Abstract
Angiocentric gliomas are pediatric low-grade gliomas (PLGGs) without known recurrent genetic drivers. We performed genomic analysis of new and published data from 249 PLGGs including 19 Angiocentric Gliomas. We identified MYB-QKI fusions as a specific and single candidate driver event in Angiocentric Gliomas. In vitro and in vivo functional studies show MYB-QKI rearrangements promote tumorigenesis through three mechanisms: MYB activation by truncation, enhancer translocation driving aberrant MYB-QKI expression, and hemizygous loss of the tumor suppressor QKI. This represents the first example of a single driver rearrangement simultaneously transforming cells via three genetic and epigenetic mechanisms in a tumor.
Introduction
Pediatric low-grade gliomas (PLGG) encompass a heterogeneous group of World Health Organization grade I and II tumors that collectively represent the most common pediatric brain tumor. PLGGs undergo frequent alterations in the MAPK pathway and in MYB family genes, including MYBL11,2 and MYB1,2. Alterations in MYB are heterogeneous; several fusion partners have been reported as rare events in PLGGs2. The frequency of specific alterations and associations with histological subtypes are unknown.
Angiocentric Gliomas arise in the temporal lobe and share histologic features with astrocytomas and ependymomas3,4. We previously identified one Angiocentric Glioma with deletion of the 3’ region of MYB1 and one other Angiocentric Glioma has been reported to harbor a MYB-QKI rearrangement2. However, the nature and incidence of MYB alterations in Angiocentric Glioma has not been determined. Furthermore, oncogenicity of MYB family transcription factors in the CNS and the mechanisms by which they contribute to gliomagenesis are yet to be defined.
To address these questions, we performed a combined analysis of newly generated and published PLGG genomic datasets1,2,5. We found MYB-QKI rearrangements to be the most common event involving a MYB family member and to be specific to Angiocentric Gliomas. We also found that this rearrangement contributes to oncogenicity through three mechanisms: generation of oncogenic MYB-QKI, enhancer translocation that establishes an auto-regulatory feedback loop selectively driving MYB-QKI expression, and partial loss of expression of QKI, a tumor suppressor gene.
Results
Angiocentric Gliomas exhibit recurrent MYB-QKI rearrangements
Previously published genomic analyses of PLGGs did not individually contain sufficient numbers of rare histologic subtypes for statistical power to detect recurrent aberrations. To address this we performed a combined genomic analysis of whole-genome sequencing (WGS) and/or RNA-sequencing (RNA-seq) data from 172 PLGGs spanning ten histologic subtypes (Supplementary Table 1), including 145 published samples2,5 and 27 rare PLGGs that are new to this study. We performed analyses of significantly recurrent somatic genetic events across all samples with WGS or RNA-seq data (See Supplementary Note 1, Supplementary Figure 1 and Supplementary Tables 2 and 3). We observed recurrent somatic alterations in 154 tumors (90%), including all 140 tumors subject to WGS. Rearrangements or structural alterations were observed in 129 tumors (83%; Figure 1a, Supplementary Table 1).
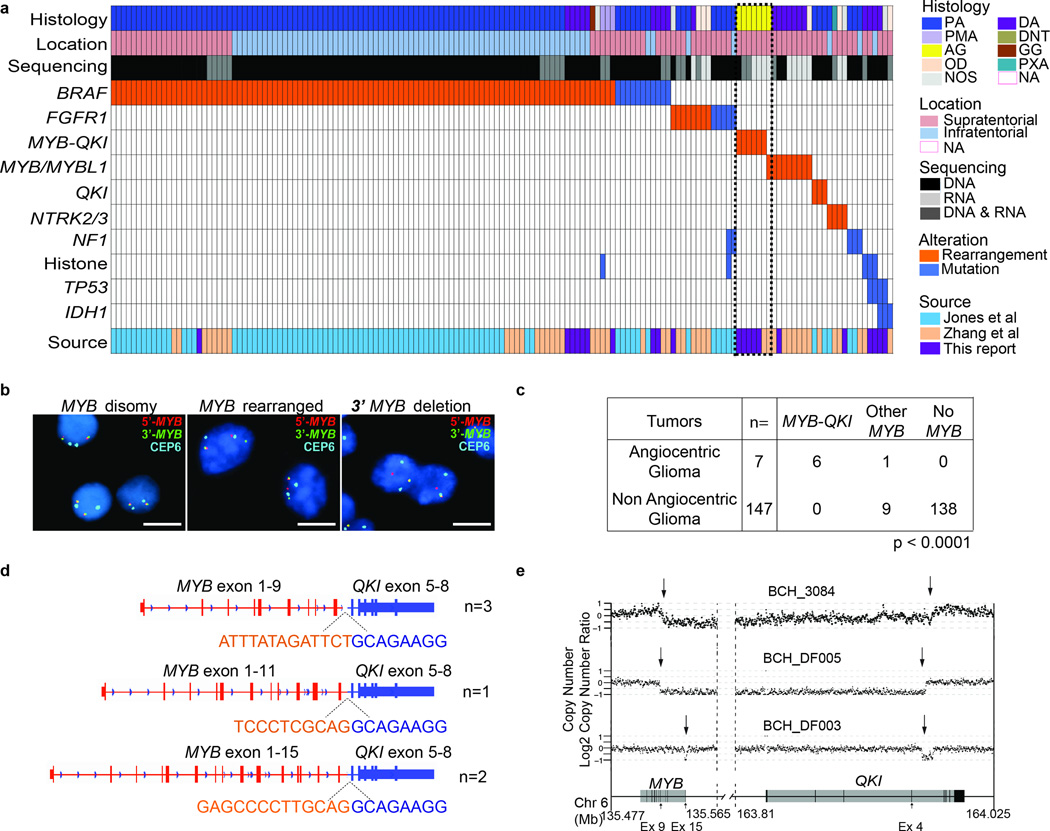
Figure 1. Genomic analysis of 172 WGS and/or RNA-seq of PLGGs reveals a recurrent rearrangement involving MYB and QKI in Angiocentric Gliomas.
a. Driver alterations were identified in 154 of 172 PLGGs profiled with WGS and/or RNA-seq. Histological subtypes include Pilocytic Astrocytoma (PA). Pilomyxoid Astrocytoma (PMA), Angiocentric Glioma (AG), Oligodendroglioma (OD), Diffuse Astrocytoma (DA), Dysembryoplastic Neuroepithelial Tumor (DNT), Ganglioglioma (GG), Pleomorphic Xanthoastrocytoma (PXA), and PLGG not otherwise specified (NOS). Tumors for which histology is unavailable are designated NA.
b. FISH using probes flanking MYB reveal three patterns in PLGG: disomy, MYB rearrangement, or 3’ MYB deletion. Scale bars = 5 microns
c. Frequency of MYB alterations or MYB-QKI rearrangements in Diffuse Astrocytoma and Angiocentric Glioma. p value represents enrichment of MYB-QKI rearrangements in Angiocentric Glioma. MYB-QKI alterations were identified with WGS alone (n=1), WGS and RNA-seq (n=2) or RNA-seq alone (n=3).
d. Breakpoints observed in MYB and QKI in Angiocentric Gliomas. Sequence across the breakpoints as determined by RNA-seq is shown for each rearrangement.
e. Copy-number profiles from WGS data of MYB and QKI in three Angiocentric Gliomas.
Rearrangements involving MYB family members (MYB, MYBL1) were the second-most recurrent alteration, affecting 16 tumors (10%), predominantly Diffuse Astrocytomas and Angiocentric Gliomas (Figure 1a and Supplementary Figure 1). Six of seven Angiocentric Gliomas, including all tumors subject to central pathology review, exhibited intra-chromosomal deletions resulting in MYB-QKI rearrangements. The other Angiocentric Glioma, which was not centrally reviewed, contained a MYB-ESR1 rearrangement.
Although MYB rearrangements have been described in PLGGs1,2, we were struck by two novel findings: QKI was the most frequent fusion partner, and MYB-QKI fusions were near-universal in Angiocentric Gliomas. For validation we identified studied 12 additional Angiocentric Gliomas with only FFPE tissue using targeted assays. Nine Angiocentric Gliomas were analyzed by FISH to detect MYB rearrangement or deletion (Figure 1b), and three Angiocentric Gliomas were analyzed by WES and/or aCGH (Supplementary Figure 2). All 12 harbored MYB aberrations.
In total, all 19 Angiocentric Gliomas profiled by WGS, RNA-seq, WES, FISH, or aCGH displayed MYB alterations, and in six of the seven cases in which its fusion partner could be detected, MYB was fused to QKI. In tumors confirmed to harbor MYB-QKI, the genetic event appeared to be present in the majority of cells, although evidence of heterogeneity (aberration in ~50% of tumor cells) was observed by FISH in 2/5 tumors with sufficient cells for quantitative scoring.
MYB-QKI rearrangements appeared specific to Angiocentric Glioma. None of the 147 non-Angiocentric Gliomas profiled with WGS or RNA-seq exhibited MYB-QKI fusions (p<0.0001, Figure 1c). We also evaluated MYB alterations in an additional 65 PLGGs from two separate cohorts: 10 non-Angiocentric Gliomas analyzed by FISH and 55 non-Angiocentric Gliomas evaluated by whole-exome sequencing (WES) and/or array CGH. Only one of these tumors exhibited alterations of MYB (vs 19/19 Angiocentric Gliomas; p<0.0001) (Supplementary Figure 2 and Supplementary Table 1). This tumor was designated not-otherwise-specified on research review but had been diagnosed as Angiocentric Glioma at the referring institution. Five tumors evaluated by WES or aCGH exhibited alterations of MYBL1; these were all Diffuse Astrocytomas. The FISH assays, aCGH, and WES, though able to detect MYB alterations, were unable to characterize its fusion partners.
All MYB-QKI rearrangements had breakpoints within intron 4 of QKI while the MYB breakpoint varied from intron 9 to 15; all were predicted to express an in-frame fusion protein MYB-QKI (Figure 1d). We identified fusion mRNA transcripts by RNA-seq (Figure 1d) and observed copy-number breakpoints in these genes from WGS data (Figure 1e).
In the WGS/RNA-seq cohort we also observed rearrangements involving QKI but not MYB in three supratentorial Pilocytic Astrocytomas (PAs), and rearrangements involving MYB or MYBL1 but not QKI in nine tumors, seven of which were Diffuse Astrocytomas. Across the entire cohort of 172 tumors profiled with WGS and/or RNA-seq, 10% harbored alterations of either MYB family members or QKI.
MYB and QKI in brain development and cancer
MYB proteins are transcription factors characterized by highly conserved DNA-binding motifs. First identified as v-myb6–8 the cellular proto-oncogene counterpart c-MYB is comprised of a N-terminus that contains helix-turn-helix (HTH) DNA binding motifs followed by a transcriptional activation domain and a C-terminal negative regulatory domain9. Full-length MYB is non-transforming or only weakly transforming in vitro10, but C-terminal MYB truncations are oncogenic10–13. MYB-QKI breakpoints in MYB intron 9 to 15 are predicted to result in C-terminal truncation of MYB.
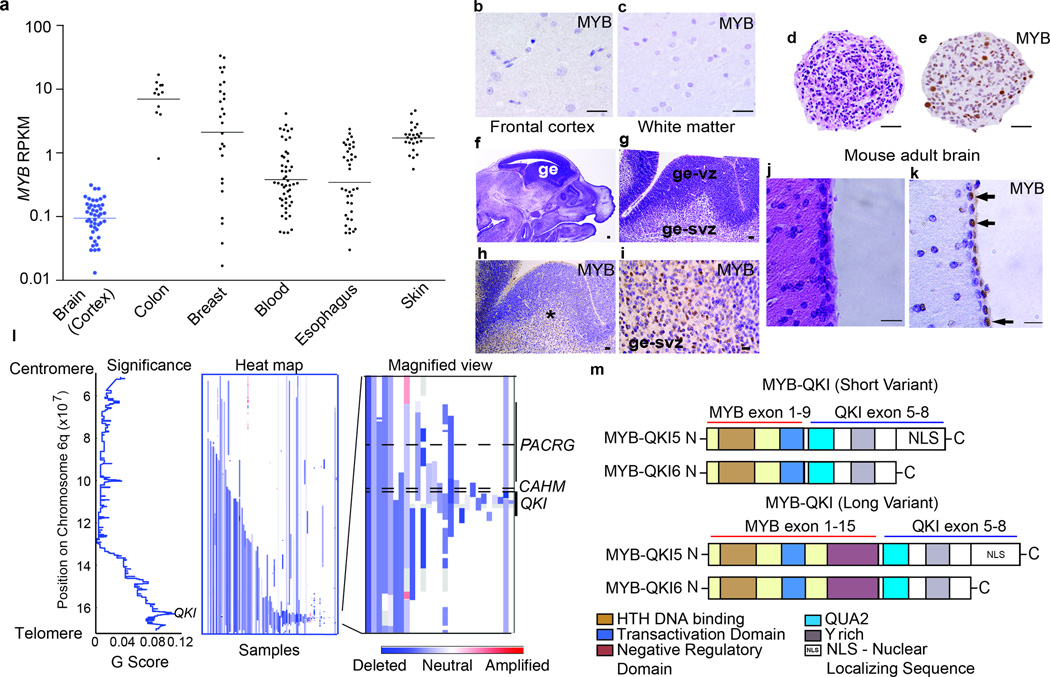
MYB is not expressed in the postnatal brain cortex, where Angiocentric Gliomas occur. We examined RNA-seq data of normal tissues14 and found MYB expression to be negligible in human brain cortex and substantially lower than MYB expression in colon, breast, blood, esophagus, or skin (Figure 2a). Likewise, immunohistochemistry of adult human frontal cortex and white matter were negative for MYB (Figure 2b and 2c); however we detected high MYB expression in human fetal neural progenitor cells generated from the ganglionic eminence at 22 weeks gestation (Figure 2d and 2e).
Figure 2. Alterations of MYB and QKI occur frequently in human cancers.
a. MYB expression (mean ± SEM) in normal human colon (n=12), breast (n=27), whole blood (n=51), esophagus (n=38), skin (n=25), and brain cortex (n=47).
b. MYB immunohistochemistry on human adult frontal cortex. Scale bar = 100 microns
c. MYB immunohistochemistry on human adult white matter. Scale bar = 100 microns
d. Hematoxylin and eosin (H&E) on human fetal neural stem cells generated from the ganglionic eminence at 22 weeks gestation. Scale bar = 100 microns
e. MYB immunohistochemistry demonstrates positive staining in a subset of cells. Scale bar = 100 microns
f. Sagittal section from embryonic 14.5 days post coitus (E14.5) mouse brain. Scale bar = 500 microns.
g. H&E of E14.5 ganglionic eminence (ge) including ventricular (ge-vz) and subventricular (ge-svz) zones. Scale bar = 50 microns.
h. MYB immunohistochemistry on the E14.5 ganglionic eminence. Scale bar = 50 microns.
i. MYB immunohistochemistry demonstrates positive staining in subventricular zone (ge-svz) but not the ventricular zone (ge-vz). Scale bars = 50 microns.
j. H&E from periventricular region of adult mouse brain. Scale bar = 100 microns.
k. Immunohistochemistry for MYB demonstrates positive cells (arrows) in the ependymal/SVZ layer. Scale bars = 100 microns.
l. (Left) Significance of deletions (x-axis) and (middle and right) heatmaps indicating copy-number profiles at 6q of individual adult Glioblastomas.
m. Structure of the MYB-QKI fusion protein. TAD denotes transactivating domain. C-terminus of QKI includes QUA2 domains. MYB-QKI5 retains a nuclear localizing sequence (NLS). Two variants of MYB-QKI are depicted corresponding to the breakpoint of MYB.
In mice MYB is expressed in E14.5 neural progenitor cells of the ganglionic eminence subventricular region (Figure 2f–i). In adult mice we detected expression in the ependyma/sub-ventricular zone (Figure 2j–k), consistent with previous reports of MYB expression in mouse progenitor cells but not in cortical brain15.
QKI encodes the STAR (Signal transduction and activation of RNA) RNA-binding protein Quaking, which plays an essential role in oligodendroglial differentiation16 and is widely expressed in the nervous system. Deletions of QKI have been suggested to be oncogenic in a number of cancers including glioblastoma17, prostate cancer18, and gastric cancer19. In copy-number analyses of 10,570 cancers within the Cancer Genome Atlas20, QKI was one of two genes in a deletion peak in adult glioblastomas (Figure 2l), renal clear cell, and cervical squamous cell carcinomas. It was also in larger peak regions of significant deletion in low-grade gliomas and bladder and adrenocortical carcinomas. Focal QKI deletions were observed in over 10% of glioblastomas.
The MYB-QKI fusion protein is expected to retain the MYB N-terminal HTH DNA binding motifs fused to the QKI C-terminus (Figure 2m). The QKI N-terminal KH RNA-binding motif is lost, while C-terminal alternative splice sites are preserved. The splice variant MYB-QKI5 retains a nuclear localizing motif which is not present in the splice variant MYB-QKI621. Fusions that contain only exons 1–9 of MYB also lose the MYB negative regulatory domain (designated short variant).
The findings that both MYB and QKI are disrupted suggest that MYB-QKI rearrangements may be oncogenic through the additive effects of alterations in both MYB and QKI. The lack of expression of MYB in normal post-natal human cortical brain regions also suggests that the rearrangement drives aberrant expression of the fusion allele. We therefore characterized mechanisms through which MYB-QKI rearrangements may contribute to aberrant MYB-QKI expression and evaluated the oncogenic potential of both genes.
MYB-QKI functions as a transcription factor
We performed genome-wide gene expression analyses of three independently-generated pools of mouse neural stem cells (mNSCs) engineered to stably over-express MYB-QKI5, MYB-QKI6, truncated MYB including exons 1–9 (MYBtrExon1–9), or eGFP. Relative to eGFP-expressing cells, those expressing MYB-QKI5 and MYB-QKI6 exhibited significantly different expression of 1621 and 1947 genes, respectively, with 1029 genes overlapping (p<0.0001; Supplementary Table 4). Gene-set enrichment analysis revealed expression of either MYBtrExon1–9 or MYB-QKI was associated with enrichment of signatures of MYB pathway activation (p<0.0001, Supplementary Table 5).
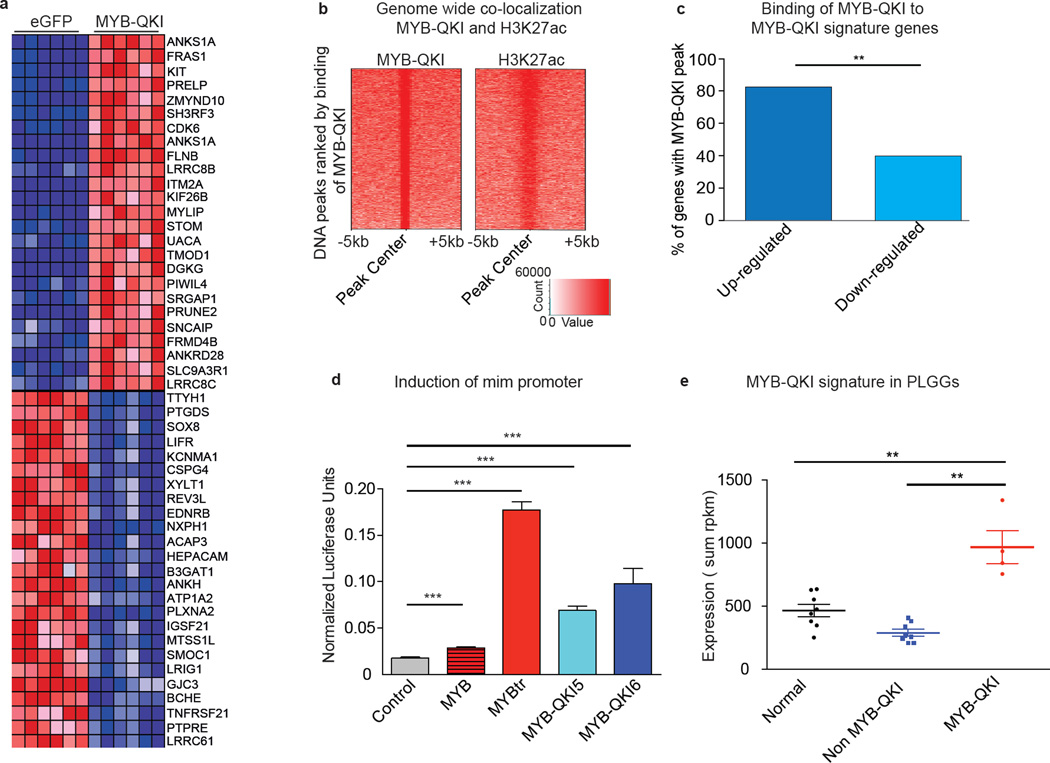
We defined a MYB-QKI gene expression signature comprising the 50 genes whose differential expression correlated most with its expression (Figure 3a). These genes include KIT and CDK6, previously reported to be associated with MYB activation22.
Figure 3. MYB-QKI functions as a transcription factor, and its molecular effects are observed in Angiocentric Gliomas.
a. MYB-QKI expression signature in mouse neural stem cells relative to cells expressing eGFP controls.
b. Heatmap of H3K27ac and MYB-QKI levels at MYB-QKI regions. Each row is centered on MYB-QKI peaks. These regions are rank-ordered by MYB-QKI signal. Scaled intensities are in units of rpm/bp
c. % of MYB-QKI signature genes with evidence of MYB-QKI ChIP-seq binding in up-regulated (n=25) and down-regulated (n=25) genes. ** depicts p<0.001 (paired t test).
d. mim-1 reporter induction following transfection of MYBtr, MYB-QKI5, MYBQKI6 or full length MYB in 293T cells. Values shown represent mean of three independent measurements ± SEM.
e. Expression of MYB-QKI signature in normal pediatric brain samples (n=8), PLGGs without MYB-QKI (n=8), or Angiocentric Gliomas with MYB-QKI (n=4). Values represent mean expression of signature in tumors ± SEM. Expression of signature within each tumor is the sum of rpkm of each gene in the signature.
We performed chromatin immunoprecipitation with parallel sequencing (ChIP-seq) in mNSCs expressing MYB-QKI, using an antibody which recognizes the N’ terminus of MYB and another antibody against H3K27ac, which defines the location of enhancer regions. We found MYB-QKI5 bound 3,672 sites (p threshold 10−7) across the genome (92% of these sites contain a MYB binding motif) and H3K27ac bound 9,122 sites, with overlap at 1,907 sites (52% of MYB binding sites, p<0.0001) (Figure 3b). These findings are consistent with reports in T-cell ALL, where MYB binding was highly correlated with H3K27ac defined enhancers23. We also identified MYB-QKI binding to the endogenous Myb promoter (Supplementary Figure 3a).
MYB-QKI5 binding sites (p threshold 10−5) were located within 100kb of 88% (22/25) of upregulated genes in the MYB-QKI signature but only 40% (10/25) of downregulated genes (p<0.001; Figure 3C). Each of the MYB-QKI binding sites associated with an upregulated gene was associated with an H3K27ac enhancer peak, while only 70% of MYB-QKI binding sites at downregulated genes overlapped enhancers (p=0.003).
The MYB-QKI fusion protein can activate transcription through binding of MYB consensus binding motifs. We generated a luciferase-reporter construct using known MYB binding sites from the target promoter mim-18, and co-transfected this reporter with MYBtrExon1–9, MYB-QKI, or full-length MYB, in 293T cells. We observed a slight induction of mim-1 promoter activity with transfection with full length MYB compared to the control vector. The greatest induction of mim-1 promoter activity was observed upon co-transfection with MYBtrExon1–9 or MYB-QKI (Figure 3d and Supplementary Figure 3b), with MYBtrExon1–9 having the highest level of activity.
Angiocentric Gliomas exhibited significantly higher expression of the MYB-QKI signature relative to normal pediatric brain (p=0.001) and PLGGs without MYB-QKI alterations (p=0.0011) (Figure 3e). PLGGs exhibited increased expression of genes associated with MYB pathway activation compared to normal brain (p=0.0003), but this was not specific to MYB-QKI rearranged tumors, and was of lower magnitude than the difference observed with the MYB-QKI signature (Supplementary Figure 3c).
MYB-QKI rearrangements drive aberrant expression of truncated MYB
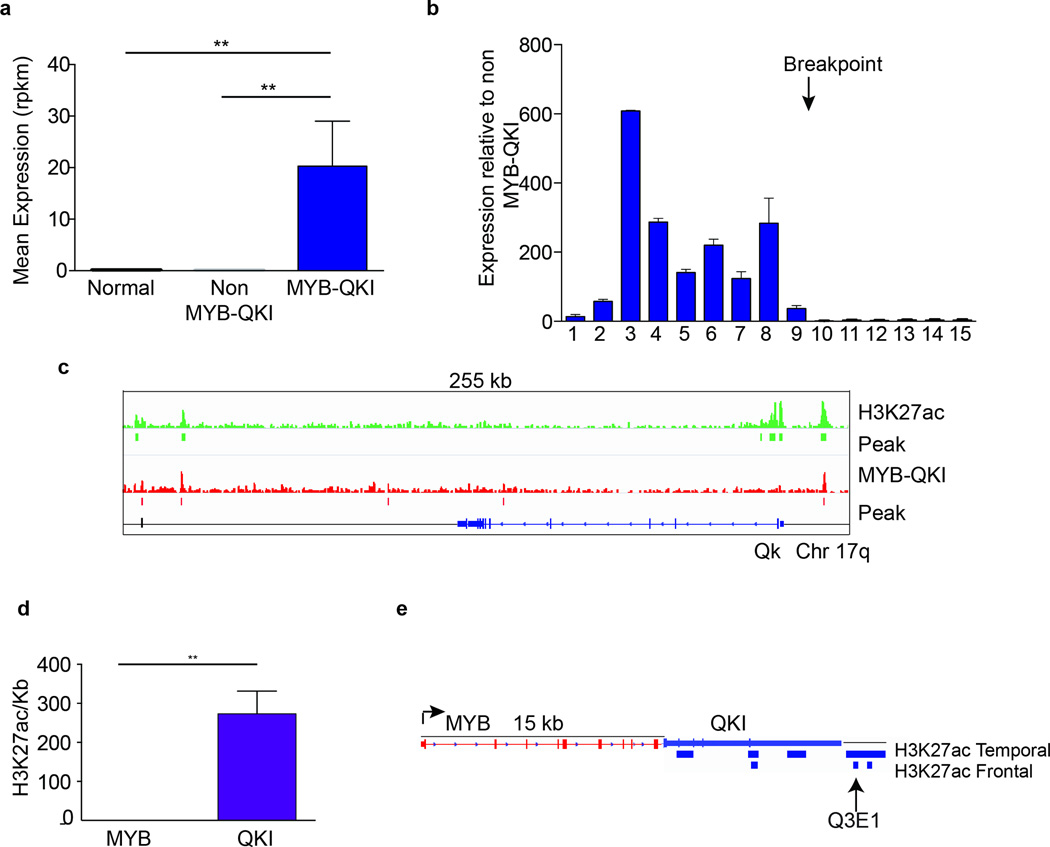
Angiocentric Gliomas with MYB-QKI exhibit significantly higher MYB expression relative to normal pediatric cortical brain (p=0.0062) or to PLGGs with BRAF or FGFR alterations (p=0.03) (Figure 4a and Supplementary Note 2). The MYB that is expressed is truncated and corresponds to the exons retained in the rearranged MYB-QKI allele. Three Angiocentric Gliomas harbored MYB-QKI rearrangement breakpoints between exons 9 and 10 of MYB. These exhibit increased expression of MYB exons 1–9 relative to PLGGs that do not harbor MYB-QKI (p<0.05), but minimal expression of the remaining exons (Figure 4b). These data support the selective, aberrant regulation of expression of truncated MYB via MYB-QKI.
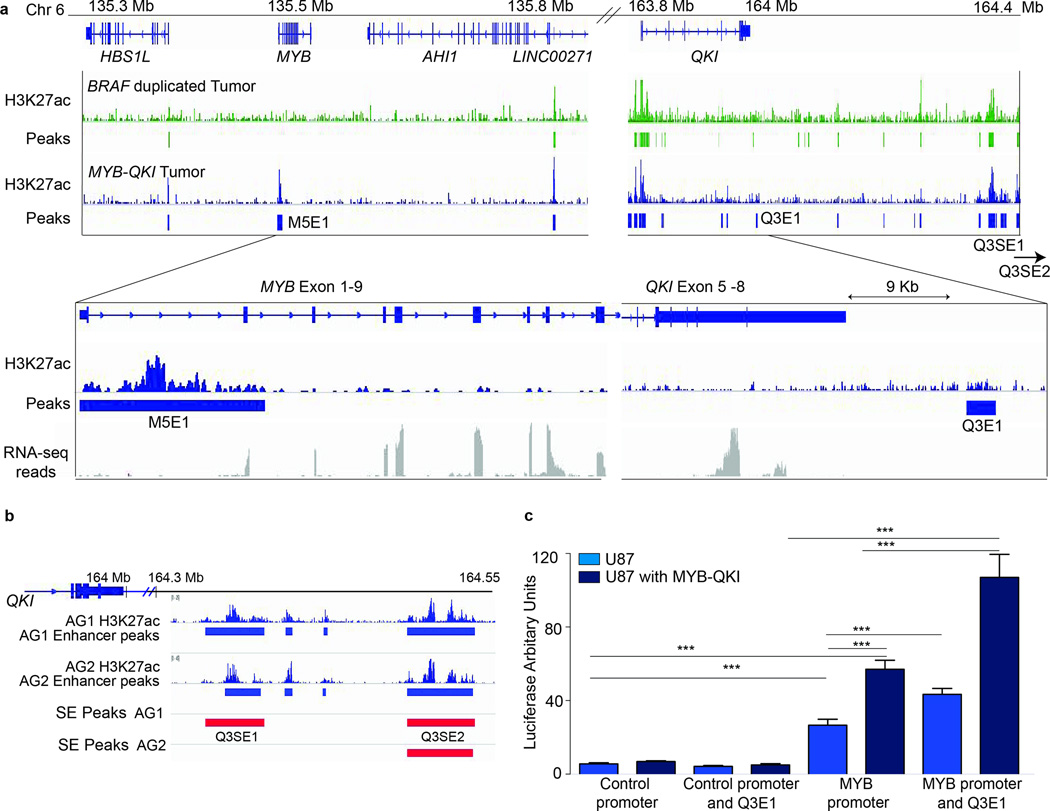
Figure 4. Angiocentric gliomas exhibit aberrant expression of MYB-QKI due to H3K27ac-associated enhancer translocation and an autoregulatory feedback circuit in which MYB-QKI binds to the MYB promoter.
a. MYB expression levels (in RPKM) of tumors with MYB-QKI rearrangement (n = 5) relative to normal brain (n = 10) or BRAF- or FGFR-driven PLGGs (n = 10). Values shown represent means ± s.e.m. **P < 0.05.
b. Exon-specific expression of MYB in angiocentric gliomas that harbor MYB-QKI rearrangement (n = 3) relative to PLGGs that harbor BRAF alterations (n = 4). Values shown represent means ± s.e.m.
c. Top track (green), H3K27ac binding within the Qk locus in mNSCs. Bottom track (red), MYB-QKI binding within the Qk locus in mNSCs. ChIP-seq binding peaks are shown.
d. H3K27ac signal within the MYB and QKI loci in human frontal and temporal lobes (Encyclopedia of DNA Elements, ENCODE). Values shown depict the mean number of nucleotides that are associated with H3K27ac (per kb) in MYB and QKI across both locations ±s.e.m. (n = 1 ChIP-seq map for each location). **P < 0.05.
e. Predicted H3K27ac-associated enhancer elements in MYB-QKI, with translocation of genomic enhancers from the 3’ region of QKI to within 15 kb of the 5’ end of MYB. The enhancer maps shown are derived from ENCODE data for normal human brain (frontal and temporal lobes). Q3E1 represents a H3K27ac-associated enhancer present in the ENCODE data from normal brain.
The MYB-QKI rearrangement results in enhancer translocation
Aberrant oncogene expression can result from enhancer translocation24. In published H3K27ac enhancer profiles from normal human cortical brain samples25, MYB is not associated with H3K27ac enhancer peaks, consistent with the finding that MYB is not expressed. In contrast, QKI, which is expressed, is associated with several H3K27ac peaks, including sequences at the 3’ end of QKI (Figure 4c, d, e). The MYB-QKI rearrangement is predicted to bring these 3’ QKI-associated H3K27ac enhancer elements to within only 15kb of the MYB promoter (Figure 4e).
H3K27ac enhancer profiling of two human Angiocentric Gliomas expressing MYB-QKI confirmed the presence of active enhancer elements that are translocated proximally towards the MYB promoter (Figure 5a and Supplementary Figure 4). ChIP-seq revealed multiple H3K27ac peaks associated with 3’ QKI, similar to the peaks observed in normal human brain, and in a BRAF-duplicated supratentorial pilocytic astrocytoma. We also observed enhancers within 10kb of 3’ QKI and a larger cluster of super-enhancers 100–500kb 3’ to QKI (Q3SE1 and Q3SE2). In Angiocentric Gliomas with MYB-QKI, these enhancers are translocated proximally towards the MYB promoter.
Figure 5. Human Angiocentric Gliomas exhibit H3K27ac enhancer translocation with an aberrant enhancer associated with the MYB promoter.
a. H3K27ac enhancer peaks in proximity to MYB and QKI in a BRAF-duplicated Pilocytic Astrocytoma (top) and MYB-QKI Angiocentric Glioma (lower). Q3E1 is an enhancer associated with the 3’UTR of QKI. Two super-enhancer clusters (Q3SE1 and Q3SE2) are located within 500kb of QKI. Angiocentric Gliomas are associated with aberrant enhancer formation at the MYB promoter (M5E1), which is not detected in the BRAF driven pilocytic astrocytoma. The breakpoints for the MYB-QKI rearrangement are between exons 1–9 MYB and 5–8 QKI. Expression as determined by RNA-sequencing is depicted for the MYB-QKI Angiocentric Glioma.
b. 3’ QKI associated super-enhancers (Q3SE1/2) presented in two Angiocentric Gliomas.
c. MYB promoter activation following transfection of the MYB-luc construct in U87 cells and U87 cells over-expressing MYB-QKI with and without Q3E1 enhancer cloned into MYB-luc construct. Changes in luciferase activity of the MYB-luc reporter is shown as mean (± SEM) of three individual replicate experiments with n=5.
We observed an aberrant enhancer associated with the MYB promoter in MYB-QKI defined Angiocentric Glioma (Figure 5a). Normal human cortical brain is not associated with H3K27ac MYB-related enhancers, and indeed we did not observe formation of H3K27ac MYB enhancer peaks in the Pilocytic Astrocytoma (Supplementary Figure 5). However, in both Angiocentric Gliomas, we observed a large H3K27ac peak associated with the MYB promoter (M5E1). RNA-seq revealed expression of the first nine exons of MYB corresponding to those retained in the rearrangement, suggesting that the aberrant M5E1 enhancer is regulating expression of truncated MYB from the rearranged allele. The lack of full-length MYB expression indicates the aberrant enhancer does not regulate the remaining wild-type MYB allele.
We examined whether MYB-QKI was able to functionally activate the MYB promoter by creating a luciferase-reporter construct possessing the human MYB-promoter (MYB-luc). We observed significant induction of MYB promoter activity in U87 cells stably expressing MYB-QKI with MYB-luc as compared to U87 cells containing MYB-luc or the promoter-less control luciferase construct alone (Figure 5c). This suggests MYB-QKI contributes to an auto-regulatory feedback loop, possibly by binding to the MYB promoter. MYB-QKI activated the MYB promoter in two additional cellular contexts (HEK 293T and NIH-3T3 cells, Supplementary Figure 6).
We predicted that enhancers in the QKI 3’ UTR could aberrantly activate the MYB promoter when translocated, thereby further driving MYB-QKI expression. We cloned the proximal QKI 3’UTR enhancer sequence (Q3E1) upstream of the human MYB promoter in the MYB-luc construct. Baseline activity of the Q3E1-MYB-luc promoter construct was higher than with MYB-luc alone in U87 glioma cells (Figure 5c), increasing activation by approximately 1.5 fold, a level of activation shown to harbor biological relevance in other diseases26. Expression of MYB-QKI with Q3E1-MYB-luc led to even higher activity, again consistent with an auto-regulatory feedback loop in the presence of the fusion protein (Figure 5c).
The MYB-QKI fusion protein is oncogenic
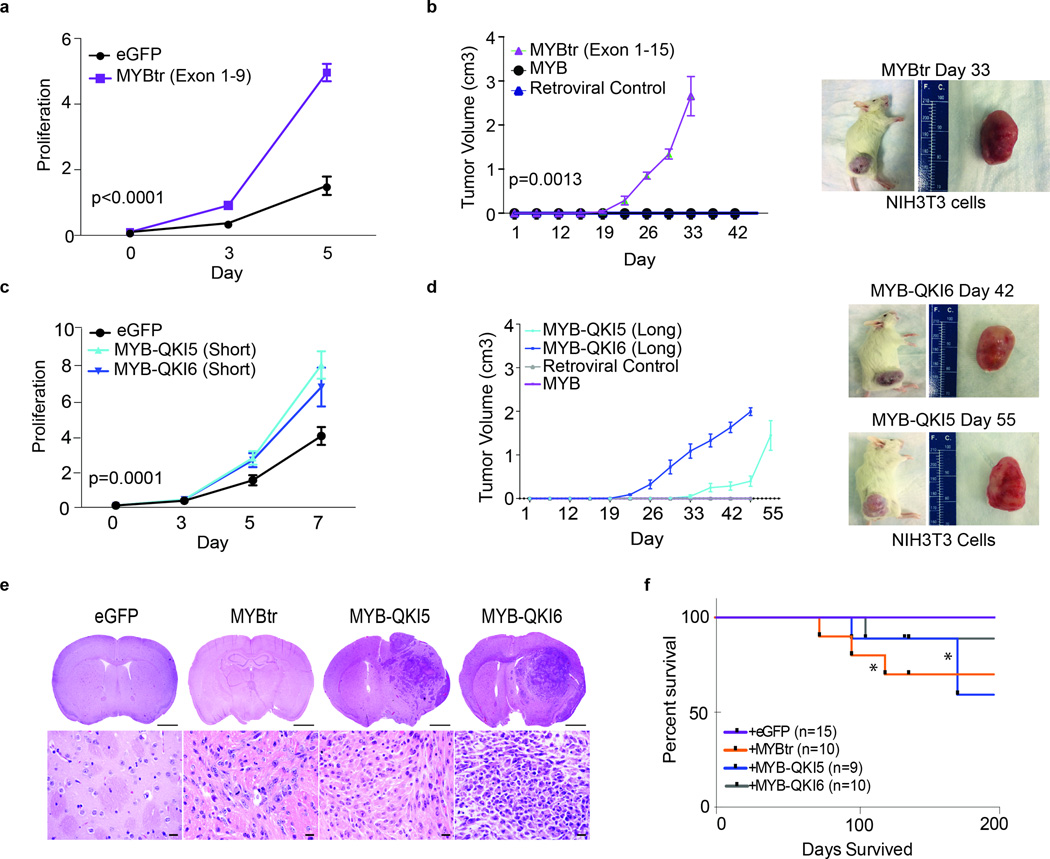
Expression of truncated MYB has previously been reported to be oncogenic10–12. In mNSCs, overexpression of MYB exons 1–9 (short variant) increased cell proliferation rates compared to eGFP controls (Figure 6a and Supplementary Figure 6b), while in NIH-3T3 cells overexpression of MYBtr (exons 1–15), but not full-length MYB, induced tumors when injected into mouse flanks (Figure 6b and Supplementary Figure 6b). Furthermore mNSCs expressing MYBtr induced diffuse gliomas on average 100 days post intracranial injection (Figure 6e, f). These tumors expressed OLIG2 and GFAP in a subset of tumor cells, a pattern similar to that observed in human diffuse gliomas (Supplementary Figure 6c).
Figure 6. MYB-QKI fusion protein and truncated MYB are oncogenic.
a. In vitro cell proliferation (number of cells relative to baseline) of mNSCs overexpressing eGFP or truncated MYBtrexons1–9. The mean values for five independent pools are depicted. Error bars, s.e.m.
b. Tumor growth following flank injections of NIH3T3 cells overexpressing MYB, MYBtrexons1–15 or a vector control. The means of five measurements are depicted. Error bars, s.e.m. Representative images are shown for intracranial mNSC-MYB-QKI6 tumors.
c. In vitro cell proliferation of mNSCs that overexpress MYB-QKI5 (short), MYB-QKI6 (short) or eGFP control. The means of five independent pools are depicted. Error bars, s.e.m.
d. Tumor growth following flank injections of NIH3T3 cells overexpressing MYB, MYB-QKI5 (long), MYB-QKI6 (long) or vector control. The mean of five measurements is depicted. Error bars, s.e.m. Representative images are shown of intracranial mNSC–truncated MYB tumors.
e. Hematoxylin and eosin analysis of severe combined immunodeficient (SCID) mouse brain after striatal injections with mNSCs expressing eGFP, truncated MYB, MYB-QKI5 or MYB-QKI6. Scale bars, 2 mm (top) and 50 ?m (bottom).
f. Kaplan-Meier survival analysis of orthotopic SCID mice injected with mNSCs overexpressing truncated MYB, MYB-QKI5 or MYB-QKI6 that develop tumors with short latency in comparison to mice injected with mNSCs expressing eGFP, which never develop tumors (**P < 0.05).
To test whether MYB-QKI fusions are oncogenic, we stably expressed MYB-QKI5 and MYB-QKI6 in mNSCs and NIH3T3 cells. In mNSCs, overexpression of either isoform led to significantly increased proliferation compared to eGFP (Figure 6c and Supplementary Figure 6b), p<0.0001. Similarly both isoforms induced anchorage-independent growth in NIH-3T3 cells (Supplementary Figure 7a); in vivo, overexpression of both MYB-QKI5 and MYB-QKI6, but not full-length MYB, led to tumor formation as flank xenografts (Figure 6b). Intracranial injections of mNSCs overexpressing MYB-QKI5 or MYB-QKI6 formed gliomas with infiltrating tumor cells with some evidence of enhanced growth around vessels and a clustered growth pattern, features similar to Angiocentric Glioma and distinct from the histology seen adult glioblastoma models (e.g. Ink4a/ARF:EGFRvIII)27. However these tumors differed from human Angiocentric Gliomas in that they had high-grade features with frequent mitoses and marked cytologic atypia (Figure 6e). Immunohistochemical analysis showed diffuse GFAP expression and a subset of OLIG2 positive tumor cells, a pattern similar to that seen in human Angiocentric Gliomas (Supplementary Figure 6c).
In total, we established flank injections in 15 mice with NIH-3T3 cells over-expressing either MYBtr or MYB-QKI (and five vector controls), and 29 intracranial injections of mNSC expressing MYBtr or MYB-QKI (15 vector controls) (Supplementary Figure 7b). We observed flank tumors in all 15 mice injected with NIH-3T3 cells over-expressing either MYBtr or MYB-QKI and five intracranial tumors from mice injected with mNSCs expressing MYBtr or MYB-QKI. We did not observe tumors in any vector controls. These data represent a significant enrichment of tumor formation in cells expressing MYBtr or MYB-QKI (p<0.0001).
The MYB-QKI rearrangement disrupts QKI, a tumor suppressor
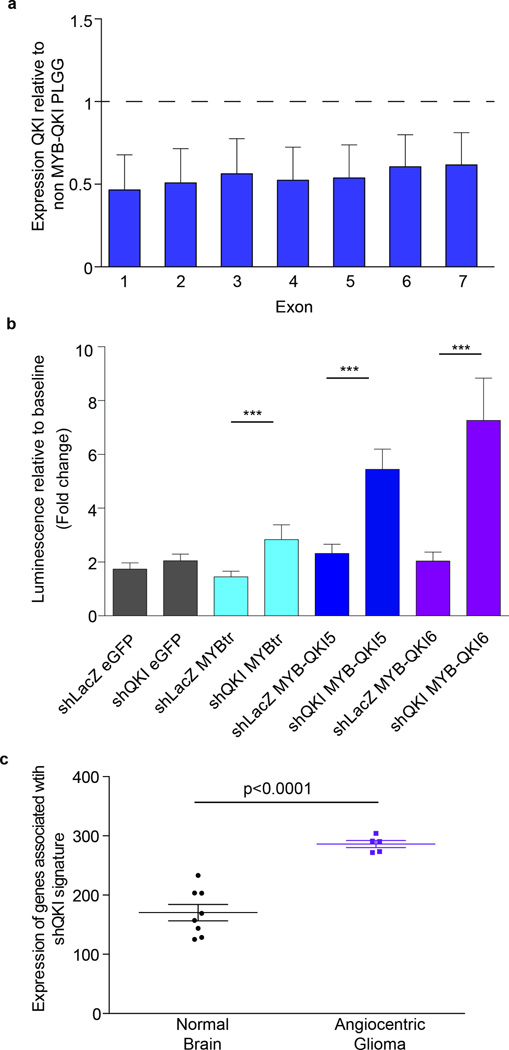
We were interested in understanding how disruption of QKI may contribute to oncogenicity in tumors that harbor MYB-QKI. Exon-specific RNA-seq analysis of Angiocentric Gliomas with MYB-QKI (n=4) showed reduced expression of QKI compared to PLGGs that harbor BRAF alterations (n=5) (Figure 7a and Supplementary Note 2). These data suggest that the MYB-QKI rearrangement may contribute to tumor formation through reduced expression of QKI, a tumor suppressor gene.
Figure 7. MYB-QKI disrupts expression of QKI, a tumor suppressor gene.
a. Exon specific expression of in Angiocentric Gliomas (n=5) relative to BRAF-driven PLGGs (n=5). Values represent mean ± SEM. RNA-sequencing data of Exon 8 of QKI revealed a high number of duplicate reads and thus is not shown.
b. Cell proliferation of mouse neural stem cells expressing MYBtr, MYB-QKI5, MYB-QKI6 or eGFP control with suppression of wild-type Qk. Values represent mean of four independent experiments ± SEM.
c. Expression of signature within each tumor is the sum of rpkm of each gene in the signature.
Indeed, suppressing wild-type Qk using shRNAs that target the first four exons of Qk led to increased proliferation of mNSCs, with the greatest increase observed in the context of pre-existing MYB-QKI expression. In mNSCs over-expressing MYBtr, MYB-QKI5 or MYB-QKI6, suppression of wild type Qk was sufficient to increase proliferation within only three days of suppression (Figure 7b and Supplementary Figure 8a). The greatest effect was observed in cells overexpressing MYB-QKI, despite a similar or lower degree of suppression of Qk in these cells compared to those over-expressing eGFP or MYBtr. We did not observe increased proliferation within three days in cells expressing eGFP, though we did observe a mild increase on day 5 (Supplementary Figure 8b). These data suggest MYB-QKI overexpression and QKI suppression exert cooperative functional effects.
Suppression of Qk by shRNAs in mNSCs expressing MYB-QKI6 led to differential expression of 309 genes relative to shLacZ (q<0.25, Supplementary Table 6). QKI has been previously reported to regulate expression of micro-RNAs28,29, and we also observed upregulation of 10 miRNAs with suppression of wild-type Qk, including Mir717 (Supplementary table 7). The mouse Qk isoform 7 is predicted to contain a miRNA regulatory element (MRE) for Mir71730.
Angiocentric Gliomas exhibit molecular effects consistent with QKI suppression. In mNSCs, we defined a signature consisting of the 50 genes whose expression was most correlated with Qk suppression (Supplementary Figure 8c). This signature was significantly enriched in Angiocentric Gliomas relative to normal brain (p<0.0001, Figure 7c).
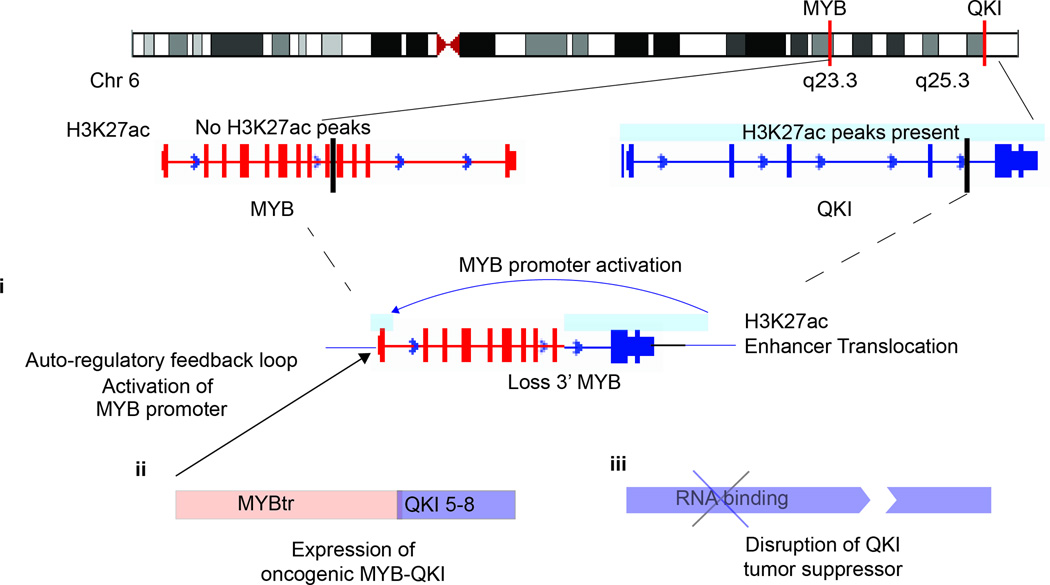
Taken together, our data suggest three mechanisms through which the MYB-QKI rearrangement contributes to oncogenicity (Figure 8). First, the alteration results in proximal translocation of H3K27ac enhancers on 3’QKI towards the MYB promoter, resulting in MYB promoter activation. Second, the MYB-QKI fusion protein that is expressed is oncogenic, functions as a transcription factor, and exhibits the ability to bind to and activate the MYB promoter, resulting in an auto-regulatory feedback loop. Third, hemizygous loss of QKI results in suppression of QKI, which functions as a tumor suppressor gene. Thus disruption of both MYB and QKI appear to contribute to tumor formation in a co-operative manner.
Figure 8. The MYB-QKI rearrangement contributes to oncogenesis through at least three mechanisms.
The MYB-QKI rearrangement disrupts both MYB and QKI, resulting in hemizygous deletion of 3’MYB and 5’ QKI. This results in proximal translocation of H3K27ac enhancers on 3’QKI towards the MYB promoter, resulting in MYB promoter activation (i). The MYB-QKI fusion protein that is expressed is oncogenic, functions as a transcription factor, and exhibits the ability to bind to and activate the MYB promoter, resulting in an auto-regulatory feedback loop (ii). Hemizygous loss of QKI results in suppression of QKI expression, which functions as a tumor suppressor gene (iii).
Discussion
We describe MYB-QKI as a novel recurrent diagnostic fusion in Angiocentric Glioma. It also represents the first example of a single driver translocation of two genes resulting in the aberrant expression of an activated oncogenic fusion protein which then participates in an auto-regulatory feedback loop, proximal translocation of enhancer elements regulating fusion-gene expression, and simultaneous functional loss of a tumor suppressor gene.
We found MYB-QKI to be a defining event in Angiocentric Glioma. This has important implications for treatment and diagnosis of this disease. The tight association of the translocation with this histology supports pathologic classification of Angiocentric Glioma as a separate biological entity. We propose that the presence of this fusion should be considered diagnostic of Angiocentric Glioma. This could aid in distinction of Angiocentric Glioma from tumors with higher potential for recurrence or require further treatment, such as IDH-mutant diffuse gliomas or ependymomas.
MYB-QKI expression was sufficient to reproducibly generate intracranial tumors. Angiocentric Gliomas are WHO grade 1 tumors and exhibit a very low mitotic index. Successful models of low-grade pediatric or adult gliomas are rare across all histologies. The low penetrance in mNSCs (5 tumors in 29 attempts) relative to high-grade glioma models (e.g. EGFRvIII and Ink4a/Arf −/− NSCs)27 suggests genetic drivers of low-grade gliomas may be borderline sufficient for transformation.
Although the development of small molecule inhibitors to target MYB directly is likely to be challenging, MYB-QKI transcriptional targets such as KIT or CDK6 can be targeted. The association of MYB-QKI with H3K27ac enhancer elements also raises the possibility of therapeutically inhibiting its effects through indirect mechanisms, such as BET-bromodomain31,32 or CDK7 inhibition33.
Adenoid cystic carcinomas harbor MYB-NFIB alterations34–36. Like MYB-QKI, MYB-NFIB also results in high levels of MYB expression, although the mechanism underlying this, the functional role of NFIB, and oncogenicity of MYB-NFIB remain undefined.
We observed an additive effect of Qk suppression with MYB-QKI over-expression, confirming QKI as a tumor suppressor and suggesting cooperation between its loss and expression of MYB-QKI. Recent studies indicate a diversity of roles for QKI in cancer, including altered splicing events37 and a role in epithelial-mesenchymal transition via regulation of circular RNAs38. QKI regulates expression of genes implicated in cancer39, and microRNA processing30. Further investigation is required to evaluate mechanisms of cooperativity between QKI suppression and MYB-QKI expression.
Angiocentric Gliomas exhibit haploinsufficiency of QKI while GBMs demonstrate biallelic loss. One explanation may be that complete loss of QKI exerts negative selection in the developing brain. This is supported by the essential role of QKI in oligodendrocytic differentiation. Haploinsufficiency may also account for the lower-grade nature of Angiocentric Glioma compared to GBM.
Pediatric tumors are characterized by simple genomes with single driver alterations40,41. Our findings that one rearrangement contributes to oncogenicity through multiple mechanisms may be applicable to a large number of pediatric tumors.
Online methods
Ethics statement
Ethics approval was granted by relevant human IRB and/or animal research committees (IACUC) of Dana-Farber Cancer Institute (DFCI), Boston Children’s Hospital, The Broad Institute and Children’s Hospital of Philadelphia (CHOP). IRB approval from all institutions was obtained, and all patients provided informed consent prior to collection of samples or were analyzed as de-identified samples with specific IRB waiver of informed consent.
Whole-genome sequencing and processing
PLGGs and normal controls from CBTTC/CHOP and DFHCC/PLGA Consortium were sequenced at BGI@CHOP, and The Broad Institute of MIT and Harvard. DNA was randomly fragmented, and libraries prepared for paired-end sequencing on an Illumina HiSeq 2000. Sequencing files from recently published PLGG datasets were accessed1,2. Read pairs were aligned to reference genome hg19 (Build 37) using the Burrows-Wheeler Aligner (bwa) with options −q 5 −l 32 −k 2 −o 13. Reads were sorted by coordinates, normalized, cleaned and duplicates were marked using SAMtools and Picard. Base quality score assignments were recalibrated to control for biases due to flow cell, lane, dinucleotide context and machine cycle using the Genome Analysis Toolkit (GATK)4. Copy-number alterations were evaluated using SegSeq5. GISTIC2 was used to identify recurrent copy-number alterations6–8. Somatic point mutations and short indels were called using Mutect9 and IndelLocator, and visual inspection in IGV10. Mutsig (version 2.0)11 was applied to detect significantly recurrent mutations. Rearrangements and breakpoints were identified using dRanger, BreakPointer12 and visual inspection. All analyses were performed within Firehose13.
RNA-sequencing and analysis pipeline
Following RNA extraction (RNeasy, Qiagen), library construction was performed using a non-strand specific Illumunia TruSeq protocol. Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocols using HiSeq 2000/2500, with a 76 bp paired-end run including an eight-base index barcode read. RNA-sequencing files were downloaded from published datasets1,2. RNA-seq bam files were transformed to fastq files using the Picard SamToFastq algorithm. Raw paired-end reads were aligned to the reference genome hg19 and preprocessed using PRADA (Pipeline for RNA-sequencing Data Analysis)14. We used PRADA within Firehose to determine gene-expression levels, exon expression levels, quality metrics, and for detection of fusion transcripts. BAM files were also assessed by visual inspection.
Array CGH
DNA was extracted from archival FFPE samples and aCGH performed as previously described15,16. GC-normalized copy-number data was cleaned of known germ-line copy-number variations and circular Binary Segmentation was used to segment the copy-number data (α = 0.001, undo.splits = sdundo, undo.SD = 1.5, minimum width = 5).
Whole exome sequencing
WES was performed from FFPE samples (without matched control). These samples were used to confirm driver alterations identified by the WGS. DNA was extracted using the QIAGEN DNA Blood and Tissue kit. Libraries with a 250 bp average insert size were prepared by Covaris sonication, followed by double-size selection (Agencourt AMPure XP beads) and ligation to specific barcoded adaptors (Illumina TruSeq) for multiplexed analysis. Exome hybrid capture was performed with the Agilent Human All Exon v2 (44 Mb) bait set.
Sequence data were aligned to the hg19 reference genome with the Burrows-Wheeler Aligner with parameters [-q 5 -l 32 -k 2 -t 4 -o 1]. Aligned data were sorted, duplicate-marked, and indexed with Picard tools. Base-quality score recalibration and local realignment around insertions and deletions was achieved with the Genome Analysis Toolkit.
Mutations were called with MuTect, filtered against a panel of normals, and annotated to genes with Oncotator. Likely germline SNPs were removed by filtering against the Exome Sequencing Project (ESP) and Exome Aggregation Consortium (ExAC).
Histological assignment
Histologic subtype assignments were according to previously published data. Samples not previously published were centrally reviewed and classified by a board-certified neuropathologist (K.L.L., S.S, or S.R.) using W.H.O. 2007 criteria.
MYB FISH
FISH was performed as previously described17 using five micron FFPE tissue sections and Homebrew probes RP11–63K22 (5’ to MYB; directly labeled in SpectrumOrange) and RP11–170P19 (3’ to MYB; directly labeled in SpectrumGreen) that map to 6q23.3. MYB status was assessed in 50 tumor nuclei per sample. A CEP6 aqua probe (Invitrogen) mapping to the centromeric region of chromosome 6 was co-hybridized as a control.
Immunohistochemistry
Diaminobenzidine (DAB), bright-field staining was performed according to standard protocols on five-micron thick paraffin sections. Heat and 10 mM sodium citrate buffer (pH 6.0) were used for antigen retrieval for the MYB (Abcam for human tissue, Bethyl Laboratories for mouse tissue), OLIG2 (Chemicon) and GFAP (Millipore) antibodies. Counterstaining for nuclei was performed using Mayer’s hematoxylin stain and coverslips were mounted with Permount (Fisher Scientific). Sections from the left occipital pole of a normal adult brain autopsy were used to assess MYB levels.
Analysis of QKI alterations in TCGA samples
GISTIC 2.0 analyses were performed across 10,570 tumor samples from 31 lineages from The Cancer Genome Atlas (TCGA), as previously described8.
Analysis of gene expression in normal tissues
RNA-sequencing of normal pediatric brain samples was accessed from the BRAINSPAN Atlas of the human developing brain and processed as previously described18. MYB expression levels from RNA-sequencing obtained from normal autopsy tissues were downloaded from the GTEx consortium19. Expression levels were compared using ANOVA and t-tests. p-values <0.05 were considered significant.
Vector construction and generation of NIH3T3 stable lines
MYB-QKI5 and MYB-QKI6 constructs were synthesized as Gateway compatible entry clones. MYBtr constructs were generated via PCR mutagenesis using MYB-QKI fusions as templates. Full-length MYB and QKI constructs were purchased as gateway entry clones from PlasmID/DF/HCC DNA Resource Core. MYB-QKI5 and MYB-QKI6, MYBtr, full-length MYB and QKI constructs were sub-cloned into a Gateway-compatible N-MYC-tagged pMXs-Puro Retroviral Vector (Cell Biolabs). Platinum-E retroviral packaging cells (Cell BioLabs) were used to generate retrovirus as per manufacturer protocols. NIH3T3 cells were infected with retrovirus containing media for 6 hours and puromycin selection commenced 48 hours post infection. Stable expression of MYC-tagged proteins was confirmed via western blot analysis (anti-MYC HRP 1:5000 (Invitrogen), anti-MYB antibody 1:5000 (Abcam) and anti-QKI 1:1000 (Bethyl Lab).
Soft Agar Colony Formation Assays and quantification
Anchorage-independent growth of NIH3T3 cells was assayed as previously described20 with the following modifications: NIH3T3 cells expressing each of the MYB-QKI5, MYB-QKI6, MYBtr, full-length MYB and full length QKI proteins and retroviral vector control were plated in 0.7% agar with DMEM and DBS in 96 well plates (in triplicates). Cell colonies were allowed to form for two weeks and images were taken. Images were analyzed using ImageJ and colonies with area greater than 500 pixels quantified.
Generation of reporter construct containing MYB promoter and enhancer constructs
To assess the effect of candidate enhancer regions on MYB promoter activity, the human MYB promoter sequence (shown below) was cloned into the pLightSwitch_Prom Vector (Active Motif) that contains a multiple cloning site upstream of a Renilla luciferase reporter gene (RenSP) without a promoter. The MluI/BglII site on pLightSwitch_Prom Vector was used to clone the MYB promoter sequence (Supplementary Table 9) and the MluI site was further used to clone candidate enhancer regions upstream of the MYB promoter. The human QKI 3’UTR enhancer sequences (hg18 chr6:163920360–163920809 and chr6:163921548–163921972) were synthesized by Invitrogen and cloned into the reporter constructs as described above. The LightSwitch™ Random Promoter Control 1 (Active Motif) containing a 1 kb non-conserved, non-genic and non-repetitive fragment from the human genome cloned upstream of the RenSP luciferase reporter gene was used as a negative control. A housekeeping gene promoter vector, LightSwitch™ ACTB Promoter Control, was used as positive control for all assays. The luciferase reporter constructs containing either the MYB promoter or MYB promoter with enhancers were transfected into U87 glioma line (or MYB-QKI stably expressing U87 line) using Lipofectamine 3000 (n=5), or co-transfected with MYB-QKI or vector control into HEK 293s via Lipofectamine 2000 (Invitrogen), or in NIH3T3/ MYB-QKI stably expressing NIH-3T3 lines using PolyFect (Qiagen). Luciferase activity was quantified 24 hours post transfection using the LightSwitch™ Luciferase Assay Reagent (Active Motif) according to the manufacturer’s protocol.
mim-1 reporter construct generation and MYB transactivation assays
Luciferase reporter constructs containing a consensus DNA-binding sequence for c-MYB were generated. The reporter construct was designed using the core MYB recognition element (MRE) consensus sequence PyAAC(G/T)G which is present in the mim-1 gene promoter, a previously described MYB target21. Double stranded oligos were generated by annealing primers mim-1 forward and mim-1 reverse (supplementary Table 9). The annealed oligo was ligated into pGL4.10[luc2] vector (Promega) digested with XhoI and HindIII. The pRL Renilla Luciferase Reporter Vector (Promega E2261) served as an internal control in all assays. The mim-1 reporter construct and pRL renilla vector (ratio 30:1) were co-transfected into HEK-293 along with indicated fusions or controls via Lipofectamine 2000. Luciferase activity was quantified 24 hours post transfection using the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer’s protocol.
Cell lines
NIH3T3, 293T and U87 MG cell lines were obtained directly from ACTT and not re-authenticated. All cell lines were routinely tested (at least every three months) for mycoplasma infection.
Generation of neural stem cells
Embryonic murine neural stem cells (mNSC) were derived from C57BL6 wild-type E14.5 dpc mouse embryos (purchased from Taconic) as previously described22. mNSCs were maintained in culture media with 1:1 Dulbecco modified Eagle medium (Gibco) and neural stem cell media (Gibco) supplemented with B27 (Gibco), EGF (02653, Stem Cell), FGF (GF003, Millipore), and Heparin (07980, Stem Cell).
Overexpression of transcripts in mouse neural stem cells
293T cells were transfected with 10 µg lentiviral pLEX307 expression vectors (gift from David Root, Addgene plasmid #41392) with packaging plasmids encoding PSPAX2 and VSVG using Lipofectamine. Lentivirus-containing supernatant was collected 48 hour after transfection, pooled and concentrated (ultrafiltration). Target neural stem cells underwent infection using a spin protocol (2000rpm for 120 minutes at 30C with no polybrene). Puromycin selection (0.5mcg/ml) commenced 48 hours after infection.
ShQk experiments and proliferation assays
Lentiviral vectors (pLKO) encoding shRNAs specific for mouse Qk, targeting sequences in the first four exons of Qk, and the control shLacZ were obtained from The RNAi Consortium (Supplementary Table 9). Lentivirus was produced by transfection of 293T cells with vectors encoding each shRNA (10 µg) with packaging plasmids encoding PSPAX2 and VSVG using Lipofectamine (Invitrogen, 56532). Lentivirus-containing supernatant was collected 48 hours after transfection, and concentrated. Target mNSC underwent infection using a spin protocol (2000rpm for 120 minutes at 30C with no polybrene). Cells were placed into proliferation assays 48 hours after infection.
Cell Proliferation Assays
1000 cells/well were plated in 96-well plates, with five replicates. Cell viability was measured by assessing ATP content using Cell Titre-Glo (Promega). Mean ± SEM was calculated.
Western immunoblotting
Cells were lysed and subjected to SDS-PAGE gradient gels as previously described23. Blots were probed with antibodies against MYB (ab45150, Abcam), QKI (A300-183A, Bethyl Laboratories) and actin (sc-1615, Santa Cruz).
RNA extraction and Real-Time RT-PCR
RNA was extracted with the RNeasy kit (Qiagen). cDNA was synthesized from 1µg RNA using High Capacity RNA to cDNA kits (Applied Biosystems). Real-time RT-PCR was performed as previously described23. Primers for MYB, QKI and β-actin are listed in Supplementary Table 9. Samples were amplified in triplicate and data analyzed using the ΔΔCT method.
Gene-expression analysis of neural stem cells expressing MYB-QKI
RNA was extracted from three independently generated pools of mNSC expressing one of eGFP, MYBtr, MYB-QKI5, MYB-QKI6 or QKItr. Gene expression profiles were assayed using Affymetrix Mouse Gene 2.0 ST microarrays (Affymetrix, Santa Clara, CA). CEL files were RMA normalized24. Comparative marker selection analysis25 was performed in GenePattern using default settings. Genes with p-value <0.05 and q-value <0.35 were considered significant. GSEA was performed using the C2 (CP) gene sets (MSigDB). Genesets with nominal p-values <0.05 were considered significant. The MYB-QKI signature was defined using the ClassNeighbours module of GenePattern (default settings).
Antibody optimization and ChIP-seq
We systematically determined the antibody and concentrations that produce the highest signal to noise ratio for MYB ChIP-seq using our automated ChIP-seq methodology26,27. We tested two MYB antibodies: abcam ab45150 and Sigma SAB4501936. Abcam45150 has been previously used to ChIP MYB28. We split the sheared chromatin between 3 ratios of antibody/chromatin (0.5µl, 1 µl and 5 µl of each antibody/1,000,000 cells), and performed ChIP-seq as previously described26. As a positive control, we included an antibody targeting H3K27ac (Cell Signaling Technologies D5E4, optimized at 1 µl/1,000,000 cells). We found 1µl ab45150/1,000,000 cells to be optimal.
Results from the MYB ChIP-seq were validated in three ways. First, we performed MYB ChIP-seq in K562 cells and confirmed enrichment at genes reported to be target genes in a prior study of these cells (Supplementary Figure 9)29. Second, we used Homer30 to perform an unbiased motif analysis across peaks identified (peak detection threshold 10−7) in mNSC over-expressing MYBQKI, and identified MYB motifs to be the most enriched motifs across all peaks (p= 1−681) (Supplementary Figure 10a). We observed 92% of all peaks (p threshold 10−7, 3392/3672 peaks) to contain a MYB motif (Supplementary Figure 10b, Supplementary Table 10). Enrichment of MYB motifs was significantly higher in data generated with the MYB antibody (p= 1−681) compared to those generated from enrichment with H3K27ac (p=1−25). Third, we compared our results from mNSCs to other published MYB ChIP-seq results. We determined whether MYB bound genes identified in our study (MYB peaks containing a MYB motif) were overlapping target genes reported in these studies, using a Chi-Square test with Yates Correction. We observed significant enrichment (p<0.0001; Supplementary Figure 10c)31,32.
ChIP libraries were indexed, pooled and sequenced on Illumina Hi-seq-2000 sequencers. Raw data was aligned to the mouse reference genome MM9 using Picard tools. Raw sequencing data was mapped to the reference genome using bowtie2 version 2.2.1 with parameters -p 4 -k 1. Peaks were called using MACS version 1.4.2 over an input control. A p-value threshold of enrichment of 10−7 was used. Density of genomic regions was calculated using bamliquidator_batch, version 1.1.0. Reads were extended 200-bp and normalized to read-density in units of reads per million mapped reads per bp (rpm/bp). To calculate genome-wide overlap, all enriched H3K27ac peaks were extended 5kb in each direction, divided into 50 bins, and read density was calculated in each bin. Density was normalized to the largest value observed in each experiment genome-wide and plotted as a heat map. Peaks and alignments were converted to TDFs by IGV tools and visualized by IGV. Bed files of published ChIP-seq data of H3K27ac chromatin maps from normal brain33 were downloaded and visualized in IGV.
ChIP-seq enriching for H3K27ac was performed on human pediatric low-grade gliomas by Active Motif as recently described34. Analysis was performed as above using a p-value threshold of enrichment of 1E-5. Super-enhancer analysis was performed as previously described35.
In vivo experiments
Mouse flank tumor studies with NIH3T3 Stable Cell Lines: NIH3T3 cell lines were injected subcutaneously into the flanks of NSG mice (5 mice for each cell line). Mice were 6–10 weeks of age, with equal representation of male and female mice. Tumor growth was measured biweekly. Ellipsoid tumor volume was calculated using the formula: volume = 1/2(length × width2).
Intracranial mouse injections: Neurospheres were dissociated and resuspended at 100,000 viable cells/µL. One microliter was injected into the right striatum of immunocompromised ICR-SCID mice. Animals were monitored and sacrificed at the onset of neurological symptoms. Brains were subjected to routine histological analysis. Tumors were scored as present based on identification of atypical cells by a neuropathologist. 4–6 week old, male IcrTac:ICRPrkdc-Scid mice from Taconic were used. A total of 44 mice were used.
Mouse injections were not randomized or blinded. Sample size was not predetermined. Qualitative assessment of tumorogenicity was the primary outcome measure. Neuropathologists were blinded to group allocation.
Statistical analysis
For statistical analysis (unless otherwise described), p values were calculated using Fisher’s, T-tests or Pearson’s as appropriate. ANOVA with correction was used for comparison of multiple groups. Log-rank (Mantel-Cox) survival analysis was performed for animal studies and Kaplan Meier curves generated. Error bars shown depict standard error of the mean.
Supplementary Material
Acknowledgments
We would like to thank and acknowledge the DFHCC/Pediatric Low-grade Astrocytoma Consortium and the Children’s Brain Tumor Tissue Consortium including additional participating sites: Ann and Robert Lurie Children’s Hospital, Seattle Children’s Hospital, and Children’s Hospital of Pittsburg for sample contribution, members of the Genomic Platform and Firehose Team at the Broad Institute for assistance with genomic sequencing and analysis, the Neuro-Histology labs of Boston Children’s Hospital and Brigham and Women’s Hospital, Heather Homer (Brigham and Women’s Hospital Department of Pathology, Center for Advanced Molecular Diagnostics, Division of Cytogenetics) for technical assistance with FISH, and members of the Ligon, Resnick and Beroukhim laboratories for useful discussions.
We acknowledge the following funding sources: A Kids’ Brain Tumor Cure Foundation Pediatric Low-Grade Astrocytoma Foundation (PB, KLL, RB, MWK, LG, CS, ACR), R01NS085336 (ACR, AJW, PBS, MS), Voices Against Brain Cancer (ACR), Children’s Brain Tumor Foundation (ACR, AJW), Stop and Shop Pediatric Brain Tumor Program (PB, MWK), Path to Cure Foundation (KLL), NIH PO1CA142536 (CS, KLL, RB), St Baldrick’s Foundation (PB), American Brain Tumor Association (LR), Team Jack Foundation (PB, MWK, RB, LG), Andrysiak Fund for LGG (MWK), Broad Institute Scientific Projects to Accelerate Research and Collaboration (SPARC) grant (AG), Jared Branfman Sunflowers For Life Fund For Pediatric Brain And Spinal Cancer Research (PB, RB, SS), Sontag Foundation (KLL, RB), Nuovo-Soldati Foundation (GB), Philippe Foundation(GB), Fondation Etoile de Martin (JG), Damon Runyon-Sohn Pediatric Fellowship Award (AJW), Hyundai Scholar Grant (AJW), Bear Necessities Pediatric Cancer Foundation (AJW, ACR) Rally Foundation for Childhood Cancer Research (AJW), K08NS087118 (SHR), Pediatric Brain Tumor Foundation (RB, PB), Thea’s Star of Hope (ACR, AJW).
Finally we would like to thank and acknowledge the many children and families affected by PLGGs for their generous contributions to this research.
Footnotes
Conflict of interest statement: The authors declare no conflicts of interest.
URLs
Samtools: samtools.sourceforge.net
Picard: picard.sourceforge.net
Bamliquidator: https://github.com/BradnerLab/pipeline/wiki/bamliquidator.
Accession codes
Microarray data have been accessioned with the Gene Expression Omnibus (GEO) under series GSE75796. The sequence reported in this paper (WES, WGS, RNA-seq and ChIP-seq) has been deposited in the database of Genotypes and Phenotypes (dbGaP) under study accession number phs001054.v1.p1.
Author Contributions
PB, LAR, PJ, GB, KLL, RB and ACR designed research, PB, LAR, PJ, GB, HJH, YZ, NC, AS, KB, REH, MRS, AMB, MS, SM, DAH, JP, AJW, PBS, JW, RZ, SES, LU, ROR, WJG, KP, SHS, YJK, DK, BRP, AGY, PMH, AF, HM, AT, SSeepo, MD, PVH, DCB, SP, CH, UT, AK, LB, PCB, CE, FJR, DAH, SSantagata, CDS, JEB, NJ, AG, JG, AHL, LG, MWK, KLL, RB, ACR completed the research, PB, LAR, PJ, GB, JW, HJH, YZ, AJW, PBS, RZ, AF, SES, WJG, SHR, SSantagata, AG, JEB, AHL, ACR, KLL, RB analyzed the data, JW, PV, MP, DCB, CG, SP, CH, UT, AK, LB, PCB, CE, MSanti, AMB, MScagnet, SM, DAH, JP, FJR, SSantagata, NJ, AG, JG, AHL, LG contributed new reagents, algorithms and/or samples, PB, LAR, PJ, GB, KLL, RB, ACR wrote the manuscript, and KLL, RB and ACR supervised the study. All authors reviewed, edited and approved the manuscript.
References – Main Text
- 1.Ramkissoon LA, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci U S A. 2013;110:8188–8193. doi: 10.1073/pnas.1300252110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M, et al. Monomorphous angiocentric glioma: a distinctive epileptogenic neoplasm with features of infiltrating astrocytoma ependymoma. J. Neuropathol. Exp. Neurol. 2005;64:875–881. doi: 10.1097/01.jnen.0000182981.02355.10. [DOI] [PubMed] [Google Scholar]
- 4.Lellouch-Tubiana A, et al. Angiocentric neuroepithelial tumor (ANET): a new epilepsy-related clinicopathological entity with distinctive MRI. Brain Pathol. 2005;15:281–286. doi: 10.1111/j.1750-3639.2005.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones DTW, et al. Recurrent somatic alterations of FGFR1 NTRK2 in pilocytic astrocytoma. Nat. Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klempnauer KH, Bonifer C, Sippel AE. Identification and characterization of the protein encoded by the human c-myb proto-oncogene. EMBO J. 1986;5:1903–1911. doi: 10.1002/j.1460-2075.1986.tb04443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klempnauer KH, Gonda TJ, Bishop JM. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell. 1982;31:453–463. doi: 10.1016/0092-8674(82)90138-6. [DOI] [PubMed] [Google Scholar]
- 8.Ness SA, Marknell A, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- 9.Sakura H, et al. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci U S A. 1989;86:5758–5762. doi: 10.1073/pnas.86.15.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonda TJ, Buckmaster C, Ramsay RG. Activation of c-myb by carboxy-terminal truncation: relationship to transformation of murine haemopoietic cells in vitro. EMBO J. 1989;8:1777–1783. doi: 10.1002/j.1460-2075.1989.tb03571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu YL, Ramsay RG, Kanei-Ishii C, Ishii S, Gonda TJ. Transformation by carboxyl-deleted Myb reflects increased transactivating capacity and disruption of a negative regulatory domain. Oncogene. 1991;6:1549–1553. [PubMed] [Google Scholar]
- 12.Press RD, Reddy EP, Ewert DL. Overexpression of C-terminally but not N-terminally truncated Myb induces fibrosarcomas: a novel nonhematopoietic target cell for the myb oncogene. Mol. Cell. Biol. 1994;14:2278–2290. doi: 10.1128/mcb.14.4.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grässer FA, Graf T, Lipsick JS. Protein truncation is required for the activation of the c-myb proto-oncogene. Mol. Cell. Biol. 1991;11:3987–3996. doi: 10.1128/mcb.11.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malaterre J, et al. c-Myb is required for neural progenitor cell proliferation and maintenance of the neural stem cell niche in adult brain. Stem Cells. 2008;26:173–181. doi: 10.1634/stemcells.2007-0293. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich VL. The myelin deficit in quacking mice. Brain Res. 1974;82:168–172. doi: 10.1016/0006-8993(74)90902-0. [DOI] [PubMed] [Google Scholar]
- 17.Yin D, et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol. Cancer. Res. 2009;7:665–677. doi: 10.1158/1541-7786.MCR-08-0270. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, et al. The tumor suppressing effects of QKI-5 in prostate cancer: a novel diagnostic prognostic protein. Cancer Biol. Ther. 2014;15:108–118. doi: 10.4161/cbt.26722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bian Y, et al. Downregulation of tumor suppressor QKI in gastric cancer its implication in cancer prognosis. Biochem. Biophys. Res. Commun. 2012;422:187–193. doi: 10.1016/j.bbrc.2012.04.138. [DOI] [PubMed] [Google Scholar]
- 20.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Zhou L, Tonissen K, Tee R, Artzt K. The quaking I-5 protein (QKI-5) has a novel nuclear localization signal shuttles between the nucleus the cytoplasm. J. Biol. Chem. 1999;274:29202–29210. doi: 10.1074/jbc.274.41.29202. [DOI] [PubMed] [Google Scholar]
- 22.Gao R, et al. A unifying gene signature for adenoid cystic cancer identifies parallel MYB-dependent and MYB-independent therapeutic targets. Oncotarget. 2014;5:12528–12542. doi: 10.18632/oncotarget.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansour MR, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014 doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menzel S, et al. The HBS1L–MYB intergenic region on chromosome 6q23.3 influences erythrocyte, platelet, and monocyte counts in humans. Blood. 2007;110:3624–3626. doi: 10.1182/blood-2007-05-093419. [DOI] [PubMed] [Google Scholar]
- 27.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Vogel G, Yu Z, Richard S. The QKI-5 QKI-6 RNA binding proteins regulate the expression of microRNA 7 in glial cells. Mol. Cell. Biol. 2013;33:1233–1243. doi: 10.1128/MCB.01604-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen A-J, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012;26:1459–1472. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji S, et al. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013;62:716–726. doi: 10.1136/gutjnl-2011-301083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandopadhayay P, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin. Cancer Res. 2014;20:912–925. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Alfonso TM, et al. MYB-NFIB gene fusion in adenoid cystic carcinoma of the breast with special focus paid to the solid variant with basaloid features. Hum. Pathol. 2014;45:2270–2280. doi: 10.1016/j.humpath.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Persson M, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens PJ, et al. Whole exome sequencing of adenoid cystic carcinoma. J. Clin. Invest. 2013;123:2965–2968. doi: 10.1172/JCI67201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danan-Gotthold M, et al. Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conn SJ, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Lu W, et al. QKI impairs self-renewal tumorigenicity of oral cancer cells via repression of SOX2. Cancer Biol. Ther. 2014;15:1174–1184. doi: 10.4161/cbt.29502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crompton BD, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 41.Kieran MW, et al. Absence of oncogenic canonical pathway mutations in aggressive pediatric rhabdoid tumors. Pediatr Blood Cancer. 2012;59:1155–1157. doi: 10.1002/pbc.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
References – Online Methods
- 1.Zhang J, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones DTW, et al. Recurrent somatic alterations of FGFR1 NTRK2 in pilocytic astrocytoma. Nat. Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drier Y, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23:228–235. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torres-García W, et al. PRADA: pipeline for RNA sequencing data analysis. Bioinformatics. 2014;30:2224–2226. doi: 10.1093/bioinformatics/btu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramkissoon LA, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci U S A. 2013;110:8188–8193. doi: 10.1073/pnas.1300252110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig JM, et al. DNA fragmentation simulation method (FSM) and fragment size matching improve aCGH performance of FFPE tissues. PLoS One. 2012;7:e38881. doi: 10.1371/journal.pone.0038881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergthold G, et al. Expression profiles of 151 pediatric low-grade gliomas reveal molecular differences associated with location and histological subtype. Neuro-oncology. 2015 doi: 10.1093/neuonc/nov045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sievert AJ, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A. 2013;110:5957–5962. doi: 10.1073/pnas.1219232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ness SA, Marknell A, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 23.Bandopadhayay P, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin. Cancer Res. 2014;20:912–925. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 25.Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22:1924–1925. doi: 10.1093/bioinformatics/btl196. [DOI] [PubMed] [Google Scholar]
- 26.Garber M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Etchegaray J-P, et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat. Cell Biol. 2015;17:545–557. doi: 10.1038/ncb3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansour MR, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014 doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bengtsen M, et al. c-Myb Binding Sites in Haematopoietic Chromatin Landscapes. PLoS One. 2015;10:e0133280. doi: 10.1371/journal.pone.0133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintana AM, Liu F, O’Rourke JP, Ness SA. Identification and regulation of c-Myb target genes in MCF-7 cells. BMC Cancer. 2011;11:30. doi: 10.1186/1471-2407-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L, et al. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.