Abstract
Phenotypic, genotypic, and antimicrobial characteristics of six phenotypically distinct human clinical isolates that most closely resembled the type strain of Streptococcus halichoeri isolated from a seal are presented. Sequencing of the 16S rRNA, rpoB, sodA, and recN genes; comparative whole-genome analysis; conventional biochemical and Rapid ID 32 Strep identification methods; and antimicrobial susceptibility testing were performed on the human isolates, the type strain of S. halichoeri, and type strains of closely related species. The six human clinical isolates were biochemically indistinguishable from each other and showed 100% 16S rRNA, rpoB, sodA, and recN gene sequence similarity. Comparative 16S rRNA gene sequencing analysis revealed 98.6% similarity to S. halichoeri CCUG 48324T, 97.9% similarity to S. canis ATCC 43496T, and 97.8% similarity to S. ictaluri ATCC BAA-1300T. A 3,530-bp fragment of the rpoB gene was 98.8% similar to the S. halichoeri type strain, 84.6% to the S. canis type strain, and 83.8% to the S. ictaluri type strain. The S. halichoeri type strain and the human clinical isolates were susceptible to the antimicrobials tested based on CLSI guidelines for Streptococcus species viridans group with the exception of tetracycline and erythromycin. The human isolates were phenotypically distinct from the type strain isolated from a seal; comparative whole-genome sequence analysis confirmed that the human isolates were S. halichoeri. On the basis of these results, a novel subspecies, Streptococcus halichoeri subsp. hominis, is proposed for the human isolates and Streptococcus halichoeri subsp. halichoeri is proposed for the gray seal isolates. The type strain of the novel subspecies is SS1844T = CCUG 67100T = LMG 28801T.
INTRODUCTION
Streptococcus halichoeri was first described in 2004 by Lawson et al. (1). The six isolates described were obtained postmortem from three gray seals in Inverness, Scotland, United Kingdom, and from three gray seals at a rehabilitation center in Cornwall, United Kingdom. Over the past 5 years, five human clinical isolates (4 blood and 1 sinus) have been received at CDC that phenotypically resembled S. halichoeri. An additional human isolate from Sweden was obtained from the Culture Collection, University of Gothenburg, Sweden (CCUG). More recently, in 2014, S. halichoeri was isolated from a 45-year-old male with empyema (2), and in 2015, a case study of S. halichoeri from a European badger with pyogranulomatous pleuropneumonia was described (3). The goal of this study was to evaluate the taxonomic status of S. halichoeri isolates associated with a human clinical source and analyze genetic and phenotypic differences for identification and potential differences in antimicrobial susceptibility patterns. Isolates with phenotypic traits resembling S. halichoeri were evaluated by 16S rRNA, rpoB, sodA, and recN gene sequencing; whole-genome sequence comparison; conventional biochemical testing; Rapid ID 32 Strep panels; and antimicrobial susceptibility patterns.
MATERIALS AND METHODS
Bacterial strains.
S. halichoeri type strain CCUG 48324T and strain CCUG 61265 were obtained from CCUG. Five human clinical isolates were received at CDC for identification. The source and associated clinical information for the seven Streptococcus halichoeri isolates used in this study are listed in Table 1.
TABLE 1.
Sources and associated clinical information for the seven Streptococcus halichoeri isolates used in this study
| Strain no. | Yr | Location | Age (yr) | Genderb | Clinical diagnosis | Source |
|---|---|---|---|---|---|---|
| CCUG 48324T | 2002 | Inverness, Scotland, UK | DNPa | DNP | Postmortem, DNP | Gray seal; airway, lung, spleen |
| SS1844 | 2006 | North Carolina, USA | 67 | M | Sepsis | Human blood |
| SS1845 | 2008 | Rhode Island, USA | 78 | M | DNP | Human blood |
| CCUG 61265 | 2011 | Kalmar, Sweden | 66 | M | Osteitis | Human wound |
| SS1846 | 2011 | Missouri, USA | 68 | F | Septicemia | Human blood |
| SS1939 | 2014 | North Carolina, USA | 52 | M | DNP | Human maxillary sinus |
| SS1940 | 2014 | Georgia, USA | 47 | F | DNP | Human blood |
DNP, data not provided.
M, male; F, female.
Phenotypic characteristics.
The S. halichoeri type strain and the six human clinical isolates were tested for their phenotypic characteristics with conventional biochemical methods as previously described (4, 5) and by the Rapid ID 32 Strep system as described in the manufacturer's instructions (bioMérieux, Inc., Durham, NC). Serogrouping was determined by the Lancefield method as previously described (5), and reactivity with group B antiserum was also evaluated using the Remel PathoDx Strep grouping latex agglutination method (Lenexa, KS) in accordance with the manufacturer's package insert.
Antimicrobial susceptibility testing.
Susceptibility testing was performed by the broth microdilution method (6) using custom-made panels manufactured by Trek Diagnostic Systems, Cleveland, OH. The following antibiotics were reported: ampicillin, cefotaxime, clindamycin, erythromycin, levofloxacin, penicillin, tetracycline, and vancomycin. The CLSI guidelines for MIC interpretive standards for Streptococcus species viridans group were used (6).
Isolation of bacterial DNA for gene sequencing.
Bacterial genomic DNA for PCR was obtained by taking a 1-μl loop of cells from an agar plate and suspending them in 200 μl dimethyl sulfoxide (DMSO) (Sigma-Aldrich, St. Louis, MO). The cell suspension was incubated at room temperature for 30 min. A 2.5-μl portion of the suspension was used in a 50-μl PCR mixture.
16S rRNA gene sequencing.
Amplification and sequencing of the 16S rRNA gene were performed as previously described (7, 8). Sequencing reaction products were purified with Sephadex G-50 (Sigma-Aldrich, St. Louis, MO). Reaction mixtures were electrophoresed on an ABI 3730 DNA analyzer using POP-7 polymer (Applied Biosystems). Chromatograms were assembled and analyzed in Geneious v7 (9) (Biomatters, Auckland, New Zealand). The consensus sequences were aligned by using Clustal W (10) and trimmed to 1,456 bp to create a phylogenetic tree. Evolutionary analyses were conducted in MEGA6 (11). The evolutionary history was inferred using the neighbor-joining method (12). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) was determined (13). The evolutionary distances were computed using the maximum composite likelihood method (11). Duplicate confirmation of 16S rRNA gene sequences was derived from whole-genome sequencing of the type strains of S. halichoeri, S. ictaluri, and S. canis and the six human clinical isolates.
rpoB gene sequencing.
A ∼1,200-bp portion of the RNA polymerase beta subunit rpoB gene from each isolate was amplified with HotStarTaq (Qiagen, Valencia, CA) using primers UnivrpoB3F (5′-ATGGGNDCGNAAYATGCA) and UnivrpoB23R (5′-GAYATGGAYGTNTGYGC) in a 50-μl PCR mixture (14). The thermal cycling conditions were 95°C for 5 min; 15 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min with a decrease of the annealing temperature by 1°C at each cycle; 25 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min; 72°C for 10 min; and a final hold at 4°C. The PCR amplicons were purified through a NucleoSpin column (Clontech) and eluted in 120 μl PCR-grade water. Cycle sequencing was performed as described for the 16S rRNA gene above but using UnivrpoB3F, UnivrpoB23R, UnivrpoBseq1 (5′-GGNGAYAARNTNKSNRR), and UnivrpoBseq2 (5′-YYNSMNANYTTRTCNCC) as sequencing primers. Raw sequence data were assembled in Geneious v7. The full-length rpoB gene sequences from the S. halichoeri, S. ictaluri, and S. canis type strains and the six clinical isolates were also determined from the whole-genome sequences. Phylogenetic trees were inferred using both neighbor-joining and maximum likelihood methods in MEGA6 using an alignment of 3,516 bp.
sodA gene sequencing.
PCR targeting the sodA gene was conducted using primers specified by Poyart et al. (15). The thermal cycler program was as follows: 94°C for 5 min followed by 10 cycles of 94°C for 15 s, 50°C for 30 s, and 72°C for 45 s, followed by 20 cycles of 94°C for 15 s, 50°C for 30 s, and 72°C for 45 s with 10 s of autoextension, and a hold at 72°C for 10 min. Raw sequence data were assembled in Geneious v7. Duplicate confirmation of sodA gene sequences was derived from whole-genome sequencing of the S. halichoeri, S. ictaluri, and S. canis type strains and the six human clinical isolates, and phylogenetic trees were inferred using both neighbor-joining and maximum likelihood methods in MEGA6 using an alignment of 417 bp.
recN gene sequencing.
PCR products were generated using primers and protocols modified from the work of Glazunova et al. (16), specifically, recNd (modified) (5′-GGA AAG TCY ATT ATY ATT GAT GC-3′) and recNr (5′-WAA CWC CNG TAT CIA CYT CAT C-3′). The thermal cycler program was as follows: 95°C for 5 min; 15 cycles of 94°C for 1 min, 55°C for 1 min (with annealing temperature dropping 1°C for each cycle), and 72°C for 1 min, followed by 25 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, and finally, a hold at 72°C for 10 min. Sequencing primers recNhalichodfin and recNphocaer were as described in the work of Glazunova et al. (16). Raw sequence data were assembled in Geneious v7. The full-length recN gene sequence was derived from the whole genome of the six isolates, and phylogenetic trees were inferred using both neighbor-joining and maximum likelihood in MEGA6 using an alignment of 1,248 bp.
Bacterial whole-genome DNA library preparation and sequencing.
Genomic DNA was prepared according to the DOE Joint Genome Institute cetyltrimethylammonium bromide (CTAB) protocol (http://jgi.doe.gov/wp-content/uploads/2014/02/JGI-Bacterial-DNA-isolation-CTAB-Protocol-2012.pdf). One microgram of DNA per sample was diluted in 50 μl of PCR-grade water and sheared on a Covaris M220 focused ultrasonicator (Covaris, Inc., Woburn, MA) for 32 s at a duty factor of 20% and 200 cycles/burst to generate 500-bp fragments according to the manufacturer's instructions. Genomic DNA libraries were constructed using SPRIworks HT fragment library kits (Beckman Coulter, Inc., Brea, CA) according to the manufacturer's instructions on a Biomek FXP laboratory automation workstation (Beckman Coulter). Libraries were labeled using Illumina indices from a TruSeq DNA sample preparation kit (Illumina, Inc., San Diego, CA) and sequenced on an Illumina MiSeq sequencer using MiSeq v2 500-cycle reagent kits. Sequencing data were assembled using CLCbio Genomics Workbench v7. The G+C content calculation is an output of the assembly program.
Analysis of whole-genome sequence.
Wet lab DNA-DNA hybridization values were estimated using the Genome-to-Genome Distance Calculator with the alignment method GGDC 2.0 BLAST+ and Formula 2 as recommended for incomplete genomes (17). Average nucleotide identity (ANI) values (9) were determined by submitting draft genome sequences to the ANI calculator (http://enve-omics.ce.gatech.edu/ani/) and EZgenome (http://www.ezbiocloud.net/ezgenome/ani). Full-length genes for 16S rRNA, rpoB, sodA, and recN were determined from whole-genome assemblies by constructing a database of each genome contig set and querying the database with the partial sequences derived from PCR as well as full-length sequences from genomic data of streptococcal species (Geneious).
Nucleotide sequence accession numbers.
The sequences described here were submitted to GenBank under the accession numbers given in Table 2.
TABLE 2.
Nucleotide sequence accession numbers
| Organism | Strain | Accession no. for sequence: |
|||
|---|---|---|---|---|---|
| 16S rRNA | sod A | rpo B | rec N | ||
| Streptococcus halichoeri subsp. hominis | SS1844T | KP851845 | KP890273 | KP890278 | KP890288 |
| SS1845 | KP851846 | KP890272 | KP890280 | KP890286 | |
| SS1846 | KP851847 | KP890271 | KP890281 | KP890290 | |
| CCUG 61265 | KP851848 | KP890270 | KP890282 | KP890287 | |
| SS1939 | KP851849 | KP890268 | KP890279 | KP890289 | |
| SS1940 | KP851850 | KP890269 | KP890277 | KP890291 | |
| Streptococcus halichoeri subsp. halichoeri | CCUG 48324T | KP851851 | KP890274 | KP890283 | KP890292 |
| Streptococcus canis | ATCC 43496T | KP851852 | KP890275 | KP890284 | KP890294 |
| Streptococcus ictaluri | ATCC BAA-1300T | KP851853 | KP890276 | KP890285 | KP890293 |
RESULTS
Gene sequence analysis.
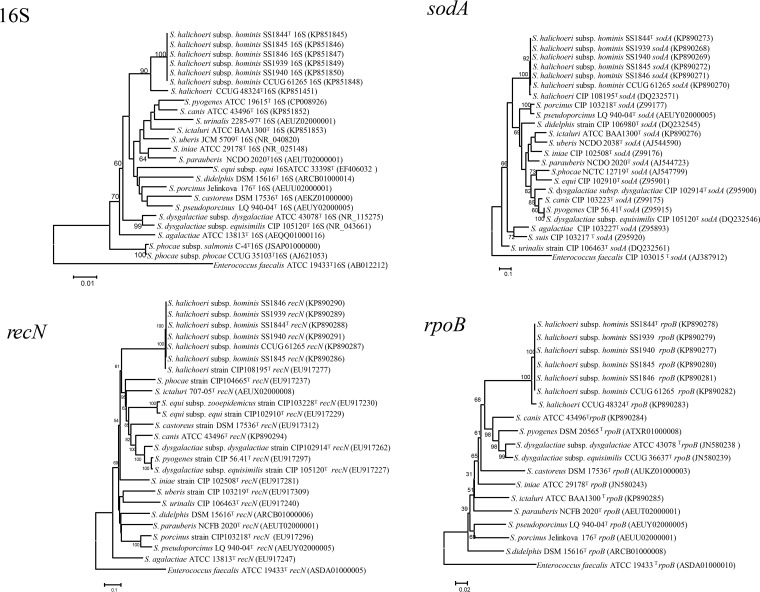
The six human clinical isolates shared 100% similarity to each other by 16S rRNA gene sequence and 98.6% similarity to S. halichoeri CCUG 48324T (seal isolate), 97.9% similarity to S. canis ATCC 43496T, and 97.8% similarity to S. ictaluri ATCC BAA-1300T. The S. halichoeri type strain sequence was 96.8% similar to S. canis and 96.6% similar to S. ictaluri. The finding of greater than 97% 16S rRNA gene sequence relatedness of the human S. halichoeri isolates with S. canis and S. ictaluri indicated that further testing was warranted to confirm the taxonomic status of the human isolates. Full-length gene sequences were determined from whole-genome sequence data. The 3,530-bp rpoB gene was identical between all six human isolates. A 3,516-bp alignment was created for phylogenetic analysis, and the rpoB sequence from the clinical isolates was 98.8% similar to the S. halichoeri type strain, 84.6% similar to the S. canis type strain, and 83.8% similar to the S. ictaluri type strain (Fig. 1). The 417-bp sodA gene fragment was identical for the 6 human clinical isolates and 98.2% similar to the S. halichoeri type strain, 77.2% similar to S. canis, and 76.7% similar to S. ictaluri. All six human isolates were identical based on recN gene sequence comparison and were 99.3% similar to the S. halichoeri CCUG 48324T seal isolate and 68.0 and 68.1% similar to S. ictaluri and S. canis, respectively. Of the four loci analyzed, the recN gene fragment of the S. halichoeri-like clinical isolates showed the greatest similarity to the type strain sequence (99.3%) (Fig. 1), while overall, the recN gene exhibited the greatest interspecies diversity (99.3 to 40.7%).
FIG 1.
Phylogenetic trees inferred from comparison of a 1,456-bp portion of the 16S rRNA gene, a 3,516-bp portion of the rpoB gene, a 417-bp portion of the sodA gene, and a 1,248-bp portion of the recN gene from Streptococcus species. The evolutionary history was inferred using the neighbor-joining method (12). Bootstrap percentage values of ≥50 from 1,000 replicates are shown at nodes (13). The evolutionary distances were computed using the maximum composite likelihood method (20) and are in units of the number of base substitutions per site. Evolutionary analyses were conducted in MEGA6 (11). No significant differences were noted in the tree topologies when maximum likelihood statistical methods were used with either Kimura 2-parameter or general-time-reversible nucleotide substitution models.
Comparative whole-genome analysis.
The comparative sequencing results for the 16S rRNA, rpoB, sodA, and recN genes showed the six human isolates to be identical. In an in silico pairwise analysis for estimation of DNA-DNA hybridization (isDDH) using the Genome-to-Genome Distance Calculator (GGDC 2.0) (17), all six clinical isolates were predicted to give an isDDH relative binding ratio of >70% to one another and to S. halichoeri CCUG 48324T (Table 3), indicating that they belong to the same species. The predicted isDDH values between the six clinical isolates and the S. canis and S. ictaluri type strains were <22. Average nucleotide identity (ANI) values were also calculated based on the assembled whole genomes. Two whole-genome sequences represent the same species if their ANI value is ≥95 to 96% (18, 19); the six clinical isolates and the S. halichoeri type strain exhibited >98% ANI. The data in Table 2 clearly demonstrate that the human isolates are not S. canis or S. ictaluri and confirm their identification as S. halichoeri. Metrics for the whole-genome sequence data are presented in Table 4.
TABLE 3.
isDDH and ANI results for Streptococcus halichoeri and closely related species
| Query genome | Reference genome | GGDCa |
ANIb | |
|---|---|---|---|---|
| Estimated isDDH | Probability of isDDH of ≥70% | |||
| SS1844T | S. halichoeri subsp. halichoeri CCUG 48324T | 84.6 | 94.4 | 98.2/98.4 |
| SS1844T | S. halichoeri subsp. hominis SS1844T | 100 | 98.3 | 100/100 |
| SS1844T | S. halichoeri subsp. hominis SS1845 | 100 | 98.2 | 100/100 |
| SS1844T | S. halichoeri subsp. hominis SS1846 | 100 | 98.3 | 98.1/100 |
| SS1844T | S. halichoeri subsp. hominis SS1939 | 98.7 | 98.0 | 99.9/99.8 |
| SS1844T | S. halichoeri subsp. hominis SS1940 | 100 | 98.2 | 100/100 |
| SS1844T | S. halichoeri subsp. hominis CCUG 61265 | 98.9 | 98.1 | 97.8/99.8 |
| SS1844T | S. canis ATCC 43496T | 21.2 | 0.00 | 72.3/79.1 |
| SS1844T | S. ictaluri ATCC BAA-1300T | 21.4 | 0.00 | 72.1/78.8 |
Genome-to-genome distance calculator with the alignment method GGDC 2.0 BLAST+ and Formula 2.
Average nucleotide identity values (9) were determined by submitting draft genome sequences to the ANI calculator (http://enve-omics.ce.gatech.edu/ani/) and EZgenome (http://www.ezbiocloud.net/ezgenome/ani), respectively, and are given as value by ANI calculator/value by EZgenome.
TABLE 4.
Whole-genome-sequence characteristics
| Genome | Assembly size (bp) | No. of contigs | G+C mol% |
|---|---|---|---|
| S. halichoeri subsp. hominis | |||
| SS1844T | 1,911,133 | 66 | 42 |
| SS1845 | 1,977,140 | 68 | 42 |
| SS1846 | 1,938,850 | 92 | 42 |
| CCUG 61265 | 2,017,305 | 89 | 42 |
| SS1939 | 1,919,301 | 68 | 42 |
| SS1940 | 1,962,910 | 65 | 42 |
| S. halichoeri subsp. halichoeri CCUG 48324T | 1,875,405 | 85 | 42 |
| S. canis ATCC 43496T | 2,022,062 | 51 | 40 |
| S. ictaluri ATCC BAA-1300T | 2,074,900 | 164 | 38 |
Phenotypic testing.
S. halichoeri CCUG 48324T and the human isolates tested catalase negative when grown on blood-free medium, and all tested positive for Lancefield serological group B and pyroglutamic acid arylamidase (PYR). Strains tested catalase negative when selection was performed correctly from non-blood-containing medium; however, a falsely catalase-positive test was observed with both the S. halichoeri type strain and the human isolates when isolates were selected from medium containing sheep blood, which may explain the discrepancy in the literature and why several isolates were initially referred to the CDC as suspected Staphylococcus species. Five human clinical isolates (four blood culture and 1 sinus) received from four different states (North Carolina [two], Rhode Island, Missouri, and Georgia) and one human wound isolate from Sweden isolated over the last 5 years were biochemically indistinguishable. Table 5 shows phenotypic characteristics that may be used to distinguish the six S. halichoeri human clinical isolates from S. halichoeri CCUG 48324T and other genetically closely related Streptococcus species based on comparative 16S rRNA sequences and Streptococcus species that may possess the group B Lancefield antigen. The six human clinical S. halichoeri isolates were phenotypically identical to one another but could be distinguished from the S. halichoeri type strain seal isolate based on a positive test for bile-esculin and esculin hydrolysis and a positive test for acid production from sucrose in conventional phenotypic tests. In the Rapid ID 32 Strep test system, the six human clinical isolates tested positive for β-glucosidase, β-glucuronidase activity, and acid production from saccharose/sucrose and for methyl-β-d-glucopyranoside, as shown in Table 5. The original species description of S. halichoeri confirms that these Rapid ID 32 Strep test reactions were negative for the six seal isolates (1).
TABLE 5.
Phenotypic characteristics for six S. halichoeri human clinical isolates, S. halichoeri type strain (gray seal isolate), and related Streptococcus species based on comparative 16S rRNA sequence identity or possession of the group B Lancefield antigen
| Characteristic | Result for species and strainb: |
|||||||
|---|---|---|---|---|---|---|---|---|
| S. halichoeri (human isolates, n = 6) | S. halichoeri CCUG 48324T | S. canis ATCC 43496T | S. ictaluri ATCC BAA-1300T | S. agalactiae ATCC 13813T | S. porcinus (n = 19) | S. pseudoporcinus (n = 72) | S. phocae ATCC 51973T | |
| 16S rRNA gene identity (%)a | 100 | 98.6 | 97.9 | 97.8 | 95.5 | 95.7 | 95.7 | 93.7 |
| Conventional tests | ||||||||
| Hemolysis | γ | γ | β | γ | β | β | β | β |
| Lancefield group | B | B | G | None | B | B (79%) | B (83%) | F |
| PYRasec | + | + | + | − | V | V | − | |
| LAPased | + | + | + | + | + | + | + | + |
| Bile-esculin | + | − | − | − | − | − | V | − |
| 6.5% NaCl | + | + | W+e | − | + | + | + | + |
| Esculin | + | − | + | − | − | + | + | − |
| Arginine | + | + | − | − | + | + | + | − |
| Hippurate | − | − | − | − | + | − | + | − |
| Acid from | ||||||||
| Lactose | − | − | + | − | + | V | − | − |
| Maltose | + | + | + | + | + | + | + | + |
| Mannitol | + | + | − | − | − | + | + | − |
| Sorbitol | − | − | − | − | − | + | + | − |
| Sucrose | + | − | + | − | + | + | + | − |
| Trehalose | − | − | − | − | + | + | + | − |
| Methyl-α-d-glucopyranoside | − | V | + | − | − | V | + | − |
| Pullulan | + | + | + | − | + | V | V | + |
| Tagatose | V | W+ | + | − | + | + | + | − |
| Rapid ID 32 | ||||||||
| Arginine dihydrolase | + | + | + | − | + | + | + | − |
| β-Glucosidase | + | − | − | − | − | + | + | − |
| β-Galactosidase | − | − | + | − | − | − | − | − |
| β-Glucuronidase | + | − | − | − | + | + | + | − |
| α-Galactosidase | − | − | + | − | − | V | V | − |
| Mannitol | + | + | − | − | − | + | + | − |
| Sorbitol | − | − | − | − | − | + | + | − |
| Lactose | − | − (V) | + | − | + | V | − | − |
| Trehalose | − | − | − | − | + | + | + | − |
| Saccharose/sucrose | + | − | + | − | + | + | + | − |
| Cyclodextrin | + | + | − | − | − | − | − | − |
| β-Galactosidase | − | − | + | − | − | − | − | − |
| Pyroglutamic acid arylamidase | + | + | − | + | − | V | − | − |
| Glycyl-tryptophan arylamidase | V | − | + | − | − | − | − | V |
| Hydrolysis of hippurate | − | − | − | − | + | V | + | − |
| Pullulan | + | + | + | − | + | V | + | − |
| Maltose | + | + | + | − | + | + | + | + |
| Methyl-β-d-glucopyranoside | + | − | + | − | − | V | − | − |
16S rRNA gene sequence percent identity of S. halichoeri human clinical isolates to S. halichoeri gray seal and related species.
+, 100% of strains positive; −, 100% of strains negative; V, variable reaction within species or repeated testing; (V), variable reaction in species description.
PYRase, pyrrolidonyl arylamidase.
LAPase, leucine aminopeptidase.
W+, weakly positive result.
Antibiotic susceptibility testing.
The CLSI guidelines for performing broth microdilution panels and MIC interpretive standards for Streptococcus species viridans group (6) were used to test ampicillin, cefotaxime, clindamycin, erythromycin, levofloxacin, penicillin, tetracycline, and vancomycin. The S. halichoeri CCUG 48324T gray seal isolate showed susceptibility to all these antimicrobials. The human clinical isolates were susceptible to all antibiotics with the exception of two isolates demonstrating resistance to tetracycline, and one of these two isolates also was resistant to erythromycin.
TAXONOMY
Emended description of Streptococcus halichoeri.
Streptococcus halichoeri (ha.lich.o′e.ri. N.L. gen. n. halichoeri, of a seal of the genus Halichoerus, systematic genus name of the gray seal). The description is essentially as proposed by Lawson et al. in 2004 (1). An additional feature is that strains grow in 6.5% sodium chloride broth.
Description of Streptococcus halichoeri subsp. halichoeri subsp. nov.
Streptococcus halichoeri (ha.lich.o′e.ri. N.L. gen. n. halichoeri, of a seal of the genus Halichoerus, systematic genus name of the gray seal). Demonstrates the characteristics previously described by Lawson et al. (1) for Streptococcus halichoeri. The type strain is M512/02/1T = CCUG 48325T = CIP 108195T.
Description of Streptococcus halichoeri subsp. hominis subsp. nov. (ho′mi.nis. L. gen. n. hominis, of a human being or from human being).
Strains conform to the species description with the following exceptions. Using the Rapid ID 32 Strep test, activity is detected for β-glucosidase and β-glucuronidase, and acid is produced from saccharose/sucrose and methyl-β-d-glucopyranoside. Activity is variable for glycyl-tryptophan arylamidase. Using the Rapid ID 32 Strep test, acid may or may not be produced from ribose, although acid is consistently produced for ribose in the longer-incubation conventional biochemical tube test. Acid is not produced from lactose in either test system. Bile-esculin and esculin are hydrolyzed. The original species description for S. halichoeri reports acid production from mannose and activity in α-glucosidase, and these tests were not performed and compared in this study. The type strain is SS1844T = CCUG 67100T = LMG 28801T isolated from human blood in 2006 in North Carolina, USA, from a 67-year-old man with sepsis. The 16S rRNA sequence accession number is KP851845.
DISCUSSION
The S. halichoeri human clinical isolates were associated with older patients whose age ranged from 47 to 78 with an average age of 63 years. Gender does not appear to be a significant clinical factor, as four isolates were from male patients and 2 were from females. The patients were from six different geographic locations. Patient contact with gray seals or ocean water was unknown. The sources of blood and wound indicate that this organism can cause serious infections in humans. Antibiotic resistance to tetracycline and/or erythromycin was observed in two isolates. Phenotypic and DNA sequence analysis of four individual loci (16S rRNA, rpoB, sodA, and recN) indicated that the six human clinical isolates were identical to each other and different from, but most closely related to, S. halichoeri. Glazunova et al. found that recN gave the most stable phylogenetic tree (16). In their study, the closest similarity between a pair of subspecies was 98% (S. gallolyticus subspecies). Our clinical isolates were 99% similar by recN to S. halichoeri CCUG 48324T. Therefore, the human clinical isolates appeared to be a subspecies or perhaps a sequence variant of S. halichoeri and not a new species. Whole-genome sequence comparison confirmed that the 6 human isolates clearly belonged to the species S. halichoeri but were genetically distinct. In addition, the six human clinical isolates were phenotypically distinct from the six S. halichoeri isolates from marine mammals. We propose the subspecies names Streptococcus halichoeri subsp. halichoeri for the marine isolates and Streptococcus halichoeri subsp. hominis for the human isolates. During the preparation of the manuscript, an additional human clinical isolate from a 45-year-old male with empyema was described in Singapore (2). That study did not provide all the biochemical data or 16S rRNA gene sequence in order to make a determination of which subspecies the isolate represented. In addition, a recently described isolate from a badger was more closely related to the gray seal isolates than to the human isolates based on comparative 16S rRNA and rpoB gene sequence analysis (3).
ACKNOWLEDGMENTS
We are grateful to Bernhard Schink for assisting with the species epithet.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Lawson PA, Foster G, Falsen E, Davison N, Collins MD. 2004. Streptococcus halichoeri sp. nov., isolated from grey seals (Halichoerus grypus). Int J Syst Evol Microbiol 54:1753–1756. doi: 10.1099/ijs.0.63082-0. [DOI] [PubMed] [Google Scholar]
- 2.Foo RM, Chan D. 2014. A fishy tale: a man with empyema caused by Streptococcus halichoeri. J Clin Microbiol 52:681–682. doi: 10.1128/JCM.03055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreno B, Bolea R, Morales M, Martín-Burriel I, González C, Badiola JJ. 2015. Isolation and phylogenetic characterization of Streptococcus halichoeri from a European badger (Meles meles) with pyogranulomatous pleuropneumonia. J Comp Pathol 152:269–273. doi: 10.1016/j.jcpa.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Facklam RR, Elliot JA. 1995. Identification, classification and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin Microbiol Rev 8:479–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Facklam RR, Washington JA II. 1991. Streptococcus and related catalase-negative, gram-positive cocci, p 238–257. In Balows A, Hausler WJ Jr Herrmann KL, Isenberg HD, Shadomy HJ (ed), Manual of clinical microbiology, 5th ed American Society for Microbiology, Washington, DC. [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing. M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Shewmaker PL, Camus AC, Bailiff T, Steigerwalt AG, Morey RE, Carvalho MGS. 2007. Streptococcus ictaluri sp. nov., isolated from channel catfish Ictalurus punctatus broodstock. Int J Syst Evol Microbiol 57:1603–1606. doi: 10.1099/ijs.0.64810-0. [DOI] [PubMed] [Google Scholar]
- 8.Morey R, Galloway R, Bragg S, Steigerwalt A, Mayer L, Levett P. 2006. Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44:3510–3516. doi: 10.1128/JCM.00670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. 2011. Geneious v5.4. Biomatters, Auckland, New Zealand. http://www.geneious.com/. [Google Scholar]
- 10.Thompson J, Higgins D, Gibson T. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 14.Glazunova OO, Raoult D, Roux V. 2009. Partial sequence comparison of the rpoB, sodA, groEL, and gyrB genes within the genus Streptococcus. Int J Syst Evol Microbiol 59:2317–2322. doi: 10.1099/ijs.0.005488-0. [DOI] [PubMed] [Google Scholar]
- 15.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol 36:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glazunova OO, Raoult D, Roux V. 2010. recN partial gene sequencing: a new tool for identification and phylogeny within the Streptococcus genus. Int J Syst Evol Microbiol 60:2140–2148. doi: 10.1099/ijs.0.018176-0. [DOI] [PubMed] [Google Scholar]
- 17.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 19.Richter M, Rosselló-Móra R. 2009. Shifting the genomic standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]