Fiber-based methods, which include direct writing and textile techniques have emerged as promising technologies for creating 3D structures with tunable mechanical and biological properties[1–3]. Fiber-based techniques also allow a precise control over the positioning of fibers[4]. These fibers can be used as carriers for drugs, factors, and micro-organisms. In the case of micro-organism laden fibrous constructs, their high porosity facilitates the transport of nutrients and waste[5]. The fibers can also serve as sacrificial structures to form vascular networks in cell-laden structures[6]. In spite of these interesting characteristics, the hydrogels that are used for fabrication of fibers and fibrous assemblies are limited, partly due to low mechanical strength of natural hydrogels[7, 8]. Another challenge that has limited the fabrication of fibers using conventional techniques such as wetspinning, microfluidic spinning, and optofluidic lithography is the need for sophisticated designs or the presence of a quick crosslinking polymer[3, 9]. To extend the applicability of these fiber fabrication techniques to other polymers and hydrogels, here we propose the use of sacrificial polymeric network templates to entrap the pre-polymer during the crosslinking process. This template can later be removed to generate pure polymeric fibers (Figure 1a). The polymeric network employed in the present study is formed from alginate, which is homogeneously mixed with a bioactive polymer.
Figure 1.
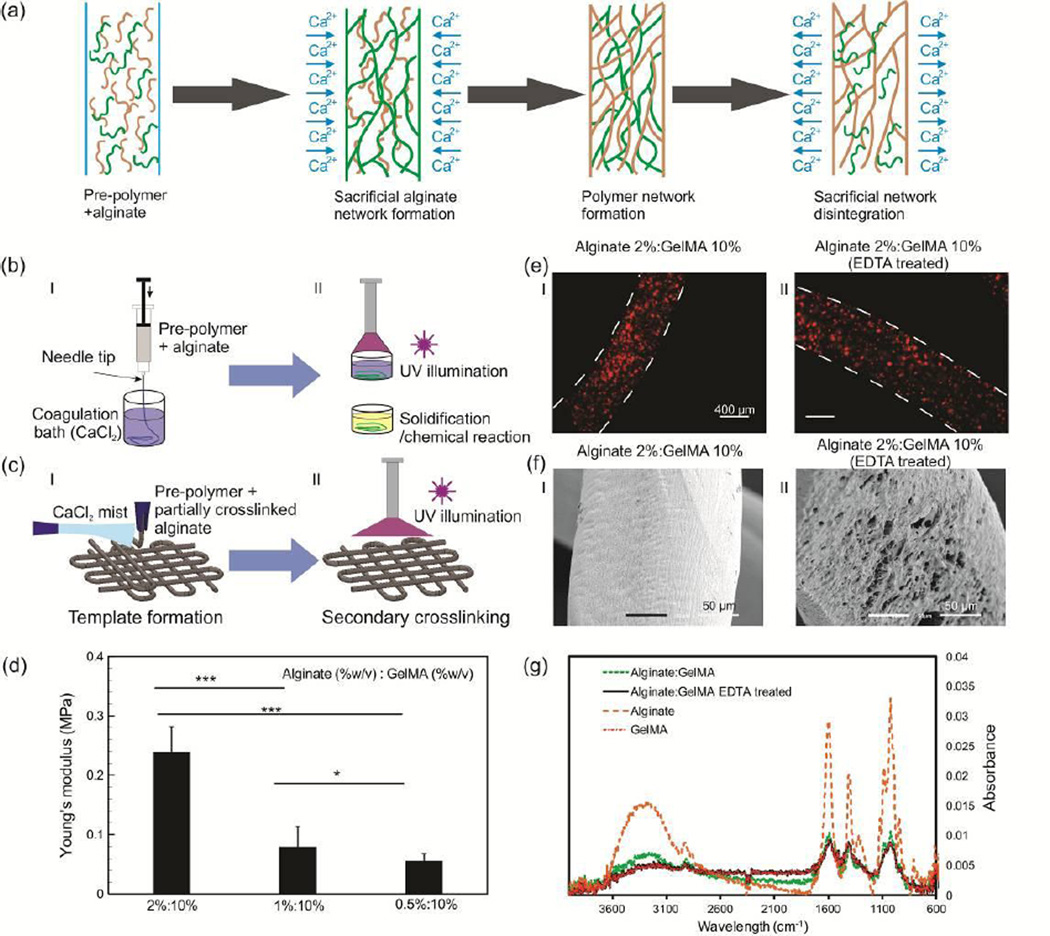
Fabrication of fibers using sacrificial polymeric networks and their characterization. a) Schematic of the process of hydrogel fibers fabrication by use of sacrificial polymeric network. b) Wetspinning of alginate-based fibers in which the process include 2 steps, (i) making a template by initial crosslinking of the mixture of pre-polymer and Na-alginate using CaCl2, (ii) a secondary crosslinking step such as solidification or UV illumination for fully crosslinking of the pre-polymer and formation of stable fibers. c) Two step bioprinting of alginate-based structures, (i) template is formed by crosslinking of Na-alginate in the mixture using CaCl2 fume; (ii) a secondary crosslinking using UV illumination or chemical treatment for fully crosslinking of the pre-polymer. d) Effect of alginate concentration on the Young’s modulus of alginate:GelMA hydrogels (* p < 0.05, ** p < 0.01, and *** p < 0.001). e) Representative micrograph of microbead laden (i) alginate:GelMA (2%:10% w/v) and (ii) GelMA (10% w/v) after alginate removal; microbeads remained entrapped after alginate removal, confirming the stability of hydrogel-based fibers. f) SEM images of (i) alginate:GelMA (2%:10% w/v) and (ii) GelMA (10% w/v) after alginate removal showing that the overall shape and geometry is preserved. g) FTIR spectra for alginate:GelMA (1%:10% w/v), alginate (1% w/v), and fibers treated with EDTA for removal of the sacrificial polymeric network, which confirmed alginate removal from the construct after the treatment.
Alginate is probably the most popular hydrogel for making fibers and 3D structures owing to its rapid and reversible ionic gelation (e.g., by the addition of calcium chloride, CaCl2) as well as its strong mechanical properties compared to many other gels[4, 10]. A common fabrication method is wetspinning, which includes the injection of the pre-polymer solution in a coagulation bath (e.g. CaCl2 solution)[11]. These fibers can also be formed through creating coaxial streams of the pre-polymer and the crosslinking solution in a microchannel[12]. In a notable study, Kang et al. developed a biomimetic microfluidic chip with multiple inlets where the pre-polymer flow rates were adjusted through the use of controllable valves. The chip allowed them to control the cellular distribution, chemical composition, and surface morphology of the fabricated alginate fibers[13]. Whereas alginate fibers alone do not promote cell adhesion, by mixing it with ECM components the desired cellular behavior can be induced[14]. Lately, Oneo et al. used double coaxial stream of ECM proteins and cells, alginate, and CaCl2 to create core shell fibers[15]. Thanks to the mechanical support of the alginate, a simple woven structure was made on a micro-loom operated with great care. However, the core-shell architecture limits the mechanical strength of the fiber, and the dissolution of the alginate can change the porosity and mechanical characteristics of the final construct. Although the mentioned techniques are capable of fabricating complex fibers and constructs, they cannot be easily applied to hydrogels with slow crosslinking process. As a result, limited successful studies are available in the literature for the fabrication of fibers from photocrosslinkable, temperature sensitive, and chemically crosslinkable materials[1, 8, 16, 17]. In a recent study, GelMA fibers were fabricated by direct injection of the solution into a cold solution, which resulted in rapid solidifacation of gelatin forming a fiber. The formed fibers were then crosslinked. However, the technique suffered from significant mixing of the injected solution with the coagulation solution and the fiber composition could not be predicted[17].
Here, alginate is used to generate a polymeric network template that physically entraps various pre-polymer solutions. The entrapped pre-polymer within the alginate template is subsequently crosslinked and forms an independent polymeric network. The use of such a sacrificial template enables extending conventional fiber fabrication techniques including wetspinning to other polymers and hydrogels. Following gelation, the polymer thus replicates the structure of the alginate template. Upon selective dissolution of the alginate by a Ca chelator, a homogeneous and polymeric structure remains (Figure 1a). It is important that the alginate does not interact or react with the target polymer to enable preserving the integrity of the construct upon its dissolution.
The technique is tested for fabrication of fibers from a number of hydrogels including gelatin, agarose, gelatin methacrylate (GelMA), poly(ethylene glycol) diacrylate (PEGDA), and poly(vinyl alcohol) (PVA) frequently used in bioengineering. A schematic of wetspinning and direct writing using a sacrificial polymeric network is presented in Figure 1b. In the wetspinning process, the pre-polymers were dissolved in Na-alginate solution mixed with cells (or beads) and were steadily injected using a syringe pump into CaCl2 solution (Figure 1b). The Na-alginate was crosslinked rapidly by exchange of Na+ with Ca2+, which resulted in the formation of hydrogel networks. Fibers were created by a secondary crosslinking step such as UV illumination for photocrosslinkable hydrogels, a secondary chemical reaction, and temperature variation for thermo-responsive polymers (Figure 1b). It is noteworthy that homogenous mixing of the pre-polymers is a precondition to faithful preservation of the shape and uniformity of the fibers and constructs following alginate dissolution. Thus, only pre-polymers that were readily dissolved in aqueous alginate solutions were used, and thorough mixing was ensured by vortexing immediately prior to spinning the fibers. In particular, we dissolved 10% (w/v) GelMA, 10% (w/v) PEGDA, 2% (w/v) agarose, and 7% (w/v) PVA in 0.5–2% (w/v) Na-alginate solution. These concentrations were selected based on recommended values found in the literature[18]. After wetspinning into 2% (w/v) CaCl2 solution, we applied UV illumination at 850 mW for 1 min to crosslink GelMA and PEGDA. For PVA, we placed the fibers in 4% (w/v) boric acid for at least 5 min. The fibers containing agarose pre-polymer were placed at 5 °C for 5 min to solidify. SEM image of the fabricated fibers are demonstrated in Figure S1. Alginate could then be removed from the fabricated constructs using 20 mM Ethylenediaminetetraacetic acid (EDTA) solution, which was previously found to be well tolerated by cells[7].
Direct writing of pre-polymers inside liquid was extremely challenging and fibers were floating. Thus, simultaneous wetspinning and direct writing was not achievable. To extend the proposed approach to direct writing, we mixed the pre-polymers with alginate and partially crosslinked it by mixing the solution with 0.3 % (w/v) of CaCl2. The partially crosslinked solution was then printed while a continuous stream of CaCl2 mist, created using an ultrasonic humidifier, was passed over the constructs during the writing process to crosslink the structure (Figure 1c). To fully crosslink the fabricated construct, we utilized a secondary crosslinking process as described previously (Figure 1c). This approach enabled us to fabricate multi-layer structures.
Upon confirming the compatibility of sacrificial hydrogel templates to a variety of hydrogels, a comprehensive physical and biological analyses were carried out using alginate:GelMA fibers as a model system. GelMA has been extensively used in tissue engineering, as it is a cost effective material with excellent cell responsive characteristics. Also, GelMA is crosslinked using widely used acrylate-based photocrossinking process. Therefore, GelMA represents a model photocrosslinkable polymer, which demonstrates the wide applicability of our proposed method for other types of hydrogels. The physical properties of fibers formed through wetspinning including the pre-polymer and crosslinking reagent viscosities, injection rate, and the nozzle diameter could be controlled. It was found that the concentration of Na-alginate in the mixture could be used to tune the size and the mechanical characteristics of the fabricated fibers. We dissolved 10% (w/v) GelMA in Na-alginate solutions of 0.5, 1, and 2% (w/v) concentrations containing fluorescent microbeads and injected them using a 25G needle in 2% CaCl2 container (Figure S2). The selected range of alginate ensured mechanically stable sacrificial polymeric networks prior to GelMA crosslinking. An inverse relationship between the Na-alginate concentration in the pre-polymer mixture and the fiber diameter was observed (Figure S2b). For example, the fiber diameter reduced from approximately 1000 µm to 550 µm when the concentration of Na-alginate increased from 0.5 to 2% (w/v). As there was no detectable interaction between alginate and GelMA, pre-polymer leakage might occur during the fabrication process. Thus, in lower alginate concentrations, where the pore sizes are larger this leakage could be more pronounced. This phenomenon might also contribute to the variation of fiber diameter by alginate concentration change. As the fabrication process using CaCl2 mist occurred in dry condition, it is expected that the leakage was less than the case with CaCl2 solution. Regardless, the entrapment within the alginate network reduces the prepolymer leakage in comparison to the method recently developed by Shi et al. for fabrication of GelMA fibers by rapidly solidifying the injected solution[17].
To characterize the compressive mechanical properties of alginate:GelMA hydrogels, circular disks were fabricated. The compressive modulus was increased from 74 kPa to 215 kPa by changing the alginate concentration from 0.5% (w/v) to 2% (w/v) (Figure 1d). To test the effect of alginate removal on the mechanical characteristics of the hybrid hydrogel, the compressive modulus of alginate:GelMA (1%:10% w/v) disks before and after their treatment with EDTA was measured. The results confirmed a reduction in the Young’s modulus as alginate was removed from the constructs (Figure S5). In addition, we noticed that increasing the concentration of alginate in the structure significantly reduced the degradation rate of the formed fibers in phosphate buffer saline (PBS) containing 1 U collagenase (Figure S2c). Following other publications, collagenase was employed to mimic the in vivo enzymatic degradation of GelMA[19].
An important factor that confirmed the success of the proposed fabrication strategy for fiber formation is the stability of the polymeric fibers after the removal of alginate from the constructs. To evaluate fiber stability, we wetspun hybrid fibers containing fluorescent microbeads as described above and treated them with 20 mM EDTA. The microscope images showed that the microbeads remained encapsulated within the fibers (Figure 1e, Figure S3). SEM images of wetspun alginate:GelMA fibers before and after EDTA treatment also illustrated that the structure of the fibers remained stable (Figure 1f, Figure S4). SEM images suggested that the structure of fibers fabricated using a lower alginate concentration (0.5% w/v) had less defects in comparison with those fabricate with a higher alginate concentration. The FTIR spectra of the alginate:GelMA fibers before and after EDTA treatment are shown in (Figure 1g). In the FTIR spectra of the fibers containing the sacrificial polymeric network, the presence of broad band at 3200–3600 cm was associated to the OH and NH component in gelatin and alginate. However, no peaks suggesting a chemical interaction between the two polymers were observed. The reduction in the peak of the alginate:GelMA samples after treatment with EDTA suggests that alginate was removed from the construct. Moreover, the magnitude of the peak at 3200–3600 cm was decreased in the treated sample compared to the non-EDTA treated fibers. This peak was related to the hydrogen bonds of the NH and hydroxyl groups. The peaks at 821 and 946 cm in pure alginate FTIR spectra were associated to the gluronic (G) and manuronic (M) acid functional groups, respectively. These peaks were observed in alginate:GelMA fibers; but the two peaks were not present in the treated fibers, which confirmed that the sacrificial polymeric network was removed from the hydrogel composition. More importantly, the similarity between the spectra of GelMA and fibers with sacrificed polymeric network through EDTA treatment confirmed the dissociation of the sacrificial alginate and the presence of pure GelMA after EDTA treatment. It should be noted that although no covalent bonding was observed between alginate and GelMA, possible non-covalent interactions might prevent complete dissociation of alginate from the fibers.
Although alginate can be removed from the fabricated fibers, its strong mechanical characteristics make fibers containing alginate network easy-to-handle during and after fiber formation. However, alginate lacks cellular binding sites which prevents the encapsulated cells from spreading, which may hinder cellular viability[4]. GelMA on the other hand allows cell attachment and spreading within 3D structures. Thus, we assessed cellular attachment to the alginate:GelMA mixture. In this manuscript we used UV light crosslinking to generate our microfibers. However, UV light illumination always carries the risk of high cell death and DNA damage which affects normal cellular function. Thus, special attention should be paid to the illumination time and intensity. Our previous studies have confirmed normal cellular function for UV (365 nm wavelength) illuminations of less than 60 s at the intensities employed in the present study[20].
To test the ability of the resulting gels to support cell encapsulation, we encapsulated NIH-3T3 fibroblasts in alginate:GelMA fibers containing 10% (w/v) GelMA and different alginate concentrations (0.5%, 1%, 2% w/v). It is known that these cells have a low proliferation rate in alginate-only gels. The cell-laden fibers were then incubated at 5% CO2 and 37 °C inside cell culture media. We assessed cellular viability and metabolic activity using a live/dead cell assay and a MTS assay, respectively (Figure 2). In the live/dead assay, viable cells appear with a green color (due to calcein) and dead cells become red (due to ethidium homodimer-1). The viability results shown in Figure 2a, b indicated that the ratio of dead cells increased more in alginate-only fibers and fibers containing higher concentration of alginate (e.g. 2% w/v). To quantify the metabolic activity of the encapsulated cells, we used MTS assay on NIH-3T3 cells that were encapsulated within approximately 3.5 cm long fibers. Since the diameter of fibers depended on the alginate concentration, the data were normalized with respect to the values for day 1. The results showed that metabolic activity was reduced in both alginate and fibers with 2% alginate concentration. However, we did not observe any reduction in the cellular metabolic activity for the cell-laden fibers containing 1% and 0.5% alginate (Figure 2d). We also investigated cellular morphology by staining for F-actin. The results shown in Figure 2c indicate that NIH-3T3 fibroblasts remained rounded after 5 days of culture in pure alginate and fibers with 2% (w/v) alginate. However, the cells retained their normal morphology inside fibers contained less than 1% (w/v) alginate. The results suggest that the use of less than 1% (w/v) alginate in the pre-polymer solution did not affect the cellular activity. Alginate:GelMA fibers containing (0.5:10% w/v) solutions were fabricated and after 5 days of culture were treated with EDTA to remove the alginate. The fibers were incubated for another day and then stained with live/dead cell assay kit. The results confirmed a high cell survival after the EDTA treatment, consistent with prior studies[7].
Figure 2.
Cellular viability, metabolic activity, and morphology in alginate:GelMA fibers. a,b) Micrographs showing the live (green) and dead (red) NIH-3T3 fibroblasts encapsulated within alginate:GelMA fibers with different compositions over 5 days of culture. c) Staining for α-actin showing the spreading of NIH-3T3 cells encapsulated within alginate:GelMA fibers after 5 days of culture. d) Cellular metabolic activity using MTS assay for 3.5 cm long fibers with the initial cell density of 106/ml. The results are normalized with respect to the values for day 1. Error bars represent standard deviation. e) Viability assay showing the high cellular viability after EDTA treatment for sacrificing the alginate network within the hydrogel fibers after 5 days of culture leaving behind a cell-laden GelMA fiber.
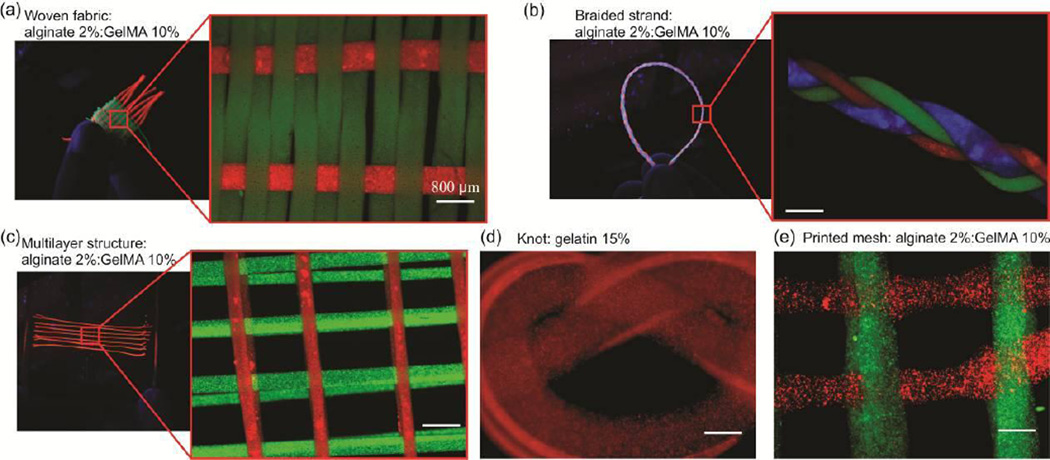
Alginate possesses high mechanical strength in comparison with other natural hydrogels; thus alginate fibers have been previously used in processes such as weaving, printing, and twisting[4, 13, 15, 21]. Textile techniques are powerful techniques that enable the fabrication of 3D constructs with a precise control over the mechanical characteristics as well as the pattern of cells or active molecules. However, textile techniques require the presence of fibers with high mechanical strength. Here, we fabricated alginate:GelMA fibers (2%:10% w/v) with a diameter of 600 µm containing fluorescent microbeads. We fabricated a custom built loom to weave the structure shown in Figure 3a. The fabricated fibers were shown to withstand the fabrication process and generated constructs that could be handled manually. Similarly, three 10 cm long fibers, were braided to create a 3D construct as shown in Figure 3b. These fibers could also be easily stacked to create multilayer structures. We also created a mold and positioned fibers in a predefined pattern as shown in Figure 3c. A fiber of alginate:gelatin (2%:15% w/v) was also created and used to form a knot. The fabricated structure was then treated with EDTA solution to remove alginate from the construct (Figure 3d). To confirm if the alginate was removed from the knot, the construct was placed in warm water (50 °C) which was melted immediately.
Figure 3.
Assembly of fibers with alginate template using various techniques. a) Microbead-laden alginate:GelMA (2%:10% w/v) fibers fabricated using a weaving machine. b) Microbead-laden alginate:GelMA (2%:10% w/v) fibers assembled into several centimeter long braided structure. c) Microbead-laden alginate:GelMA (2%:10% w/v) fibers stacked to form a 3D mesh. d) Alginate:gelatin (2%:15% w/v) fibers were used to form a knot; alginate was dissolved using EDTA to leave behind a gelatin (15% w/v) knot. This knot was instantaneously dissolved in warm water (50 °C). e) Alginate:GelMA (2%:10% w/v) hydrogels printed to form a 3D mesh.
To generate biofabricated structures using this approach, an alginate:GelMA solution with the concentration of 2%:10% (w/v) was mixed with 0.3% (w/v) CaCl2 solution at the ratio of 7 to 3 and was loaded into a syringe connected to a syringe pump. The pre-polymer was then injected through a 21G needle at a flow rate of 1 ml/hr. The needle was moved in a predefined pattern to deposit the fibers parallel to each other (Figure 3e). To crosslink the printed fibers, 2% (w/v) CaCl2 mist was created using an ultrasonic humidifier to crosslink the fibers. After creating the first layer, the pre-polymer solution was changed and fibers were deposited in the perpendicular direction. This procedure was repeated several times to form 3D constructs. A direct written construct with 4 layers is shown in Figure 3e.
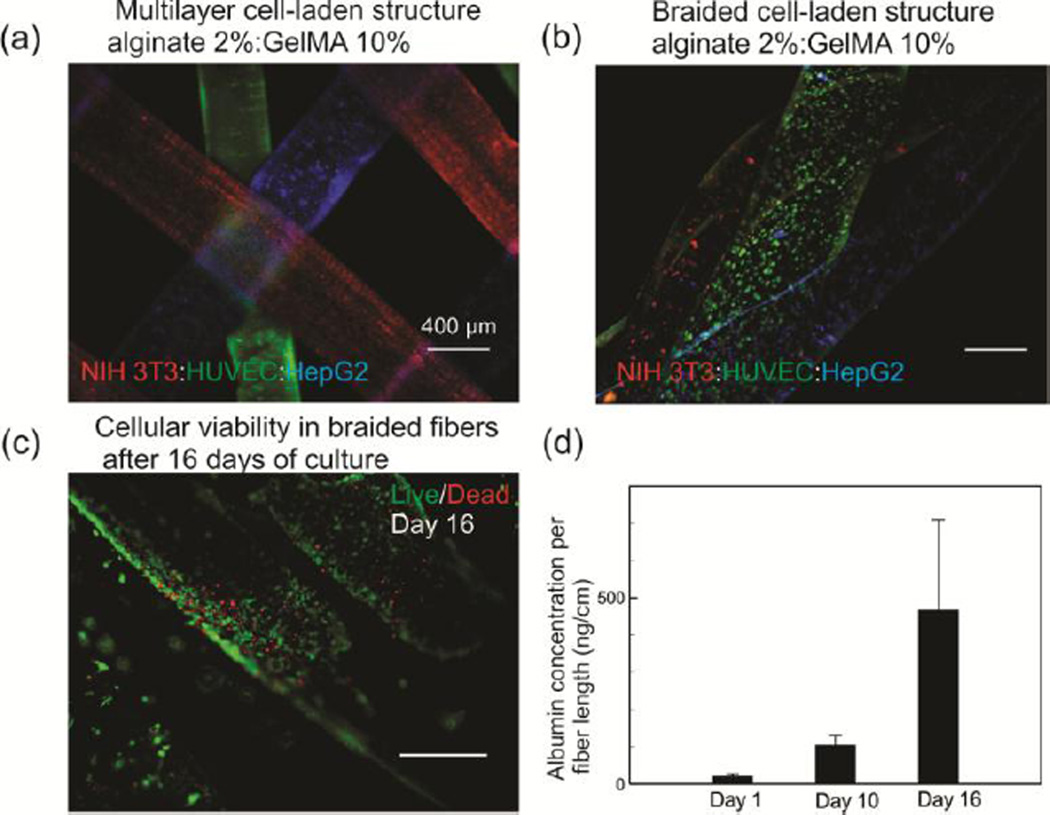
Hydrogel fibers containing NIH-3T3 fibroblasts, human umbilical vein endothelial cells (HUVEC), and HepG2 liver cells with a concentration of 2×106 cell/ml were fabricated. Due to significant mechanical forces applied during the assembly process, cell-laden fibers with alginate concentration of 2% were utilized in the analysis despite the slight lower cellular viability. The cells were stained with different florescent dyes and the cell-laden fibers were assembled by stacking and braiding to form liver models (Figure 4a,b). We fabricated 10 cm of braided structures with fibers containing NIH-3T3 fibroblasts, HUVECs, and HepG2s. The constructs were placed in T25 tissue culture flasks with 5 ml of media and 50% of media was refreshed every 5 days. We measured the albumin concentration in the media over 16 days of culture. The results showed an increasing trend in the albumin concentration, which is an indication of the cellular activity (Figure 4d). The value of albumin secretion per cell in the first day was also estimated to be 1.7 pg/cell/day, which was comparable to the value of 0.6 pg/cell/day reported by Guzzardi et al.[22]. In the analysis we considered a fiber diameter of 600 µm and the concentration of 2×106 cell/ml. After 16 days of culture, the fibers which were very fragile were stained by live/dead assay kit and the results showed a high viability ratio in the fibers (Figure 4c).
Figure 4.
Fabrication and assessment of 3D constructs formed from separate HUVEC-, HepG2-, and NIH-3T3-laden alginate:GelMA (2%:10% w/v) fibers as a model of liver tissue. a) A micrograph showing a multilayer construct form from different cell types (concentration of cells is 2×106 /ml for each type). Cells are stained with cell trackers (NIH-3T3-red; HUVEC-green; HepG2-blue) to assist visualization. b) A micrograph showing a typical braided cell-laden structure from three different cell-laden alginate:GelMA (2%:10% w/v) fibers. The concentration of each cell type is 2×106 /ml for each type. c) High cellular viability in the braided fibers after 16 days of culture. d) Albumin secretion from a braided construct containing three different fibers with HepG2s, HUVECs, and fibroblasts which is normalized to the length of the fibers over 16 days of culture. Error bars represent standard deviation.
In summary, a robust approach for fabricating fibers and fibrous structures from hydrogels was proposed in which alginate was used to create a sacrificial template. The entrapped pre-polymer was then crosslinked and alginate could be removed from the construct. We also proposed an alternative approach for direct writing of hydrogels containing a sacrificial polymeric network, where we used CaCl2 mist to form the fiber templates during the deposition process. A secondary polymerization step created hydrogels. We confirmed that after alginate removal polymeric fibers stayed stable. Alginate can be removed from the fiber or construct using a calcium chelator to manufacture pure polymeric constructs. However, alginate presence improves the mechanical properties of the engineered constructs from protein-based hydrogel with low mechanical properties. Thus, if the alginate concentration is low enough that the biological characteristics of the protein-based network will not be affected significantly, these hybrid fibers can be used for engineered complex fibrous constructs. For example, hybrid gels containing more than 1% (w/v) alginate inhibited cells from spreading in 3D environment, but their morphology became normal following the alginate dissolution.
The hybrid hydrogel fibers were mechanically stronger than those treated with EDTA, which greatly facilitated the assembly and handling of complex 3D structures by direct writing, stacking, or by using textile techniques such as weaving, knotting and braiding. To illustrate the potential of this method, alginate:GelMA fibers containing NIH-3T3 fibroblasts, HepG2s, and HUVECs were formed and assembled into a 3D liver model. The cells remained active over 16 days of culture and the level of albumin in the media increased continuously.
The proposed approach for fabrication of fibers and 3D constructs is easy-to-implement and can be achived using common lab equipment. This approach may be expanded by making finer fibers, exploring novel material combinations, possibly including tri-gels, and assembling them into even more complex structures.
Material and methods
Materials
Chemical reagents were purchased from Sigma-Aldrich St. Louis, MO, USA) unless mentioned otherwise. GelMA was prepared using the method and conditions described in our previous work[23]. Briefly, type-A porcine skin gelatin was dissolved in PBS at 60 °C. Methacrylic anhydride (MA) was added drop wise to the gelatin solution under continuous stirring. The solution was dialyzed against deionized water using 12–14 kDa cut-off dialysis tubes at 50 °C for 7 days to remove unreacted MA. The solution was then freeze-dried and stored at room temperature for further use. GelMA pre-polymer solution was mixed with 0.25% (w/v) photoinitiator (PI) and exposed to UV light (850 mW) for 30 s to crosslink. Cell culture reagents and assays were purchased from Invitrogen (Carlsbad, CA, USA).
Wetspinning of fibers with sacrificial polymeric network
The mixed solution of pre-polymers was prepared in a way that the final alginate concentration was higher than 0.5% (w/v). The solution was loaded in a 3 ml plastic syringe and was connected to a syringe pump (Harvard Apparatus PHD 4400, Holliston, MA, USA). The syringe was connected to a needle using a flexible tube. The solution was directly injected in a bath of 2% CaCl2, where the fiber template was formed immediately and settled at the bottom of the bath. The fibers were then collected and a secondary crosslinking step was performed, depending on the mixed pre-polymer. The secondary crosslinking steps include 30 s UV exposure at 850 mW for GelMA, 60 s UV exposure at 850 mW for PEGDA, 5 min treatment with 4% (w/v) boric acid for PVA, and for 5 min at 5 °C for agarose and gelatin. For alginate network from the fabricated fibers, they were placed in 20 mM EDTA solution for 5 minutes.
Direct writing of hydrogels
To direct-write 3D hydrogel structures, a alginate:GelMA mixture at the final concentration of 2:10% (w/v) was prepared. PI was added to the solution at the concentration of 0.25% (w/v). This concentration of PI has been shown not to affect cellular viability[24]. A solution of 0.3% (w/v) CaCl2 was added this mixture to form a highly viscous and semi-crosslinked solution. The resulting liquid was then loaded on a syringe connected to a needle via a flexible tube. The needle was placed in the slots of a stencil-like pattern and was moved slowly to deposit fibers. The mist crosslinking approach reported by Ahn et al. was followed[25]. Briefly, 2% (w/v) CaCl2 solution was added to an ultrasonic humidifier, so aerosolized CaCl2 would be flown over the bioprinted fibers during the fabrication process. The structures were then fully crosslinked with 2% (w/v) CaCl2 followed by UV illumination for 30 s UV at 850 mW.
Supplementary Material
Acknowledgements
A.K. acknowledges funding from the National Science Foundation (EFRI-1240443), the office of Naval Research Young National Investigator Award, and the National Institutes of Health (HL092836, DE019024, EB012597, AR057837, DE021468, HL099073, EB008392). D.J. acknowledges the financial support of NSERC, CIHR, CHRP, CFI, Genome Canada, Canada Research Chair, and Genome Quebec. M.A. and A.T. acknowledge NSERC Postdoctoral fellowships. N.A. acknowledges the support from the National Health and Medical Research Council.
Footnotes
The authors declare no conflict of interests in this work.
Contributor Information
Dr. Ali Tamayol, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA.
Dr. Alireza Hassani Najafabadi, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA.
Dr. Bahar Aliakbarian, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA.
Dr. Elmira Arab-Tehrany, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA.
Dr. Mohsen Akbari, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, 02115, USA.
Dr. Nasim Annabi, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, 02115, USA.
Prof. David Juncker, Department of Biomedical Engineering, McGill University and Genome Quebec Innovation Centre, McGill University, Montreal, H3A 0G1, Canada; McGill University and Genome Quebec Innovation Centre, Montreal, H3A 2B4, Canada; Department of Neurology and Neurosurgery, McGill University, Montreal, H3A 0G1, Canada.
Prof. Ali Khademhosseini, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, 02115, USA; Department of Physics, King Abdulaziz University, Jeddah 21569, Saudi Arabia.
References
- 1.Tamayol A, Akbari M, Annabi N, Paul A, Khademhosseini A, Juncker D. Biotechnology Advances. 2013;31:669. doi: 10.1016/j.biotechadv.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang CM, Khademhosseini A, Park Y, Sun K, Lee S-H. Langmuir. 2008;24:6845. doi: 10.1021/la800253b. [DOI] [PubMed] [Google Scholar]
- 3.Chung BG, Lee K-H, Khademhosseini A, Lee S-H. Lab on a Chip. 2012;12:45. doi: 10.1039/c1lc20859d. [DOI] [PubMed] [Google Scholar]
- 4.Ghorbanian S, Qasaimeh M, Akbari M, Tamayol A, Juncker D. Biomed Microdevices. 2014;1 doi: 10.1007/s10544-014-9842-8. [DOI] [PubMed] [Google Scholar]
- 5.Tamayol A, Wong K, Bahrami M. Physical Review E. 2012;85:026318. doi: 10.1103/PhysRevE.85.026318. [DOI] [PubMed] [Google Scholar]
- 6.Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Khademhosseini A. Lab on a Chip. 2014;14:2202. doi: 10.1039/c4lc00030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbari M, Tamayol A, Laforte V, Annabi N, Hassani A, Khademhosseini A, Juncker D. Advanced Functional Materials. 2014;24:4060. doi: 10.1002/adfm.201303655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Annabi N, Tamayol A, Uquillas JA, Akbari M, Bertassoni LE, Cha C, Camci-Unal G, Dokmeci MR, Peppas NA, Khademhosseini A. Advanced Materials. 2014;26:85. doi: 10.1002/adma.201303233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho S, Shim TS, Yang S-M. Lab on a Chip. 2012;12:3676. doi: 10.1039/c2lc40439g. [DOI] [PubMed] [Google Scholar]; Habasaki S, Yoshida S, Lee WC, Takeuch S. Vertical continuous flow lithography for fabricating long 3D structures. presented at Micro Electro Mechanical Systems (MEMS), 2013 IEEE 26th International Conference on; 20–24 Jan. 2013.2013. [Google Scholar]; Cheng Y, Zheng F, Lu J, Shang L, Xie Z, Zhao Y, Chen Y, Gu Z. Advanced Materials. 2014 doi: 10.1002/adma.201400798. [DOI] [PubMed] [Google Scholar]; Panda P, Ali S, Lo E, Chung BG, Hatton TA, Khademhosseini A, Doyle PS. Lab on a Chip. 2008;8:1056. doi: 10.1039/b804234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takei T, Kishihara N, Sakai S, Kawakami K. Biochemical Engineering Journal. 2010;49:143. [Google Scholar]; Kang E, Choi YY, Chae S-K, Moon J-H, Chang J-Y, Lee S-H. Advanced Materials. 2012;24:4271. doi: 10.1002/adma.201201232. [DOI] [PubMed] [Google Scholar]; Augst AD, Kong HJ, Mooney DJ. Macromolecular Bioscience. 2006;6:623. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]; Chae S-K, Kang E, Khademhosseini A, Lee S-H. Advanced Materials. 2013;25:3071. doi: 10.1002/adma.201300837. [DOI] [PubMed] [Google Scholar]; Grigoryev A, Sa V, Gopishetty V, Tokarev I, Kornev KG, Minko S. Advanced Functional Materials. 2013;23:5903. [Google Scholar]
- 11.Qiu W, Teng W, Cappello J, Wu X. Biomacromolecules. 2009;10:602. doi: 10.1021/bm801296r. [DOI] [PubMed] [Google Scholar]
- 12.Jun Y, Kang E, Chae S-K, Lee S-H. Lab on a Chip. 2014 doi: 10.1039/c3lc51414e. [DOI] [PubMed] [Google Scholar]
- 13.Kang E, Jeong GS, Choi YY, Lee KH, Khademhosseini A, Lee S-H. Nature Materials. 2011;10:877. doi: 10.1038/nmat3108. [DOI] [PubMed] [Google Scholar]
- 14.Gillette BM, Jensen JA, Wang M, Tchao J, Sia SK. Advanced Materials. 2010;22:686. doi: 10.1002/adma.200902265. [DOI] [PubMed] [Google Scholar]
- 15.Onoe H, Okitsu T, Itou A, Kato-Negishi M, Gojo R, Kiriya D, Sato K, Miura S, Iwanaga S, Kuribayashi-Shigetomi K. Nature Materials. 2013 doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- 16.Lewitus DY, Landers J, Branch JR, Smith KL, Callegari G, Kohn J, Neimark AV. Advanced Functional Materials. 2011;21:2624. doi: 10.1002/adfm.201002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi X, Ostrovidov S, Zhao Y, Liang X, Kasuya M, Kurihara K, Nakajima K, Bae H, Wu H, Khademhosseini A. Advanced Functional Materials. 2015;25:2250. [Google Scholar]
- 18.Chen YC, Lin RZ, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A. Advanced Functional Materials. 2012;22:2027. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sonnet C, Simpson CL, Olabisi RM, Sullivan K, Lazard Z, Gugala Z, Peroni JF, Weh JM, Davis AR, West JL. Journal of Orthopaedic Research. 2013;31:1597. doi: 10.1002/jor.22407. [DOI] [PubMed] [Google Scholar]; Wirawan F, Cheng C-L, Kao W-C, Lee D-J, Chang J-S. Applied Energy. 2012;100:19. [Google Scholar]
- 19.Zhao X, Lang Q, Yildirimer L, Lin ZY, Cui W, Annabi N, Ng KW, Dokmeci MR, Ghaemmaghami AM, Khademhosseini A. Advanced healthcare materials. 2015 doi: 10.1002/adhm.201500005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen YC, Lin RZ, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A. Advanced Functional Materials. 2012 doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annabi N, Tsang K, Mithieux SM, Nikkhah M, Ameri A, Khademhosseini A, Weiss AS. Advanced Functional Materials. 2013 doi: 10.1002/adfm.201300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song S-J, Choi J, Park Y-D, Hong S, Lee JJ, Ahn CB, Choi H, Sun K. Artificial Organs. 2011;35:1132. doi: 10.1111/j.1525-1594.2011.01377.x. [DOI] [PubMed] [Google Scholar]
- 22.Guzzardi MA, Vozzi F, Ahluwalia AD. Tissue Engineering Part A. 2009;15:3635. doi: 10.1089/ten.TEA.2008.0695. [DOI] [PubMed] [Google Scholar]
- 23.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancock MJ, Piraino F, Camci-Unal G, Rasponi M, Khademhosseini A. Biomaterials. 2011;32:6493. doi: 10.1016/j.biomaterials.2011.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doggui S, Sahni JK, Arseneault M, Dao L, Ramassamy C. Journal of Alzheimer's Disease. 2012;30:377. doi: 10.3233/JAD-2012-112141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.