Abstract
Klebsiella pneumoniae, a Gram-negative bacillus that causes life-threatening infections in both hospitalized patients and ambulatory persons, can be classified into nine lipopolysaccharide (LPS) O-antigen serotypes. The O-antigen type has important clinical and epidemiological significance. However, K. pneumoniae O serotyping is cumbersome, and the reagents are not commercially available. To overcome the limitations of conventional serotyping methods, we aimed to create a rapid and accurate PCR method for K. pneumoniae O genotyping. We sequenced the genetic determinants of LPS O antigen from serotypes O1, O2a, O2ac, O3, O4, O5, O8, O9, and O12. We established a two-step genotyping scheme, based on the two genomic regions associated with O-antigen biosynthesis. The first set of PCR primers, which detects alleles at the wzm-wzt loci of the wb gene cluster, distinguishes between O1/O2, O3, O4, O5, O8, O9, and O12. The second set of PCR primers, which detects alleles at the wbbY region, further differentiates between O1, O2a, and O2ac. We verified the specificity of O genotyping against the O-serotype reference strains. We then tested the sensitivity and specificity of O genotyping in K. pneumoniae, using the 56 K-serotype reference strains with known O serotypes determined by an inhibition enzyme-linked immunosorbent assay (iELISA). There is a very good correlation between the O genotypes and classical O serotypes. Three discrepancies were observed and resolved by nucleotide sequencing—all in favor of O genotyping. The PCR-based O genotyping, which can be easily performed in clinical and research microbiology laboratories, is a rapid and accurate method for determining the LPS O-antigen types of K. pneumoniae isolates.
INTRODUCTION
Klebsiella pneumoniae is an encapsulated Gram-negative bacillus that frequently causes outbreaks of nosocomial infections in hospitalized patients (1–3). It can also affect ambulatory persons and cause community-acquired invasive diseases, including pyogenic liver abscess, endophthalmitis, meningitis, empyema, lung abscess, and necrotizing fasciitis (4–9). The worldwide emergence of multidrug-resistant strains and hypervirulent strains of K. pneumoniae has become an increasing clinical challenge and public health concern (10, 11).
K. pneumoniae strains can be distinguished by their capsular polysaccharide (CPS) K-antigen types (77 serotypes) and lipopolysaccharide (LPS) O-antigen types (9 serotypes) (2). The K type and O type of K. pneumoniae both have important clinical and epidemiological significance (8, 12, 13). K1 clonal complex 23 is a newly emerged hypervirulent clade, which causes pyogenic liver abscesses with septic ocular or central nervous system complications (8, 14, 15). Likewise, O1 is associated with hypervirulent strains that cause pyogenic liver abscess (12). The O:K typing also helps establish the clonality of K. pneumoniae in nosocomial outbreak investigations (13). K-genotyping methods are now available for quickly and correctly determining the K types in K. pneumoniae (8, 16, 17). On the other hand, there is still a lack of genotyping techniques for identifying the O types of K. pneumoniae strains.
A rapid and accurate O-genotyping method for K. pneumoniae is desirable, because O-serotyping methods have several notable limitations. Traditional O serotyping such as tube agglutination is interfered with by the masking effect of heat-stable CPS (18), which necessitates the use of capsule-free mutants (19). The introduction of an inhibition enzyme-linked immunosorbent assay (iELISA) has obviated the need for mutants (18, 20, 21). Nevertheless, the procedure is still cumbersome and the reagents are not commercially available. Furthermore, serological cross-reactivity exists between biochemically distinctive O types due to similarities in their O-antigen structures (19, 21, 22). For example, serological cross-reactions occur between O1, O2(2a), and O2(2a,2c) because the O-antigen polysaccharides in O1 (d-galactan I and d-galactan II), O2(2a) (d-galactan I alone), and O2(2a,2c) (d-galactan I and 2c polysaccharide) have a common element: d-galactan I (23, 24). The technical difficulties in O serotyping have hindered the clinical and epidemiological applications of K. pneumoniae O typing.
To overcome the limitations of conventional O-serotyping methods, we designed a novel PCR-based O-genotyping approach. The main genetic determinant of K. pneumoniae O antigen is the wb gene cluster (25–28), which contains the wzm and wzt genes, which encode an ATP-binding cassette transporter for O-polysaccharide biosynthesis (29). The wbbY region, which contains the wbbY gene, which encodes a glycosyltransferase, is also involved in the O-antigen synthesis (30). In K. pneumoniae O1, d-galactan I synthesis requires the wb gene cluster only (25), while d-galactan II synthesis further requires the wbbY region (30). Therefore, we hypothesized that O1, O2(2a), and O2(2a,2c) have identical wb gene clusters for d-galactan I synthesis, yet they can be distinguished by the existence or absence of specific alleles at the wbbY region for synthesis of a second O-polysaccharide (d-galactan II or 2c polysaccharide); meanwhile, each of the other O types (O3, O4, O5, O8, O9, and O12) has a unique allele at the wb gene cluster for generating type-specific O-polysaccharides. In other words, one assay (wb) would allow us to separate the O1/O2a/O2ac strains from the other O types, and the second assay (wbbY) would further differentiate between O1, O2a, and O2ac.
This study aimed to sequence and analyze the genetic determinants of the nine O serotypes (O1, O2a, O2ac, O3, O4, O5, O8, O9, and O12) that have been found in clinical K. pneumoniae isolates (13, 18, 20, 21, 31) and to create and validate a novel genotyping scheme for determining the O-antigen types in K. pneumoniae.
MATERIALS AND METHODS
Bacterial strains.
Nine Klebsiella O-serotype reference strains and 77 Klebsiella K-serotype reference strains (of which 56 are K. pneumoniae) were purchased from Statens Serum Institut (Copenhagen, Denmark) (see Tables S1 and S2 in the supplemental material). The O serotypes of the 77 K-serotype reference strains were previously determined using an iELISA by Hansen et al. (18). The O2ac reference strain 5053 and six K-serotype reference strains (K4, K27, K28, K35, K43, and K59) are the prototype strains of the serotype O2 subgroups established by Ørskov (32). The taxonomic designation of the 77 K reference strains was reevaluated and updated using rpoB gene sequencing by Brisse et al. (16, 33). An epidemiological sample of 87 nonduplicate K. pneumoniae strains, which were consecutively collected from routine cultures in patients with community-acquired K. pneumoniae infections at National Taiwan University Hospital (Taipei, Taiwan), were used to evaluate the performance of our O-genotyping method in clinical settings. The study procedure was reviewed and approved by National Taiwan University Hospital's Institutional Review Board (no. 201506057RINA). All strains were stored at −80°C before use.
Sequencing the wb gene cluster.
We sequenced the wb gene clusters of O1, O2ac, O3, O4, O5, O8, and O12 reference strains, using PCR primers designed from published nucleotide sequences of the following strains: K. pneumoniae NTUH-K2044 (serotype O1; complete genome: GenBank accession number NC012731) (34), K. variicola 342 (O serotype uncertain; complete genome: accession no. NC011283) (33, 35), K. pneumoniae KPNIH31 (O serotype uncertain; complete genome: accession no. CP009876) (36), K. pneumoniae KT769 (serotype O5; partial wb gene cluster: accession no. AF189151) (27), K. terrigena 889 (serotype O8; partial wb gene cluster: accession no. L41518) (16, 37), and K. pneumoniae KT776 (serotype O12; partial wb gene cluster: accession no. AF097519) (28). The PCR products were generally 1 to 3 kb in size. When the standard PCR was not successful, we used the long and accurate PCR (LAPCR) method (38), which yielded products of up to 8 kb. Nucleotide sequencing was performed using an ABI 3730 DNA sequencer at the Sequencing Laboratory of the National Taiwan University Hospital.
Sequencing the wbbY region.
The wbbY region was originally characterized and sequenced by Hsieh et al. (30) in an O1 serotype strain: K. pneumoniae NTUH-K2044 (GenBank accession no. KJ451390). We sequenced the wbbY region in an O2ac serotype strain: K. pneumoniae subsp. ozaenae D5050 (the K4 reference strain) (see Table S2 in the supplemental material), using PCR primers designed from the whole-genome shotgun sequence of K. pneumoniae subsp. ozaenae ATCC 11296 (accession no. NZ_CDJH00000000.1).
Extraction of genomic DNA.
The pellet from the overnight culture preparation was lysed by cell lysis solution (1% sodium dodecyl sulfate, 200 mM Tris-HCl buffer [pH 8.5], and 100 mM EDTA [pH 8.0]). The protein components were then precipitated by 10 M ammonium acetate. The genomic DNA in the supernatant was extracted by phenol-chloroform.
Genotyping procedure.
Genomic DNA (100 ng) extracted from the strains tested was used as the template. Standard Taq polymerase (DreamTaq; Thermo Fisher Scientific, Waltham, MA) was used for PCR, with 0.4 μM primers and 2.5 mM Mg2+ in a final reaction volume of 50 μl. The cycle was set as 96°C for 3 min and then 30 cycles of the following steps: 96°C for 30 s, 56°C for 15 s, and 72°C for 1 min. Afterwards, the samples were incubated at 72°C for 10 min.
Sensitivity and specificity of O genotyping.
We examined the specificity of O genotyping against the nine O-serotype reference strains. We then assessed the sensitivity and specificity of O genotyping in K. pneumoniae strains, using the 56 K. pneumoniae K-serotype reference strains with known O serotypes (see Table S2 in the supplemental material) (18). The sensitivity and specificity of O genotyping (against the iELISA) are assessed by the true-positive rate (i.e., the proportion of strains with a given O serotype that did have the corresponding O genotype) and the true-negative rate (i.e., the proportion of strains without a given O serotype that did not have the corresponding O genotype), respectively (39). For strains with inconsistent results between the iELISA and O genotyping, we sequenced the PCR products to resolve the discrepancy.
Nucleotide sequence accession numbers.
We deposited complete nucleotide sequence of the wb gene clusters from the O1, O2ac, O3, O4, O5, O8, and O12 reference strains in GenBank under accession no. AB819964, AB795943, AB795941, KU310493, AB819962, AB819963, and AB795942, respectively. We deposited the nucleotide sequences of the wzm and wzt genes from the O9 reference strain in GenBank under accession no. LC107950. We also deposited the complete nucleotide sequence of the wbbY region from the O2ac serotype strain D5050 in GenBank under accession no. LC074715. We performed sequence analysis and annotation using Sequencher (Gene Codes, Ann Arbor, MI) and the National Center for Biotechnology Information (NCBI) BLAST. The phylogenetic trees were constructed using Molecular Evolutionary Genetics Analysis (MEGA) (http://megasoftware.net/) (40), based on the maximum likelihood method with 2,000 bootstrap replicates.
RESULTS
Nucleotide sequence diversity in the wb gene clusters of O1, O3, O4, O5, O8, O9, and O12.
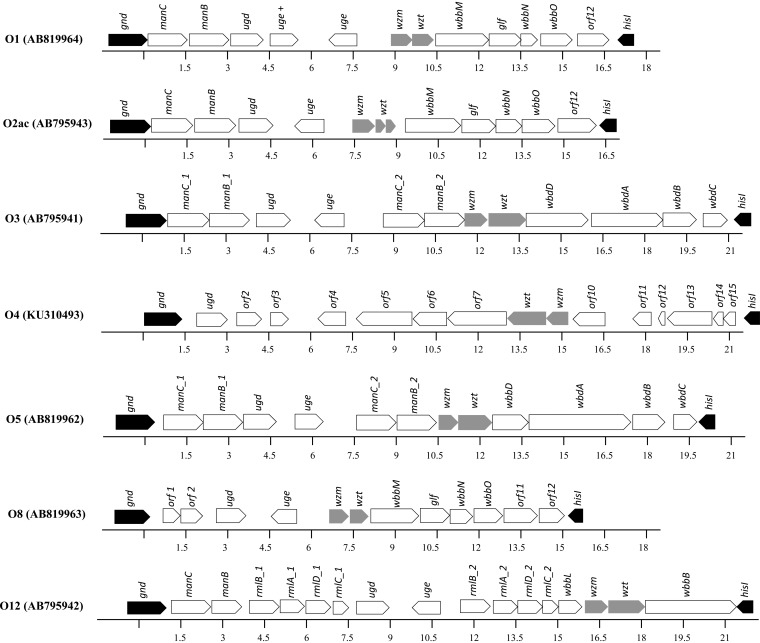
Figure 1 shows the organization of the wb gene clusters in O1, O2ac, O3, O4, O5, O8, and O12 reference strains, all of which contain the wzm and wzt genes (shown in gray). The nucleotide sequences at the wzm-wzt loci are <70% similar between the O1, O3, O4, O5, O8, and O12 reference strains. In contrast, the sequences are nearly identical between the O1 and O2ac reference strains at wzm (99.6% similar) and wzt (97.2% similar).
FIG 1.
Organization of the wb gene clusters in the O1, O2ac, O3, O4, O5, O8, and O12 reference strains. The wb gene clusters are located between the highly conserved gnd and hisI genes (black color). The nucleotide position (in kilobases) is relative to the first nucleotide of the complete sequence submitted to GenBank. The wzm and wzt loci are shown in gray. The O1 reference strain has an additional uge gene, which is not present in the other O1 strains. The O2ac reference strain has a truncated wzt gene because of a frameshift mutation (secondary to the addition of two DNA bases, adenine and cytosine, at nucleotides 324 and 325, respectively), which is not present in the other O2ac strains.
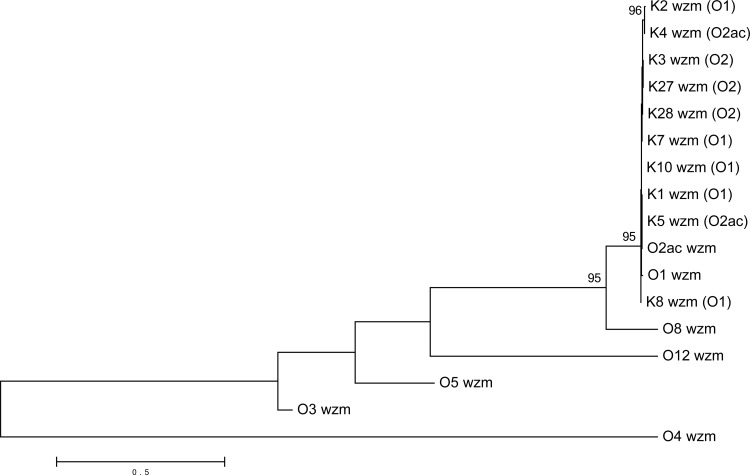
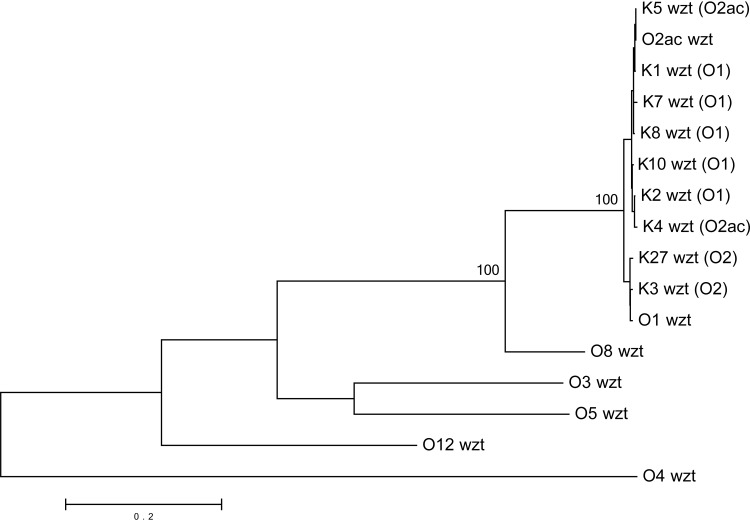
To confirm the identity in the nucleotide sequences at the wzm-wzt loci between O1, O2a, and O2ac, we sequenced 10 K reference strains of a known O1 serotype (K1, K2, K7, K8, K10), O2 serotype (K3, K27, K28) (for which the principal antigen inhibited in the O2 iELISA system is O2a) (18), or O2ac serotype (K4, K5) (see Table S2 in the supplemental material). The wzm and wzt genes are nearly identical among the 10 O1, O2, and O2ac strains (see File S1 in the supplemental material). The phylogenetic analysis of wzm and wzt shows that all of the sequenced O1, O2, and O2ac strains (n = 12) form a single clade that is separate from the O3, O4, O5, O8, and O12 strains (Fig. 2 and 3).
FIG 2.
Phylogenetic relationships among the wzm genes from O1, O2ac, O3, O4, O5, O8, and O12 reference strains, as well as from the 10 K reference strains of an O1, O2, or O2ac serotype by iELISA (for which the principal antigen inhibited in the O2 iELISA system is O2a) (18). Bootstrap percentages of remarkable nodes are shown (distance scale, 0.1 represents 10% difference between sequences).
FIG 3.
Phylogenetic relationships among the wzt genes from the O1, O2ac, O3, O4, O5, O8, and O12 reference strains and from the 10 K reference strains of an O1, O2, or O2ac serotype by iELISA (for which the principal antigen inhibited in the O2 iELISA system is O2a) (18). Bootstrap percentages of remarkable nodes are shown.
Since the previous iELISA studies showed that O9 is closely related to O2 (18), we sequenced the wzm-wzt region of the O9 reference strain (see Table S3 and File S2 in the supplemental material) and compared it with that of the O2ac reference strain. We found that the nucleotide sequences are only 78 to 79% similar between O9 and O2ac.
Based on the diversity in nucleotide sequences, we designed the first set of PCR primers to detect the O1/O2, O3, O4, O5, O8, O9, and O12 alleles at the wzm-wzt loci of the wb gene clusters (Tables 1 and 2).
TABLE 1.
Klebsiella pneumoniae O-genotyping scheme
| Genotype |
Serotype |
|||||
|---|---|---|---|---|---|---|
| Designation | wb allele | wbbY allele | Designationa | Biochemical component(s)b | Antigen factor(s)a | Reference strain(s) |
| O1 | O1/O2 | O1 | O1 | d-Galactan I, d-galactan II | O2a, O1 | Friedländer 204 |
| O2 | ||||||
| O2a | O1/O2 | None | O2(2a) | d-Galactan I | O2a | 5758, 2482 |
| O2(2a,2f,2g) | d-Galactan I (with d-Galp side group) | O2a, O2f, O2g | 6613c | |||
| O2ac | O1/O2 | O2ac | O2(2a,2c) O2(2a,2c,2d) |
d-Galactan I, 2c polysaccharide | O2a, O2c O2a, O2c, O2d |
5053d D5050e |
| O3 | O3 | NAf | O3 | O3 polysaccharide | O3 | 636/52 |
| O4 | O4 | NA | O4 | O4 polysaccharide | O4 | Mich.61 |
| O5 | O5 | NA | O5 | O5 polysaccharide | O5 | 5710/52 |
| O7g | NSg | NA | O7 | O7 polysaccharide | O7 | 264(1)h |
| O8 | O8 | O1 | O8 | O-acetylated d-galactan I, d-galactan II | O2a, O1 | 889 |
| O9 | O9 | NA | O9 | O-acetylated d-galactan I with d-Galp side group | O2a, O9 | 1205 |
| O2(2a,2e) | O2a, O2e | 7444i | ||||
| O12 | O12 | NA | O12 | O12 polysaccharide | O12 | 708 |
From Ørskov and Ørskov (19), Ørskov (32), Trautmann et al. (21), and Hansen et al. (18). O2b is not a true O-antigenic factor (24). O2(2a,2e,2h) is biochemically closely related to O9 and O2(2a,2e), with minor differences in the stoichiometry of the d-Galp side chain addition and O-acetylation of the d-Gal residue (43). We found that the prototype O2(2a,2e,2h) strain (K. michiganensis 2212/52) does not react with the O9, O1/O2, or other O-type wb primers. O6 is identical to O1 (19). O10 is not Klebsiella (21). O11 (for which the biochemical components have not yet been elucidated) is serologically closely related to O4 (21).
From Ørskov and Ørskov(19), Whitfield et al. (23, 24), Kelly et al. (41, 43), and Vinogradov et al. (22).
The d-galactan I in strain 6613 has a d-Galp side group (43). The genetic determinant for addition of the d-Galp side group is located outside the wb gene cluster (43). Therefore, O2(2a,2f,2g) has the same O1/O2 wb allele as O2(2a). The genes responsible for the addition of the d-Galp side group have not yet been identified.
Strain 5053 has a truncated wzt gene, which retains a certain degree of function that allows the expression of O antigen (24, 47). Compared with strains D5050 (K4 reference strain) and E5051 (K5 reference strain), both of which have a normal wzt gene, strain 5053 produces significantly more low-molecular-weight d-galactan I in its SDS-PAGE LPS profile (24).
Strain D5050 has a normal wzt gene. Compared with strain 5053, which has a truncated wzt gene, strain D5050 produces predominantly high-molecular-weight 2c polysaccharide in its SDS-PAGE LPS profile (24).
NA, not applicable.
Currently nontypeable. NS, not sequenced yet.
Taxonomic reevaluation using rpo sequencing in 2013 found that strain 264(1) is of the species K. (Raoultella) terrigena (16).
The structure of O-polysaccharide in strain 7444, O2(2a,2e), is identical to that of O9 (43). Strain 7444 reacts with the O9-specific wb primers, but not with the O1/O2 wb primers.
TABLE 2.
Primers used for Klebsiella pneumoniae lipopolysaccharide O genotyping by allele-specific PCR
| O type detected | Primer | Nucleotide sequence | Primer positiona | PCR product size (kb) |
|---|---|---|---|---|
| Allele at the wzm-wzt loci of the wb gene clusterb | ||||
| O1/O2 | wb O1/O2-A-F | 5′-CGCTATAAGAGCAGCATGCTAG-3′ | 7470–7491 | 1.3 |
| wb O1/O2-A-R | 5′-CGATATCACCTACTGCCAGA-3′ | 8703–8722 | ||
| wb O1/O2-B-F | 5′-TTGTTGAGCCTGACAGGATC-3′ | 7317–7336 | 1.6 | |
| wb O1/O2-B-R | 5′-GCCATTGCTTGCTTGTACAG-3′ | 8890–8909 | ||
| O3 | wb O3-A-F | 5′-CTATCGCTACCGTGGCTTTA-3′ | 11093–11112 | 0.8 |
| wb O3-A-R | 5′-TCTCGTCCACAATATCAGCG-3′ | 11840–11859 | ||
| wb O3-B-F | 5′-GCCTACAGTATCTACCTCTG-3′ | 11271–11290 | 0.9 | |
| wb O3-B-R | 5′-CGGTAAAGTCAGGATGGAAG-3′ | 12154–12173 | ||
| O4 | wb O4-A-F | 5′-CAGAAGCGCGAGTTAATCTG-3′ | 15212–15231 | 0.7 |
| wb O4-A-R | 5′-GGTCCAGTTAGGCTCAATTC-3′ | 14554–14573 | ||
| wb O4-B-F | 5′-GTCAGCGGGAATTATTGGAC-3′ | 14258–14277 | 1.2 | |
| wb O4-B-R | 5′-CTTGAGATCCAGAATGCCAC-3′ | 13077–13096 | ||
| O5 | wb O5-A-F | 5′-GCTACCAAACCAGTATGCTG-3′ | 10564–10583 | 1.8 |
| wb O5-A-R | 5′-AGGTGCGTACTGGAAGTATG-3′ | 12365–12384 | ||
| wb O5-B-F | 5′-GGTGATGAAAGCCAGAATGC-3′ | 10649–10668 | 1.4 | |
| wb O5-B-R | 5′-CAGTGCCTGAAACAGTTTGC-3′ | 12052–12071 | ||
| O8 | wb O8-A-F | 5′-CGTGGCAATGGTTTGCTAGT-3′ | 7784–7803 | 1.2 |
| wb O8-A-R | 5′-TCAATCCACACAACTCGGTC-3′ | 8994–9013 | ||
| wb O8-B-F | 5′-GCTAGTTCGGCAACTAACTCAC-3′ | 7798–7819 | 0.8 | |
| wb O8-B-R | 5′-AGTTCCAGCATCGAAGCAACTC-3′ | 8617–8638 | ||
| O9 | wb O9-A-F | 5′-CGCGCTCAGTTATTCCATTG-3′ | 305–324 | 1.0 |
| wb O9-A-R | 5′-CTGGCTGATGACAGAGAATC-3′ | 1258–1277 | ||
| wb O9-B-F | 5′-GCATTCCTGTTCGTGTATGG-3′ | 379–398 | 0.9 | |
| wb O9-B-R | 5′-ATGTCACCGACAGCAAGTAC-3′ | 1308–1327 | ||
| O12 | wb O12-A-F | 5′-CTGCAGATGGCTAAACGTGA-3′ | 16269–16288 | 0.6 |
| wb O12-A-R | 5′-CCGTTCGGGCTTGTTCAATA-3′ | 16877–16896 | ||
| wb O12-B-F | 5′-GAAGTCGACTTTGCTGCAGA-3′ | 17238–17257 | 1.0 | |
| wb O12-B-R | 5′-ACGTTGATCAAGCTCCTCTC-3′ | 18227–18246 | ||
| Allele at the wbbY loci of the wbbY regionc | ||||
| O1 | wbb O1-F | 5′-GATTTCACTTTCCGGGCAAC-3′ | 2191–2210 | 1.1 |
| wbb O1-R | 5′-GGCTTGCTGAATCACAAGAC-3′ | 1136–1155 | ||
| O2ac | wbb O2ac-F | 5′-AAACATCGCTGACTCGAGTC-3′ | 1905–1924 | 1.0 |
| wbb O2ac-R | 5′-CGACTATGATCGTACCAACG-3′ | 879–898 |
For the wb gene cluster, the primer position is relative to the first nucleotide of the corresponding complete wb gene cluster sequence submitted to GenBank. For the primers targeting O1/O2, O3, O4, O5, O8, and O12 alleles, the reference sequences are the following: O2ac, AB795943; O3, AB795941; O4, KU310493; O5, AB819962; O8, AB819963; and O12, AB795942, respectively (Fig. 1; the positions of the corresponding genes, wzm and wzt, are shown in gray). For O9, the primer position is relative to the first nucleotide of the wzm-wzt region sequence submitted to GenBank (accession no. LC107950). The primer position is relative to the first nucleotide of the wbbY alleles. For the primers targeting the O1 and O2ac wbbY allele, the reference sequences are the following: O1, KJ451390; and O2ac, LC074715, respectively (see Fig. 4, the positions of the corresponding gene, wbbY, are shown in gray color).
To enhance specificity, we designed two PCR primer pairs for each allele at the wb gene cluster. Only the strains that tested positive for both primer pairs are classified as having a specific genotype.
The pair of primers wbb O1-F and wbb O1-R, which detects the allele for d-galactan II, also reacts with O8 (which contains d-galactan II and O-acetylated d-galactan I).
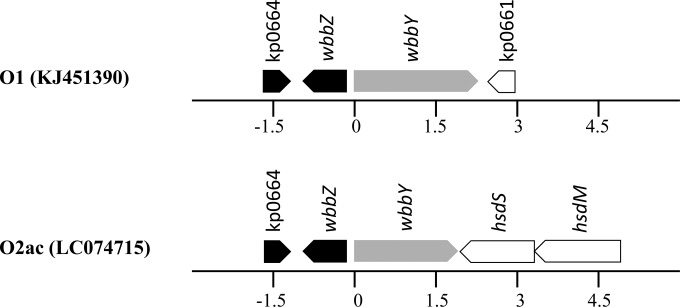
Nucleotide sequence diversity at wbbY loci of O1 and O2ac.
Figure 4 shows the organization of the wbbY region in O1 and O2ac serotype strains, both of which contain the wbbY gene (shown in gray). Nucleotide sequences of the O1 allele (2,223 nucleotides) and the O2ac allele (1,926 nucleotides) are nearly identical up to nucleotide 1896, after which the sequence similarity is <60%. Based on this difference, we designed a second set of PCR primers to detect the O1 and O2ac alleles at the wbbY locus (Tables 1 and 2).
FIG 4.
Organization of the wbbY regions in Klebsiella pneumoniae NTUH-K2044 (serotype O1) and K. pneumoniae subsp. ozaenae D5050 (serotype O2ac). wbbZ is highly conserved (black). The wbbY locus is shown in gray. The nucleotide position (in kilobases) is relative to the first nucleotide of the wbbY gene.
Two-step O-genotyping scheme.
We initially tested all strains using the first set of PCR primers, which detect the O1/O2, O3, O4, O5, O8, O9, and O12 alleles at the wzm-wzt loci of the wb gene clusters. To enhance the specificity, we designed two primer pairs for each allele at the wb gene cluster. Only the strains that tested positive for both primer pairs are classified as having a specific genotype. Strains that did not belong to the above genotypes are considered nontypeable. The strains with the O1/O2 allele were further tested using the second set of PCR primers, which detect the O1 and O2ac alleles at the wbbY locus. The strains with the O1/O2 allele that did not react with the O1- or O2ac-specific primers are considered the O2a genotype.
Specificity against O reference strains.
We verified that O genotyping is specific against the nine O-serotype reference strains. In the wb assay, each of the PCR primer pairs targeting the O3, O4, O5, O8, O9, and O12 alleles reacts with its corresponding reference strain, without cross-reacting with the other reference strains; those targeting the O1/O2 allele react only with the O1 and O2ac reference strains. In the wbbY assay of the O1 and O2ac reference strains, the PCR primers are also specific, without cross-reactions.
Sensitivity and specificity in 56 K. pneumoniae K reference strains.
There is a very good correlation between the iELISA O serotypes and the PCR-based O genotypes (Table 3). The sensitivities and specificities, respectively, of O genotyping (against iELISA) are 100% (24/24) and 94% (30/32) for O1, 80% (4/5) and 96% (49/51) for O2, 100% (2/2) and 100% (54/54) for O2ac, 86% (6/7) and 100% (49/49) for O3, 100% (3/3) and 98% (52/53) for O4, 100% (3/3) and 100% (53/53) for O5, 100% (1/1) and 98% (54/55) for O8, and 100% (1/1) and 100% (55/55) for O12. The four strains (K9, K13, K17, and K38) that did not express O antigen and were thus nontypeable by iELISA (18) were found to be genotypes O2a, O1, O4, and O1, respectively. There were another three strains that expressed O antigen and had discordant results between iELISA and O genotyping, including K6 (iELISA nontypeable versus genotype O2a), K50 (serotype O3 versus O-genotyping nontypeable), and K68 (serotype O2 versus genotype O8). These discrepancies were resolved by nucleotide sequencing, all in favor of O genotyping. With nucleotide sequencing as the gold standard, O genotyping has 100% sensitivity and 100% specificity.
TABLE 3.
Comparison of O-typing results between iELISA and genotyping, with discrepancies resolved by nucleotide sequencing, in 56 Klebsiella pneumoniae K-serotype reference strains
| K type | Speciesa | Strain designation | iELISAb | Genotyping | Sequencing |
|---|---|---|---|---|---|
| 1 | K. pneumoniae | A 5054 | O1 | O1 | |
| 2 | K. pneumoniae | B 5055 | O1 | O1 | |
| 3 | K. pneumoniae | C 5046 | O2 | O2a | |
| 4 | K. pneumoniae subsp. ozaenae | D 5050 | O2ac | O2ac | |
| 5 | K. pneumoniae subsp. ozaenae | E 5051 | O2ac | O2ac | |
| 6 | K. pneumoniae subsp. ozaenae | F 5052 | NTc O+ | O2a | O2a |
| 7 | K. pneumoniae | Aerogenes 4140 | O1 | O1 | |
| 8 | K. pneumoniae | Klebs. 1015 | O1 | O1 | |
| 9 | K. pneumoniae | Klebs. 56 | NT O− | O2a | O2a |
| 10 | K. pneumoniae | Klebs. 919 | O1 | O1 | |
| 11 | K. pneumoniae | Klebs. 390 | O3 | O3 | |
| 12 | K. pneumoniae | Klebs. 313 | O1 | O1 | |
| 13 | K. pneumoniae | Klebs. 1470 | NT O− | O1 | O1 |
| 14 | K. pneumoniae | Klebs. 1193 | O5 | O5 | |
| 15 | K. pneumoniae | Mich. 61 | O4 | O4 | |
| 16 | K. pneumoniae | 2069/49 | O1 | O1 | |
| 17 | K. pneumoniae | 2005/49 | NT O− | O4 | O4 |
| 18 | K. pneumoniae | 1754/49 | O1 | O1 | |
| 19 | K. pneumoniae | 293/50 | O1 | O1 | |
| 20 | K. pneumoniae | 889/50 | O1 | O1 | |
| 21 | K. pneumoniae | 1702/49 | O1 | O1 | |
| 22 | K. pneumoniae | 1996/49 | O1 | O1 | |
| 23 | K. pneumoniae | 2812/50 | O1 | O1 | |
| 24 | K. pneumoniae | 1680/49 | O1 | O1 | |
| 25 | K. pneumoniae | 2002/49 | O3 | O3 | |
| 27 | K. pneumoniae | 6613 | O2 | O2a | |
| 28 | K. pneumoniae | 5758 | O2 | O2a | |
| 30 | K. pneumoniae | 7824 | O1 | O1 | |
| 31 | K. pneumoniae | 6258 | O3 | O3 | |
| 33 | K. pneumoniae | 6168 | O3 | O3 | |
| 34 | K. pneumoniae | 7522 | O1 | O1 | |
| 36 | K. pneumoniae | 8306 | O4 | O4 | |
| 37 | K. pneumoniae | 8238 | O1 | O1 | |
| 38 | K. pneumoniae | 8414 | NT O− | O1 | O1 |
| 39 | K. pneumoniae | 7749 | O1 | O1 | |
| 40 | K. pneumoniae | 8588 | NT O− | NT | |
| 42 | K. pneumoniae | 1702 | O4 | O4 | |
| 43 | K. pneumoniae | 2482 | O2 | O2a | |
| 45 | K. pneumoniae | 8464 | O1 | O1 | |
| 46 | K. pneumoniae | 5281 | O1 | O1 | |
| 47 | K. pneumoniae | 9682 | O1 | O1 | |
| 50 | K. quasipneumoniae subsp. similipneumoniae | 1303/50 | O3 | NT | NTd |
| 51 | K. pneumoniae | 4715/50 | O3 | O3 | |
| 52 | K. pneumoniae | 5759/50 | NT O− | NT | |
| 55 | K. pneumoniae | 3985/51 | O3 | O3 | |
| 60 | K. quasipneumoniae subsp. similipneumoniae | 4463/52 | O5 | O5 | |
| 61 | K. pneumoniae | 5710/52 | O5 | O5 | |
| 62 | K. pneumoniae | 5711/52 | O1 | O1 | |
| 63 | K. pneumoniae | 5845/52 | O1 | O1 | |
| 64 | K. pneumoniae | NCTC 8172 | O1 | O1 | |
| 67 | K. terrigena (previously identified as K. pneumoniae) | 264 (1) | O7 | NT | |
| 68 | K. terrigena (previously identified as K. pneumoniae) | 265 (1) | O2 | O8 | O8 |
| 69 | K. terrigena (previously identified as K. pneumoniae) | 889 | O8 | O8 | |
| 80 | K. quasipneumoniae subsp. similipneumoniae | 708 | O12 | O12 | |
| 81 | K. pneumoniae | 370 | NT O+ | NT | |
| 82 | K. pneumoniae | 3454-70 | NT O+ | NT |
Taxonomic designation updated in 2013 (16). K. quasipneumoniae subsp. similipneumoniae, previously K. pneumoniae phylogroup II-B (33).
iELISA results reported by Hansen et al. (18). The principal antigen inhibited in the O2 iELISA system is O2a (18).
NT, nontypeable.
Nucleotide sequencing was unsuccessful (i.e., all of the PCR primer pairs designed from the wb sequence of the O3 reference strain yielded negative results) (see Table S4 in the supplemental material), which indicates that this strain was incorrectly assigned to the O3 serotype.
Nucleotide diversity between Klebsiella species.
To explore whether the O-genotyping method primarily developed for K. pneumoniae may also be applied to other Klebsiella species, we sequenced wzm and wzt in the nine K reference strains with an O3 serotype; these included four K. pneumoniae strains (K31, K33, K51, K55), four K. variicola strains (K48, K49, K53, K54), and one K. oxytoca strain (K74) (see File S3 in the supplemental material). At wzm, the sequences are 98 to 99% similar among the different strains of a given species but are only 80 to 81% similar between K. pneumoniae and K. oxytoca. At wzt, substantial diversity (nucleotide sequence similarity of 77 to 81%) between the two species is also observed. The significant sequence variations across species indicate a need to design the O-genotyping PCR primers separately for different Klebsiella species.
O-genotype distribution in 87 clinical K. pneumoniae strains.
The 87 strains are isolates from blood (n = 82), liver abscesses (n = 4), or urine (n = 1). The O1 genotype is the most common (n = 56; 64%), followed by the O2a (n = 17; 20%), O3 (n = 5; 6%), and O5 (n = 4; 5%) genotypes (Table 4). Overall, 83 (95%) of the 87 strains are typable.
TABLE 4.
O-genotype distribution of 87 clinical Klebsiella pneumoniae strains
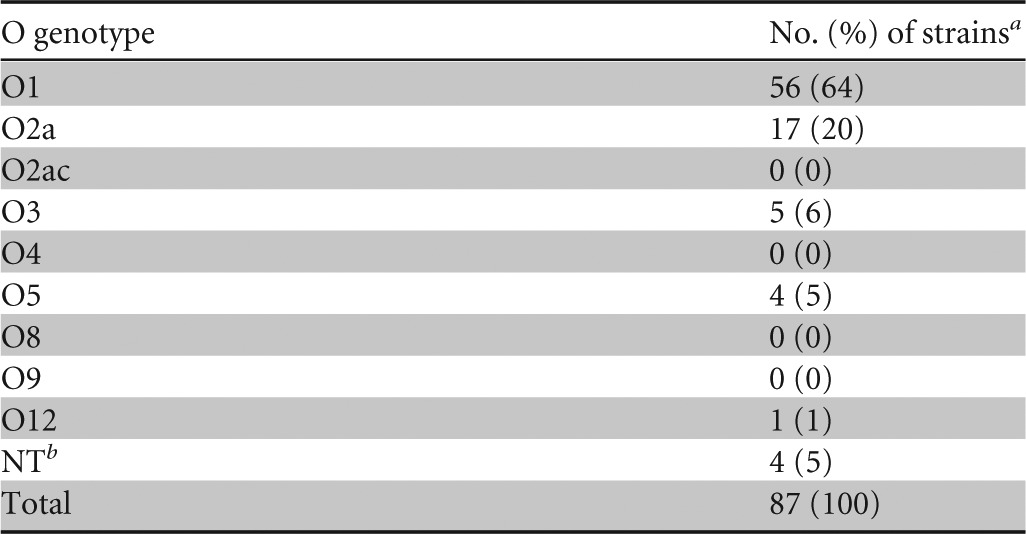
Consecutively collected from the patients with community-acquired K. pneumoniae infections diagnosed at the National Taiwan University Hospital from 1 July 2006 to 31 July 2007. For each patient, only one isolate was included. The 87 strains are isolates from blood (n = 82), liver abscesses (n = 4), or urine (n = 1).
NT, nontypeable.
DISCUSSION
We have successfully developed and validated a PCR-based O-genotyping method for the rapid and accurate determination of O-antigen types in K. pneumoniae. This new technique overcomes several limitations of conventional serotyping methods. First, O genotyping directly detects the genetic determinants of O antigen and thus avoids the problems of the capsular masking effect and serological cross-reaction. Second, unlike O antisera, PCR primers can be commercially synthesized with high quality at a low cost, which allows the O-genotyping procedure to be easily performed in clinical and research microbiology laboratories.
In addition to the technical convenience, our O-genotyping method accurately identifies the O-antigen types of K. pneumoniae isolates. There is a very good correlation between the O genotypes and classical O serotypes, with a complete match in 100% (9/9) of the O reference strains and 88% (49/56) of the K reference strains. For the three strains that express O antigen and had discordant results between iELISA and O genotyping, nucleotide sequencing evidently supports the accuracy of O genotyping in all of the cases. With nucleotide sequencing as the gold standard, the O-genotyping method has 100% sensitivity and 100% specificity in K. pneumoniae.
The nine O types (O1, O2, O2ac, O3, O4, O5, O8, O9, and O12) that are typable by our method together represent the great majority (83/87, 95%) of the K. pneumoniae clinical isolates in an epidemiologically representative sample in the current study. This validates the applicability and good performance of our O-genotyping method in the clinical setting. Similarly, based on a large survey in Denmark, Spain, and the United States (18), these nine O types account for up to 99.6% (528/530) of all O-typable K. pneumoniae clinical isolates. In another classical seroepidemiological study in Germany, the nine O types also accounted for up to 93.8% (315/336) of all O-typable Klebsiella clinical isolates (21). Both studies show that O7, the type not included in our current O-genotyping scheme, is extremely rare among K. pneumoniae clinical isolates. In fact, a reevaluation using phylogenetic analysis of the rpoB gene sequence in 2013 found that K. pneumoniae strain 264(1), the strain in which the O7 serotype was originally described, is actually K. (Raoultella) terrigena, rather than K. pneumoniae (16).
It should be emphasized that O-genotyping complements, rather than replaces, iELISA. Since mutations within the wb and wbbY gene cluster(s) might abolish O-antigen expression on the cell surface, strains of a given O genotype may not exhibit the corresponding phenotype. This is shown by the four K. pneumoniae strains that have a detectable O genotype yet are nontypeable by iELISA because they did not produce O antigen. As bacterial virulence depends not only on the genotype but also on the phenotype actually expressed, iELISA remains an essential phenotyping tool for K. pneumoniae.
With the novel O-genotyping technology, our study provides new and important evidence for previously unresolved questions. There have been controversies about the status of O8 and O9 in the Klebsiella O-type classification system (18, 21). Although O8 is biochemically discernible from O1 by the O-acetylation of d-galactan I (41), serological methods are unable to separate O8 from O1 (18, 21). It has been proposed that genetic methods might be able to distinguish between these two types (18), since the wb gene cluster of O8 is substantially different from that of O1 (37, 41). In the present study, using allele-specific PCR primers targeting the wzm-wzt loci of the wb gene cluster, we demonstrate that O8 can be explicitly discriminated from O1. This finding confirms that O8 is a distinct O type. Likewise, O9 is biochemically discernible from O2(2a) (by the O-acetylation of d-galactan I and the α-d-Galp residues attached to alternate d-galactan I-repeating units) (22, 42), yet there is serological cross-reactivity between O9 and O2(2a) (18, 21). Hansen et al. considered O9 as part of the O2 group (18, 43). Conversely, Trautmann et al. have described O9 as a distinct serotype, based on its possession of an additional, non-O2 antigenic factor (21). In the current study, we found substantial diversity in the nucleotide sequences of the wzm-wzt loci between the O9 and O2ac reference strains. Moreover, the successful development and validation of the O9-specific wb primers also support the classification of O9 as a discrete O type.
Our two-step genotyping scheme precisely corresponds to the structural composition of the O antigen. The O1 and O2(2a,2c) antigens contain both d-galactan I and another O polysaccharide (23, 24), while the O2(2a) antigen contains only d-galactan I. Accordingly, we defined the O1 and O2ac genotypes as having not only the genes (at the wb gene cluster) for d-galactan I but also that (at the wbbY region) for the second O polysaccharide and defined the O2a genotype as having the genes for d-galactan I only. Both the wb and wbbY assays are required to distinguish between O1, O2a, and O2ac. In contrast, the first assay (wb) alone is sufficient to specifically identify genotypes O3, O4, O5, O9, and O12, each of which has a unique O polysaccharide (22) and thus is defined as having a specific allele at the wzm-wzt loci. Interestingly, the O8 antigen also has two polysaccharides: O-acetylated d-galactan I and d-galactan II (41). We did find that the O8 reference strain reacts with the second set of PCR primers targeting the allele for d-galactan II (Tables 1 and 2). However, since the wb gene cluster of O8 is markedly different from that of O1, the first assay (wb) alone is adequate to unequivocally identify O8. Another example of a good correlation between biochemical structure and O genotyping is our result for O9 and O2(2a,2e). Studies revealed that O9 and O2(2a,2e) polysaccharides have identical structures that are different from that of the O2(2a) antigen (43). In keeping with previous biochemical findings, we show that both the O9 and O2(2a,2e) reference strains react with the O9 wb primers but not with the O1/O2 wb primers (Table 1).
O genotyping can provide important information regarding the pathogenic ability of K. pneumoniae strains to cause invasive infections. Studies have suggested that O1 is a major virulence factor in K. pneumoniae (12, 30, 44). Compared with non-O1 strains, the O1 strains are generally more resistant to the bactericidal effect of human serum (12), which has been attributed to the d-galactan II component in O1 antigen (30, 44). One study further shows that the association of O1 with invasive K. pneumoniae strains causing pyogenic liver abscesses (90.5% [38/42]) is greater than that with noninvasive strains (28.1% [9/32]) (12). Nevertheless, two classical O-seroepidemiological studies found a lack of significant differences in the distribution of O1 between invasive and noninvasive strains (18, 21). This seeming inconsistency between studies may be due to the fact that K. pneumoniae virulence also depends on the K-antigen type; K1 is the most virulent, followed by K2, K5, K20, and K54 (8). To characterize the significance of the O-antigen types in K. pneumoniae pathogenicity, analysis of both the O type and K type would be required in future studies. Our O-genotyping technique facilitates studies on the role of the O type in the pathogenesis of invasive K. pneumoniae diseases.
When combined with K genotyping (8, 16), O genotyping also helps determine the clonality of K. pneumoniae isolates. The O:K-typing system (with nine O types and 77 K types) has a discriminative power comparable to that of pulsed-field gel electrophoresis (PFGE) (13). Unlike PFGE, which still lacks an international database for real-time pattern recognition of K. pneumoniae strains, the O:K genotyping method yields definitive O:K types and thus facilitates the direct discrimination of K. pneumoniae strains isolated from different geographical locations or time periods (13). Furthermore, by targeting the two major virulent factors (LPS and CPS) in K. pneumoniae, O:K typing can also complement multilocus sequence typing (MLST) (which targets housekeeping genes in the genomic background) (45) for the purposes of tracing the source and transmission of K. pneumoniae in large-scale epidemiological investigations (46). Hence, the O:K genotyping method helps to investigate outbreaks, evaluate trends, and detect emerging strains of K. pneumoniae.
Given the greater convenience and high accuracy, our PCR-based O-genotyping technique represents an important breakthrough in the methodology of K. pneumoniae LPS O-antigen typing. The O:K genotyping system provides a useful tool for the clinical and epidemiological investigations of K. pneumoniae and its associated diseases.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Statens Serum Institut (Copenhagen, Denmark) for providing Klebsiella O-serotype reference strains and K-serotype reference strains.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02494-15.
REFERENCES
- 1.Jarvis WR, Munn VP, Highsmith AK, Culver DH, Hughes JM. 1985. The epidemiology of nosocomial infections caused by Klebsiella pneumoniae. Infect Control 6:68–74. [DOI] [PubMed] [Google Scholar]
- 2.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asensio A, Oliver A, Gonzalez-Diego P, Baquero F, Perez-Diaz JC, Ros P, Cobo J, Palacios M, Lasheras D, Canton R. 2000. Outbreak of a multiresistant Klebsiella pneumoniae strain in an intensive care unit: antibiotic use as risk factor for colonization and infection. Clin Infect Dis 30:55–60. doi: 10.1086/313590. [DOI] [PubMed] [Google Scholar]
- 4.Wang JH, Liu YC, Lee SS, Yen MY, Chen YS, Wann SR, Lin HH. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis 26:1434–1438. doi: 10.1086/516369. [DOI] [PubMed] [Google Scholar]
- 5.Fang CT, Chang SC, Hsueh PR, Chen YC, Sau WY, Luh KT. 2000. Microbiologic features of adult community-acquired bacterial meningitis in Taiwan. J Formos Med Assoc 99:300–304. [PubMed] [Google Scholar]
- 6.Chen KY, Hsueh PR, Liaw YS, Yang PC, Luh KT. 2000. A 10-year experience with bacteriology of acute thoracic empyema: emphasis on Klebsiella pneumoniae in patients with diabetes mellitus. Chest 117:1685–1689. doi: 10.1378/chest.117.6.1685. [DOI] [PubMed] [Google Scholar]
- 7.Wang JL, Chen KY, Fang CT, Hsueh PR, Yang PC, Chang SC. 2005. Changing bacteriology of adult community-acquired lung abscess in Taiwan: Klebsiella pneumoniae versus anaerobes. Clin Infect Dis 40:915–922. doi: 10.1086/428574. [DOI] [PubMed] [Google Scholar]
- 8.Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. 2007. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 45:284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 9.Cheng NC, Yu YC, Tai HC, Hsueh PR, Chang SC, Lai SY, Yi WC, Fang CT. 2012. Recent trend of necrotizing fasciitis in Taiwan: focus on monomicrobial Klebsiella pneumoniae necrotizing fasciitis. Clin Infect Dis 55:930–939. doi: 10.1093/cid/cis565. [DOI] [PubMed] [Google Scholar]
- 10.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shon AS, Russo TA. 2012. Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol 7:669–671. doi: 10.2217/fmb.12.43. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh PF, Lin TL, Yang FL, Wu MC, Pan YJ, Wu SH, Wang JT. 2012. Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PLoS One 7:e33155. doi: 10.1371/journal.pone.0033155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen DS, Skov R, Benedi JV, Sperling V, Kolmos HJ. 2002. Klebsiella typing: pulsed-field gel electrophoresis (PFGE) in comparison with O:K-serotyping. Clin Microbiol Infect 8:397–404. doi: 10.1046/j.1469-0691.2002.00411.x. [DOI] [PubMed] [Google Scholar]
- 14.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, Andersen PS, Driebe EM, Keim P, Krogfelt KA. 2015. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6:e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decre D. 2013. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol 51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan YJ, Lin TL, Lin YT, Su PA, Chen CT, Hsieh PF, Hsu CR, Chen CC, Hsieh YC, Wang JT. 2015. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob Agents Chemother 59:1038–1047. doi: 10.1128/AAC.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen DS, Mestre F, Alberti S, Hernandez-Alles S, Alvarez D, Domenech-Sanchez A, Gil J, Merino S, Tomas JM, Benedi VJ. 1999. Klebsiella pneumoniae lipopolysaccharide O typing: revision of prototype strains and O-group distribution among clinical isolates from different sources and countries. J Clin Microbiol 37:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ørskov I, Ørskov F. 1984. Serotyping of Klebsiella, p 143–164. In Bergan T. (ed), Methods in microbiology. Academic Press, London, United Kingdom. [Google Scholar]
- 20.Alberti S, Hernandez-Alles S, Gil J, Reina J, Martinez-Beltran J, Camprubi S, Tomas JM, Benedi VJ. 1993. Development of an enzyme-linked immunosorbent assay method for typing and quantitation of Klebsiella pneumoniae lipopolysaccharide: application to serotype O1. J Clin Microbiol 31:1379–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trautmann M, Ruhnke M, Rukavina T, Held TK, Cross AS, Marre R, Whitfield C. 1997. O-antigen seroepidemiology of Klebsiella clinical isolates and implications for immunoprophylaxis of Klebsiella infections. Clin Diagn Lab Immunol 4:550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinogradov E, Frirdich E, MacLean LL, Perry MB, Petersen BO, Duus JØ, Whitfield C. 2002. Structures of lipopolysaccharides from Klebsiella pneumoniae: elucidation of the structure of the linkage region between core and polysaccharide O chain and identification of the residues at the non-reducing termini of the O chains. J Biol Chem 277:25070–25081. doi: 10.1074/jbc.M202683200. [DOI] [PubMed] [Google Scholar]
- 23.Whitfield C, Richards J, Perry M, Clarke B, MacLean L. 1991. Expression of two structurally distinct D-galactan O antigens in the lipopolysaccharide of Klebsiella pneumoniae serotype O1. J Bacteriol 173:1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitfield C, Perry MB, MacLean LL, Yu SH. 1992. Structural analysis of the O-antigen side chain polysaccharides in the lipopolysaccharides of Klebsiella serotypes O2(2a), O2(2a,2b), and O2(2a,2c). J Bacteriol 174:4913–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke BR, Whitfield C. 1992. Molecular cloning of the rfb region of Klebsiella pneumoniae serotype O1:K20: the rfb gene cluster is responsible for synthesis of the d-galactan I O polysaccharide. J Bacteriol 174:4614–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugiyama T, Kido N, Kato Y, Koide N, Yoshida T, Yokochi T. 1998. Generation of Escherichia coli O9a serotype, a subtype of E. coli O9, by transfer of the wb* gene cluster of Klebsiella O3 into E. coli via recombination. J Bacteriol 180:2775–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merino S, Altarriba M, Izquierdo L, Nogueras MM, Regue M, Tomas JM. 2000. Cloning and sequencing of the Klebsiella pneumoniae O5 wb gene cluster and its role in pathogenesis. Infect Immun 68:2435–2440. doi: 10.1128/IAI.68.5.2435-2440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izquierdo L, Merino S, Regué M, Rodriguez F, Tomás JM. 2003. Synthesis of a Klebsiella pneumoniae O-antigen heteropolysaccharide (O12) requires an ABC 2 transporter. J Bacteriol 185:1634–1641. doi: 10.1128/JB.185.5.1634-1641.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenfield LK, Whitfield C. 2012. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr Res 356:12–24. doi: 10.1016/j.carres.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh PF, Wu MC, Yang FL, Chen CT, Lou TC, Chen YY, Wu SH, Sheu JC, Wang JT. 2014. d-Galactan II is an immunodominant antigen in O1 lipopolysaccharide and affects virulence in Klebsiella pneumoniae: implication in vaccine design. Front Microbiol 5:608. doi: 10.3389/fmicb.2014.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trautmann M, Cross AS, Reich G, Held H, Podschun R, Marre R. 1996. Evaluation of a competitive ELISA method for the determination of Klebsiella O antigens. J Med Microbiol 44:44–51. doi: 10.1099/00222615-44-1-44. [DOI] [PubMed] [Google Scholar]
- 32.Ørskov I. 1954. O antigens in the Klebsiella group. Acta Pathol Microbiol Scand 34:145–156. [DOI] [PubMed] [Google Scholar]
- 33.Brisse S, Passet V, Grimont PA. 2014. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol 64:3146–3152. doi: 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- 34.Wu KM, Li LH, Yan JJ, Tsao N, Liao TL, Tsai HC, Fung CP, Chen HJ, Liu YM, Wang JT, Fang CT, Chang SC, Shu HY, Liu TT, Chen YT, Shiau YR, Lauderdale TL, Su IJ, Kirby R, Tsai SF. 2009. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191:4492–4501. doi: 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fouts DE, Tyler HL, DeBoy RT, Daugherty S, Ren Q, Badger JH, Durkin AS, Huot H, Shrivastava S, Kothari S. 2008. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet 4:e1000141. doi: 10.1371/journal.pgen.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, Program NCS, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly RF, Whitfield C. 1996. Clonally diverse rfb gene clusters are involved in expression of a family of related d-galactan O antigens in Klebsiella species. J Bacteriol 178:5205–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng S, Fockler C, Barnes WM, Higuchi R. 1994. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci U S A 91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fletcher RH, Fletcher SW. 2005. Sensitivity and specificity, p 38–40. In Clinical epidemiology: the essentials, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 40.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly RF, Severn WB, Richards JC, Perry MB, MacLean LL, Tomas JM, Merino S, Whitfield C. 1993. Structural variation in the O-specific polysaccharides of Klebsiella pneumoniae serotype O1 and O8 lipopolysaccharide: evidence for clonal diversity in rfb genes. Mol Microbiol 10:615–625. doi: 10.1111/j.1365-2958.1993.tb00933.x. [DOI] [PubMed] [Google Scholar]
- 42.MacLean LL, Whitfield C, Perry MB. 1993. Characterization of the polysaccharide antigen of Klebsiella pneumoniae O:9 lipopolysaccharide. Carbohydr Res 239:325–328. doi: 10.1016/0008-6215(93)84231-T. [DOI] [PubMed] [Google Scholar]
- 43.Kelly RF, Perry MB, MacLean LL, Whitfield C. 1995. Structures of the O-antigens of Klebsiella serotypes O2 (2a,2e), O2 (2a,2e,2h), and O2 (2a,2f,2g), members of a family of related d-galactan O-antigens in Klebsiella spp. J Endotoxin Res 2:131–140. [Google Scholar]
- 44.McCallum KL, Schoenhals G, Laakso D, Clarke B, Whitfield C. 1989. A high-molecular-weight fraction of smooth lipopolysaccharide in Klebsiella serotype O1:K20 contains a unique O-antigen epitope and determines resistance to nonspecific serum killing. Infect Immun 57:3816–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis GS, Waits K, Nordstrom L, Weaver B, Aziz M, Gauld L, Grande H, Bigler R, Horwinski J, Porter S, Stegger M, Johnson JR, Liu CM, Price LB. 2015. Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin Infect Dis 61:892–899. doi: 10.1093/cid/civ428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar V, Jayasuryan N, Kumar R. 1996. A truncated mutant (residues 58-140) of the hepatitis B virus X protein retains transactivation function. Proc Natl Acad Sci U S A 93:5647–5652. doi: 10.1073/pnas.93.11.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.