome-profiling libraries, respectively, under the supervision of D.P.B. D.E.W. performed initial data analysis. P.S. performed detailed data analyses and simulations with help from J.B.P. J.A.H. contributed new analytical tools.
REFERENCES
- Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Codon preferences in free-living microorganisms. Microbiological reviews. 1990;54:198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Gilbert WV. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome research. 2013;23:977–987. doi: 10.1101/gr.150342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB. Evolution at two levels of gene expression in yeast. Genome research. 2014;24:411–421. doi: 10.1101/gr.165522.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic acids research. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentele K, Saffert P, Rauscher R, Ignatova Z, Bluthgen N. Efficient translation initiation dictates codon usage at gene start. Molecular systems biology. 2013;9:675. doi: 10.1038/msb.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beznoskova P, Cuchalova L, Wagner S, Shoemaker CJ, Gunisova S, von der Haar T, Valasek LS. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLoS genetics. 2013;9:e1003962. doi: 10.1371/journal.pgen.1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski CA, Hurst LD. Positively charged residues are the major determinants of ribosomal velocity. PLoS biology. 2013;11:e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AK, Kahn LE, Bourne CM. Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. The American journal of anatomy. 1987;178:1–10. doi: 10.1002/aja.1001780102. [DOI] [PubMed] [Google Scholar]
- Chu D, von der Haar T. The architecture of eukaryotic translation. Nucleic acids research. 2012;40:10098–10106. doi: 10.1093/nar/gks825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, Franks A, Choi DS, Airoldi EM, Drummond DA. Accounting for experimental noise reveals that mRNA levels, amplified by post-transcriptional processes, largely determine steady-state protein levels in yeast. PLoS genetics. 2015;11:e1005206. doi: 10.1371/journal.pgen.1005206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Ding Y, Shah P, Plotkin JB. Weak 5'-mRNA secondary structures in short eukaryotic genes. Genome biology and evolution. 2012;4:1046–1053. doi: 10.1093/gbe/evs082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villen J, Bartel DP. mRNA Destabilization Is the Dominant Effect of Mammalian MicroRNAs by the Time Substantial Repression Ensues. Molecular cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, Futcher B. Measurement of average decoding rates of the 61 sense codons in vivo. eLife. 2014;3 doi: 10.7554/eLife.03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic acids research. 2014 doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17394–17399. doi: 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godefroy-Colburn T, Ravelonandro M, Pinck L. Cap accessibility correlates with the initiation efficiency of alfalfa mosaic virus RNAs. European journal of biochemistry / FEBS. 1985;147:549–552. doi: 10.1111/j.0014-2956.1985.00549.x. [DOI] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome biology. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS computational biology. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3' untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nature reviews Genetics. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic acids research. 2011;39:e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, May GE, Spealman P, Shteyman A. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome research. 2014;24:422–430. doi: 10.1101/gr.164996.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova DD, Leidel SA. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nature structural & molecular biology. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. Widespread Co-translational RNA Decay Reveals Ribosome Dynamics. Cell. 2015;161:1400–1412. doi: 10.1016/j.cell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nature reviews Genetics. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Molecular systems biology. 2014;10:770. doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS genetics. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nature chemical biology. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Gilchrist MA. Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10231–10236. doi: 10.1073/pnas.1016719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic acids research. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Pais H, Hall AE, Kozomara A, Griffiths-Jones S, Moulton V, Dalmay T. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence. 2012;3:4. doi: 10.1186/1758-907X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. Journal of molecular biology. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalova L, Munzarova V, Herrmannova A, Nielsen KH, Burela L, Hinnebusch AG, Valasek L. eIF3a cooperates with sequences 5' of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes & development. 2008;22:2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj TA, Argos P. Ribosome-mediated translational pause and protein domain organization. Protein science : a publication of the Protein Society. 1996;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Tuller T, Veksler-Lublinsky I, Gazit N, Kupiec M, Ruppin E, Ziv-Ukelson M. Composite effects of gene determinants on the translation speed and density of ribosomes. Genome biology. 2011;12:R110. doi: 10.1186/gb-2011-12-11-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. Journal of molecular biology. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. SUPERFAMILY--sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic acids research. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ma P, Shah P, Rokas A, Liu Y, Johnson CH. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature. 2013;495:116–120. doi: 10.1038/nature11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495:111–115. doi: 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Gilbert WV. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS genetics. 2013;9:e1003675. doi: 10.1371/journal.pgen.1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur H, Tuller T. New universal rules of eukaryotic translation initiation fidelity. PLoS computational biology. 2013;9:e1003136. doi: 10.1371/journal.pcbi.1003136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.