Abstract
Currently, there is no noninvasive test that can reliably diagnose early invasive pulmonary aspergillosis (IA). An electronic nose (eNose) can discriminate various lung diseases through an analysis of exhaled volatile organic compounds. We recently published a proof-of-principle study showing that patients with prolonged chemotherapy-induced neutropenia and IA have a distinct exhaled breath profile (or breathprint) that can be discriminated with an eNose. An eNose is cheap and noninvasive, and it yields results within minutes. We determined whether Aspergillus fumigatus colonization may also be detected with an eNose in cystic fibrosis (CF) patients. Exhaled breath samples of 27 CF patients were analyzed with a Cyranose 320. Culture of sputum samples defined the A. fumigatus colonization status. eNose data were classified using canonical discriminant analysis after principal component reduction. Our primary outcome was cross-validated accuracy, defined as the percentage of correctly classified subjects using the leave-one-out method. The P value was calculated by the generation of 100,000 random alternative classifications. Nine of the 27 subjects were colonized by A. fumigatus. In total, 3 subjects were misclassified, resulting in a cross-validated accuracy of the Cyranose detecting IA of 89% (P = 0.004; sensitivity, 78%; specificity, 94%). Receiver operating characteristic (ROC) curve analysis showed an area under the curve (AUC) of 0.89. The results indicate that A. fumigatus colonization leads to a distinctive breathprint in CF patients. The present proof-of-concept data merit external validation and monitoring studies.
INTRODUCTION
The diagnosis of invasive pulmonary aspergillosis (IA) in patients with prolonged chemotherapy-induced neutropenia (PCIN) remains a frequently occurring and difficult challenge (1–3). The signs and symptoms are nonspecific (4, 5). Mortality is high, especially in case of a diagnostic delay (3, 6, 7). The Platelia galactomannan assay (Bio-Rad) performed on bronchoalveolar lavage fluid samples is the only test with sufficient sensitivity and specificity to demonstrate or rule out IA (8). Its major drawback is the invasive and highly uncomfortable nature of bronchoalveolar lavage (9–13).
The Platelia galactomannan assay was originally developed to be used on serum. Although diagnostic accuracy is significantly lower in this setting, it creates the opportunity to serially monitor patients who are at high risk, such as acute myeloid leukemia (AML) patients treated with intensive induction chemotherapy (14). This might enable earlier diagnosis and thus a better prognosis. This is the topic of a number of ongoing studies. Unfortunately, two already-published studies did not show that serial monitoring of galactomannan leads to an earlier diagnosis (15, 16). Previously, we showed that patients with PCIN and IA have a distinct exhaled breath profile that can be discriminated by an electronic nose (eNose) (17). Very recently, this was confirmed by using gas chromatography-mass spectrometry (GC-MS), demonstrating exhaled metabolite signatures of Aspergillus fumigatus in vitro and IA in vivo (18).
An eNose is a cheap and noninvasive instrument with real-time measurement capacity. It can discriminate complex mixtures of volatile organic compounds (VOCs) by measuring these VOCs by pattern recognition using a limited number of cross-reactive sensors. This is achievable, because VOCs react differently with each sensor (19, 20). Exhaled breath can contain about 2,000 to 3,000 VOCs, and numerous disease states may be diagnosed by their exhaled breath profile (or breathprint, the composite pattern in exhaled VOCs) (21–30). For asthma and chronic obstructive pulmonary disease (COPD), this has been corroborated by external validation (23).
In cystic fibrosis (CF), proper chloride and sodium channel functioning is hampered, leading to an altered rheology of airway secretions, among other manifestations (31). The secretions become thick and difficult to clear, leading to chronic colonization with microorganisms. The reported incidence of A. fumigatus colonization varies between 6 and 58% (32–38). It is unknown whether colonization by A. fumigatus in CF results in a change in exhaled breath profile. If so, it would extend the proof-of-principle study that showed that an eNose may be used to diagnose invasive pulmonary aspergillosis (17). However, if an eNose can detect A. fumigatus colonization, this might also form the basis of false-positive test results in detecting invasive pulmonary aspergillosis.
We hypothesized that A. fumigatus colonization in CF patients leads to a distinct exhaled breath profile. Therefore, we aimed to determine whether an eNose can discriminate CF patients with and without A. fumigatus colonization.
MATERIALS AND METHODS
Design.
The study had a cross-sectional design in which exhaled breath analysis was performed once at a regular outpatient visit for each subject for pulmonary function assessment and sputum culture. Within 5 min of the exhaled breath analysis, subjects were asked to expectorate sputum for culture. Subjects with a culture positive for A. fumigatus were considered to be cases, and subjects with a negative culture were controls. We compared the breathprints between cases and controls.
Subjects.
Twenty-seven patients (age range, 17 to 50 years) from the CF clinic of the Academic Medical Center, Amsterdam, the Netherlands, with a confirmed diagnosis of CF (defined by 2 known CF mutations and/or a sweat chloride level of >60 mEq/liter) (39, 40), participated in this study (Table 1). Due to the noninvasive nature of merely collecting exhaled breath samples during routine lung function testing, the ethics review board of the Academic Medical Center considered that the protocol did not require ethical review under Dutch legislation. We calculated the P values in Table 1 using a two-sample t test (for age and the median forced expiratory volume in 1 s [FEV1]) and a Pearson's chi-square test with Yates' continuity correction (for gender).
TABLE 1.
Study subject characteristics, stratified according to A. fumigatus colonization status
| Characteristic | Colonized with A. fumigatus | Not colonized with A. fumigatus | P value |
|---|---|---|---|
| No. of subjects | 9 | 18 | >0.2 |
| Age (mean [SD]) (yr) | 28.0 (23.0) | 30.3 (9.6) | >0.2 |
| Male gender (%) | 67 | 67 | >0.2 |
| % of predicted FEV1 (mean [SD]) | 46.0 (28.3) | 63.5 (23.6) | >0.2 |
Exhaled breath analysis.
Exhaled breath analysis was performed as described previously (17, 22, 24). Subjects had to breathe through a mouthpiece with their nostrils closed by a clip for 5 min. The mouthpiece was connected by a three-way nonrebreathing valve to a VOC filter (A2; North Safety, Middelburg, the Netherlands) at the inspiratory port and a silica reservoir at the expiratory port. Without interruption, the subject was then asked to inspire deeply and exhale a vital capacity volume. The exhaled breath was stored in a 10-liter Tedlar bag attached to the silica reservoir. Analysis was performed within 30 min with a Cyranose 320 (Smith Detections, Pasadena, CA), a hand-held eNose with 32 nanocomposite sensors (19). The electrical resistance differential in all sensors was recorded. The procedure of sampling the Tedlar bag was done twice, using the first measurement solely to prime the sensors for the exhaled VOC mixture, as previously described (22), and as recommended by the manufacturer, in order to avoid a “first smell effect.” The first measurement was not used in the statistical analysis.
Index culture.
Subjects were asked to expectorate sputum once within 5 min of the exhaled breath maneuver. The sputum samples were processed and cultured according to the standard operating procedure of our hospital. The sputum samples were inoculated for bacterial culture on the following media: sheep blood agar (COS; bioMérieux), chromogenic agar (CPS; bioMérieux), chocolate agar containing thymidine, Columbia red agar containing thymidine, and N-acetyl-d-glucosamine agar. For the fungal cultures, samples were inoculated on two Sabouraud agars containing gentamicin and chloramphenicol (SGC2; bioMérieux). These were incubated for 14 days at 10°C and 36°C, respectively, and assessed twice a week. If growth was observed, fungi were identified based on morphology (both macroscopically and microscopically) and growth characteristics (41). We reviewed all sputum cultures from a study subject that were performed during the 2 years before the study culture. If one of those cultures showed a nonconsistent result with regard to Aspergillus spp., the subject was excluded from analysis. Sputum samples from our CF patients are routinely cultured twice per year, and often, more frequently.
Analysis.
As a primary analysis, we compared the breathprints of subjects with and without colonization by A. fumigatus, using the method of analysis from our previous study (17). We used the R statistical package (version 2.11.1). Principal component analysis (PCA) was executed to reduce the raw data of the 32 sensors to a smaller number of principal components. The number of components was chosen to include at least 99.9% of the variance within the data set. With a t test (equal variance assumed depending on an F-test), all PCA factors were categorized as either discriminatory between the two groups or not. A two-sided P value of 0.10 was chosen as a cutoff. Next, a diagnostic algorithm was constructed by using the discriminatory PCA factors as input for a linear canonical discriminant analysis, assuming an equal chance of being a member of one of the two groups. The discriminant function was optimized to best distinguish between categories. The percentage of correctly classified subjects (combining cases and controls) was cross-validated using the leave-one-out method. This resulted in cross-validated accuracy. The P value of our cross-validated accuracy was derived by generating 100,000 random classifications of our subjects (whether or not colonized by A. fumigatus) and calculating the chance that a random classification would have resulted in the same cross-validated accuracy or better, constructing a new pattern recognition algorithm for each of the random classifications. Receiver operating characteristic (ROC) curve analysis was performed.
RESULTS
All measurements were performed according to the predefined protocol. No patients had an exacerbation at the time of the measurement. None of the subjects were treated with antifungal therapy in the 2 years preceding the eNose measurement, either systemically or as an inhalant. All subjects were able to produce sputum. None of the subjects had a sputum culture in the previous 2 years that was nonconsistent with respect to Aspergillus spp., i.e., all cultures were either positive or negative. The entire eNose analysis took about 45 min in our experimental setup, including data handling. Principal component analysis resulted in 7 PCA factors that described >99.9% of the variance. In 9 of the 27 subjects, A. fumigatus was cultured from sputum. Of the 7 PCA factors, 3 discriminated between subjects with and without A. fumigatus colonization. Canonical discriminant analysis based on these 3 principal components yielded a cross-validated accuracy of 88.9%. Figure 1 shows the individual discriminant scores of subjects with and without colonization. Three subjects were misclassified in the cross-validation, two false positively and one false negatively, corresponding to a sensitivity of 78% and a specificity of 94%. The area under the curve of the ROC curve was 0.89. ROC analysis of the cross-validated posterior probabilities of group membership showed an area under the curve of 80.2% (Fig. 2). In our simulation, 0.4% of the random classifications resulted in a cross-validated accuracy of ≥88.9% (Fig. 3). The raw eNose data are presented in Fig. 4.
FIG 1.
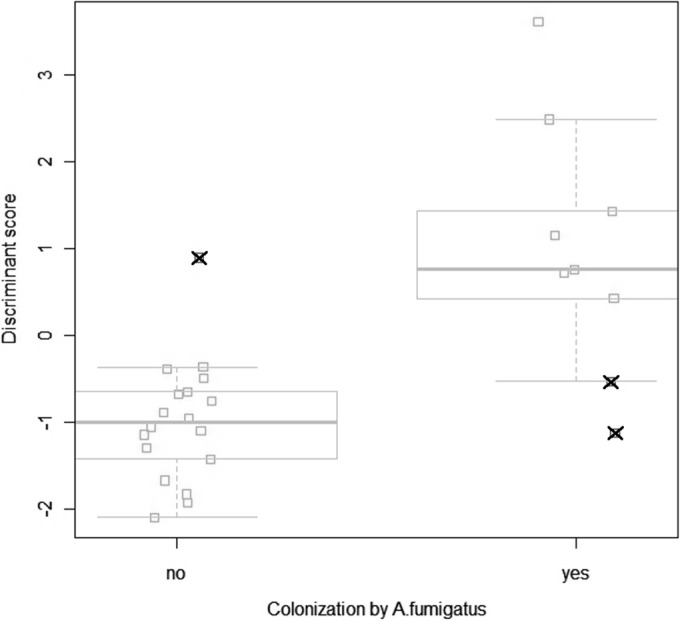
Breathprints of CF patients with and without A. fumigatus colonization, in a one-dimensional “jittered” canonical discriminant analysis plot. Squares with a cross represent subjects that were misclassified in the cross-validation. Box-and-whisker plots (with the median shown as a line through the box, the first and third quartiles shown as the lower and upper edges of the box, respectively, and the 95% confidence interval of the median shown as whiskers) are superimposed.
FIG 2.
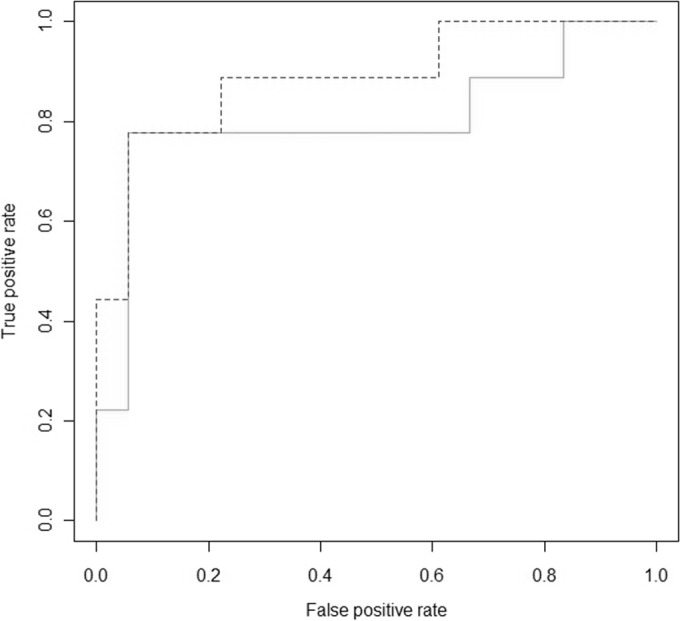
Receiver operating characteristic (ROC) curves showing the true-positive and false-positive rates in detecting A. fumigatus colonization with an eNose as a function of the cutoff value. Dashed line, ROC curve of the non-cross-validated measurements; solid line, the ROC curve of the cross-validated measurements.
FIG 3.
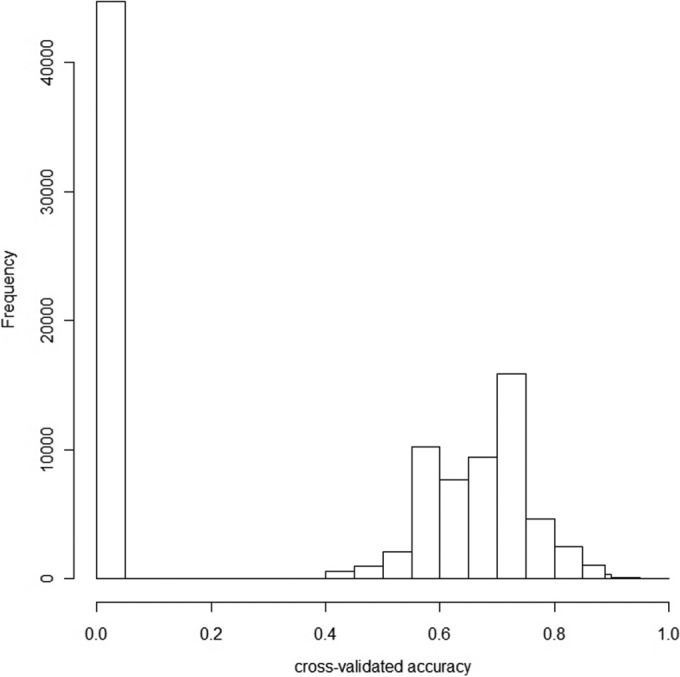
Histogram of the cross-validated accuracies produced by the random simulation.
FIG 4.
Spider graph of the raw eNose data showing a circular representation of the 32 sensor signals from all 27 samples, zoomed in to demonstrate between-sample differences. As the sensors are exposed to the breath sample, the electrical resistance over the sensors changes. On the axes of the spider graph, the quotient (calculated as [Rmax − Rmin]/[Rmin]) is displayed (Rmax, greatest amount of resistance; Rmin, smallest amount of resistance). The data have not been normalized.
We explored the effect of cocolonization by other microorganisms, for example, to exclude the possibility that the eNose had merely “smelled” another microorganism with a coinciding colonization pattern. Pseudomonas spp., Staphylococcus aureus, and Haemophilus influenzae were found in the index culture of more than one subject (see Table 2). The extent to which the colonization pattern of these microorganisms coincided with colonization by A. fumigatus is also shown in Table 2, as we calculated the percentage of cases in which a subject was colonized by both A. fumigatus and the other microorganism, or by neither of these two. We determined the cross-validated accuracy with which an eNose could detect those microorganisms by repeating the statistical method of the primary analysis afresh. Only P. aeruginosa colonization was associated with a PCA factor that differed statistically significantly between groups. The cross-validated accuracy of an eNose for detecting P. aeruginosa colonization was 63%, only slightly >50%.
TABLE 2.
Incidence and pattern of cocolonization
| Factor | No. of subjects | Coincidence with A. fumigatus colonization (%) | Cross-validated accuracy (%) |
|---|---|---|---|
| Pseudomonas spp. colonization | 17 | 67 | 63.0 |
| S. aureus colonization | 14 | 52 | NSa |
| H. influenzae colonization | 6 | 67 | NS |
| Nebulizing pulmonary medication | 15 | 56 | NS |
NS, no statistically significant principal component was found.
DISCUSSION
To the best of our knowledge, this study is the first to examine whether an eNose can detect A. fumigatus colonization with moderate to good accuracy. It shows that A. fumigatus colonization in CF patients leads to a distinct breathprint. The low P value obtained by comparison with 100,000 random classifications indicates that there is only a minor chance that this represents a false-positive outcome merely due to coincidence.
Our study has a number of strengths. It is novel, and we were able to examine a unique cohort of CF patients according to a strict protocol, using both electronic nose technology and microbiological techniques. The study also has limitations. First and foremost, the sample size was too small to allow split-half analysis. However, the 100,000 random classifications and internal validation using the leave-one-out method strengthened the validity of our results. Furthermore, our data indicate that it is highly unlikely that another microorganism was mistaken for A. fumigatus by our algorithm. We explored this possibility by repeating the statistical method of our analysis for every bacterial species that was encountered in the sputum culture results of more than one subject. Only P. aeruginosa colonization was associated with one or more PCA factors that differed statistically significantly between groups. The cross-validated accuracy was only 63.0%. Eventually, only external validation, i.e., the confirmation of our diagnostic algorithm in a separate group of subjects, would definitively establish the ability of an eNose to diagnose A. fumigatus colonization (42). Such validation has already been performed successfully in a similar eNose study on differentiating between COPD and asthma (23).
The second shortcoming of our study is that there is no gold standard reference test available for A. fumigatus colonization. As the sensitivity and specificity of a single culture are insufficient to conclude airway colonization by Aspergillus spp., we reviewed the sputum cultures of all subjects during the 2 years prior to the study. Sputum samples from CF patients are routinely cultured twice per year at our outpatient clinic, and much more often in practice. A subject was excluded from the study if one of the sputum cultures during the previous 2 years showed a result that was different from the index culture with respect to Aspergillus species. No patient had to be excluded from our study.
The present data add to the findings of a number of earlier studies. First of all, our in vivo findings extend the in vitro data of a number of research groups (43–45). These studies showed that A. fumigatus can be discriminated in vitro from multiple other microorganisms by an analysis of the metabolites they produce. Second, it was already suggested that the analysis of an exhaled metabolite profile may diagnose aspergillosis by use of gas chromatography-mass spectrometry identification of the exhaled biomarker 2-pentylfuran, which is specific for colonization and invasive disease by A. fumigatus. This biomarker was demonstrated to be exhaled by patients and to be produced by A. fumigatus in vitro (46, 47). Similarly, recent GC-MS and gas chromatography–solid-phase microextraction studies of A. fumigatus in vitro and in vivo demonstrated other candidate VOCs for IA, such as monoterpenes and sesquiterpenes (18, 48). Last, the current study strengthens the suggestion that eNose technology can detect such a distinct profile of metabolites produced by A. fumigatus in vivo by means of exhaled breath analysis. Our previous study already showed that invasive aspergillosis is associated with a distinctive breathprint, and as most subjects in the control group of that study had a pulmonary infection other than invasive aspergillosis, the distinct breathprint was unlikely to be caused by inflammation in general (17).
eNose technology has been designed to capture and compare complex molecular mixtures (20). Like human olfaction, this is extremely effective but does not allow the identification of the responsible VOCs, as it does not measure individual molecules or their concentrations (49). Therefore, eNoses are highly suitable for probabilistic diagnostic assessment, whereas gas chromatography-mass spectrometry (GC-MS) is required for pathophysiological research into the roles of individual molecular compounds (50). The combined output of all cross-reactive eNose sensors provides a pattern that can be subjected to pattern recognition algorithms. The VOCs that yielded the distinct breathprint in our study might of course be produced by the mold itself, as suggested above. 2-Pentylfuran and sesquiterpenes might be examples of this. However, differences in exhaled breath profiles may also be due to other factors than A. fumigatus metabolites. First, there might be an unknown confounding factor predisposing to (a positive culture suggesting) A. fumigatus colonization, such as a higher fungal load, more-severe CF, or more exposure to antibiotics or steroids, etc. A more likely possibility is the inflammatory host response, as Aspergillus spp. can trigger an immune response in the airways (37, 51, 52). Interestingly, asthma, CF and COPD, all inflammatory airway diseases, can be discriminated fairly accurately using eNose technology (22, 24, 25, 53, 54). More pathophysiological research on this topic is warranted, as the question is of more than theoretical interest: eNoses can be fine-tuned to react to specific VOCs. A comparison of the VOCs produced by molds in vitro and VOCs exhaled by patients with colonization or invasive disease by A. fumigatus will shed more light on this issue. This was recently done in patients with ventilator-associated pneumonia (VAP), demonstrating that the VOCs of tracheal aspirates were not influenced by colonization (55). Taken together, the success of the eNose can most likely be explained by a combination of VOCs originating in the specific inflammatory host response and/or VOCs originating from the mold itself.
The present results raise the hypothesis that in the future, an eNose could be used to diagnose invasive pulmonary aspergillosis, which frequently occurs during or after intensive high-risk treatment of acute leukemia or stem cell transplantation (1–3). Mortality due to invasive pulmonary aspergillosis is high, and while diagnosis is challenging, it is dependent on the amount of diagnostic delay (3–7). An eNose could be used to test patients regularly (e.g., twice a week) for the development of invasive pulmonary aspergillosis, instead of the current practice in some institutions of performing a serum galactomannan assay twice a week in high-risk populations (56, 57). The resulting reduction in diagnostic delay will hopefully improve prognosis. If accurate enough, exhaled breath analysis might also obviate bronchoscopy, an uncomfortable procedure associated with complications (9–13). As this study shows that colonization by A. fumigatus leads to a distinct breathprint, future research on exhaled breath analysis as a tool to diagnose invasive pulmonary mycoses should examine the possibility of colonization as a cause of false-positive test results.
In summary, our study shows that electronic nose technology can detect A. fumigatus colonization with moderate to good accuracy. These proof-of-concept data merit external validation and justify research to clarify the responsible compounds. Longitudinal monitoring studies are warranted to examine whether an electronic nose can detect developing invasive pulmonary aspergillosis at an earlier stage than the current state-of-the-art diagnostic strategies.
ACKNOWLEDGMENTS
We thank the 27 patients who made this study possible.
This study was supported by the Egbers Foundation. The funder had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Dettenkofer M, Wenzler-Röttele S, Babikir R, Bertz H, Ebner W, Meyer E, Rüden H, Gastmeier P, Daschner FD, Hospital Infection Surveillance System for Patients with Hematologic/Oncologic Malignancies Study Group . 2005. Surveillance of nosocomial sepsis and pneumonia in patients with a bone marrow or peripheral blood stem cell transplant: a multicenter project. Clin Infect Dis 40:926–931. doi: 10.1086/428046. [DOI] [PubMed] [Google Scholar]
- 2.Cornillet A, Camus C, Nimubona S, Gandemer V, Tattevin P, Belleguic C, Chevrier S, Meunier C, Lebert C, Aupe M, Caulet-Maugendre S, Faucheux M, Lelong B, Leray E, Guiguen C, Gangneux JP. 2006. Comparison of epidemiological, clinical, and biological features of invasive aspergillosis in neutropenic and nonneutropenic patients: a 6-year survey. Clin Infect Dis 43:577–584. doi: 10.1086/505870. [DOI] [PubMed] [Google Scholar]
- 3.Robenshtok E, Gafter-Gvili A, Goldberg E, Weinberger M, Yeshurun M, Leibovici L, Paul M. 2007. Antifungal prophylaxis in cancer patients after chemotherapy or hematopoietic stem-cell transplantation: systematic review and meta-analysis. J Clin Oncol 25:5471–5489. doi: 10.1200/JCO.2007.12.3851. [DOI] [PubMed] [Google Scholar]
- 4.Barth PJ, Rossberg C, Koch S, Ramaswamy A. 2000. Pulmonary aspergillosis in an unselected autopsy series. Pathol Res Pract 196:73–80. doi: 10.1016/S0344-0338(00)80036-9. [DOI] [PubMed] [Google Scholar]
- 5.Groll AH, Shah PM, Mentzel C, Schneider M, Just-Nuebling G, Huebner K. 1996. Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J Infect 33:23–32. doi: 10.1016/S0163-4453(96)92700-0. [DOI] [PubMed] [Google Scholar]
- 6.Aisner J, Wiernik PH, Schimpff SC. 1977. Treatment of invasive aspergillosis: relation of early diagnosis and treatment to response. Ann Intern Med 86:539–543. doi: 10.7326/0003-4819-86-5-539. [DOI] [PubMed] [Google Scholar]
- 7.von Eiff M, Roos N, Schulten R, Hesse M, Zuhlsdorf M, van de Loo J. 1995. Pulmonary aspergillosis: early diagnosis improves survival. Respiration 62:341–347. doi: 10.1159/000196477. [DOI] [PubMed] [Google Scholar]
- 8.Guo YL, Chen YQ, Wang K, Qin SM, Wu C, Kong JL. 2010. Accuracy of BAL galactomannan in diagnosing invasive aspergillosis: a bivariate metaanalysis and systematic review. Chest 138:817–824. doi: 10.1378/chest.10-0488. [DOI] [PubMed] [Google Scholar]
- 9.Tukey MH, Wiener RS. 2012. Population-based estimates of transbronchial lung biopsy utilization and complications. Respir Med 106:1559–1565. doi: 10.1016/j.rmed.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr IM, Koegelenberg CFN, von Groote-Bidlingmaier F, Mowlana A, Silos K, Haverman T, Diacon AH, Bolliger CT. 2012. Blood loss during flexible bronchoscopy: a prospective observational study. Respiration 84:312–318. doi: 10.1159/000339507. [DOI] [PubMed] [Google Scholar]
- 11.Facciolongo N, Patelli M, Gasparini S, Lazzari Agli L, Salio M, Simonassi C, Del Prato B, Zanoni P. 2009. Incidence of complications in bronchoscopy. Multicentre prospective study of 20,986 bronchoscopies. Monaldi Arch Chest Dis 71:8–14. [DOI] [PubMed] [Google Scholar]
- 12.Jin F, Mu D, Chu D, Fu E, Xie Y, Liu T. 2008. Severe complications of bronchoscopy. Respiration 76:429–433. doi: 10.1159/000151656. [DOI] [PubMed] [Google Scholar]
- 13.Pue CA, Pacht ER. 1995. Complications of fiberoptic bronchoscopy at a university hospital. Chest 107:430–432. doi: 10.1378/chest.107.2.430. [DOI] [PubMed] [Google Scholar]
- 14.Leeflang MM, Debets-Ossenkopp YJ, Visser CE, Scholten RJ, Hooft L, Bijlmer HA, Reitsma JB, Bossuyt PM, Vandenbroucke-Grauls CM. 2008. Galactomannan detection for invasive aspergillosis in immunocompromized [sic] patients. Cochrane Database Syst Rev (4):CD007394. [DOI] [PubMed] [Google Scholar]
- 15.Hebart H, Klingspor L, Klingebiel T, Loeffler J, Tollemar J, Ljungman P, Wandt H, Schaefer-Eckart K, Dornbusch HJ, Meisner C, Engel C, Stenger N, Mayer T, Ringden O, Einsele H. 2009. A prospective randomized controlled trial comparing PCR-based and empirical treatment with liposomal amphotericin B in patients after allo-SCT. Bone Marrow Transplant 43:553–561. doi: 10.1038/bmt.2008.355. [DOI] [PubMed] [Google Scholar]
- 16.Cordonnier C, Pautas C, Maury S, Vekhoff A, Farhat H, Suarez F, Dhdin N, Isnard F, Ades L, Kuhnowski F, Foulet F, Kuentz M, Maison P, Bretagne S, Schwarzinger M. 2009. Empirical versus preemptive antifungal therapy for high-risk, febrile, neutropenic patients: a randomized, controlled trial. Clin Infect Dis 48:1042–1051. doi: 10.1086/597395. [DOI] [PubMed] [Google Scholar]
- 17.de Heer K, van der Schee MP, Zwinderman K, van den Berk IA, Visser CE, van Oers R, Sterk PJ. 2013. Electronic nose technology for detection of invasive pulmonary aspergillosis in prolonged chemotherapy-induced neutropenia: a proof-of-principle study. J Clin Microbiol 51:1490–1495. doi: 10.1128/JCM.02838-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koo S, Thomas HR, Daniels SD, Lynch RC, Fortier SM, Shea MM, Rearden P, Comolli JC, Baden LR, Marty FM. 2014. A breath fungal secondary metabolite signature to diagnose invasive aspergillosis. Clin Infect Dis 59:1733–1740. doi: 10.1093/cid/ciu725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Röck F, Barsan N, Weimar U. 2008. Electronic nose: current status and future trends. Chem Rev 108:705–725. doi: 10.1021/cr068121q. [DOI] [PubMed] [Google Scholar]
- 20.Wilson AD. 2015. Advances in electronic-nose technologies for the detection of volatile biomarker metabolites in the human breath. Metabolites 5:140–163. doi: 10.3390/metabo5010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pauling L, Robinson AB, Teranishi R, Cary P. 1971. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proc Natl Acad Sci U S A 68:2374–2376. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fens N, Zwinderman AH, van der Schee MP, de Nijs SB, Dijkers E, Roldaan AC, Cheung D, Bel EH, Sterk PJ. 2009. Exhaled breath profiling enables discrimination of chronic obstructive pulmonary disease and asthma. Am J Respir Crit Care Med 180:1076–1082. doi: 10.1164/rccm.200906-0939OC. [DOI] [PubMed] [Google Scholar]
- 23.Fens N, Roldaan AC, van der Schee MP, Boksem RJ, Zwinderman AH, Bel EH, Sterk PJ. 2011. External validation of exhaled breath profiling using an electronic nose in the discrimination of asthma with fixed airways obstruction and chronic obstructive pulmonary disease. Clin Exp Allergy 41:1371–1378. doi: 10.1111/j.1365-2222.2011.03800.x. [DOI] [PubMed] [Google Scholar]
- 24.Dragonieri S, Schot R, Mertens BJ, Le Cessie S, Gauw SA, Spanevello A, Resta O, Willard NP, Vink TJ, Rabe KF, Bel EH, Sterk PJ. 2007. An electronic nose in the discrimination of patients with asthma and controls. J Allergy Clin Immunol 120:856–862. doi: 10.1016/j.jaci.2007.05.043. [DOI] [PubMed] [Google Scholar]
- 25.Dragonieri S, Annema JT, Schot R, van der Schee MP, Spanevello A, Carratu P, Resta O, Rabe KF, Sterk PJ. 2009. An electronic nose in the discrimination of patients with non-small cell lung cancer and COPD. Lung Cancer 64:166–170. doi: 10.1016/j.lungcan.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 26.van der Schee MP, Paff T, Brinkman P, van Aalderen WMC, Haarman EG, Sterk PJ. 2015. Breathomics in lung disease. Chest 147:224–231. doi: 10.1378/chest.14-0781. [DOI] [PubMed] [Google Scholar]
- 27.Machado RF, Laskowski D, Deffenderfer O, Burch T, Zheng S, Mazzone PJ, Mekhail T, Jennings C, Stoller JK, Pyle J, Duncan J, Dweik RA, Erzurum SC. 2005. Detection of lung cancer by sensor array analyses of exhaled breath. Am J Respir Crit Care Med 171:1286–1291. doi: 10.1164/rccm.200409-1184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Natale C, Macagnano A, Martinelli E, Paolesse R, D'Arcangelo G, Roscioni C, Finazzi-Agro A, D'Amico A. 2003. Lung cancer identification by the analysis of breath by means of an array of non-selective gas sensors. Biosens Bioelectron 18:1209–1218. doi: 10.1016/S0956-5663(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 29.Peng G, Hakim M, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A, Tisch U, Haick H. 2010. Detection of lung, breast, colorectal, and prostate cancers from exhaled breath using a single array of nanosensors. Br J Cancer 103:542–551. doi: 10.1038/sj.bjc.6605810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedrich MJ. 2009. Scientists seek to sniff out diseases: electronic “noses” may someday be diagnostic tools. JAMA 301:585–586. doi: 10.1001/jama.2009.90. [DOI] [PubMed] [Google Scholar]
- 31.Collins FS. 1992. Cystic fibrosis: molecular biology and therapeutic implications. Science 256:774–779. doi: 10.1126/science.1375392. [DOI] [PubMed] [Google Scholar]
- 32.Milla CE, Wielinski CL, Regelmann WE. 1996. Clinical significance of the recovery of Aspergillus species from the respiratory secretions of cystic fibrosis patients. Pediatr Pulmonol 21:6–10. doi:. [DOI] [PubMed] [Google Scholar]
- 33.Skov M, McKay K, Koch C, Cooper PJ. 2005. Prevalence of allergic bronchopulmonary aspergillosis in cystic fibrosis in an area with a high frequency of atopy. Respir Med 99:887–893. doi: 10.1016/j.rmed.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 34.Valenza G, Tappe D, Turnwald D, Frosch M, Konig C, Hebestreit H, Abele-Horn M. 2008. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros 7:123–127. doi: 10.1016/j.jcf.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Nelson LA, Callerame ML, Schwartz RH. 1979. Aspergillosis and atopy in cystic fibrosis. Am Rev Respir Dis 120:863–873. [DOI] [PubMed] [Google Scholar]
- 36.Bakare N, Rickerts V, Bargon J, Just-Nubling G. 2003. Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses 46:19–23. doi: 10.1046/j.1439-0507.2003.00830.x. [DOI] [PubMed] [Google Scholar]
- 37.Mroueh S, Spock A. 1994. Allergic bronchopulmonary aspergillosis in patients with cystic fibrosis. Chest 105:32–36. doi: 10.1378/chest.105.1.32. [DOI] [PubMed] [Google Scholar]
- 38.Skov M, Koch C, Reimert CM, Poulsen LK. 2000. Diagnosis of allergic bronchopulmonary aspergillosis (ABPA) in cystic fibrosis. Allergy 55:50–58. doi: 10.1034/j.1398-9995.2000.00342.x. [DOI] [PubMed] [Google Scholar]
- 39.Boyle MP. 2003. Nonclassic cystic fibrosis and CFTR-related diseases. Curr Opin Pulm Med 9:498–503. doi: 10.1097/00063198-200311000-00009. [DOI] [PubMed] [Google Scholar]
- 40.De Boeck K, Wilschanski M, Castellani C, Taylor C, Cuppens H, Dodge J, Sinaasappel M, Diagnostic Working Group . 2006. Cystic fibrosis: terminology and diagnostic algorithms. Thorax 61:627–635. doi: 10.1136/thx.2005.043539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Hoog GS, Guarro J, Gené J, Figueras MJ. 2001. Atlas of clinical fungi, 2nd edition ASM Press, Washington, DC. [Google Scholar]
- 42.Broadhurst D, Kell DB. 2006. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2:171–196. [Google Scholar]
- 43.Moens M, Smet A, Naudts B, Verhoeven J, Ieven M, Jorens P, Geise HJ, Blockhuys F. 2006. Fast identification of ten clinically important micro-organisms using an electronic nose. Lett Appl Microbiol 42:121–126. doi: 10.1111/j.1472-765X.2005.01822.x. [DOI] [PubMed] [Google Scholar]
- 44.Fend R, Kolk AH, Bessant C, Buijtels P, Klatser PR, Woodman AC. 2006. Prospects for clinical application of electronic-nose technology to early detection of Mycobacterium tuberculosis in culture and sputum. J Clin Microbiol 44:2039–2045. doi: 10.1128/JCM.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dutta R, Hines EL, Gardner JW, Boilot P. 2002. Bacteria classification using Cyranose 320 electronic nose. Biomed Eng Online 1:4. doi: 10.1186/1475-925X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chambers ST, Syhre M, Murdoch DR, McCartin F, Epton MJ. 2009. Detection of 2-pentylfuran in the breath of patients with Aspergillus fumigatus. Med Mycol 47:468–476. doi: 10.1080/13693780802475212. [DOI] [PubMed] [Google Scholar]
- 47.Syhre M, Scotter JM, Chambers ST. 2008. Investigation into the production of 2-pentylfuran by aspergillus fumigatus and other respiratory pathogens in vitro and human breath samples. Med Mycol 46:209–215. doi: 10.1080/13693780701753800. [DOI] [PubMed] [Google Scholar]
- 48.Heddergott C, Calvo AM, Latge JP. 2014. The volatome of Aspergillus fumigatus. Eukaryot Cell 13:1014–1025. doi: 10.1128/EC.00074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bushdid C, Magnasco MO, Vosshall LB, Keller A. 2014. Humans can discriminate more than 1 trillion olfactory stimuli. Science 343:1370–1372. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boots AW, Bos LD, van der Schee MP, van Schooten FJ, Sterk PJ. 2015. Exhaled molecular fingerprinting in diagnosis and monitoring: validating volatile promises. Trends Mol Med 21:633–644. doi: 10.1016/j.molmed.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 51.Peetermans M, Goeminne P, De Boeck C, Dupont LJ. 2014. IgE sensitization to Aspergillus fumigatus is not a bystander phenomenon in cystic fibrosis lung disease. Chest 146:e99–e100. doi: 10.1378/chest.14-0635. [DOI] [PubMed] [Google Scholar]
- 52.Baxter CG, Moore CB, Jones AM, Webb AK, Denning DW. 2013. IgE-mediated immune responses and airway detection of Aspergillus and Candida in adult cystic fibrosis. Chest 143:1351–1357. doi: 10.1378/chest.12-1363. [DOI] [PubMed] [Google Scholar]
- 53.Joensen O, Paff T, Haarman EG, Skovgaard IM, Jensen P, Bjarnsholt T, Nielsen KG. 2014. Exhaled breath analysis using electronic nose in cystic fibrosis and primary ciliary dyskinesia patients with chronic pulmonary infections. PLoS One 9:e115584. doi: 10.1371/journal.pone.0115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paff T, van der Schee MP, Daniels JMA, Pals G, Postmus PE, Sterk PJ, Haarman EG. 2013. Exhaled molecular profiles in the assessment of cystic fibrosis and primary ciliary dyskinesia. J Cyst Fibros 12:454–460. doi: 10.1016/j.jcf.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 55.Bos LDJ, Martin-Loeches I, Kastelijn JB, Gili G, Espasa M, Povoa P, Kolk AHJ, Janssen HG, Sterk PJ, Artigas A, Schultz MJ. 2014. The volatile metabolic fingerprint of ventilator-associated pneumonia. Intensive Care Med 40:761–762. doi: 10.1007/s00134-014-3260-5. [DOI] [PubMed] [Google Scholar]
- 56.Maertens J, Theunissen K, Verhoef G, Verschakelen J, Lagrou K, Verbeken E, Wilmer A, Verhaegen J, Boogaerts M, Van Eldere J. 2005. Galactomannan and computed tomography-based preemptive antifungal therapy in neutropenic patients at high risk for invasive fungal infection: a prospective feasibility study. Clin Infect Dis 41:1242–1250. doi: 10.1086/496927. [DOI] [PubMed] [Google Scholar]
- 57.Maertens J, Van Eldere J, Verhaegen J, Verbeken E, Verschakelen J, Boogaerts M. 2002. Use of circulating galactomannan screening for early diagnosis of invasive aspergillosis in allogeneic stem cell transplant recipients. J Infect Dis 186:1297–1306. doi: 10.1086/343804. [DOI] [PubMed] [Google Scholar]