Abstract
The nontuberculous mycobacteria (NTM) cause miscellaneous disorders in humans, especially in the lungs, which present with a variety of radiological features. To date, knowledge of the pathogenic role of the Mycobacterium avium-intracellulare complex (MAC) in the human lung and the definitive criteria for initiating multidrug therapy are still lacking. However, there is little doubt that clarithromycin is the most efficacious drug among the various treatment regimens for lung NTM. In this study, with the use of a bridged nucleic acid (BNA) probe a detection system based on a real-time PCR (BNA-PCR) for the identification of the point mutations at position 2058 or 2059 in domain V of the 23S rRNA gene responsible for clarithromycin resistance was developed and has been assessed using MAC isolates from clinical samples. Out of 199 respiratory specimens, the drug susceptibility test demonstrated 12 strains resistant to clarithromycin, while the BNA-PCR showed 8 strains carrying the point mutation at position 2058 or 2059 of the 23S rRNA gene. This system revealed that there were mycobacterial strains resistant to clarithromycin which do not carry previously identified resistance genes. This paper documents a novel system for detecting clarithromycin-resistant strains and demonstrates that although these mutations are tacitly assumed to account for >90% of the reported resistant mutants, there is a significant fraction of resistant mutants that do not harbor these mutations. Therefore, unknown mechanisms affecting clarithromycin resistance remain to be elucidated.
INTRODUCTION
The nontuberculous mycobacteria (NTM) are ubiquitous microorganisms found in various environments. They are capable of causing miscellaneous disorders in humans, especially in the lungs, which present with a variety of radiological features. It is noteworthy that the prevalence of lung NTM is increasing throughout the world (1, 2). Whereas NTM diversity in clinical manifestations and geographic distribution has been well documented, Mycobacterium avium and Mycobacterium intracellulare, collectively referred to as the Mycobacterium avium-intracellulare complex (MAC), are frequently isolated from such patients in most countries (3).
To date, knowledge of the pathogenic role of MAC strains in the human lung and the definitive criteria for initiating multidrug therapy are still lacking. However, there is little doubt that clarithromycin is the most efficacious drug among the various treatment regimens for lung NTM. Clarithromycin is a ribosome-targeting macrolide antibiotic that interacts with a bacterial 23S rRNA hairpin-like structure in domain II and the peptidyl transferase loop in domain V (4, 5). Additionally, clarithromycin is the only agent for which susceptibility testing has high clinical efficacy. Therefore, management of clarithromycin-resistant MAC isolates from advanced lung NTM is an emerging concern. It is worth noting that multidrug therapy containing clarithromycin as the key drug is now a mandatory implementation to prevent resistance.
Other studies have concluded that the mechanism responsible for clarithromycin-resistant MAC emergence, although it has not been fully unraveled, is point mutations in domain V of the 23S rRNA gene (6, 7, 8). It has been also reported that 4.0 to 10% of MAC organisms causing lung NTM are capable of acquiring clarithromycin resistance (6, 7). Meanwhile, a great deal of attention has been paid to finding alternative ways to identify the clarithromycin-resistant genes in MAC isolates. Currently, culture-based drug susceptibility testing is the only means to evaluate the bioactivity of antibiotics against MAC strains despite the guideline recommendation that clarithromycin susceptibility testing be performed for all patients before therapy is initiated (3). To this end, with a bridged nucleic acid (BNA) probe which has improved hybridizing capability between base pairs, a detection system for clarithromycin resistance mutations based on a real-time PCR (BNA-PCR) was developed and has been assessed using MAC isolated from clinical samples. In addition, the characteristics of patients with lung NMT who have the resistant organism were also studied.
MATERIALS AND METHODS
Clinical sample.
Expectorated sputum was submitted to the microbiology laboratory at the Saitama Medical University Hospital, and an equal volume of Sputazyme (Kyokuto Pharmaceuticals Industrial Co., Ltd., Tokyo, Japan) was added for sputum lysis. The samples were centrifuged at 3,000 × g for 20 min, and the pellet was first examined with Ziehl-Neelsen staining. Following N-acetyl-l-cysteine-sodium hydroxide (NALC-NaOH) digestion and decontamination, the pellet of the sample was inoculated into BBL MGIT broth medium (BD, Tokyo, Japan), which was incubated in the Bactec MGIT 960 system (BD). After this incubation, M. avium or M. intracellulare was identified by the Cobas TaqMan 48 analyzer (Roche Diagnostics K.K., Tokyo, Japan). Two milliliters of the MGIT broth was analyzed by PCRs, and 500 μl was transferred for drug susceptibility tests.
Drug susceptibility tests.
The clarithromycin susceptibility of isolated M. avium or M. intracellulare was measured by BrothMIC NTM (Kyokuto Pharmaceuticals Industrial Co., Ltd., Tokyo, Japan), which employed a broth microdilution susceptibility panel containing 96 reagent wells and was in accordance with the current Clinical and Laboratory Standards Institute guidelines (9). A week after 500 μl of the MGIT broth was transferred to Myco broth (Kyokuto Pharmaceuticals Industrial Co., Ltd.), the Myco broth was split into each well (see the manufacturer's instructions for details). Strains with MICs of ≤8 μg/ml at pH 7.4 were considered susceptible to clarithromycin, those with MICs of ≥32 μg/ml were considered resistant, and those with MICs between 8 and 32 μg/ml were considered intermediate.
DNA preparation.
Two milliliters of MGIT broth containing M. avium or M. intracellulare was centrifuged at 3,000 × g for 10 min, the pellet was mixed with 200 μl of AL buffer (Qiagen, Tokyo, Japan) containing 20 μl of proteinase K (TaKaRa Bio Inc., Shiga, Japan), and the resultant mixture was incubated at 56°C for 1 h. DNA was purified into 100 μl of Tris-EDTA (TE) buffer with the QIAamp DNA blood minikit (Qiagen), according to the manufacturer's instructions (10).
PCR.
The final solution of the PCR mixture contained 12.5 μl of the TaKaRa Premix Ex Taq (TaKaRa Bio Inc., Shiga, Japan), 300 nM each primer, 300 nM fluorescence-labeled TaqMan probe, 1.0 μl of purified DNA from MGIT broth, and deionized distilled water up to 25.0 μl. Two real-time PCRs were simultaneously performed to detect the clarithromycin resistance gene using the BNA-PCR and to identify M. avium or M. intracellulare as a control using the TaqMan probe (11). The PCR was performed by starting at 95°C for 30 s followed by 40 cycles at 95°C for 5 s and 66°C for 40 s using the SmartCycler II (Cepheid, Sunnyvale, CA). The sequences of primers and probes are described in Table 1. The electrophoresis was carried out with a 100-bp DNA ladder (TaKaRa Bio Inc.) in a 1.5% agarose gel containing 0.5 μg/ml ethidium bromide.
TABLE 1.
Sequences for primers and probes
| Primer/probea | Oligonucleotide name | Target gene (GenBank accession no.) |
Oligonucleotide conc (μM) | Sequence | PCR product (bp) |
|---|---|---|---|---|---|
| BNA-PCR for clarithromycin resistance gene | |||||
| FW primer | MAV_Fw2485 | 23S rRNA (X74494) | 0.3 | 5′-GTAACGACTTCCCAACTGTCTC-3′ | 146 |
| RV primer | MAV_Rv2631 | 0.3 | 5′-ACCTATCCTACACAAACCGTACC-3′ | ||
| BNA probeb | MAV_BNA2555 | 0.3 | Alex532-CGCGGCAGGACGAAAAGAC-BHQ1 | ||
| Real-time PCR for M. avium identification | |||||
| FW primer | MAV_Fw0021 | 16S rRNA (X52918) | 0.3 | 5′-CAAGTCGAACGGAAAGGCCTCT-3′ | 269 |
| RV primer | MAV_Rv0269 | 0.3 | 5′-GCCGTATCTCAGTCCCAGTGTG-3′ | ||
| TaqMan probe | MAV_Pb0129 | 0.3 | FAM-TACCGGATAGGACCTCAAGACGC-TAMRA | ||
| Real-time PCR for M. intracellulare identification | |||||
| FW primer | MINT_Fw0068 | ITSc 16–23S rRNA (AM709724) | 0.3 | 5′-AGCACCACGAAAAGCACTCCAATT-3′ | 243 |
| RV primer | MINT_Rv0284 | 0.3 | 5′-CGAACGCATCAGCCCTAAGGACTA-3′ | ||
| TaqMan probe | MINT_Pb0181 | 0.1 | FAM-CCTGAGACAACACTCGGTCGATCC-TAMAR | ||
| Sequence of 23S rRNA in M. avium or M. intracellulare | |||||
| Sequence analysis | MAV_Fw1073 | 23S rRNA (X74494) | 0.3 | 5′-ACCCGAAGCGGAGTGATCTACCCA-3′ | |
| Sequence analysis | MAV_Fw1533 | 0.3 | 5′-ATGTAGCGGGGCTCAAGCACACC-3′ | ||
| Sequence analysis | MAV_Fw2938 | 0.3 | 5′-GATAAAAGGTACCCCGGGGATAAC-3′ |
FW, forward; RV, reverse.
Two BNA bases introduced at positions 2058 and 2059 corresponding to E. coli numbering AF053966.1 are underlined.
ITS, internal transcribed spacer.
Resistance gene synthesis.
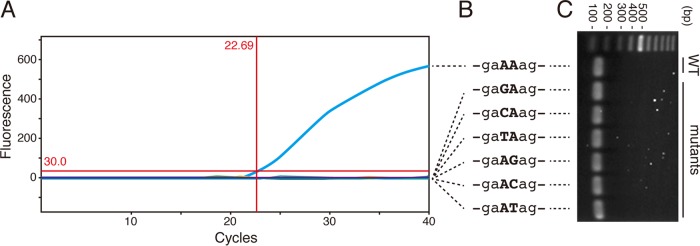
Template DNA for the resistance gene, a 237-bp fragment from position 2415 through 2650 in the 23S rRNA gene of Mycobacterium avium (GenBank accession number X74494) harboring the sequences of both the MAV_Fw2485 and MAV_Rv2631 regions, was generated at TaKaRa Bio Inc. A point mutation (A→G, C, or T) was inserted at position 2058 or 2059 equivalent to Escherichia coli (GenBank accession number AF053966.1). A set of resistance genes showing 2 bp before or after position 2058 and 2059 are depicted in Fig. 1.
FIG 1.
Development of the BNA-PCR system to identify the clarithromycin resistance gene in Mycobacterium avium complex (MAC). (A) BNA-PCR on SmartCycler. The seven real-time PCRs were simultaneously carried out using a wild type (WT) template and six patterns of mutant templates. The BNA-PCR was able to identify only the WT sequence at a 22.69 threshold cycle value but not the mutants. (B) Sequences of the synthesized genes. A 237-bp fragment DNA of the WT as well as six forms of mutants between positions 2415 and 2650 in 23S rRNA of Mycobacterium avium were generated. At position 2058 or 2059 corresponding to Escherichia coli numbering the adenine (A) was replaced by a mutant (G, C, or T), as shown by the capital letters. (C) Agarose gel electrophoresis. The WT and six mutant sequences in panels A and B were identical as amplified by MAV_Fw2485 and MAV_Rv2631.
Nucleotide sequencing.
PCR products from the BNA-PCR were purified by a StrataPrep PCR purification kit (Agilent Technologies, Santa Clara, CA, USA) and then subjected to sequence analysis using MAV_Fw2485 at Fasmac Co., Ltd. (Kanagawa, Japan). The sequence analysis of the domain II and domain V regions in 23S rRNA was done with MAV_Fw1073, MAV_Fw1533, and MAV_Fw12938 (Table 1).
Ethical considerations.
The research protocol involving patients' samples was reviewed by the institutional review boards of the Saitama Medical University Hospital (institutional review board [IRB] number 11-006, 9 June 2011).
RESULTS
Development of a detection system for the clarithromycin resistance gene by BNA-PCR.
The primary factor resulting in clarithromycin resistance in M. avium or M. intracellulare has been reported to be point mutations at position 2058 or 2059 in domain V of the 23S rRNA gene; the position number corresponds to the 23S rRNA gene of E. coli (6, 7, 12). Current clinical laboratory techniques have enabled the detection of gene mutations readily and effortlessly. A variety of techniques have now been employed for the detection of mutations in a clinical setting (13, 14). BNAs are artificial nucleic acids with a high binding affinity to a specific sequence and enhanced biochemical stability (15, 16). With this technique, even a single mismatch sequence was able to be discriminated from the amplified product. Hence, we designed a real-time PCR-based system employing BNA probes, in which two BNA bases were introduced at positions 2058 and 2059 (-gaAAag-) to detect the clarithromycin resistance gene of MAC isolates (BNAs are underlined) (Fig. 1). If a point mutation exists, the resulting sequence mismatch leads to a decrease in the melting temperature (Tm) value. This prevents the BNA probe from annealing to the PCR product and consequently reduces the emitted fluorescence level. Additionally, the established real-time PCR using a TaqMan probe to identify M. avium or M. intracellulare was simultaneously performed as a positive control (10, 11) (Fig. 1A).
Analyzing the specificity of the BNA-PCR for MAC.
In order to assess the analytical specificity of this system, first the BNA-PCR and the TaqMan PCR were both tested against a cross-reactivity panel. The panel contains DNA from 76 organisms (14) and showed that both the BNA-PCR and the TaqMan-PCR for M. avium and M. intracellulare amplified the M. avium and M. intracellulare strains specifically, respectively, with no cross-reaction with other organisms. Next, 237-bp DNA of wild type (WT) (-gaAAag-) and six forms of point mutations (-gaACag-, -gaAGag-, -gaATag-, -gaCAag-, -gaGAag-, and -gaTAag-) were synthesized (Fig. 1B). With these oligonucleotides, BNA-PCR was capable of reacting to the amplicon of WT strains but not the mutants. Additionally, the target sequences were well amplified by primer pairs using MAV_Fw2485 and MAV_Rv2631 and were confirmed by gel electrophoresis (Fig. 1C). These data show that BNA-PCR was able to discriminate the WT from the mutant oligonucleotides.
Features of patients with lung NTM from whom clarithromycin-resistant MAC strains were isolated.
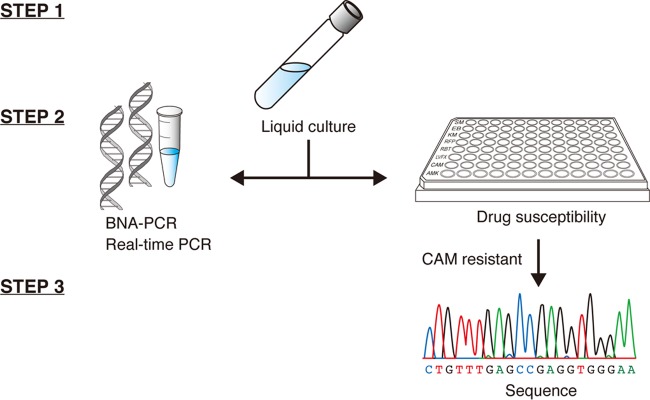
In order to confirm whether this system worked with clinical samples and to assess the prevalence of clarithromycin-resistant MAC infections, respiratory samples were collected from patients who visited the Saitama Medical University Hospital and were suspected of or diagnosed as having lung NTM. The study was performed between 9 June 2011 and 9 June 2015, and a total of 199 sputum samples were analyzed. The sputum samples were digested and then aliquoted for a drug susceptibility test and BNA-PCR as shown in the schematic diagram in Fig. 2. Out of 199 strains, 12 strains were demonstrated to be resistant to clarithromycin by the drug susceptibility test, while the BNA-PCR showed 8 strains carrying point mutations at position 2058 of the 23S rRNA gene (Table 2). The characteristics of the patients who had clarithromycin-resistant MAC infections are summarized in Table 3. This shows that a female of lighter build had a greater tendency than a male to be affected by a clarithromycin-resistant MAC infection even in the absence of a smoking habit or other underlying conditions as previously indicated (6); however, all physical features resembled those of individuals who had clarithromycin-susceptible MAC infections (17). In addition, although the data from patients were gathered retrospectively, they indicate that macrolide monotherapy might result in the development of the resistant strain: half of the patients with clarithromycin-resistant MAC infections had received monotherapy. Indeed, 3 out of 12 patients who had resistant MAC infections died due to the progression of lung NTM and subsequent deterioration of lung function.
FIG 2.
Procedure for the BNA-PCR and drug susceptibility test. In step 1, M. avium or M. intracellulare was isolated from the respiratory specimens using liquid culture (BBL MGIT). In step 2, the culture media were divided into two tests. One was submitted for two real-time PCRs, which were simultaneously performed to detect the clarithromycin (CAM) resistance gene using the BNA probe and to identify M. avium or M. intracellulare using the TaqMan probe, and the other was submitted for drug susceptibility testing. In step 3, sequence analysis was conducted for the clarithromycin-resistant strains.
TABLE 2.
Summary of drug susceptibility testing, BNA-PCR, and sequence analysis of M. avium and M. intracellulare from clinical isolates
| Clinical isolate | Drug susceptibility (clarithromycin MIC [μg/ml]) | Real-time PCR | BNA-PCR | Sequence analysisa |
|||||
|---|---|---|---|---|---|---|---|---|---|
| A2058 | A2059 | A752 | A2062 | G2505 | 1198–1247 | ||||
| Sensitive strain | |||||||||
| NTM 0019 | 0.5 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0122 | 0.25 | M. intracellulare | Negative | A | A | A | A | G | No mutations |
| NTM 0123 | 1 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0124 | 1 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0176 | 2 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0177 | 1 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0178 | 0.5 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0179 | 0.5 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0180 | 0.5 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0181 | 0.5 | M. avium | Negative | A | A | A | A | G | No mutations |
| Resistant strains | |||||||||
| NTM 0036 | >32 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0130 | >32 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0142 | >32 | M. avium | Positive | C | A | A | A | G | No mutations |
| NTM 0146 | >32 | M. intracellulare | Positive | C | A | A | A | G | No mutations |
| NTM 0164 | >32 | M. avium | Negative | A | A | A | A | G | No mutations |
| NTM 0165 | >32 | M. avium | Positive | A | C | A | A | G | No mutations |
| NTM 0169 | >32 | M. avium | positive | A | C | A | A | G | No mutations |
| NTM 0184 | >32 | M. avium | Positive | A | C | A | A | G | No mutations |
| NTM 0186 | >32 | M. avium | Positive | A | C | A | A | G | No mutations |
| NTM 1006 | >32 | M. avium | Positive | G | A | A | A | G | No mutations |
| NTM 1014 | >32 | M. avium | Positive | A | C | A | A | G | No mutations |
| NTM 1033 | >32 | M. intracellulare | Negative | A | A | A | A | G | No mutations |
The mutated positions at 2058 and 2059 correspond to E. coli GenBank accession number AF053966.1, and positions at 752, 2062, and 2505 of the 23S rRNA gene of E. coli correspond to NR_103073.1. The sequence 1198 to 1247 in clarithromycin-resistant M. avium and M. intracellulare refers to those of sensitive strains.
TABLE 3.
Characteristics of patients who have clarithromycin-resistant MAC infections and the features of their lung NTM (n = 12)
| Characteristic/featurea | Value |
|---|---|
| Age at diagnosis (mean [minimum–maximum]) (yr) | 62.8 (44–79) |
| Body wt (mean ± SD) (kg) | 42.0 ± 7.6 |
| Sex (male/female) | 1/11 |
| Smoking status (no. [%]) | |
| Current/former | 1 (8.3) |
| Never | 11 (91.7) |
| Underlying condition other than lung NTM (no. [%]) | |
| Chronic respiratory disorder | 2 (16.7) |
| Chronic other organ dysfunction | 1 (8.3) |
| Identified species | |
| M. avium/M. intracellulare (no.) | 10/2 |
| Microbiology result at diagnosis (no. [%]) | |
| AFB smear positive | 9 (75.0) |
| Radiological feature (no. [%]) | |
| Nodular bronchiectatic form | 5 (41.7) |
| Upper lobe cavitary form | 2 (16.7) |
| Mixed/advanced form | 5 (41.7) |
| Treatment history (no. [%]) | |
| Macrolide monotherapy | 6 (50.0) |
| Multidrug therapy (CAM + RFP + EB) | 10 (83.3) |
| SM administration | 3 (25.0) |
| Surgical resection | 1 (8.3) |
AFB, acid-fast bacilli; CAM, clarithromycin; RFP, rifampin; EB, ethambutol; SM, streptomycin.
Detection of other mutations causing clarithromycin resistance in MAC.
Some data were inconsistent with previous reports that mutations at position 2058 or 2059 of the 23S rRNA gene accounted for most of clarithromycin-resistant MAC strains (6, 7, 8, 12). Therefore, sequence analysis of the PCR product that showed clarithromycin resistance was implemented and showed that the clarithromycin-resistant samples, except for eight mutant specimens, were WT (-gaAAag-). Based on the this result, other possibilities for the development of clarithromycin resistance were subsequently examined. It has been reported that point mutations at position 752 in the hairpin-like structure in domain II as well as at positions 2062 and 2505 at the peptidyl transferase loop in domain V of 23S rRNA were involved in the clarithromycin resistance of E. coli (18). Therefore, the sequence of domain II covering the position 752 was scrutinized using primer MAV_Fw1073 (Table 2). Similarly, the sequences of positions 2062 and 2505 were examined using primers MAV_Fw2485 and MAV_Fw2938, respectively (Table 2). The 12 clarithromycin-resistant MAC strains and a randomly selected 10 clarithromycin-sensitive MAC strains were analyzed and showed no mutations in domain II and domain V of 23S rRNA. Likewise, the mutations of nucleotides 1198 through 1247 at the hairpin-like structure in domain II have been reported to introduce macrolide resistance in E. coli (19). Thus, the sequence between 1198 and 1247 was also analyzed using primer MAV_Fw1533 (Table 2). No mutations in this sequence were detected.
DISCUSSION
In this work, we set out to develop a real-time PCR-based clarithromycin resistance gene detection system using BNAs. An additional goal was to assess its efficacy using clinical samples from patients with lung NTM. Although our technique should be validated against the various techniques currently available, one of the advantages in applying BNAs is that the BNA-PCR can be simultaneously implemented with most conventional PCR systems since all of the reaction steps for the BNA-PCR are the same as those of ordinary PCR. Therefore, this technique not only enables the detection of mutations within the target gene but also allows for the use of existing devices. Moreover, the BNA-PCR was readily applicable to clinical practice; the consumables cost $20 US per sample, and the results can be delivered within 4 h after Mycobacterium species are grown. Thus, considering the increase in the number of patients with lung NTM and the ease of implementation of this system using existing devices, it is practical and perhaps advisable that clinical laboratories employ this system to address the growing demand for the identification of clarithromycin-resistant MAC strains.
In prokaryotic cells, rRNAs that serve as the sites for protein synthesis are composed of three distinct size species of rRNA (23S, 16S, and 5S). The ribosomal peptidyl transferase activity resides in domain V of 23S rRNA. Domain V is also the most common binding site for antibiotics and accounts for macrolide resistance in a variety of bacteria (5, 20, 21). Indeed, an abundance of literature reports suggest that point mutations at either position 2058 or 2059 in the 23S rRNA gene are the most responsible for macrolide-resistant MAC strain (6, 7, 8, 12, 22, 23). However, our results indicate that some of the resistance did not arise from the mutations in domain V. Given the inherent diversity in the mechanisms of pathogenicity displayed by MAC strains, it is conceivable and likely that there are other mutations that can result in the emergence of resistance. In line with this, it has been reported that the mutations of nucleotides 1198 through 1247 or of position 752 in domain II or point mutations at positions 2062 and 2505 in domain V of 23S rRNA are involved in macrolide resistance in E. coli. With this in mind, those mutations were scrutinized using clarithromycin-resistant NTM. However, this investigation demonstrated no further mutants. At least one strain showing clarithromycin resistance but carrying no mutation at position 2058 or 2059 has been observed in separate reports using clinical samples (6, 7, 8). This implies that other underlying mechanisms causing clarithromycin resistance undoubtedly exist in MAC strains.
Clarithromycin-resistant MAC disease emerges as a threat to treatment and requires a more complicated strategy. This is in part due to the fact that there has been no reliable in vitro drug susceptibility evaluation yet. Surgical intervention should be considered first if the patient can tolerate such an invasive procedure. The majority of patients, however, are already compromised due to the progressive disorder and destruction of lung function. Lung transplant, although still somewhat of a contraindication, may be one potential treatment for lung NTM infection (24). Indeed, lung transplantation has been performed in patients with cystic fibrosis who carried MAC (25, 26). Although it should be noted that posttransplant immunosuppressive management together with antimycobacterials lacking competent macrolide ability raises the possibility of reemergence of resistant MAC infections in the donor lung. Despite a paucity of reliable studies, treatment against MAC strains resistant to clarithromycin should be initiated using medication that has not been previously used in the patient. The regimen should be composed of at least three or four antimycobacterials, including a parenteral aminoglycoside, high-dose ethambutol (6), rifabutin (3), fluoroquinolones acting against DNA gyrase and topoisomerase IV (27, 28), and clofazimine (28, 29). Although somewhat controversial, continuation of macrolide treatment, even at a lower dose, if well tolerated, may also be considered due to its broad mechanism of action.
Our experiment has shown that the BNA-PCR system, established to identify the clarithromycin resistance gene, also elucidated the existence of clarithromycin-resistant MAC strains carrying no mutation at position 2508 or 2509. The disease-causing processes of Mycobacterium species are still poorly understood, and these bacteria also show an abundance of diversity in their regional characteristics and in the individuals more likely to be affected. Since clarithromycin is a final weapon against lung disorders due to MAC, the exact mechanism for the clarithromycin resistance requires further investigation.
ACKNOWLEDGMENTS
We thank Johnathan Canton, Program in Cell Biology, Hospital for Sick Children, Toronto, ON, Canada, for proofreading of the manuscript draft.
The manuscript has been reviewed by all of the authors and is not being submitted to any other journals.
We declare no conflict of interest.
Funding Statement
This work was supported by Grant-in-Aid for Young Scientists (B) (no. 23790918) from Japan Society for the Promotion of Science and A Grant Fostering Young Physician (no. 24-C-1-15) from Saitama Medical University Hospital. No additional external funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Hoefsloot W, Van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, Beylis N, Boeree MJ, Cacho J, Chihota V, Chimara E, Churchyard G, Cias R, Daza R, Daley CL, Dekhuijzen PNR, Domingo D, Drobniewski F, Esteban J, Fauville-Dufaux M, Folkvardsen DB, Gibbons N, Gómez-Mampaso E, Gonzalez R, Hoffmann H, Hsueh PR, Indra A, Jagielski T, Jamieson F, Jankovic M, Jong E, Keane J, Koh WJ, Lange B, Leao S, Macedo R, Mannsåker T, Marras TK, Maugein J, Milburn HJ, Mlinkó T, Morcillo N, Morimoto K, Papaventsis D, Palenque E, Paez-Peña M, Piersimoni C, Polanová M, Rastogi N, Richter E, et al. 2013. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J 42:1604–1613. doi: 10.1183/09031936.00149212. [DOI] [PubMed] [Google Scholar]
- 2.Marras TK, Daley CL. 2002. Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin Chest Med 23:553−567. doi: 10.1016/S0272-5231(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 3.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, Von Reyn CF, Wallace RJ, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 4.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 5.Schlünzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 6.Griffith DE, Brown-Elliott BA, Langsjoen B, Zhang Y, Pan X, Girard W, Nelson K, Caccitolo J, Alvarez J, Shepherd S, Wilson R, Graviss EA, Wallace RJ. 2006. Clinical and molecular analysis of macrolide resistance in Mycobacterium avium complex lung disease. Am J Respir Crit Care Med 174:928–934. doi: 10.1164/rccm.200603-450OC. [DOI] [PubMed] [Google Scholar]
- 7.Inagaki T, Yagi T, Ichikawa K, Nakagawa T, Moriyama M, Uchiya K, Nikai T, Ogawa K. 2011. Evaluation of a rapid detection method of clarithromycin resistance genes in Mycobacterium avium complex isolates. J Antimicrob Chemother 66:722–729. doi: 10.1093/jac/dkq536. [DOI] [PubMed] [Google Scholar]
- 8.Jamal MA, Maeda S, Nakata N, Kai M, Fukuchi K, Kashiwabara Y. 2000. Molecular basis of clarithromycin-resistance in Mycobacterium avium-intracellulare complex. Tuber Lung Dis 80:1–4. doi: 10.1054/tuld.1999.0227. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2012. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, 2nd ed Document M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 10.Hirama T, Mogi H, Egashira H, Yamamoto E, Kukisaki S, Hagiwara K, Takei O. 2015. A pressure-driven column-based technique for the efficient extraction of DNA from respiratory samples. Clin Chim Acta 445:122–126. doi: 10.1016/j.cca.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 11.Hirama T, Yamaguchi T, Miyazawa H, Tanaka T, Hashikita G, Kishi E, Tachi Y, Takahashi S, Kodama K, Egashira H, Yokote A, Kobayashi K, Nagata M, Ishii T, Nemoto M, Tanaka M, Fukunaga K, Morita S, Kanazawa M, Hagiwara K. 2011. Prediction of the pathogens that are the cause of pneumonia by the battlefield hypothesis. PLoS One 6:e24474. doi: 10.1371/journal.pone.0024474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meier A, Kirschner P, Springer B, Steingrube VA, Brown BA, Wallace RJ, Bottger EC. 1994. Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob Agents Chemother 38:381−384. doi: 10.1128/AAC.38.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyazawa H, Tanaka T, Nagai Y, Matsuoka M, Sutani A, Udagawa K, Zhang J, Hirama T, Murayama Y, Koyama N. 2008. Peptide nucleic acid-locked nucleic acid polymerase chain reaction clamp-based detection test for gefitinib-refractory T790M epidermal growth factor receptor mutation. Cancer Sci 99:595–600. doi: 10.1111/j.1349-7006.2007.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirama T, Minezaki S, Yamaguchi T, Kishi E, Kodama K, Egashira H, Kobayashi K, Nagata M, Ishii T, Nemoto M, Tanaka M, Fukunaga K, Kanazawa M, Hagiwara K. 2014. HIRA-TAN: a real-time PCR-based system for the rapid identification of causative agents in pneumonia. Respir Med 108:395. doi: 10.1016/j.rmed.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Imanishi T, Obika S. 2002. BNAs: novel nucleic acid analogs with a bridged sugar moiety. Chem Commun (Camb.) 16:1653–1659. [DOI] [PubMed] [Google Scholar]
- 16.Rahman SA, Seki S. 2008. Design, synthesis, and properties of 2′,4′-BNANC: a bridged nucleic acid analogue. J Am Chem Soc 130:4886–4896. doi: 10.1021/ja710342q. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka E, Kimoto T, Tsuyucuchi K, Watanabe I, Matsumoto H, Niimi A, Suzuki K, Murayama T, Amitani R, Kuze F. 1999. Effect of clarithromycin regimen for Mycobacterium avium complex pulmonary disease. Am J Respir Crit Care Med 160:866–872. doi: 10.1164/ajrccm.160.3.9811086. [DOI] [PubMed] [Google Scholar]
- 18.Hansen LH, Mauvais P, Douthwaite S. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol Microbiol 31:623–631. doi: 10.1046/j.1365-2958.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- 19.Dam M, Douthwaite S, Tenson T, Mankin AS. 1996. Mutations in domain II of 23 S rRNA facilitate translation of a 23 S rRNA-encoded pentapeptide conferring erythromycin resistance. J Mol Biol 259:1–6. doi: 10.1006/jmbi.1996.0296. [DOI] [PubMed] [Google Scholar]
- 20.Morozumi M, Hasegawa K, Kobayashi R, Inoue N, Iwata S, Kuroki H, Kawamura N, Nakayama E, Tajima T, Shimizu K, Ubukata K. 2005. Emergence of macrolide-resistant Mycoplasma pneumoniae with a 23S rRNA gene mutation. Antimicrob Agents Chemother 49:2302–2306. doi: 10.1128/AAC.49.6.2302-2306.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Versalovic J, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, Tanaka SK, Graham DY, Go MF. 1996. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother 40:477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nash KA, Inderlied CB. 1995. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob Agents Chemother 39:2625−2630. doi: 10.1128/AAC.39.12.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier a. Heifets L, Wallace RJ, Zhang Y, Brown BA, Sander P, Böttger EC. 1996. Molecular mechanisms of clarithromycin resistance in Mycobacterium avium: observation of multiple 23S rDNA mutations in a clonal population. J Infect Dis 174:354–360. doi: 10.1093/infdis/174.2.354. [DOI] [PubMed] [Google Scholar]
- 24.Qvist T, Pressler T, Thomsen VO, Skov M, Iversen M, Katzenstein TL. 2013. Nontuberculous mycobacterial disease is not a contraindication to lung transplantation in patients with cystic fibrosis: a retrospective analysis in a Danish patient population. Transplant Proc 45:342–345. doi: 10.1016/j.transproceed.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 25.Knoll BM, Kappagoda S, Gill RR, Goldberg HJ, Boyle K, Baden LR, Fuhlbrigge AL, Marty FM. 2012. Non-tuberculous mycobacterial infection among lung transplant recipients: a 15-year cohort study. Transpl Infect Dis 14:452–460. doi: 10.1111/j.1399-3062.2012.00753.x. [DOI] [PubMed] [Google Scholar]
- 26.Lobo LJ, Noone PG. 2014. Respiratory infections in patients with cystic fibrosis undergoing lung transplantation. Lancet Respir Med 2:73–82. doi: 10.1016/S2213-2600(13)70162-0. [DOI] [PubMed] [Google Scholar]
- 27.Fujita M, Kajiki A, Tao Y, Miyazaki M, Ouchi H, Harada E, Ikegame S, Matsumoto T, Uchino J, Watanabe K, Nakanishi Y. 2012. The clinical efficacy and safety of a fluoroquinolone-containing regimen for pulmonary MAC disease. J Infect Chemother 18:146–151. doi: 10.1007/s10156-011-0303-5. [DOI] [PubMed] [Google Scholar]
- 28.Jo K-W, Kim S, Lee JY, Lee S-D, Kim WS, Kim DS, Shim TS. 2014. Treatment outcomes of refractory MAC pulmonary disease treated with drugs with unclear efficacy. J Infect Chemother 20:602–606. doi: 10.1016/j.jiac.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Jarand J, Davis JP, Cowie RL, Field SK, Fisher DA. Long term follow up of Mycobacterium avium complex lung disease in patients treated with regimens including clofazimine and/or rifampin. Chest, in press. [DOI] [PubMed] [Google Scholar]