Abstract
Enterobacter cloacae strain G6809 with reduced susceptibility to carbapenems was identified from a patient in a long-term acute care hospital in Kentucky. G6809 belonged to sequence type (ST) 88 and carried two carbapenemase genes, blaKPC-18 and blaVIM-1. Whole-genome sequencing localized blaKPC-18 to the chromosome and blaVIM-1 to a 58-kb plasmid. The strain was highly resistant to ceftazidime-avibactam. Insidious coproduction of metallo-β-lactamase with KPC-type carbapenemase has implications for the use of next-generation β-lactam–β-lactamase inhibitor combinations.
TEXT
Carbapenem-resistant Enterobacteriaceae (CRE) has become one of the most urgent threats facing hospitals worldwide due to the limited options available for treatment once infection develops. In the United States, the most common mechanism of carbapenem resistance in Enterobacteriaceae is the production of KPC-type carbapenemases (1). Treatment of infections caused by KPC-producing Enterobacteriaceae has largely relied on combinations of polymyxins, tigecycline, and other agents, yielding less than optimal clinical outcomes (2). To address this gap, a series of novel β-lactamase inhibitors are in clinical development in combination with various partner β-lactams (3), including avibactam and relebactam, which are diazabicyclooctanes, and RPX7009, which is a cyclic boronic acid compound. They inhibit class A β-lactamases (including KPC-type carbapenemases) and class C β-lactamases (AmpCs) and variably inhibit class D β-lactamases (OXAs), but they do not inhibit class B β-lactamases (metallo-β-lactamases [MBLs]). The ceftazidime-avibactam combination has been approved for the treatment of complicated intra-abdominal infections and complicated urinary tract infections in the United States. Ceftazidime-avibactam is highly active in vitro against KPC-producing Enterobacteriaceae and has become the standard therapy for CRE infections in some hospitals (4). However, no practical susceptibility testing method has been approved for clinical use; therefore, the agent is used for treatment of CRE infections without susceptibility data in most instances.
Carbapenem resistance due to the production of MBL is still relatively rare in the United States, but hospital outbreaks due to Pseudomonas aeruginosa producing VIM-2 MBL and Klebsiella pneumoniae and Escherichia coli producing NDM-type MBL have been reported (5–7). Since these newer β-lactamase inhibitors do not inhibit MBLs, the β-lactam–β-lactamase inhibitor combinations (BLBLIs) that contain them are not active against MBL-producing bacteria. Here, we report a case of suspected ventilator-associated pneumonia caused by Enterobacter cloacae coproducing KPC-type carbapenemase and MBL to highlight this potential caveat.
E. cloacae strain G6809 was isolated from a respiratory specimen of a 58-year-old man who was admitted to a long-term acute care hospital (LTACH) in Kentucky. The strain was subsequently referred to the University of Louisville Hospital for KPC confirmation testing. MICs were determined by a commercial broth microdilution plate (Sensititre GNX2F; Thermo Fisher Scientific, Oakwood Village, OH), except for those of carbapenems, which were tested with Etest (bioMérieux, Durham, NC) (Table 1). E. cloacae G6809 was resistant to conventional BLBLIs, including ticarcillin-clavulanic acid and piperacillin-tazobactam and cephalosporins and aztreonam, and to ertapenem and imipenem when applying the latest Clinical and Laboratory Standards Institute breakpoints (8). Notably, the strain was highly resistant to ceftazidime-avibactam with an MIC of >256/4 μg/ml by Etest (bioMérieux). Phenotypic tests with imipenem and Tris-EDTA disks (BD, Franklin Lakes, NJ) were positive for production of both an MBL and a class A or class D carbapenemase (9, 10). The strain was prepared as a 0.5 McFarland standard suspension in saline and was loaded onto a cartridge of a Verigene Gram-negative blood culture test (GN-BC) (Nanosphere, Northbrook, IL) (11). This assay was positive for blaKPC and blaVIM carbapenemase genes. Coproduction of KPC-type carbapenemase and VIM-type MBL has been reported on occasion but is unusual in the United States (12–16). We therefore conducted whole-genome sequencing to elucidate the genetic context of the two carbapenemase genes. The genomic DNA was extracted from the E. cloacae G6809 isolate by a DNeasy blood and tissue kit (Qiagen, Valencia, CA), sequenced by RSII (Pacific Biosciences, Menlo Park, CA) at the Yale Center for Genome Analysis, and assembled using SMRT analysis 2.1 (Pacific Biosciences). De novo assembly yielded 10 contigs, with an average genome-wide coverage of 172×.
TABLE 1.
MICs obtained in tests with E. cloacae G6809 isolate (blaKPC-18 and blaVIM-1 positive) and its blaVIM-1-positive transformant
| Drug | MICa with: |
||
|---|---|---|---|
| E. cloacae G6809 | E. coli TOP10 (pG6809-2) | E. coli TOP10 | |
| Ticarcillin-clavulanic acid | >128/2 | >128/2 | ≤16/2 |
| Piperacillin-tazobactam | >64/4 | >64/4 | ≤8/4 |
| Cefotaxime | >32 | 32 | ≤1 |
| Ceftazidime | >16 | >16 | ≤1 |
| Cefepime | >16 | 8 | ≤2 |
| Aztreonam | >16 | ≤2 | ≤2 |
| Ertapenem | 1.5 | 0.19 | 0.004 |
| Imipenem | 3 | 2 | 0.38 |
| Meropenem | 0.75 | 0.5 | 0.032 |
| Ceftazidime-avibactam | >256/4 | >256/4 | 0.25/4 |
| Aztreonam-avibactam | 0.5/4 | ≤0.06/4 | ≤0.06/4 |
| Gentamicin | ≤1 | ≤1 | ≤1 |
| Tobramycin | 8 | 8 | ≤1 |
| Amikacin | 8 | 8 | ≤4 |
| Levofloxacin | >8 | ≤1 | ≤1 |
| Ciprofloxacin | >2 | ≤1 | ≤1 |
| Trimethoprim-sulfamethoxazole | >4/76 | 2/38 | ≤0.5/9.5 |
| Minocycline | 8 | ≤2 | ≤2 |
| Doxycycline | 8 | ≤2 | ≤2 |
| Tigecycline | 0.5 | ≤0.25 | ≤0.25 |
| Polymyxin B | ≤0.25 | ≤0.25 | ≤0.25 |
| Colistin | ≤0.25 | ≤0.25 | ≤0.25 |
The MICs were obtained with broth microdilution (Sensititre GNX2F), except for the carbapenems and ceftazidime-avibactam, which were obtained with Etest, and aztreonam-avibactam, which were obtained with standard broth microdilution.
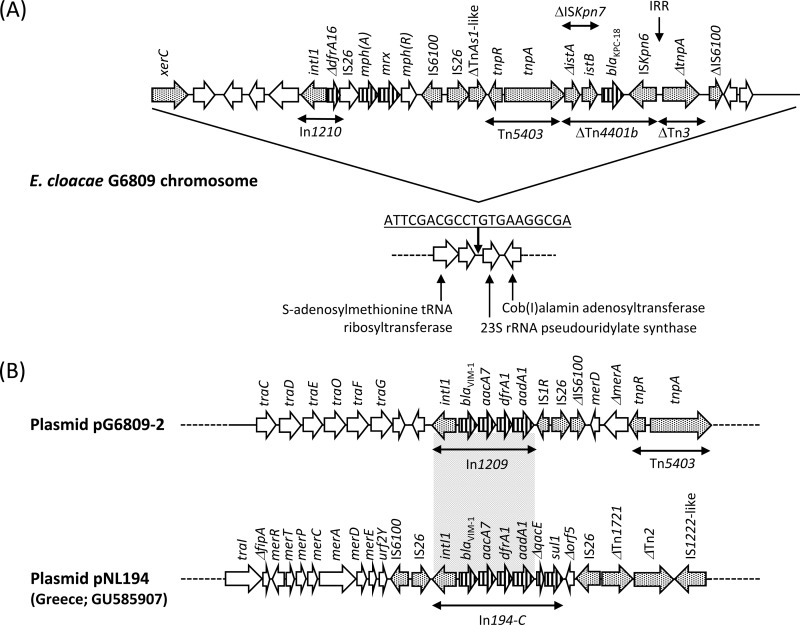
E. cloacae G6809 belonged to sequence type (ST) 88. ST88 was initially assigned to a VIM-1-producing strain found in Greece as a singleton (17). The KPC gene blaKPC was located on a 3.3-Mb contig representing a chromosomal sequence and encoded KPC-18 (GenBank accession no. KP681699; an E. coli urinary strain from the United States), which differs from KPC-2 by a single amino acid substitution, V8I. This substitution was located in the signal peptide and was unlikely to affect the kinetic properties of the enzyme. Indeed, when the coding regions of blaKPC-18 and blaKPC-2 were cloned into pBCSK− (18), the ertapenem MIC of E. coli TOP10 harboring blaKPC-18 and blaKPC-2 was 0.12 μg/ml for both, compared with 0.003 μg/ml for E. coli TOP10 alone. Using E. cloacae strain 34977 as the reference genome (GenBank accession no. CP010376), it was apparent that a 29,874-bp sequence was inserted in an intergenic region upstream of the 23S rRNA pseudouridylate synthase gene located on the chromosome, generating 21-bp direct repeats (Fig. 1A). Here, blaKPC-18 was located on a Tn4401b structure that was truncated at the 5′ end in the istA component of ISKpn7 by insertion of a Tn5403-like transposon. However, the 3′ end of Tn4401b was conserved with an intact 32-bp right-inverted repeat sequence (IRR), which in turn truncated Tn3. This downstream sequence has been observed in blaKPC-2-carrying IncN plasmids from various Enterobacteriaceae species that were involved in an outbreak at the NIH Clinical Center (19), but the upstream sequence consisting of Tn5403-like was unique to E. cloacae G6809. Chromosomal integration of blaKPC appears to be a relatively rare event overall but has been reported in K. pneumoniae (20) and Acinetobacter baumannii (21).
FIG 1.
(A) Genetic context of blaKPC-18 and blaVIM-1 in Enterobacter cloacae G6809. Antimicrobial resistance genes are indicated by vertical stripes. Genes involved in transposition are shaded. (B) The region surrounding the class 1 integron in pG6809-2 is compared with that of pNL194, a blaVIM-1-carrying plasmid reported from K. pneumoniae in Greece.
E. cloacae G6809 possessed two large plasmids, pG6809-1 (108,462 bp; GenBank accession no. KT345945) and pG6809-2 (58,120 bp; GenBank accession no. KT345946). pG6809-1 was related to p34399, which is a 121-kb plasmid registered from an E. cloacae clinical strain from the United States (GenBank accession no. CP010386). It carried a dihydrofolate reductase gene as the only antimicrobial resistance determinant, and the function of the rest of the plasmid was unclear. pG6809-2 was an IncN plasmid that most resembled pNL194, another IncN plasmid that was reported in K. pneumoniae from Greece and carried blaVIM-1 (22). The structure of the class I integron was similar to that of pNL194 and contained blaVIM-1, aacA7 [encoding aminoglycoside acetyltransferase AAC(6′)-Ib], dfrA1b (encoding dihydrofolate reductase DHFR), and aadA1b [encoding aminoglycoside adenylyltransferase ANT(3″)-Ia], whereas the 3′-conserved segment of this integron was missing due to insertion of IS1R and IS26 (Fig. 1B).
E. coli TOP10 transformant carrying pG6809-2 was resistant to BLBLIs, including ceftazidime-avibactam and cephalosporins, and had reduced susceptibility to carbapenems but remained susceptible to aztreonam, which reflected the spectrum of β-lactam hydrolysis by VIM-1 (Table 1) (23). However, E. cloacae G6809 was resistant to aztreonam due to the additional production of KPC-18 and the chromosomal AmpC, which together masked this unique susceptibility phenotype that is helpful in identifying MBL-producing organisms. As expected, addition of avibactam restored the susceptibility of E. cloacae G6809 to aztreonam with an MIC of 0.5/4 μg/ml, as this combination inhibits MBL, KPC, and AmpC (24).
The advent of a series of novel BLBLIs that inhibit KPC-type carbapenemases has the potential to transform the management of infections caused by KPC-producing Enterobacteriaceae for the better. However, this report illustrates the caveat that their activity is lost by coproduction of MBLs, such as VIM-1 in the KPC background, which is difficult to detect in the absence of sophisticated molecular diagnostics. Further complicating this issue is the lack of approved susceptibility testing methods for ceftazidime-avibactam so far, as well as inconsistent breakpoints from CLSI (for ceftazidime, MIC of ≤4 μg/ml for susceptibility) and the FDA (for ceftazidime-avibactam, MIC of ≤8/4 μg/ml for susceptibility). As more novel BLBLI combinations approach late-stage clinical development, we insist that due attention be paid to timely development and validation of the susceptibility testing method along with development of the new agents themselves.
Nucleotide sequence accession numbers.
The 100-kb region surrounding blaKPC-18 has been annotated and submitted to GenBank under accession number KT884517. The sequences of pG6809-1 and pG6809-2 have been submitted under accession numbers KT345945 and KT345946, respectively.
ACKNOWLEDGMENTS
Y.D. has served on advisory boards for Shionogi, Meiji Seika Pharma, and Tetraphase Pharmaceuticals; consulted for Melinta Therapeutics; and received research funding from Merck and The Medicines Company for studies unrelated to this work. All other authors declare no conflicts of interest.
REFERENCES
- 1.Pitout JD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. doi 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzouvelekis LS, Markogiannakis A, Piperaki E, Souli M, Daikos GL. 2014. Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 20:862–872. doi: 10.1111/1469-0691.12697. [DOI] [PubMed] [Google Scholar]
- 3.Drawz SM, Papp-Wallace KM, Bonomo RA. 2014. New β-lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother 58:1835–1846. doi: 10.1128/AAC.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasoo S, Cunningham SA, Cole NC, Kohner PC, Menon SR, Krause KM, Harris KA, De PP, Koh TH, Patel R. 2015. In vitro activities of ceftazidime-avibactam, aztreonam-avibactam, and a panel of older and contemporary antimicrobial agents against carbapenemase-producing Gram-negative bacilli. Antimicrob Agents Chemother 59:7842–7846. doi: 10.1128/AAC.02019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lolans K, Queenan AM, Bush K, Sahud A, Quinn JP. 2005. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob Agents Chemother 49:3538–3540. doi: 10.1128/AAC.49.8.3538-3540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epson EE, Pisney LM, Wendt JM, MacCannell DR, Janelle SJ, Kitchel B, Rasheed JK, Limbago BM, Gould CV, Kallen AJ, Barron MA, Bamberg WM. 2014. Carbapenem-resistant Klebsiella pneumoniae producing New Delhi metallo-β-lactamase at an acute care hospital, Colorado, 2012. Infect Control Hosp Epidemiol 35:390–397. doi: 10.1086/675607. [DOI] [PubMed] [Google Scholar]
- 7.Epstein L, Hunter JC, Arwady MA, Tsai V, Stein L, Gribogiannis M, Frias M, Guh AY, Laufer AS, Black S, Pacilli M, Moulton-Meissner H, Rasheed JK, Avillan JJ, Kitchel B, Limbago BM, MacCannell D, Lonsway D, Noble-Wang J, Conway J, Conover C, Vernon M, Kallen AJ. 2014. New Delhi metallo-β-lactamase-producing carbapenem-resistant Escherichia coli associated with exposure to duodenoscopes. JAMA 312:1447–1455. doi: 10.1001/jama.2014.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Kim SY, Hong SG, Moland ES, Thomson KS. 2007. Convenient test using a combination of chelating agents for detection of metallo-β-lactamases in the clinical laboratory. J Clin Microbiol 45:2798–2801. doi: 10.1128/JCM.02486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson GK, AbdelGhani S, Snyder JW, Thomson KS. 2015. Indirect Tris-EDTA disk testing using imipenem and meropenem for detection of OXA-48 carbapenemase production. J Clin Microbiol 53:3705–3706. doi: 10.1128/JCM.02094-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelhamed AM, Bajaksouzian S, Bonomo RA, Jacobs MR. 2015. Evaluation of Verigene BC-GN for identification of Gram negative bacteria and detection of resistance to β-lactams, including carbapenems from solid media, poster D-1153. Abstr 55th Intersci Conf Antimicrob Agents Chemother, San Diego, CA. [Google Scholar]
- 12.Porres-Osante N, Azcona-Gutierrez JM, Rojo-Bezares B, Undabeitia E, Torres C, Saenz Y. 2014. Emergence of a multiresistant KPC-3 and VIM-1 carbapenemase-producing Escherichia coli strain in Spain. J Antimicrob Chemother 69:1792–1795. doi: 10.1093/jac/dku055. [DOI] [PubMed] [Google Scholar]
- 13.Steinmann J, Kaase M, Gatermann S, Popp W, Steinmann E, Damman M, Paul A, Saner F, Buer J, Rath P. 2011. Outbreak due to a Klebsiella pneumoniae strain harbouring KPC-2 and VIM-1 in a German university hospital, July 2010 to January 2011. Euro Surveill 16: pii=19944. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19944. [PubMed]
- 14.Rojas LJ, Mojica MF, Blanco VM, Correa A, Montealegre MC, De La Cadena E, Maya JJ, Camargo RD, Quinn JP, Villegas MV. 2013. Emergence of Klebsiella pneumoniae coharboring KPC and VIM carbapenemases in Colombia. Antimicrob Agents Chemother 57:1101–1102. doi: 10.1128/AAC.01666-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meletis G, Tzampaz E, Protonotariou E, Sofianou D. 2010. Emergence of Klebsiella pneumoniae carrying blaVIM and blaKPC genes. Hippokratia 14:139–140. doi: 10.3109/23744235.2015.1094822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markovska R, Schneider I, Stoeva T, Bojkova K, Boyanova L, Bauernfeind A, Mitov I. 2013. First identification of KPC-2 and VIM-1 producing Klebsiella pneumoniae in Bulgaria. Diagn Microbiol Infect Dis 77:252–253. doi: 10.1016/j.diagmicrobio.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 17.Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, MOSAR WP2, WP3, and WP5 Study Groups . 2015. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother 70:48–56. doi: 10.1093/jac/dku359. [DOI] [PubMed] [Google Scholar]
- 18.Adams-Haduch JM, Potoski BA, Sidjabat HE, Paterson DL, Doi Y. 2009. Activity of temocillin against KPC-producing Klebsiella pneumoniae and Escherichia coli. Antimicrob Agents Chemother 53:2700–2701. doi: 10.1128/AAC.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, Program NCS, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Chavda KD, DeLeo FR, Bryant KA, Jacobs MR, Bonomo RA, Kreiswirth BN. 2015. Genome sequence of a Klebsiella pneumoniae sequence type 258 isolate with prophage-encoded K. pneumoniae carbapenemase. Genome Announc 3:pii=e00659-15. doi: 10.1128/genomeA.00659-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez T, Vazquez GJ, Aquino EE, Martinez I, Robledo IE. 2014. ISEcp1-mediated transposition of blaKPC into the chromosome of a clinical isolate of Acinetobacter baumannii from Puerto Rico. J Med Microbiol 63:1644–1648. doi: 10.1099/jmm.0.080721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miriagou V, Papagiannitsis CC, Kotsakis SD, Loli A, Tzelepi E, Legakis NJ, Tzouvelekis LS. 2010. Sequence of pNL194, a 79.3-kilobase IncN plasmid carrying the blaVIM-1 metallo-β-lactamase gene in Klebsiella pneumoniae. Antimicrob Agents Chemother 54:4497–4502. doi: 10.1128/AAC.00665-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franceschini N, Caravelli B, Docquier JD, Galleni M, Frere JM, Amicosante G, Rossolini GM. 2000. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob Agents Chemother 44:3003–3007. doi: 10.1128/AAC.44.11.3003-3007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazmierczak KM, Rabine S, Hackel M, McLaughlin RE, Biedenbach DJ, Bouchillon SK, Sahm DF, Bradford PA. 2015. Multi-year, multi-national survey of the incidence and global distribution of metallo-β-lactamase-producing Enterobacteriaceae and P. aeruginosa. Antimicrob Agents Chemother. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]