Abstract
A persistent 8-year infection by a Beijing Mycobacterium tuberculosis strain from a previous outbreak after importation from West Africa obliged us to investigate secondary cases. We developed a multiplex PCR method based on whole-genome sequencing to target strain-specific single nucleotide polymorphisms (SNPs). In 1 week, we analyzed 868 isolates stored over 6 years. Only 2 cases (immigrants from Guinea Conakry) harbored the strain, which ruled out transmission—despite opportunities—and challenged some of the advantages associated with Beijing strains.
TEXT
Extensive application of genotyping strategies to characterize Mycobacterium tuberculosis has enabled us to identify strains that are advantageous because of their virulence, ability to acquire resistance, or efficiency in causing severe outbreaks.
Once such strains are identified, their fingerprints can be used as a reference to closely monitor their emergence in populations where they remain undetected (1). However, this approach requires universal population-based systematic fingerprinting programs.
Alternatively, strain-specific PCRs can be used to track the emergence of theoretically advantageous strains (2, 3). Whole-genome sequencing (WGS) has simplified the identification of genotypic peculiarities (based on specific single nucleotide polymorphisms [SNPs]) from which we can design strain-specific PCRs (4, 5).
We developed an allele-specific oligonucleotide (ASO)-multiplex PCR method based on WGS data to track the presence of a susceptible Beijing strain in Madrid, Spain. This strain was responsible for a severe outbreak on Gran Canaria Island (GCI) in the 1990s, and it was eventually shown to be the cause of around one-quarter of the total number of cases on the island (6). The GCI strain was still found in 2007 and 2008 as shown after the application of a specific PCR (7). In Madrid, we identified a nonadherent case with persistently active infectious tuberculosis (8 years) caused by the Gran Canaria strain (8), which obliged us to fast track potential secondary cases.
(The results of this study were partially presented at the 2015 ESM Meeting, Riga, Latvia, 28 June to 1 July 2015.)
The persistent case was diagnosed in our institution in 2006. The mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) pattern of this isolate coincided with the pattern described for the GCI strain (7). Using fingerprinting data from 2006 to 2008, we only detected 3 patients, all of whom were linked to the GCI outbreak. Given that the population-based genotyping program running in our area was discontinued at the end of 2008, we were unable to identify potential secondary cases that occurred from 2009 to 2014, when previously exposed cases were more likely to develop the disease. To update the 2009 to 2014 gap, we developed an ASO-PCR method to retrospectively fast track the GCI strain by directly analyzing stored isolates.
WGS of 4 isolates from our case was performed as indicated elsewhere (4). We followed standard library preparation protocols. Using a HiSeq 2000 device generating 51- to 101-bp paired-end reads, we were able to identify 132 GCI-strain-specific SNPs after comparing the SNPs extracted from WGS data with those from an in-house database of 219 strains representing the geographic and phylogenetic diversity of the M. tuberculosis complex (9). To improve the specificity of our selection, our list of 132 candidate-specific SNPs underwent a second filter by comparison with a database containing 358 unique Beijing genomes (10). Eighty-two SNPs were found in the database, but 50 continued to be GCI strain specific. We selected the 16 synonymous SNPs to ensure their stability as genetic markers (11).
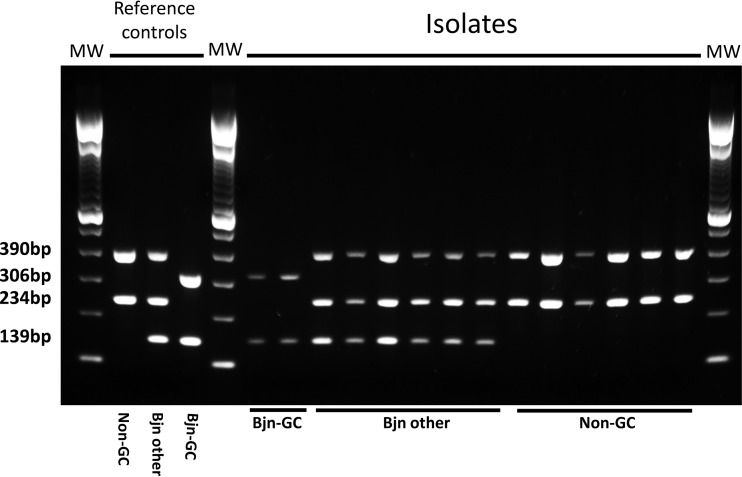
In order to rule out false positives and negatives, we decided to simultaneously target several GCI-strain-specific SNPs as recommended elsewhere (5). We designed a multiplex ASO-PCR method to target 3 of these SNPs (GCI-SNP1 [Rv2524, C1398T], GCI-SNP2 [Rv3869, G1347C], and GCI-SNP3 [Rv0926c, G162A]). We also targeted a fourth SNP that was specific for the Beijing lineage (Rv2952, G526A) (5). Our PCR method was designed to ensure amplification products for any strain in order to be able to identify inhibitions. To do so, we designed 2 selective primers (GCI-SNP3 and Beijing-SNP) (Table 1) to target the alleles found in the GCI strain, whereas the remaining 2 primers (GCI-SNP1 and GCI-SNP2) targeted the alleles from the non-GCI strains. Our design would thus generate 3 different patterns depending on whether the strain tested was non-GCI, Beijing GCI, or Beijing other (Fig. 1).
TABLE 1.
Primers involved in the multiplex ASO-PCR
| SNP | Primer | Sequencea |
|---|---|---|
| GCI-SNP 1 | GCI-2524Fw | 5′-CCGCATCGAACAAATGGCC-3′ |
| GCI-SNP 1 | GCI-2524R | 5′-TCCGAGTAGGTAGCCAGCAG-3′ |
| GCI-SNP 2 | GCI-3869F | 5′-ATCAAGCAGATCCACGGGAC-3′ |
| GCI-SNP 2 | GCI-3869Rw | 5′-GGCATCTTTCGACAGCACC-3′ |
| GCI-SNP 3 | GCI-0926Fm | 5′-GGCTGGCGGATTCGACA-3′ |
| GCI-SNP 3 | GCI-0926R | 5′-CCATCGGCAACAGGTCGTT-3′ |
| Beijing-SNP | Bjn-2952F | 5′-GCCGAAAACCTGCCCTTC- 3′ |
| Beijing-SNP | Bjn-2952Rm | 5′-CGGCGTATGGGAAGTACCT-3′ |
The SNPs targeted by the selective primers are highlighted in bold.
FIG 1.
Amplification patterns of the quadruplex allele-specific PCR. At the left side of the gel, the amplification patterns are shown for non-Beijing (Bjn) strains (390 and 234 bp), non-GCI Beijing strains (Bjn other; 390, 234, and 139 bp), and Beijing-Gran Canaria Island (Bjn-GC) strains (306 and 139 bp). The remaining lanes correspond to the 2 Beiijng GCI isolates and a selection of representative isolates for Beijing other and non-Beijing strains.
The ASO-PCR method was applied to all of the 2009 to 2014 strains isolated at the Hospital General Universitario Gregorio Marañón (HGUGM) and at the Hospital Doce de Octubre (HDO), which cover 25.9% of the population of Madrid. The assay conditions were as follows: 1.8 mM MgCl2, 0.1 μM of each GCI primer, 0.3 μM Bejing primers, 200 μM deoxynucleoside triphosphates (dNTPs), and 0.4 μl AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA). The PCR conditions were 95°C for 10 min followed by 30 cycles of 95°C for 1 min, 65°C for 1 min, and 72°C for 1 min followed by 72°C for 10 min.
The 868 isolates (364 from HGUGM and 504 from HDO) were interrogated directly using a boiled aliquot from the stored collection. All but 17 samples (98.04%) produced a result, which shows the efficiency of the test without the delays caused by DNA purification and subculture. Despite the long-term infectivity of our case, only 2 cases (1 from each institution) were caused by the GCI strain. Infection by the GCI strain in these 2 new cases was confirmed by MIRU-VNTR 24 analysis (7) and Sanger sequencing of the SNPs. They corresponded to recent isolations (2014) and involved 2 immigrants (Guinea Conakry). Both patients denied having lived on GCI, and contacts with our case were ruled out. These data suggest an unexpected lack of transmission in Madrid by the strain causing the severe outbreak on GCI. The index case of the outbreak was from Liberia, which shares borders with the country of origin of the 2 new cases. The findings indicate that the GCI strain is prevalent in that area of West Africa (6) and seems to have been recently reimported into Spain following a new path that is not linked to GCI.
In addition to the 2 GCI cases, we identified 15 (1.72%) new cases infected by non-GCI Beijing strains (10 from HGUGM and 5 from HDO), all of which were confirmed by Sanger sequencing of another specific SNP, namely, A191C in Rv2629 (12). Most were pansusceptible (12/15), but we identified 2 strains that were resistant to rifampin, isoniazid, and streptomycin and 1 strain that was resistant to isoniazid, streptomycin, and ethionamide. Most of the isolates were from immigrants (12/15) (China [4], Peru [3], Ukraine [2], Ecuador [1], Bangladesh [1], and Mongolia [1]).
The ASO-PCR-based screening strategy proved to be highly efficient. The turnaround time for complete analysis of the two collections at each hospital was distributed as follows: 4 and 6 h to take the aliquots and 3 and 4 consecutive days for PCR, electrophoresis, and analysis of patterns. Therefore, within 1 week, we updated information from around 900 strains covering a 6-year period and 26% of the population of Madrid. The estimated cost of the procedure was €1.50 per isolate. These data indicate that our strategy offers accuracy and is also cost-effective and time efficient. Although WGS would have offered a complete analysis of the entire genome, it would have been less cost-effective and less efficient.
Our data suggest an absence of transmission of the GCI strain in Madrid despite the presence of circumstances favoring it due to the existence of a long-term infectious TB case, an intravenous drug user (IVDU), infected by this strain. Consequently, the cause of the GCI outbreak seems more likely to be epidemiology related than it is to be strain related. Nevertheless, we must continue to monitor the dynamics of the GCI strain in Madrid. Our specific multiplex ASO-PCR method may prove to be a suitable tool for fast tracking this strain.
The combination of the knowledge derived from universal genotyping programs with the in-depth analysis provided by WGS and the design of high-risk strain-specific PCRs using these data can optimize and simplify the procedure, reduce costs, and completely transform the way in which we track TB transmission.
Nucleotide sequence accession number.
All sequences are available at EBI under project number PRJEB9158.
ACKNOWLEDGMENTS
We thank Thomas O'Boyle for proofreading the manuscript. We thank Pedro Montilla from the Unidad de Valoración y Apoyo a la Atención Domiciliaria (UVAAD) in HGUGM for obtaining the epidemiological data of the 2 new cases infected by the GCI strain. We greatly appreciate the help of Tao Luo (Key Laboratory of Medical Molecular Virology, Institute of Medical Microbiology, Fudan University, Shanghai, China) for purging our list of SNPs by comparison with his Beijing database.
The authors declare that they have no conflicts of interest.
REFERENCES
- 1.Kremer K, Glynn JR, Lillebaek T, Niemann S, Kurepina NE, Kreiswirth BN, Bifani PJ, van Soolingen D. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J Clin Microbiol 42:4040–4049. doi: 10.1128/JCM.42.9.4040-4049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mokrousov I, Vyazovaya A, Zhuravlev V, Otten T, Millet J, Jiao WW, Shen AD, Rastogi N, Vishnevsky B, Narvskaya O. 2014. Real-time PCR assay for rapid detection of epidemiologically and clinically significant Mycobacterium tuberculosis Beijing genotype isolates. J Clin Microbiol 52:1691–1693. doi: 10.1128/JCM.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plikaytis BB, Marden JL, Crawford JT, Woodley CL, Butler WR, Shinnick TM. 1994. Multiplex PCR assay specific for the multidrug-resistant strain W of Mycobacterium tuberculosis. J Clin Microbiol 32:1542–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Lago L, Martinez-Lirola M, Herranz M, Comas I, Bouza E, Garcia-de-Viedma D. Fast and low-cost decentralized surveillance of transmission of tuberculosis based on strain-specific PCRs tailored from whole genome sequencing data: a pilot study. Clin Microbiol Infect 21:249.e1–249.e9. [DOI] [PubMed] [Google Scholar]
- 5.Stucki D, Ballif M, Bodmer T, Coscolla M, Maurer AM, Droz S, Butz C, Borrell S, Längle C, Feldmann J, Furrer H, Mordasini C, Helbling P, Rieder HL, Egger M, Gagneux S, Fenner L. 2015. Tracking a tuberculosis outbreak over 21 years: strain-specific single-nucleotide polymorphism typing combined with targeted whole-genome sequencing. J Infect Dis 211:1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caminero JA, Pena MJ, Campos-Herrero MI, Rodríguez JC, García I, Cabrera P, Lafoz C, Samper S, Takiff H, Afonso O, Pavón JM, Torres MJ, van Soolingen D, Enarson DA, Martin C. 2001. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am J Respir Crit Care Med 164:1165–1170. doi: 10.1164/ajrccm.164.7.2101031. [DOI] [PubMed] [Google Scholar]
- 7.Millán-Lou MI, Alonso H, Gavín P, Hernández-Febles M, Campos-Herrero MI, Copado R, Cañas F, Kremer K, Caminero JA, Martín C, Samper S. 2012. Rapid test for identification of a highly transmissible Mycobacterium tuberculosis Beijing strain of sub-Saharan origin. J Clin Microbiol 50:516–518. doi: 10.1128/JCM.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-Lago L, Navarro Y, Montilla P, Comas I, Herranz M, Rodríguez-Gallego C, Ruiz Serrano MJ, Bouza E, García de Viedma D. 2015. Persistent infection by a Mycobacterium tuberculosis strain that was theorized to have advantageous properties, as it was responsible for a massive outbreak. J Clin Microbiol 53:3423–3429. doi: 10.1128/JCM.01405-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, Yeboah-Manu D, Bothamley G, Mei J, Wei L, Bentley S, Harris SR, Niemann S, Diel R, Aseffa A, Gao Q, Young D, Gagneux S. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo T, Comas I, Luo D, Lu B, Wu J, Wei L, Yang C, Liu Q, Gan M, Sun G, Shen X, Liu F, Gagneux S, Mei J, Lan R, Wan K, Gao Q. 2015. Southern East Asian origin and coexpansion of Mycobacterium tuberculosis Beijing family with Han Chinese. Proc Natl Acad Sci U S A 112:8136–8141. doi: 10.1073/pnas.1424063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stucki D, Malla B, Hostettler S, Huna T, Feldmann J, Yeboah-Manu D, Borrell S, Fenner L, Comas I, Coscollà M, Gagneux. 2012. Two new rapid SNP-typing methods for classifying Mycobacterium tuberculosis complex into the main phylogenetic lineages. PLoS One 7:e41253. doi: 10.1371/journal.pone.0041253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homolka S, Koser C, Archer J, Rusch-Gerdes S, Niemann S. 2009. Single-nucleotide polymorphisms in Rv2629 are specific for Mycobacterium tuberculosis genotypes Beijing and Ghana but not associated with rifampin resistance. J Clin Microbiol 47:223–226. doi: 10.1128/JCM.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]