Abstract
Our previous publications and the data presented here provide evidences on the ability of plant-based culture media to optimize the cultivability of rhizobacteria and to support their recovery from plant-soil environments. Compared to the tested chemically-synthetic culture media (e.g. nutrient agar and N-deficient combined-carbon sources media), slurry homogenates, crude saps, juices and powders of cactus (Opuntia ficus-indica) and succulent plants (Aloe vera and Aloe arborescens) were rich enough to support growth of rhizobacteria. Representative isolates of Enterobacter spp., Klebsiella spp., Bacillus spp. and Azospirillum spp. exhibited good growth on agar plates of such plant-based culture media. Cell growth and biomass production in liquid batch cultures were comparable to those reported with the synthetic culture media. In addition, the tested plant-based culture media efficiently recovered populations of rhizobacteria associated to plant roots. Culturable populations of >106–108 cfu g−1 were recovered from the ecto- and endo-rhizospheres of tested host plants. More than 100 endophytic culture-dependent isolates were secured and subjected to morphophysiological identification. Factor and cluster analyses indicated the unique community structure, on species, genera, class and phyla levels, of the culturable population recovered with plant-based culture media, being distinct from that obtained with the chemically-synthetic culture media. Proteobacteria were the dominant (78.8%) on plant-based agar culture medium compared to only 31% on nutrient agar, while Firmicutes prevailed on nutrient agar (69%) compared to the plant-based agar culture media (18.2%). Bacteroidetes, represented by Chryseobacterium indologenes, was only reported (3%) among the culturable rhizobacteria community of the plant-based agar culture medium.
Keywords: Plant-based culture media, Rhizobacteria, Cultivability, Community structure of rhizobacteria, Cactus, Succulent plants
Introduction
Prokaryotic taxonomy is nowadays based on genome data, which allows classification of non-culturable bacteria, and greatly contributes to our understanding of the microbial diversity in the plant-soil system [1], [2]. However, culturable methods are far from redundant but required, and present a challenge to environmental microbiology specialists [3]. They provide information about communities that cannot be obtained directly from sequencing efforts and/or culture-independent methods alone [4], [5], [6], and remain important for the physiological and genetical characterization of specific bacterial species containing functionally important traits. In addition, the isolation of individual bacterial species in pure cultures allows full assessment of environmental impacts and further manipulation for the benefit of natural ecosystems. Developments over the last decade have led to the recovery of unculturables from various populated habitats, e.g. the use of dilute nutrient media, long-term incubation, encapsulating individual cells into gel microdroplets (GMD), diffusion chambers, and the soil substrate membrane system [6], [7], [8].
Through rhizodeposition, small-molecular weight secondary metabolites, amino acids, secreted enzymes, mucilage, and cell lysates, plants may induce selective pressure on the microbial composition in the root region [9], [10], [11], [12]. In an effort to better reproduce the natural environment, plant-based culture media have been introduced for the culturing of rhizobacteria as a sole growth milieu [13]. From a theoretical point of view, plant juices and/or extracts are well suited as culture media for microbial growth and fermentations, as they contain all the necessary nutrients as well as growth factors such as amino acids, vitamins and minerals. Representatives of pathogenic fungi and human pathogens were successfully grown on the extracts/juices of a variety of plants as well as legume seeds-protein [14], [15]. Recovery and isolation of fungal endophytes of Hordeum murinum were significantly improved by supplementing commercial culture media with the whole host plant extract [16]. Furthermore, microbial metabolites were productively recovered from culture media based on plant substrates, especially the by-products of agro-industries [17], [18], [19], [20]. Green biorefinery of brown and green juices produces varieties of organic acids, amino acids, feed additives, enzymes, proteins, peptides or fungal and bacterial biomass [21].
Data presented here provide further support for our original approach of the sole use of plant-based culture media to replace the chemically-synthetic standard ones, traditionally used for culturing of rhizobacteria [13]. A number of cacti (Opuntia. ficus-indica; prickly pears) and succulent (Aloe vera and Aloe arborescens) plants were used to obtain the nutrient juices, saps, slurry homogenates and powders for the preparation of plant-based culture media. Such media were tested for culturing rhizobacteria present in pure cultures (in vitro) and for recovering the rhizobacteria population associated with roots of homologous and heterologous host plants. Secured isolates of culturable endophytic rhizobacteria were subjected to morphophysiological identification, to compare the effect of culture media tested on their community structure.
Material and methods
Tested plants
The tested plants, the cactus Opuntia ficus-indica (prickly pears) and the succulent plants Aloe vera and Aloe arborescens, are cultivated in Orman Botanical Garden, Giza- Egypt, as ornamental plants for display and research. Such plants were chosen for their availability in arid and semi-arid environments as well as their copious juicy nature.
Samples of the full-grown plants were obtained by first insertion and separation of the vegetative part of plant into plastic bags. Then, the root system (intact roots with closely adhering soil) was carefully removed and transferred to plastic bags. Free soil samples were secured from the soil nearby the roots. Samples were kept in the refrigerator until analyses, which were conducted within few days of sampling.
Preparation of plant-based culture media
The succulent leaves of A. vera and A. arborescens and mature stem pads of O. ficus-indica were washed, sliced, and then blended with equal aliquots of distilled water (w/v) for ca. 5 min in a Waring blender. The resulting slurry homogenate was used as such or coarse-filtered through cheesecloth to obtain plant juice; ca. 73–82% of the plant fresh weight was recovered as juice. To obtain plant saps, the succulent leaves of Aloe vera and Aloe arborescens were washed, sliced and manually pressed by a squeezer; the sap recovered represented ca. 24–26% of the plant fresh weight. The pH for saps, juices and slurry homogenates was in the range of 3.6–5.2. All plant substrates were stored at −20 °C. In addition, a dehydrated powder was prepared from cactus (O. ficus-indica): stem pads were sliced and sun-dried (>30 °C) for 3–4 days, then further oven-dried in a hot air (70 °C) for 48 h and mechanically-grinded to pass through a 2-mm sieve.
The plant homogenates, juices and saps obtained from the tested plants were further diluted with distilled water (v/v); 1:10, 1:20, 1:40, 1:80, and 1:100. Exclusively, such diluted juices and saps were used as such to prepare the plant-based agar culture media (2% agar, w/v). For the dehydrated powder of cactus the liquid and agar (2% agar) culture media were prepared by dissolving 4 g in 1 L of distilled water. All media were adjusted to pH 7.0 and autoclaved at 121 °C for 20 min.
Chemically-synthetic standard culture media
The rich nutrient agar [22] and N-deficient combined carbon-sources medium (CCM) [23] were used.
Nutrient agar [22]: It contains (g l−1): beef extract, 3.0; peptone, 5.0; glucose, 1.0; yeast extract, 0.5; agar, 15; pH, 7.2.
N-deficient combined carbon sources medium, CCM [23]: It comprises of (g l−1): glucose, 2.0; malic acid, 2.0; mannitol, 2.0; sucrose, 1.0; K2HPO4, 0.4; KH2PO4, 0.6; MgSO4, 0.2; NaCl, 0.1; MnSO4, 0.01; yeast extract, 0.2; fermentol (a local product of corn-steep liquor), 0.2; KOH, 1.5; CaCl2, 0.02; FeCl3, 0.015; Na2 MoO4, 0.002. In addition, CuSO4, 0.08 mg; ZnSO4, 0.25 mg; sodium lactate, 0.6 ml (50% v/v) were added per litre.
Growth of rhizobacteria isolates on agar plates
Representative pure isolates of rhizobacteria (Azospirillum brasilense, Bacillus circulans, Bacillus macerans, Bacillus polymyxa, Enterobacter agglomerans, and Klebsiella oxytoca) were obtained from the culture collection of the Department of Microbiology, Faculty of Agriculture, Cairo University, Giza [24], [25]. They were initially inoculated into semi-solid CCM test tubes, and the bacterial batch cultures were microscopically examined for growth and purity. Aliquots of 100 μl were carefully spread on the surfaces of agar plates representing plant-based agar plates, prepared from successive dilutions of various plant materials, as well as the standard nutrient agar and CCM. With incubation at 30 °C for 7 days, the growth index recorded was: 1, scant (discontinued bacterial lawn, with scattered colonies); 2–3, good (continued bacterial lawn); and 4–5, very good (continued and more dense bacterial lawn).
Growth and biomass production of rhizobacteria isolates in liquid batch cultures
The growth of Enterobacter agglomerans and Klebsiella oxytoca was monitored in liquid plant-based culture media, using cactus powder (4 g/litre) and slurry homogenate (diluted with distilled water 1:20, v/v). For comparisons, the standard liquid combined carbon sources medium (CCM) was included. The liquid culture media were prepared (100 ml in 250 ml-capacity Erlenmeyer flasks), inoculated with tested isolates (2%, v/v), and incubated at 30 °C in a rotary shaker (100 rpm) for up to 45 days. Periodic samples from the resulting batch cultures were surface inoculated on CCM agar plates, in triplicate, for cfu counting. Growth curves were plotted and doubling times were calculated [26].
Cultivability and recovery of rhizobacteria associated with plant roots on plant-based culture media
The ecto-rhizosphere samples, representing the root surfaces together with closely-adhering soil, were prepared [24], [25] from the tested plants, A. vera and A. arborescens. For the endo–rhizosphere samples, roots were surface-sterilized with 95% ethanol for 5–10 s followed by 3% sodium hypochlorite for 30 min, and then carefully washed with sterilized distilled water before crushing in Waring blender with adequate volume of basal salts of CCM [27]. Further serial dilutions were prepared for each of the ecto- and endo-rhizosphere samples. For each sample, aliquots of 200 μl of suitable dilutions were used to surface-inoculate 21 agar plates, 3 replicates from each dilution, representing plant-based culture media prepared from the tested crude juices/saps (diluted with distilled water; 1:20 and/or 1:40, v/v), as well as the standard nutrient agar and CCM culture media. Incubation took place at 30 °C for 2–7 days and cfu were counted. Dry weights for suspended roots (70 °C) and rhizosphere soil (105 °C) were determined.
Morphophysiological identification of endophytic rhizobacteria developed on agar plates
Representative agar plates, having 30–70 cfu plate−1, were selected to represent various culture media: standard nutrient agar, N-deficient combined carbon sources medium (CCM) and plant juice/sap-based culture media. By single colony isolation, the secured pure isolates of all developed colonies were examined for growth, colony and cell morphology, Gram stain and cultural characteristics including catalase and oxidase production. Then, biochemical test kits (bioMerieux API) were used for bacterial identification [28]: API 20E for Enterobacteriaceae, API 20NE for non-Enterobacteriaceae and API 50CHB for bacilli. Test results and constructed numerical profiles were entered into the online database [29] to determine bacterial identification.
Chemical analysis of the dehydrated powder of cactus (O. ficus-indica)
The chemical compositions and nutritional contents of the tested succulent plants (A. vera and A. arborescens) are available in the literature [30]. Therefore, special attention was given to the analysis of the tested powder of cactus (O. ficus-indica), for possible future application in biomass production required for formulation of bio-preparates (biofertilizers and biopesticides). Macro- and micro-nutrients were detected by atomic absorption analysis, total protein by TruSpec N instrument, amino acids by performic oxidation method and vitamins by GC/MS/MS analyses. Total crude fibre and ash were also determined (Table 1).
Table 1.
Nutritional profile of the dehydrated powder of cactus (O. ficus-indica) as determined by chemical analysis.
| Parameters | O. ficus-indica (oven dried powder) | Parameters | O. ficus-indica (oven dried powder) |
|---|---|---|---|
| Macro nutrients (ppm)a | Micronutrients (ppm)a | ||
| Ca++ | 0.5625 | Cu | 2.06 |
| Mg++ | 0.0143 | Zn | 0.3393 |
| K+ | 1.736 | Fe | 1.955 |
| Na+ | 1.246 | Mn | 1.16 |
| Se (ppb) | 41.08 | ||
| Pb (ppb) | 0.1066 | ||
| Total P (%) | 0.09 | Total carbohydrate (%) | 52.44 |
| Total ash (%)f | 21.4 | Total crude protein (%)b,c | 8.5 |
| Total crude fibre (%)f | 9.16 | ||
| Amino acids (%)d | Amino acids (%) | ||
| Aspartic | 0.65 | Tyrosine | 0.25 |
| Threonine | 0.31 | Phenylalanine | 0.39 |
| Serine | 0.32 | Histidine | 0.16 |
| Glutamic | 0.8 | Lysine | 0.28 |
| Glycine | 0.37 | Arginine | 0.34 |
| Alanine | 0.4 | Proline | 0.48 |
| Valine | 0.35 | Cysteine | 0.14 |
| Isoleucine | 0.29 | Methionine | 0.08 |
| Leucine | 0.51 | ||
| Vitamin A (ppm)e | 7.55 | ||
| Vitamin B (ppm) | 556 | ||
Baker, A.S., & Smith, R.L. (1974). Preparation of solutions for atomic absorption analyses of iron, manganese, zinc, and copper in plant tissue. J. Agric. Food Chem. 22, 103–107.
Truspec Nitrogen Determinator Instruction Manual. March (2006) Part number 200–289.
AOAC International, & Latimer, G. W. (2012a). Official Methods of Analysis of AOAC International. AOAC International. Chapter 4, P. 25–26.
AOAC International, & Latimer, G. W. (2012b). Official Methods of Analysis of AOAC International. AOAC International. Chapter 4, P. 9–13.
Lehotay, S. J., & Hajšlová, J. (2002). Application of gas chromatography in food analysis. TrAC Trends in Analytical Chemistry, 21(9), 686–697.
AOAC International. “Official Methods of Analysis.” (1998).
Statistical analysis
STATISTICA 10.0 [31] was used for the analysis of variance (ANOVA) to examine the significant effects of culture media, root spheres and incubation periods at the level of p < 0.05. Culture media clustering was also done by the principle components extraction.
Results and discussion
Growth of rhizobacteria isolates on agar plates
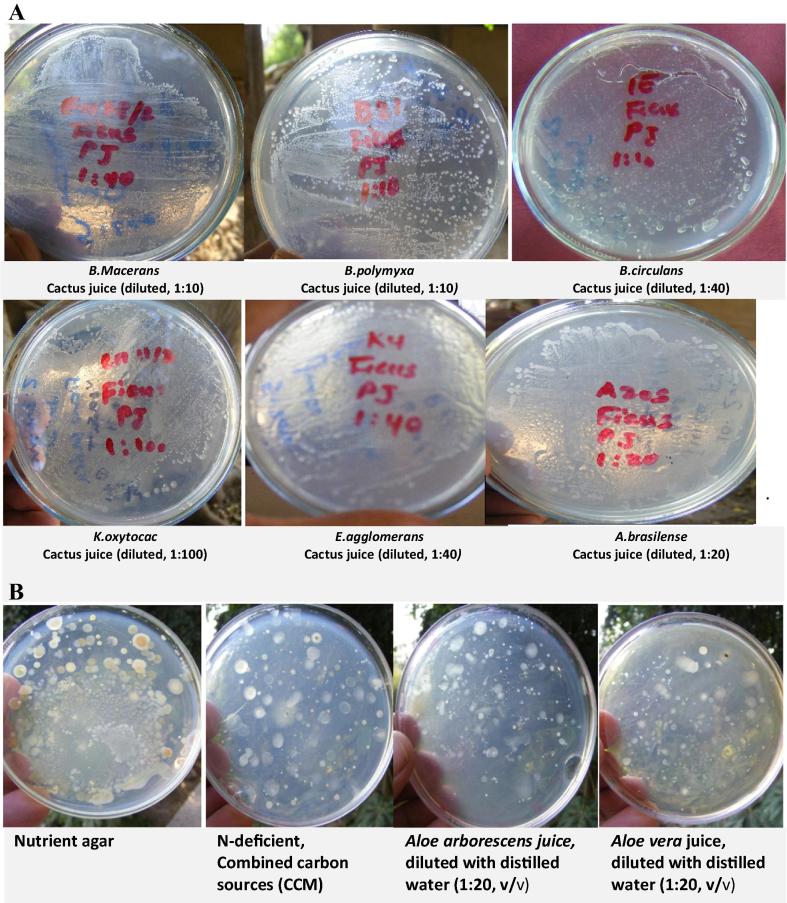
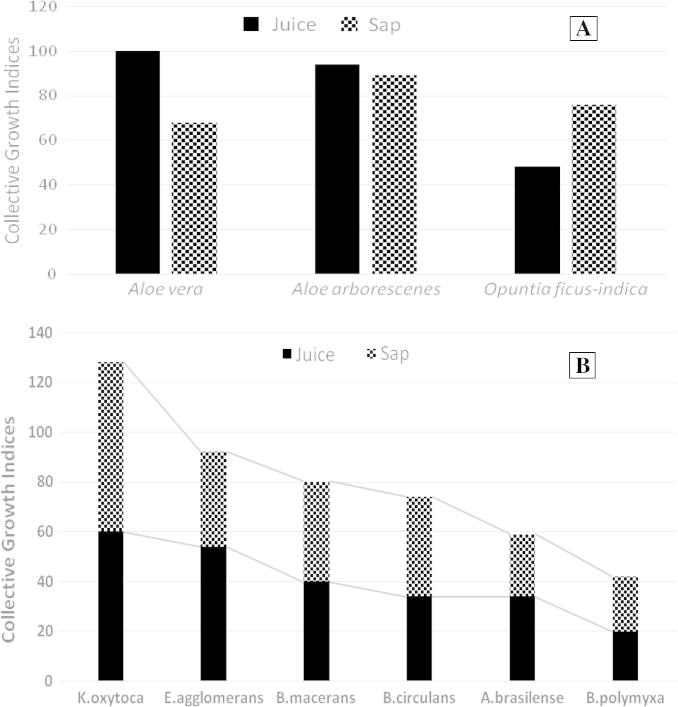
The growth of representative isolates of rhizobacteria was tested on agar plates prepared from various plant-based and synthetic standard culture media. Results indicated that both the juices and saps of all tested plants were nutritionally rich enough to support good growth of the majority of tested rhizobacteria isolates. Good bacterial growth was even obtained with further dilutions up to 1:80 (juice or sap: distilled water, v/v) (Fig. 1A). Such positive dilution effect very possibly attributed to decreasing the osmotic effect of concentrated nutrients as well as minimizing the inhibitory effect of antimicrobial compounds present in the juices/saps of tested plants [30]. The tested cactus (Table 1) and succulent plants [30] are reported to contain >75 active constituents: vitamins, enzymes, minerals, sugars, lignins, saponins, salicylic acid and amino acids. The plant effect was demonstrated; the sap of Opuntia ficus-indica was relatively richer to support better growth of rhizobacteria compared to its juice, Aloe vera sap was not as supportive of growth as its juice, while both juice and sap of Aloe arborescens were of about the same nutritional reserve to support good growth of tested rhizobacteria (Fig. 2A). Irrespective of plant type and material (juice/sap), Klebsiella oxytoca exhibited the highest overall growth on the plant-based culture media, followed, in a descending order, by E. agglomerans, B. macerans, B. circulans, A. brasilense and B. polymyxa (Fig. 2B). Several isolates representing other genera of rhizobacteria, e.g. Pseudomonas pp. (Ps. putida, Ps. luteola and Ps. cepacia), Azotobacter spp. (A. chroococcum), Enterobacter spp. (E. cloacae and E. sakazakii) and yeasts (Saccharomyces spp.) were nicely developed on a wide variety of plant-based culture media (unpublished data of the graduation projects of Rahma Nemr and Dina ElSabagh, personal communication).
Fig. 1.
(A) Growth of pure isolates of rhizobacteria on agar plates prepared from the diluted crude juice (1:10–1:100, v/v) of the cactus O. ficus-indica. (B) Recovery of endophytic rhizobacteria associated with plant roots on various culture media. Normal distinctive colonies of rhizobacteria associated with the roots of Aloe vera developed on agar plates of plant-based culture media (inoculated with similar aliquots of the same root dilution, 10–2), prepared from diluted juices (1:20, v/v) of A. vera and A. arborescens in comparison with those developed on synthetic standard culture media (nutrient agar and CCM).
Fig. 2.
Growth of pure isolates of rhizobacteria on agar plates. Collective, i.e. the aggregate of growth indices scored for all tested isolates of rhizobacteria on plant-based culture media: (A) Growth on plant juices and saps irrespective of independent tested rhizobacteria isolates and/or juice/sap dilutions, (B) Growth of individual rhizobacteria isolates, irrespective of plant type or juice/sap dilutions.
Growth and biomass production of rhizobacteria isolates in liquid batch cultures
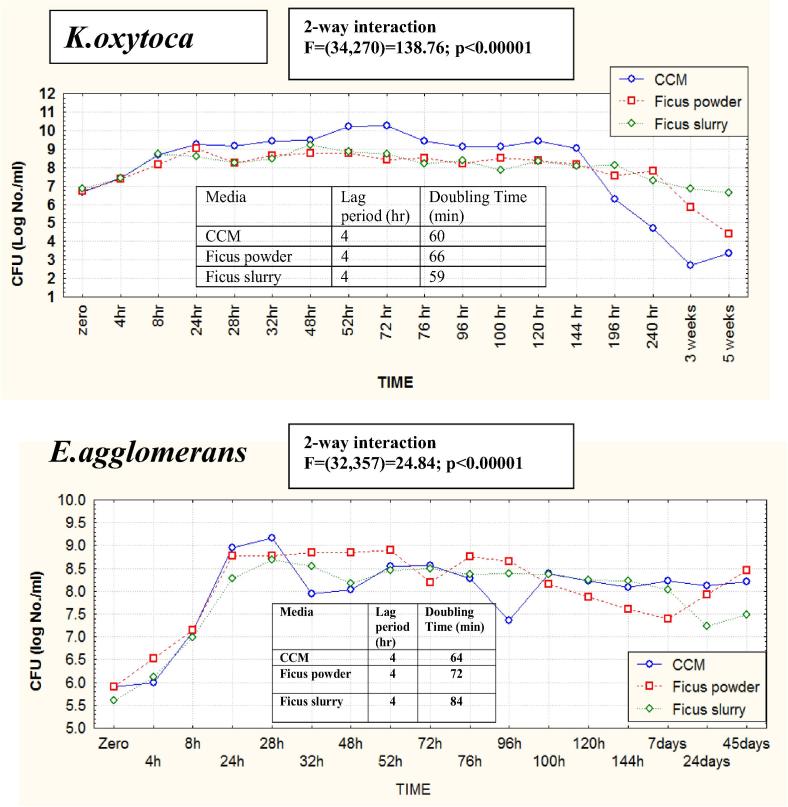
When grown in liquid batch cultures, Klebsiella oxytoca exhibited excellent growth in culture media prepared from O. ficus-indica slurry homogenate and powder, with growth velocity very much comparable to the synthetic standard CCM culture medium (doubling times of 59–66 min) (Fig. 3). Relatively, the plant-based culture media supported longer cell viability that extended to >3 weeks. This is probably due to the nutrient complexity and diversity of the plant nutrients together with the limited development of acidity and suppressive metabolites. Similarly, cells of Enterobacter agglomerans were sufficiently produced in the plant-based culture (Fig. 3). These results strongly recommend the use of plant-based culture media for rhizobacterial biomass production required for the formulation of bio-preparates (biofertilizers/biopesticides) [32].
Fig. 3.
The normal growth curves and cell biomass production of Klebsiella oxytoca (above) and Enterobacter agglomerans (below) in liquid batch cultures prepared from the slurry homogenate and dehydrated powder of cactus (O. ficus-indica), compared to the synthetic standard CCM culture medium. Inserted are tables showing calculated doubling times and lag periods.
Cultivability and recovery of rhizobacteria associated with plant roots on plant-based culture media
The tested plant-based culture media successfully supported the culturing of rhizobacteria present in the root theatre, free soil, ecto- and endo-rhizospheres of A. vera. The nutrient store in the tested plant juices as such (Table 1) was rich enough to support growth of rhizobacteria, very much comparable to the chemically-synthetic standard culture media (nutrient agar and CCM). Developed colonies were distinct, easily distinguished and of confined not spread over growth (Fig. 1B). Statistically, significant differences were attributed to the independent effects of incubation period, plant sphere and culture medium (Table 2). Higher recovery of rhizobacteria was reported, for the plant-based culture media in particular, by extending the incubation period up to 7 days, as differences among tested culture media were diminished. The ecto-rhizosphere accommodated the highest population densities of rhizobacteria (>107 g−1 root) compared to the endo-rhizosphere (>106 g−1 root). In fact, the ecto-rhizosphere represents the most bioactive interface of roots with the adjacent soil, and often reported to be the richest sphere in populations of rhizobacteria. With rice, further metagenomic and proteomic approaches [33] have clearly identified not two but three distinct compartments, rhizosphere, rhizoplane and endosphere, with a decreasing gradient in microbial richness and diversity from the rhizosphere to the endosphere.
Table 2.
Mean values and ANOVA analysis of culturable rhizobacteria (g−1 root) associated with roots of A. vera developed on agar plates of plant-based culture media (prepared from juices of A. vera and A. arborescens diluted 1:20 and 1:40, v/v) and standard culture media (NA, nutrient agar; CCM, N-deficient combined carbon sources). Means followed by the same letter are not statistically different.
| Treatments | (Log No g−1 root) |
|---|---|
| Factor (A): time of incubation | |
| 1–3 days | 6.51B |
| 2–7 days | 7.02A |
| L.S.D. (p ⩽ 0.05) | 0.15 |
| Factor (B): root sphere | |
| 1-Free soil | 6.69B |
| 2-Endo-rhizosphere | 6.16C |
| 3-Ecto-rhizosphere | 7.44A |
| L.S.D. (p ⩽ 0.05) | 0.18 |
| Factor (C): culture media | |
| 1-NA | 7.15A |
| 2-CCM | 6.56C |
| 3-A. vera juice (diluted 1:20) | 6.74BC |
| 4-A. vera juice (diluted 1:40) | 6.63BC |
| 5-A. arbo juice (diluted 1:20) | 6.87B |
| 6-A. arbo juice (diluted 1:40) | 6.64BC |
| L.S.D. (p ⩽ 0.05) | 0.26 |
| 2-way interactions: root sphere × culture media (B × C) | |
| 1. Free soil × NA | 6.74CD |
| 2. Free soil × CCM | 6.50DEF |
| 3. Free soil × A. vera 1:20 | 6.59DE |
| 4. Free soil × A. vera 1:40 | 6.48DEF |
| 5. Free soil × A. arbo 1:20 | 7.21B |
| 6. Free soil × A. arbo 1:40 | 6.63D |
| 1. Endo-rhizosphere × NA | 6.75CD |
| 2. Endo-rhizosphere × CCM | 6.01G |
| 3. Endo-rhizosphere × A. vera 1:20 | 6.08FG |
| 4. Endo-rhizosphere × A. vera 1:40 | 5.94G |
| 5. Endo-rhizosphere × A. arbo 1:20 | 6.01G |
| 6. Endo-rhizosphere × A. arbo 1:40 | 6.16EFG |
| 1. Ecto-rhizosphere × NA | 7.95A |
| 2. Ecto -rhizosphere × CCM | 7.15BC |
| 3. Ecto -rhizosphere × A. vera 1:20 | 7.56AB |
| 4. Ecto -rhizosphere × A. vera 1:40 | 7.47B |
| 5. Ecto -rhizosphere × A. arbo 1:20 | 7.39B |
| 6. Ecto -rhizosphere × A. arbo 1:40 | 7.13BC |
| L.S.D. (p ⩽ 0.05) | 0.45 |
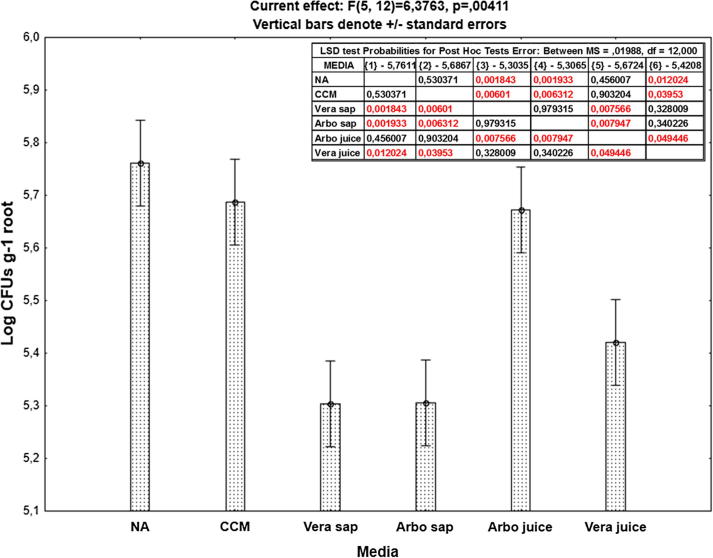
As to culture media, the plant juice-based culture media yielded populations in the range of >106–107 g−1 root, being higher than those reported by the use of the N-deficient combined carbon sources medium (ca. 106 g−1 root) and very much comparable to those of the rich nutrient agar (ca. 107 g−1 root). The two-way interactions of culture media and root spheres indicated the ability of plant juice-based culture media to support cultivability of rhizobacteria present in both ecto- and endo-rhizospheres, very much similar to the synthetic standard culture media (Table 2).
Similarly, endophytic rhizobacteria of Aloe arborescens were successfully recovered on agar plates prepared from plant juices and saps of homologous (Aloe arborescens) and heterologous (Aloe vera) plants. The homologous not the heterologous juice/sap supported the recovery of higher populations of endophytic rhizobacteria (>105–106 g−1 root), very much similar to those developed on the synthetic standard culture media (Fig. 4).
Fig. 4.
The CFUs numbers of rhizobacteria recovered from the endo-rhizosphere of Aloe arborescens on different culture media: NA, nutrient agar; CCM, N-deficient combined carbon sources medium; and plant-based culture media prepared from diluted (1:20, v/v) juices and saps of A. vera, and A. arborescens. Inserted is statistical analysis indicating levels of significance (p = <0.01).
As to culturing of rhizobacteria, pH of the culture medium is among the critical factors, and the pH is adjusted to near the sampled soil values [7]. The plant-soil environment under investigation is of neutral pH (pH 7.2–8.0), and therefore, the tested plant-based culture medium was adjusted to a corresponding neutral scale. However, in case of endophytic rhizobacteria it would be of great interest to investigate the implication of other pH values close to those of the plant sap/juice used for culture media preparation, a possibility that might facilitate culturing a fraction of the uncultivable endophytes.
Morphophysiological identification of endophytic rhizobacteria developed on agar plates
Isolates of endophytic rhizobacteria, associated with roots of tested plants and developed on representative agar plates of various culture media, were further grown and identified with the objective of defining the community structure of culturable endophytic rhizobacteria. In general, the composition of culturable rhizobacteria developed on juice/sap-based culture media differed to that grown on the standard nutrient agar and CCM.
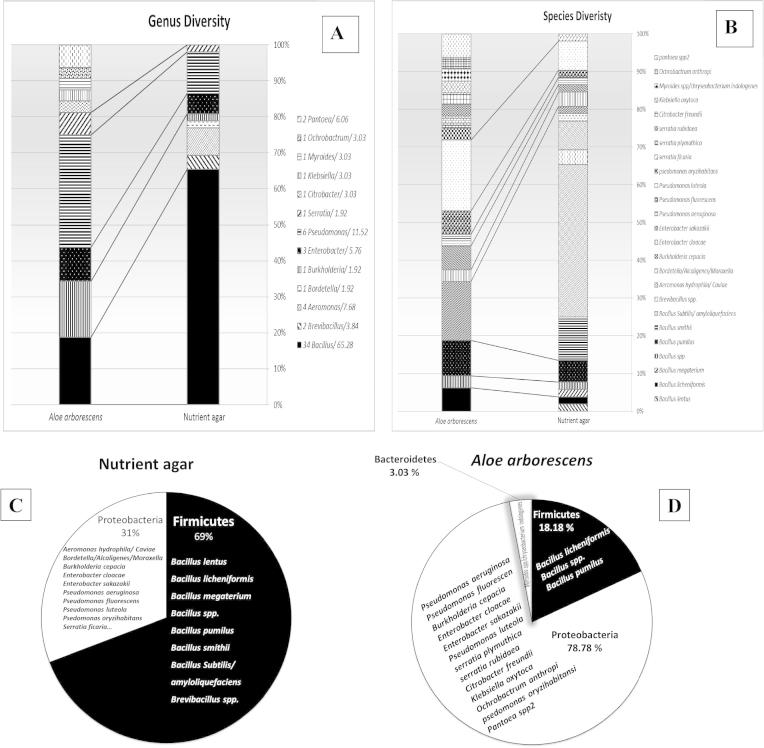
Regarding rhizobacteria of Aloe vera, while all of the 52 single discrete colonies developed on nutrient agar plates were successfully sub-cultured, only 32 out of total 42 colonies grown on the plant juice of A. arborescens were able to sustain sub-culturing. The comparative distribution of all isolates identified was noticeably different. Among the eight genera identified, five genera (Bacillus, Burkholderia, Enterobacter, Mycoides and Serratia) commonly developed on both culture media (Fig. 5A). The genera of Brevibacillus, Aeromonas, and Bordetella only developed on nutrient agar, while Citrobacter, Klebsiella, Ochrobactrum, Pantoea and Chryseobacterium were confined to the plant-based agar culture medium. The compositional differences were also evident at the species level where all tested culture media supported nine species, but differed in the occurrence of the remaining eight species (Fig. 5B). On the phylum level, Proteobacteria were the dominant (78.8%) on plant-based agar culture medium (Fig. 5D) compared to only 31% on nutrient agar (Fig. 5C). To the contrary, Firmicutes were prevailing on nutrient agar (69%) compared to the plant-based agar culture media (18.2%). Such prevalence of Proteobacteria in association with the roots of plants, e.g. maize, was also reported [34], where 68% of total CFUs belonged to Betaproteobacteria (Achromobacter), 30% to Firmicutes (Bacillus) and only 2% as Gammaproteobacteria. The phylum Bacteroidetes, represented by Chryseobacterium indologenes, was only reported (3%) among the rhizobacteria community of the plant-based agar culture medium. Members of the genus Chryseobacterium are considered an important bacterial group associated with plants, and currently there is enough evidence to show that strains of plant-associated species of the genus exhibit plant-growth promoting activities [35].
Fig. 5.
Community composition of endophytic rhizobacteria associated with roots of Aloe vera, based on API biochemical identification of isolates developed on nutrient agar compared to the plant-based culture medium (prepared from the juice of A. arborescens): (A) Species; (B) Genera; (C) and (D) phyla levels.
We also examined the diversity of culturable endophytic rhizobacteria associated with roots of A. arborescens recovered on both the N-deficient combined carbon sources medium (CCM) and the culture medium prepared from the homologous sap of A. arborescens (Table 3). Out of total 41 colonies developed on representative agar plates, 20 colonies (ca. 49%) failed to grow further and very possibly entered the phase of ‘viable but not culturable’ (VBNC or VNC). Only 21 colonies sustainably grew and were identified by API system. The plant-sap based culture media supported the development of 3 phyla, Firmicutes (22%), Proteobacteria (10%) and Bacteriodetes (2%). Nine species belonged to 8 genera were identified, with the majority of the species Paenibacillus macerans (40%) and the genus Paenibacillus (75%) (Table 3). The community structure of rhizobacteria on the synthetic standard medium (N-deficient combined carbon sources, medium, CCM) was different, where only two phyla (Firmicutes, 34% and Proteobacteria 27%) were distinguished. Specifically, 6 genera (majority for Bacillus spp. and Pseudomonas spp., 32%) and 12 species (majority for Pseudomonas cepacia, Bacillus licheniformis and Bacillus megaterium, 33%) were identified. Common to the plant-based and standard CCM culture media were the genera Bacillus spp. and Paenibacillus spp., and the species Bacillus circulans, Paenibacillus macerans and Pseudomonas luteola. All other genera and species were different. In agreement with these results was the culturable community composition of rhizobacteria identified from both root and inner tissues of maize as well as rice seedlings [33], [34]. Bacillus species were the most common within Firmicutes, and Pseudomonas and Enterobacter within Gammaproteobacteria.
Table 3.
Community structure of endophytic rhizobacteria associated with roots of A. arborescens developed on its plant sap-based culture medium compared to the synthetic standard CCM culture medium.
| Culture media | Phyla | Class | Genera | Species |
|---|---|---|---|---|
| CCM | (2) | (3) | (6) | (12) |
| Firmicutes | Bacillus | Aeromonas, Bacillus, Paenibacillus, Pseudomonas, Rhizobium, Sphingomonas | Aeromonas hydrophila/caviae | |
| Proteobacteria | Gammaproteobacteria | Bacillus circulans | ||
| Alphaproteobacteria | Bacillus licheniformis | |||
| Bacillus megaterium | ||||
| Bacillus pumilus | ||||
| Paenibacillus macerans | ||||
| Pseudomonas aeruginosa | ||||
| Pseudomonas luteola | ||||
| Burkholderia cepacia | ||||
| Pseudomonas fluorescens | ||||
| Rhizobium radiobacter | ||||
| Sphingomonas paucimobilis | ||||
| Plant-based | (3) | (3) | (8) | (9) |
| Firmicutes | Bacillus | Aneurinibacillus, Bacillus, Chryseobacterium, EnterobacterPaenibacillus, Pantoea Pasteurella, Pseudomonas | Aneurinibacillus aneurinilyticus | |
| Proteobacteria. Bacteriodetes | Gammaproteobacteria | Bacillus smithii | ||
| Flavobacterium | Bacillus circulans | |||
| Chryseobacterium indologenes | ||||
| Enterobacter cloacae | ||||
| Paenibacillus macerans | ||||
| Pantoea spp. 3 | ||||
| Pasteurella pneumotropica | ||||
| Pseudomonas luteola | ||||
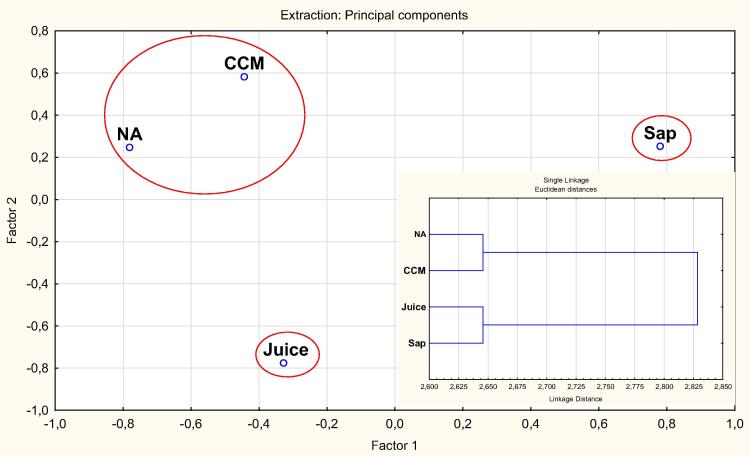
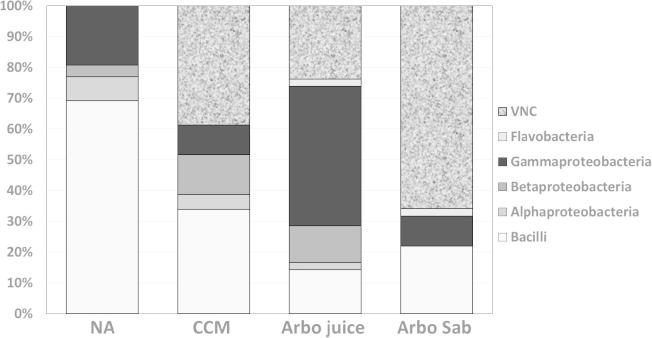
In accordance with our previous results [13], the tested plant-based culture media successfully supported the culturing of rhizobacteria associated with the plant roots. Furthermore, the community structure of the culturable population is different among all tested culture media. Factor analysis (Fig. 6) indicated the unique community structure revealed with culture media based on plant juices or saps, being distinct from that obtained with the chemically-synthetic culture media, rich (nutrient agar) or N-deficient combined carbon-sources medium CCM). Fig. 7 presents as well variations in class distribution of rhizobacteria developed on various culture media. The phylum Firmicutes with its major class Bacilli ranged from 22 to 69%, being predominant among populations recovered on the synthetic standard culture media, nutrient agar in particular. The predominance of Gammaproteobacteria, ranging from 10% to 45%, was evident on plant-based culture media, followed by Betaproteobacteria (4–13%) being highest in CCM and juice, then Alpha proteobacteria (2–8%). Flavobacteria were only reported on plant-based culture media.
Fig. 6.
Factor analysis-species level for the community structure of endophytic rhizobacteria of Aloe vera and Aloe arborescens as revealed by the use of various culture media, illustrating the different community structures of rhizobacteria species recovered on plant-based culture media (sap and juice) compared to those grown on the chemically-synthetic standard media of nutrient agar and CCM. Inserted is cluster distribution on the genus level, supporting the clear distinction between genera of rhizobacteria developed on plant-based culture media and those grown on chemically-synthetic standard media.
Fig. 7.
Variations in class distribution of rhizobacteria recovered from the endo-rhizosphere of Aloe vera and Aloe arborescens using various culture media tested: NA, nutrient agar; CCM, N-deficient combined carbon sources medium; plant-based culture media prepared from diluted (1:20, v/v) juices and saps of A. arborescens. VNC; viable non-culturable rhizobacteria.
The particular predominance of Proteobacteria on the tested plant-based culture media was also reported using a number of culture-independent techniques, e.g. ITS sequencing, as well as morphophysiological analyses. Perira et al. [34], [36], and Peiffer et al. [37] found that the largest fraction of the clones of root endophytic bacteria, for maize, belonged to Proteobacteria (50%) and the remaining clones belonged to Bacillus. Their results as well as of others [38] point to the somewhat agreement with both genomic and morphophysiological analyses, and that culture dependent and independent approaches are complementary. They also support the general conclusion of the particular adaptation of Proteobacteria to the plant rhizosphere generally and cross-diverse plant species, because of their response to labile carbon sources, and are generally r-selected [39]. Similar findings were reported with other host plants, e.g. rice roots [33], as the relative abundance of Proteobacteria is increased in the endosphere compared with soil, while the relative abundance of Acidobacteria decreases from soil to the endosphere. Our results further indicated that the chemically synthetic standard media, nutrient agar in particular, favour the growth of fast growing colonies, the majority of which are Bacilli, that were 100% easily cultivable further on. However, ⩾40% of colonies that grew on plant-based culture media were somewhat fastidious and difficult to sustain cultivability, i.e. viable but not culturable-VBNC. Very possibly, they require more defined endophytic growth conditions, e.g. nutrient complexity [40], long-term incubation [41], adjustable gas phases, and/or coculturing conditions [8], [42], [43]. It is reported as well that the gelling agent is a crucial factor for the growth of rhizobacteria on plate culture media, and that alternatives to agar, e.g. gellan gum, are very important for increasing the culturability of VBNC and/or yet-to-be cultured populations [44].
Conclusions
The presented results provide additional clues that plant materials in the form of crude slurry homogenates, juices, saps and/or dehydrated powder are rich enough, as such without any supplement, to support culturability of rhizobacteria. The tested plant-based culture media supported good in vitro growth of representative isolates of rhizobacteria, and in situ recovery of rhizobacteria associated with plant roots. The culturable population of rhizobacteria developed on plant-based culture media with densities very much comparable to that developed on chemically-synthetic culture media. Furthermore, the community structure of existing rhizobacteria differed among culture media tested, a conclusion that is based on morphophysiological identification of CFUs developed on agar plates, and remains to be confirmed by genomic analysis. If verified, the sole use of plant-based culture media is confirmed to be a methodological breakthrough. In addition, the need arises to revise the long-established information on the ecology of rhizobacteria solely based on the use of chemically-synthetic culture media. Further research is also encouraged to investigate how far the plant-based culture media significantly increase culturability of rhizobacteria, and effectively mirror their community structure in the root spheres.
Conflict of Interest
The authors have declared no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Acknowledgements
The present work was supported by the Research Grant of the Egyptian Ministry of Agriculture and Land Reclamation. The technical support and cooperation of colleagues at IGZ-Grossbeeren, Germany, during the Alexander von Humboldt Stiftung-research visit of NA Hegazi, is very much appreciated. IGZ support was generously extended for co-authors of this publication during their DAAD-training on “molecular biological techniques for studying microbial ecology”.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Rosselló-Mora R., Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 2.Konstantinidis K.T., Tiedje J.M. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102(2567–2572):99. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols D. Cultivation gives context to the microbial ecologist. FEMS Microbiol Ecol. 2007;60:351–357. doi: 10.1111/j.1574-6941.2007.00332.x. [DOI] [PubMed] [Google Scholar]
- 4.Janssen P.H., Yates P.S., Grinton B.E., Taylor P.M., Sait M. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol. 2002;68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis R.J., Morgan P., Weightman A.J., Fry J.C. Cultivation-dependent and independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl Environ Microbiol. 2003;69:3223–3230. doi: 10.1128/AEM.69.6.3223-3230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vartoukian S.R., Palmer R.M., Wade W.G. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 7.Pham V.H.T., Kim J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012;30:475–484. doi: 10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Stewart E.J. Growing unculturable bacteria. J Bacteriol. 2012;194:4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis P.G., Miller A.J., Hirsch P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol. 2010;72:313–327. doi: 10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed] [Google Scholar]
- 10.Doornbos R.F., van Loon L.C., Bakker P.A. Impact of root exudates and plant defence signalling on bacterial communities in the rhizosphere. A review. Agronom Sustain Dev. 2012;32:227–243. [Google Scholar]
- 11.Bais H.P., Broeckling C.D., Vivanco J.M. Root exudates modulate plant-microbe interactions in the rhizosphere. In: Karlovsky P., editor. Secondary Metabolites in Soil Ecology. vol. 14. Springer-Verlag; Berlin, Heidelberg: 2008. pp. 241–252. (Soil Biology). [Google Scholar]
- 12.Grayston S.J., Wang S., Campbell C.D., Edwards A. CSelective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem. 1998;30:369–378. [Google Scholar]
- 13.Nour E.H., Hamza M.A., Fayez M., Monib M., Ruppel S., Hegazi N.A. The crude plant juices of desert plants as appropriate culture media for the cultivation of rhizospheric microorganisms. J Adv Res. 2012;3:35–43. [Google Scholar]
- 14.Arulanantham R., Pathmanathan S., Ravimannan N., Niranjan K. Alternative culture media for bacterial growth using different formulation of protein sources. J Nat Prod Plant Resour. 2012;2:697–700. [Google Scholar]
- 15.Osman Z.A., Elsanousi S.M., Elsheikh E.A.E. Plant materials as probable growth promoters for certain fungi. Euro J Exp Biol. 2012;2:1785–1791. [Google Scholar]
- 16.Murphey B., Batke S.P., Doohan F.M., Hodkinson T.R. Media manipulation and the culture of beneficial fungal root endophytes. Int J Biol. 2015;7:94–102. [Google Scholar]
- 17.de Oliveira D.W., França I.W., Félix A.K., Martins J.J., Giro M.E., Melo V.M. Kinetic study of biosurfactant production by Bacillus subtilis LAMI005 grown in clarified cashew apple juice. Colloids Surf, B. 2013;101:34–43. doi: 10.1016/j.colsurfb.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Kurosumi A., Sasaki C., Yamashita Y., Nakamura Y. Utilization of various fruit juices as carbon source for production of bacterial cellulose by Acetobacter xylinum NBRC 13693. Carbohydr Polym. 2009;76:333–335. [Google Scholar]
- 19.Kosugi A., Tanaka R., Magara K., Murata Y., Arai T., Sulaiman O. Ethanol and lactic acid production using sap squeezed from old oil palm trunks felled for replanting. J Biosci Bioeng. 2010;110:322–325. doi: 10.1016/j.jbiosc.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Wang B., Sharma-Shivappa R.R., Olson J.W., Khan S.A. Production of polyhydroxybutyrate (PHB) by Alcaligenes latus using sugarbeet juice. Indust Crop Prod. 2013;43:802–811. [Google Scholar]
- 21.Thomsen M.H. Complex media from processing of agricultural crops for microbial fermentation. Appl Microbiol Biotechnol. 2005;68:598–606. doi: 10.1007/s00253-005-0056-0. [DOI] [PubMed] [Google Scholar]
- 22.Jensen V. Studies on the microflora of Danish beech forest soils. I. The dilution plate count technique for the enumeration of bacteria and fungi in soil. Zentbl Bakteriol Parasitenkd. 1962;Abt 2:13–32. [Google Scholar]
- 23.Hegazi N.A., Hamza M.A., Osman A., Ali S., Sedik M.Z., Fayez M. Modified combined carbon N-deficient medium for isolation, enumeration and biomass production of diazotrophs. In: Malik K.A., Mirza M.S., Ladha J.K., editors. Nitrogen fixation with non-legumes. Kluwer Academic Publishers; Dordrecht: 1998. pp. 247–253. [Google Scholar]
- 24.Othman A.A., Amer W.M., Fayez M., Monib M., Hegazi N.A. Biodiversity of diazotrophs associated to the plant cover of north sinai deserts. Arch Agron Soil Sci. 2003;49:683–705. [Google Scholar]
- 25.Othman A.A., Amer W.M., Fayez M., Hegazi N.A. Rhizosheath of Sinai desert plants is a potential repository for associative diazotrophs. Microbiol Res. 2004;159:285–293. doi: 10.1016/j.micres.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Wistreich G.A. Prentice Hall; Upper Saddle River, NJ: 2003. Microbiology laboratory fundamentals and applications, 2/E. [Google Scholar]
- 27.Youssef H.H., Fayez M., Monib M., Hegazi N. Gluconacetobacter diazotrophicus: a natural endophytic diazotroph of Nile Delta sugarcane capable of establishing an endophytic association with wheat. Biol Fert Soils. 2004;39:391–397. [Google Scholar]
- 28.Logan N.A., Berkeley R.C.W. Identification of Bacillus strains using the API system. J Gen Microbiol. 1984;130:1871–1882. doi: 10.1099/00221287-130-7-1871. [DOI] [PubMed] [Google Scholar]
- 29.API web™ version: 1.2.1. <http://apiweb.biomerieux.com>.
- 30.Pellizzoni M., Ruzickova G., Kalhotka L., Lucini L. Antimicrobial activity of different Aloe barbadensis Mill. and Aloe arborescens Mill. Leaf fractions. J. Appl. Med. Plants Res. 2012;6:1975–1981. [Google Scholar]
- 31.Statictica 10.0. StatSoft Inc, Tusla, USA.
- 32.Hegazi N.A., Fayez M., Hamza A. LAP Lambert Academic Publishing; Saarbrücken: 2013. Biofertilizers for organic farming. [Google Scholar]
- 33.Edwards J, Johnson C, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, et al. Structure, variation, and assembly of the root-associated microbiome of rice. <www.pnas.org/cgi/doi/10.1073/pnas.1414592112>. [DOI] [PMC free article] [PubMed]
- 34.Pereira P., Ibáñez F., Rosenblueth M., Etcheverry M., Martinéz-Romero E. Analysis of the bacterial diversity associated with the roots of maize (Zea mays L.) through culture-dependent and culture-independent methods. ISRN Ecol. 2011:1–10. [Google Scholar]
- 35.Montero-Calasanz M.C., Göker M., Rohde M., Spröer C., Schumann P., Busse H., Schmid M. Chryseobacterium hispalense sp. nov., a plant growth-promoting bacterium isolated from a rainwater pond in an olive plant nursery, and emended descriptions of Chryseobacterium defluvii, Chryseobacterium indologenes, Chryseobacterium wanjuense and Chryseobacterium gregarium. Int J Sys Evol Microbiol. 2013;63:4386–4395. doi: 10.1099/ijs.0.052456-0. [DOI] [PubMed] [Google Scholar]
- 36.Pereira P., Nesci A., Etcheverry M. Impact of two bacterial biocontrol agents on bacterial and fungal culturable groups associated with the roots of field-grown Maize. Lett Appl Microbiol. 2009;48:493–499. doi: 10.1111/j.1472-765X.2009.02558.x. [DOI] [PubMed] [Google Scholar]
- 37.Peiffer J.A., Spor A., Koren O., Jin Z., Tringe S.G., Dangl J.L. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci. 2013;110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardoim P.R., von Overbeek L.S., Elsas J.D.V. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Fierer N., Bradford M.A., Jackson R.B. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 40.Connon S.A., Giovannoni S.J. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol. 2002;68:3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis K.E., Joseph S.J., Janssen P.H. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol. 2005;71:826–834. doi: 10.1128/AEM.71.2.826-834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Onofrio A., Crawford J.M., Stewart E.J., Witt K., Gavrish E., Epstein S. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 2010;17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nichols D., Lewis K., Orjala J., Mo S., Ortenberg R., O’connor P.B. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl Environ Microbiol. 2008;74:4889–4897. doi: 10.1128/AEM.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamaki H., Hanada S., Sekiguchi Y., Tanaka Y. Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ Microbiol. 2009;11:1827–1834. doi: 10.1111/j.1462-2920.2009.01907.x. [DOI] [PubMed] [Google Scholar]