Abstract
Pneumonia due to Pneumocystis jirovecii (PCP) is a frequent infection among HIV-positive or other immunocompromised patients. In the past several years, PCR on pulmonary samples has become an essential element for the laboratory diagnosis of PCP. Nevertheless, very few comparative studies of available PCR assays have been published. In this work, we evaluated the concordance between four real-time PCR assays, including three commercial kits, AmpliSens, MycAssay, and Bio-Evolution PCR, and an in-house PCR (J. Fillaux et al. 2008, J Microbiol Methods 75:258–261, doi:http://dx.doi.org/10.1016/j.mimet.2008.06.009), on 148 pulmonary samples. The results showed concordance rates ranging from 81.6% to 96.6% (kappa, 0.64 to 0.93). Concordance was excellent between three assays: the in-house assay, AmpliSens, and the MycAssay PCR (kappa, >0.8). The performances of these PCR assays were also evaluated according to the classification of the probability of PCP (proven, probable, possible, or no final diagnosis of PCP) based on clinical and radiological signs as well as on the direct examination of bronchoalveolar lavage samples. In the proven PCP category, Pneumocystis jirovecii DNA was detected with all four assays. In the probable PCP category, the in-house PCR, AmpliSens, and the MycAssay PCR were positive for all samples, while the Bio-Evolution PCR failed to detect Pneumocystis jirovecii DNA in two samples. In the possible PCP category, the percentage of positive samples according to PCR varied from 54.5% to 86.4%. Detection of colonized patients is discussed. Finally, among the four evaluated PCR assays, one was not suitable for colonization detection but showed good performance in the proven and probable PCP groups. For the three other assays, performances were excellent and allowed detection of a very low fungal burden.
INTRODUCTION
Pneumocystis jirovecii is an opportunistic pathogen responsible for Pneumocystis pneumonia (PCP) in immunocompromised patients. PCP often occurs in HIV-infected patients, but frequency of PCP in other immunocompromised populations, such as patients with kidney transplantation, is increasing (1–3). Common clinical manifestations and outcomes of PCP between HIV-positive and HIV-negative patients are well known; in non-HIV-infected patients symptomatology is more acute, mortality is generally higher, and fungal burden is lower (2, 4). In any case, biological diagnosis of PCP is required, and the microscopic identification of P. jirovecii (cysts and trophic forms) by staining bronchoalveolar lavage (BAL) samples continues to be the most widely used method. However, microscopic diagnosis lacks sensibility for low fungal burdens, especially in non-HIV-infected patients and when noninvasive pulmonary samples are used. The development of PCR methods, particularly that of real-time PCR, has improved the diagnosis of PCP and seems to be essential with a negative predictive value close to 100% (5, 6). The first assays used for molecular diagnosis of PCP used “in-house” PCR. In recent years, commercial kits were made available by several companies, but few studies evaluated their performances (5, 7–10) and none compared them. The main objective of this study was to evaluate the concordance between four real-time PCR assays, three commercial kits and the in-house PCR (11), for the detection of P. jirovecii DNA in pulmonary samples. Performances of these assays were then evaluated according to the probability of PCP based on clinical and radiological data and the result of direct examination (DE) of bronchoalveolar lavage fluid.
(Preliminary data were presented at the 6th Trends in Medical Mycology Meeting, Copenhagen, Denmark, 11 to 14 October 2013 [12].)
MATERIALS AND METHODS
Patients and biological samples.
A total of 148 pulmonary samples (75 BAL samples, 1 pulmonary biopsy specimen, 12 bronchial aspirates [BA], and 60 sputum samples [Spu]) were collected between March 2009 and February 2013 in Nîmes University Hospital. All patients presented pulmonary clinical symptoms, and P. jirovecii detection in lung samples was required to support or refute the diagnosis of PCP.
Direct examination (DE) was systematically performed on BAL fluid (after cytospin procedure) and on pulmonary biopsy specimens by using May-Grünwald-Giemsa and Gomori-Grocott staining.
DNA extraction.
All DNA were extracted by using the EZ1 DNA tissue kit (Qiagen) and following the manufacturer's instructions. Briefly, after the centrifugation of 2 ml of BAL fluid, Spu, and BA (10,000 rpm for 5 min), the pellet (resuspended in 190 μl lysis buffer) was incubated with proteinase K (10 μl) for 10 min at 56°C. DNA was then automatically extracted (BioRobot EZ1; Qiagen) and was resuspended in 100 μl. DNA samples were stored at −20°C.
PCR assays.
Four real-time PCR assays were used: an in-house PCR according to Fillaux et al. protocol (11) and three commercial kits, including the real-time PCR Pneumocystis jirovecii (Bio-Evolution), the AmpliSens Pneumocystis jirovecii (carinii)-FRT PCR kit, and the MycAssay Pneumocystis (Myconostica). Commercial kits were used according to the manufacturer's instructions. Five-microliter DNA samples were added in each reaction tube for the in-house and Bio-Evolution PCR, and 10-μl samples were added for the MycAssay and AmpliSens PCR. PCRs were performed on the LightCycler 480 (Roche). The in-house PCR primers target major surface glycoprotein genes, while the three commercial kits target the mitochondrial large subunit rRNA (mtLSU). For each PCR run, negative (water) and positive (DNA of P. jirovecii) controls were included. For each sample, DNA extraction and inhibition controls, consisting of the amplification of a fragment of the tissue plasminogen activator (TPA) gene, were also tested. For disparate results, reactions were repeated at least one time with each PCR.
PCR results analysis.
PCR results were given as positive or negative. For positive samples, threshold cycle (CT) values were also recorded. Nevertheless, as no standard curve was generated with a positive control, CT values were not interpretable for quantification.
Classification of the probability of PCP.
Clinical, radiological, and treatment data were collected retrospectively for all patients included. The clinical history of each patient was analyzed by two specialists (a mycologist and an infectious diseases specialist) to classify PCP as proven, probable, possible, or no final diagnosis of PCP. This classification was established blindly to the PCR results. Our classification was based on the four following criteria: at least one factor of immunosuppression, clinical signs of progressive pneumonia, radiological signs of PCP (diffuse ground glass opacities and/or diffuse infiltration), and resolution of symptoms after P. jirovecii-specific treatment. The categories were determined as follow: (i) proven PCP, presence of four criteria and positive DE; (ii) probable PCP, presence of four criteria and negative DE; (iii) possible PCP, three out of four criteria, negative DE, and no other cause for pneumonia; (iv) no final diagnosis of PCP, less than three criteria, negative DE, and other cause for pneumonia (see Table S1 in the supplemental material).
Statistical analysis.
Quantitative data are expressed as median with range. Qualitative variables are expressed as frequency with percentage. The concordance of four real-time PCR assays was assessed by the kappa concordance coefficient for qualitative results (13) with its 95% confidence interval (95% CI). All analyses were performed using SAS version 9.3 (SAS Institute Inc., Cary, NC) with a 2-sided type 1 error rate of 5% as the threshold for statistical significance.
Sample size.
The sample size was previously determined considering a minimal concordance of 0.6 (minimal concordance that is considered a good accord between two measures), i.e., a kappa coefficient equal to 0.8 with a half precision of the confidence interval equal to 0.2. A minimum sample size of 138 subjects was calculated as necessary to estimate this parameter with a 5% type 1 error rate, but a bigger number was considered necessary to minimize the risk of nonanalyzable variables.
Ethics.
The study was approved by the Ethics Committee of Nîmes University Hospital (IRB 13/02.03). All the participants were informed of the study and its objectives.
RESULTS
Patient characteristics.
The main characteristics of the 148 included patients are presented in Table 1. Among the 148 patients, 27.4% were HIV positive, 30.8% had a hematologic malignancy, and 48.6% received an immunosuppressive treatment. Most patients (79.6%) had radiological signs compatible with PCP.
TABLE 1.
Main characteristics of the 148 patients included in the study (according to available clinical data)
| Characteristic | Value | MVa |
|---|---|---|
| Median age (range) (yr) | 57 (0–90) | 0 |
| Sex ratio (male/female) | 2.9 | 0 |
| Underlying disease, no. (%) | ||
| HIV infection | 40 (27.4) | 2 |
| CD4+ T-cell, mean/mm3 (range) | 56 (1–1,240) | 2 |
| Solid cancer | 25 (17.1) | 2 |
| Solid organ transplantation | 9 (6.2) | 2 |
| Hematologic malignancy | 45 (30.8) | 2 |
| Bone marrow transplant | 3 (2.1) | 2 |
| Autoimmune or inflammatory disease | 11 (7.8) | 7 |
| COPDb | 19 (13.0) | 2 |
| Other | 10 (6.8) | |
| Concomitant treatment, no. (%) | ||
| Anti-tumor chemotherapy | 48 (32.7) | 1 |
| Immunosuppressive agents (other than chemotherapy or corticosteroids) | 18 (12.2) | 0 |
| Long-term corticosteroid therapy | 6 (4.1) | 0 |
| Clinical signs, no. (%) | ||
| Cough | 74 (54.8) | 13 |
| Fever | 88 (65.2) | 13 |
| Dyspnea | 92 (67.6) | 12 |
| Radiological findings (chest radiography or computed tomography scan), no. (%) | ||
| Abnormalities consistent with PCP | 113 (79.6) | 6 |
| No abnormality or abnormalities not consistent with PCP | 29 (20.4) | 6 |
| Curative treatment of PCP | 78 (52.7) | 0 |
| Other documented etiology for the pneumonia | 33 (26.2) | 22 |
MV, missing value.
COPD, chronic obstructive pulmonary disease.
Concordance of the four molecular methods (n = 147 samples, one missing result for the MycAssay PCR).
The quality of DNA extraction was assessed by TPA gene amplification. For all of the samples, amplification of the TPA gene was positive with CT values of <24. Table 2 presents the number of positive (presence of P. jirovecii DNA) and negative (no detection of P. jirovecii DNA) results obtained with the four assays. The Bio-Evolution PCR detected the fewest positive samples (55 samples), and the AmpliSens PCR detected the most positive samples (82 samples). The concordance rate of the four assays varied from 81.6% to 96.6% (kappa, 0.64 to 0.93). For all samples, concordance was excellent between the following three assays: the in-house assay, the AmpliSens assay, and the MycAssay PCR (kappa, >0.8) (13). The Bio-Evolution PCR was only moderately concordant with the three other assays (0.64 < kappa < 0.75), and this kit detected the fewest positive samples.
TABLE 2.
PCR results of the 147 samplesa and concordance rate between the four PCR methods
| PCR method and result | Amplisens PCR |
MycAssay PCR |
Bio-Evolution PCR |
|||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | |
| In-house PCR (11) | ||||||
| No. positive | 77 | 1 | 73 | 5 | 55 | 23 |
| No. negative | 5 | 64 | 0 | 69 | 0 | 69 |
| Kappa concordance coefficient (95% CI) | 0.92 (0.85–0.98) | 0.93 (0.87–0.99) | 0.69 (0.58–0.80) | |||
| Concordance, % | 95.9 | 96.6 | 84.4 | |||
| Bio-Evolution PCR | ||||||
| No. positive | 55 | 0 | 55 | 0 | ||
| No. negative | 27 | 65 | 18 | 74 | ||
| Kappa concordance coefficient (95% CI) | 0.64 (0.53–0.76) | 0.75 (0.65–0.86) | ||||
| Concordance, % | 81.6 | 87.8 | ||||
| MycAssay PCR | ||||||
| No. positive | 72 | 1 | ||||
| No. negative | 10 | 64 | ||||
| Kappa concordance coefficient (95% CI) | 0.85 (0.77–0.93) | |||||
| Concordance, % | 92.5 | |||||
There is one missing result for the MycAssay PCR.
Patient classification for probability of PCP (n = 139 patients).
Clinical data were available for 139/148 patients and allowed for the classification of these patients based on the probability of PCP. Thirteen patients were classified as proven PCP (available samples: 12 BAL samples/biopsy specimens and 1 BA/Spu), 32 were classified as probable PCP (6 BAL samples and 26 BA/Spu), and 31 were classified as possible PCP (18 BAL samples and 13 BA/Spu). For 63 patients, final diagnosis of PCP was excluded (37 BAL samples and 26 BA/Spu). Patients with proven or probable PCP received specific P. jirovecii treatment (41 received sulfamethoxazole and trimethoprim, 3 received atovaquone, and 1 received atovaquone plus pentamidine isethionate). In the possible PCP group, 21 patients were treated with sulfamethoxazole and trimethoprim and 3 were treated with atovaquone. For the no final diagnosis of PCP group, the classification data were presented in Table S1 in the supplemental material. Among these 139 patients, 25 received P. jirovecii treatment at the time of pulmonary sampling and were not included in the analysis of PCR results according to the classification of PCP to avoid bias of interpretation.
PCR results for patients without P. jirovecii treatment at the time of pulmonary sampling according to their classification (n = 113 patients).
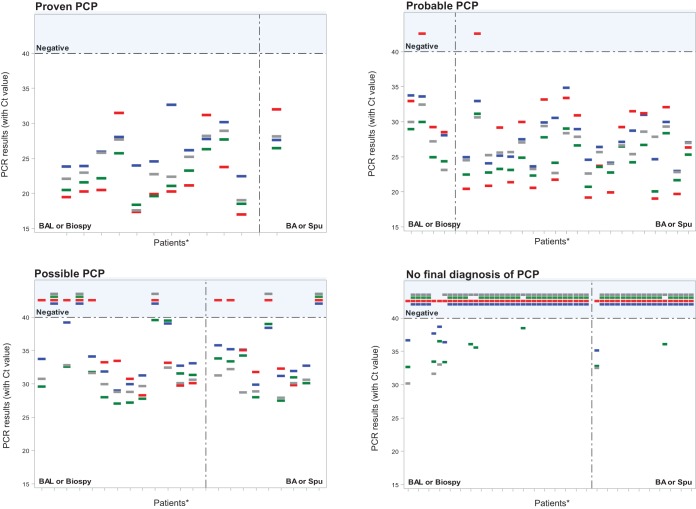
Table 3 presents global PCR results according to patient classification and the type of pulmonary sample. The results obtained for each patient with the four assays (negative or positive PCR with CT value) according to classification and pulmonary sample are shown in Fig. 1.
TABLE 3.
Results of the four PCR assays according to patient classification and type of samplea
| PCR method and result | Proven PCP (n = 12) |
Probable PCP (n = 25) |
Possible PCP (n = 22) |
No final diagnosis of PCP (n = 54) |
||||
|---|---|---|---|---|---|---|---|---|
| BAL or biopsy specimen (n = 11) | BA or Spu (n = 1) | BAL or biopsy specimen (n = 4) | BA or Spu (n = 21) | BAL or biopsy specimen (n = 13) | BA or Spu (n = 9) | BAL or biopsy specimen (n = 35) | BA or Spu (n = 19) | |
| In-house PCR (11) | ||||||||
| No. negative (%) | 0/11 (0) | 0/1 (0) | 0/4 (0) | 0/21 (0) | 3/13 (23.1) | 1/9 (11.1) | 31/35 (88.6) | 18/19 (94.7) |
| No. positive (%) | 11/11 (100) | 1/1 (100) | 4/4 (100) | 21/21 (100) | 10/13 (76.9) | 8/9 (88.9) | 4/35 (11.4) | 1/19 (5.3) |
| Median CT (range) | 25.91 (22.45–32.63) | 27.62b | 31.4 (28.03–33.75) | 27.03 (22.97–34.83) | 32.88 (28.96–39.20) | 33.92 (29.89–38.38) | 37.14 (36.32–38.63) | 35.12b |
| AmpliSens PCR | ||||||||
| No. negative (%) | 0/11 (0) | 0/1 (0) | n = 0/4 (0) | 0/21 (0) | 2/13 (15.4) | 1/9 (11.1) | 28/35 (80) | 17/19 (89.5) |
| No. positive (%) | 11/11 (100) | 1/1 (100) | 4/4 (100) | 21/21 (100) | 11/13 (84.6) | 8/9 (88.9) | 7/35 (20) | 2/19 (10.5) |
| Median CT (range) | 21.55 (18.32–27.67) | 26.41b | 26.94 (24.31–29.96) | 24.12 (20.00–31.08) | 31.28 (27.05–39.54) | 32.16 (27.47–38.94) | 35.54 (32.60–38.48) | 34.39 (32.76–36.01) |
| MycAssay PCR | ||||||||
| No. negative (%) | 0/11 (0) | 0/1 (0) | 0/4 (0) | 0/21 (0) | 3/13 (23.1) | 2/9 (22.2) | 32/35 (91.4) | 18/19 (94.7) |
| No. positive (%) | 11/11 (100) | 1/1 (100) | 4/4 (100) | 21/21 (100) | 10/13 (76.9) | 7/9 (77.8) | 3/35 (8.6) | 1/19 (5.3) |
| Median CT (range) | 22.94 (17.53–28.88) | 28.13b | 28.54 (23.11–32.42) | 25.67 (22.56–30.61) | 30.33 (28.77–32.76) | 30.1 (27.91–32.21) | 31.62 (30.18–33.00) | 32.47b |
| Bio-Evolution PCR | ||||||||
| No. negative (%) | 0/11 (0) | 0/1 (0) | 1/4 (25) | 1/21 (4.8) | 6/13 (46.2) | 4/9 (44.4) | 35/35 (100) | 19/19 (100) |
| No. positive (%) | 11/11 (100) | 1/1 (100) | 3/4 (75) | 20/21 (95.2) | 7/13 (53.9) | 5/9 (55.6) | 0/35 (0) | 0/19 (0) |
| Median CT (range) | 20.26 (16.97–31.46) | 31.99b | 29.17 (28.45–32.95) | 24.96 (18.98–33.33) | 30.72 (28.26–33.43) | 31.75 (29.76–35.00) | c | c |
Only untreated patients at the time of pulmonary sampling were included (n = 113).
Only one positive value was available.
No positive samples were available.
FIG 1.
PCR results obtained for each patient with the four assays (negative or positive with CT value) according to the probability of PCP. Results were also presented depending on the type of pulmonary samples. *, Patients without P. jirovecii treatment at the time of pulmonary sampling. Each patient is represented by a dash. Blue dash, in house PCR; red dash, Bio-Evolution PCR; green dash, AmpliSens PCR; grey dash, MycAssay PCR.
In the proven PCP category, P. jirovecii DNA was detected with all four assays in all samples. In the probable PCP category, the in-house, AmpliSens, and MycAssay PCR assays were positive for all samples, including BA/Spu samples. The Bio-Evolution PCR failed to detect P. jirovecii DNA in two samples (1 BAL fluid and 1 BA/Spu). In the possible PCP category, the percentage of positive samples according to PCR varied from 54.5% (Bio-Evolution PCR) to 86.4% (AmpliSens PCR). In the no final diagnosis of PCP category, only the Bio-Evolution kit was negative for all of the samples. With the three other assays, the percentage of P. jirovecii DNA detection ranged from 8.6% (MycAssay PCR) to 20.0% (AmpliSens PCR) for BAL samples and from 5.3% (MycAssay and in-house PCR) to 10.5% (AmpliSens PCR) for BA/Spu.
Although CT values were not interpretable for an absolute quantification, as no standard curve was generated, the distribution of a positive CT for each method and clinical category was reported. Regarding the CT results of positive samples, the median was slightly lower for the proven PCP category independently of the assay performed (Table 3). Figure 1 shows that, depending on the PCR assay used, a wide range of positive CT values within a category may be obtained. For example, in the proven PCP category, CT values ranged from 16.97 to 32.63. We also observed that for a given patient, CT values were different with the four assays (often superior to 3 cycles).
DISCUSSION
PCP is an opportunistic and frequent infection among HIV-infected patients, but its incidence in non-HIV immunocompromised patients is increasing. Although microscopic detection of trophic forms or cysts in pulmonary samples continues to be the gold standard, the use of highly sensitive techniques, such as real-time PCR, has largely improved PCP diagnosis. Indeed, real-time PCR is currently considered an essential method for PCP diagnosis (14, 15).
The main objective of this study was to evaluate the concordance of four real-time PCR assays (three commercial kits and an in-house PCR) for the detection of P. jirovecii DNA (11). Only two commercial kits, MycAssay and Bio-Evolution PCR, have been previously evaluated in the literature (5, 7–9). To our knowledge, no comparison of these four assays is available. Our data showed that concordance calculated with interpreted results (positive/negative PCR) is excellent between 3 assays: the in-house assay, AmpliSens, and MycAssay. Performances of real-time PCR were also evaluated in a clinical and a radiological context.
Several groups have proposed to classify patients according to the probability of having PCP (5, 16–18). Criteria analyzed included the presence of risk factors, clinical and radiological signs, outcomes of anti-P. jirovecii treatment, and the results of DE of respiratory samples. In our study, patients were classified into four categories according to the criteria described in the Materials and Methods. In the proven PCP group, all samples were PCR positive independently of the PCR assay. Considering the probable PCP group only, the AmpliSens, in-house, and MycAssay PCR assays confirmed the diagnosis of PCP in 100% of cases while the Bio-Evolution PCR confirmed the diagnosis of PCP in 92% of cases. This highlights the better sensitivity of PCR and its interest for PCP diagnosis confirmation using BAL fluid and BA/Spu. For the possible PCP group, the rate of PCR-positive samples varied from 54.5% (Bio-Evolution PCR) to 86.4% (AmpliSens PCR). Interpretation of PCR results in this group is more complex and must take into account the patient's clinical condition, radiological data, the type of sample tested, and the CT value (see below). Finally, for the no final diagnosis of PCP group, the rate of positive PCR ranged from 0% (Bio-Evolution PCR) to 16.7% (AmpliSens PCR), and patients with a positive PCR were considered colonized. Indeed, real-time PCR has allowed for the introduction of the concept of colonized patients, corresponding to detection of P. jirovecii DNA in respiratory specimens in patients without clinical and radiological signs of PCP (19). The proportion of patients colonized by P. jirovecii varies among studies, PCR assays, and underlying risk factors (17, 19, 20). The fungal burden was lower in colonized patients compared to that in patients with PCP (5, 6, 16–18, 21–23). To date, the clinical course of colonized patients and their involvement in epidemiology are not clear. Colonization may promote the occurrence of PCP or play a role as reservoir in the transmission of P. jirovecii; therefore, treatment and isolation of hospitalized colonized patients need to be evaluated (reviewed in reference 4). In this work, among the four PCR assays evaluated, the Bio-Evolution kit was not suitable for colonization detection but showed good performance in the proven and probable groups (94.6% of positive samples). For the three other methods, performances were excellent and allowed the detection of very low fungal burdens. To facilitate the interpretation of PCR results and to differentiate PCP from colonization, cutoff values of P. jirovecii DNA copy number or CT values were suggested (6, 11, 16, 17, 21–24). However, as the fungal load during PCP can vary according to the population of the patients tested (e.g., HIV infected versus non-HIV infected) and is often higher in HIV-infected patients (5, 17, 18), cutoffs need to be adapted to the population. Nevertheless, the lack of standardization of pulmonary samples (volume, number of cells, etc.) and the variability among the PCR assays does not allow for the comparison of cutoff values among studies. In our study, the wide dispersion in the distribution of CT values for the four PCR assays and for each category and the lack of a standard curve does not allow for the establishment of cutoffs (Fig. 1). Nevertheless, quantification or semiquantification of PCR results remains important for bioclinical interpretation; in addition, some authors have proposed to associate the detection of β-d-glucans in serum to refine the diagnosis of PCP and guide therapeutic decision making (17, 21, 22).
Conclusion.
Finally, three out of four PCR assays had a high concordance rate and similar performance; these were relevant for the confirmation of PCP or for the detection of colonized patients. The fourth assay evaluated showed a lower concordance rate but remains effective for PCP diagnosis. This work highlights once again the need for the implementation of a molecular diagnostic method when DE is negative. Commercial kits show excellent performances for PCP diagnosis. Nevertheless, a positive PCR result must necessarily be interpreted in clinical context. Standardized rules of PCR results interpretation are required and must include type and quality of lung sampling, PCR technique, and the entire clinical and radiological context.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mariella Lomma for revision of the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02876-15.
REFERENCES
- 1.Catherinot E, Lanternier F, Bougnoux M-E, Lecuit M, Couderc L-J, Lortholary O. 2010. Pneumocystis jirovecii pneumonia. Infect Dis Clin North Am 24:107–138. doi: 10.1016/j.idc.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Roux A, Canet E, Valade S, Gangneux-Robert F, Hamane S, Lafabrie A, Maubon D, Debourgogne A, Le Gal S, Dalle F, Leterrier M, Toubas D, Pomares C, Bellanger AP, Bonhomme J, Berry A, Durand-Joly I, Magne D, Pons D, Hennequin C, Maury E, Roux P, Azoulay ´ E. 2014. Pneumocystis jirovecii pneumonia in patients with or without AIDS, France. Emerg Infect Dis 20:1490–1497. doi: 10.3201/eid2009.131668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maini R, Henderson KL, Sheridan EA, Lamagni T, Nichols G, Delpech V, Phin N. 2013. Increasing Pneumocystis pneumonia, England, UK, 2000–2010. Emerg Infect Dis 19:386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris A, Norris KA. 2012. Colonization by Pneumocystis jirovecii and its role in disease. Clin Microbiol Rev 25:297–317. doi: 10.1128/CMR.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauser PM, Bille J, Lass-Flörl C, Geltner C, Feldmesser M, Levi M, Patel H, Muggia V, Alexander B, Hughes M, Follett SA, Cui X, Leung F, Morgan G, Moody A, Perlin DS, Denning DW. 2011. Multicenter, prospective clinical evaluation of respiratory samples from subjects at risk for Pneumocystis jirovecii infection by use of a commercial real-time PCR assay. J Clin Microbiol 49:1872–1878. doi: 10.1128/JCM.02390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mühlethaler K, Bögli-Stuber K, Wasmer S, von Garnier C, Dumont P, Rauch A, Mühlemann K, Garzoni C. 2012. Quantitative PCR to diagnose Pneumocystis pneumonia in immunocompromised non-HIV patients. Eur Respir J 39:971–978. doi: 10.1183/09031936.00095811. [DOI] [PubMed] [Google Scholar]
- 7.Orsi CF, Gennari W, Venturelli C, La Regina A, Pecorari M, Righi E, Machetti M, Blasi E. 2012. Performance of 2 commercial real-time polymerase chain reaction assays for the detection of Aspergillus and Pneumocystis DNA in bronchoalveolar lavage fluid samples from critical care patients. Diagn Microbiol Infect Dis 73:138–143. doi: 10.1016/j.diagmicrobio.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 8.McTaggart LR, Wengenack NL, Richardson SE. 2012. Validation of the MycAssay Pneumocystis kit for detection of Pneumocystis jirovecii in bronchoalveolar lavage specimens by comparison to a laboratory standard of direct immunofluorescence microscopy, real-time PCR, or conventional PCR. J Clin Microbiol 50:1856–1859. doi: 10.1128/JCM.05880-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montesinos I, Brancart F, Schepers K, Jacobs F, Denis O, Delforge M-L. 2015. Comparison of 2 real-time PCR assays for diagnosis of Pneumocystis jirovecii pneumonia in human immunodeficiency virus (HIV) and non-HIV immunocompromised patients. Diagn Microbiol Infect Dis 82:143–147. doi: 10.1016/j.diagmicrobio.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Orsi CF, Bettua C, Pini P, Venturelli C, La Regina A, Morace G, Luppi M, Forghieri F, Bigliardi S, Luppi F, Codeluppi M, Girardis M, Blasi E. 2015. Detection of Pneumocystis jirovecii and Aspergillus spp. DNA in bronchoalveolar lavage fluids by commercial real-time PCR assays: comparison with conventional diagnostic tests. New Microbiol 38:75–84. [PubMed] [Google Scholar]
- 11.Fillaux J, Malvy S, Alvarez M, Fabre R, Cassaing S, Marchou B, Linas M-D, Berry A. 2008. Accuracy of a routine real-time PCR assay for the diagnosis of Pneumocystis jirovecii pneumonia. J Microbiol Methods 75:258–261. doi: 10.1016/j.mimet.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Lachaud L, Sasso M, Dumas-Chastang E, Boutet-Dubois A, Bourgeois A, Lechiche C. 2013. Abstr 6th Trends in Medical Mycology, abstr P061. [Google Scholar]
- 13.Landis JR, Koch GG. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 14.Fan L-C, Lu H-W, Cheng K-B, Li H-P, Xu J-F. 2013. Evaluation of PCR in bronchoalveolar lavage fluid for diagnosis of Pneumocystis jirovecii pneumonia: a bivariate meta-analysis and systematic review. PLoS One 8(9):e73099. doi: 10.1371/journal.pone.0073099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summah H, Zhu Y-G, Falagas ME, Vouloumanou EK, Qu J-M. 2013. Use of real-time polymerase chain reaction for the diagnosis of Pneumocystis pneumonia in immunocompromised patients: a meta-analysis. Chin Med J (Engl) 126:1965–1973. [PubMed] [Google Scholar]
- 16.Maillet M, Maubon D, Brion JP, François P, Molina L, Stahl JP, Epaulard O, Bosseray A, Pavese P. 2014. Pneumocystis jirovecii (Pj) quantitative PCR to differentiate Pj pneumonia from Pj colonization in immunocompromised patients. Eur J Clin Microbiol Infect Dis 33:331–336. doi: 10.1007/s10096-013-1960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alanio A, Desoubeaux G, Sarfati C, Hamane S, Bergeron A, Azoulay E, Molina JM, Derouin F, Menotti J. 2011. Real-time PCR assay-based strategy for differentiation between active Pneumocystis jirovecii pneumonia and colonization in immunocompromised patients. Clin Microbiol Infect 17:1531–1537. doi: 10.1111/j.1469-0691.2010.03400.x. [DOI] [PubMed] [Google Scholar]
- 18.Robert-Gangneux F, Belaz S, Revest M, Tattevin P, Jouneau S, Decaux O, Chevrier S, Le Tulzo Y, Gangneux J-P. 2014. Diagnosis of Pneumocystis jirovecii pneumonia in immunocompromised patients by real-time PCR: a 4-year prospective study. J Clin Microbiol 52:3370–3376. doi: 10.1128/JCM.01480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris A, Wei K, Afshar K, Huang L. 2008. Epidemiology and clinical significance of Pneumocystis colonization. J Infect Dis 197:10–17. doi: 10.1086/523814. [DOI] [PubMed] [Google Scholar]
- 20.Gutiérrez S, Respaldiza N, Campano E, Martínez-Risquez MT, Calderón EJ, De La Horra C. 2011. Pneumocystis jirovecii colonization in chronic pulmonary disease. Parasite 18:121–126. doi: 10.1051/parasite/2011182121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damiani C, Le Gal S, Da Costa C, Virmaux M, Nevez G, Totet A. 2013. Combined quantification of pulmonary Pneumocystis jirovecii DNA and serum (1→3)-β-d-glucan for differential diagnosis of Pneumocystis pneumonia and Pneumocystis colonization. J Clin Microbiol 51:3380–3388. doi: 10.1128/JCM.01554-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumura Y, Ito Y, Iinuma Y, Yasuma K, Yamamoto M, Matsushima A, Nagao M, Takakura S, Ichiyama S. 2012. Quantitative real-time PCR and the (1→3)-β-d-glucan assay for differentiation between Pneumocystis jirovecii pneumonia and colonization. Clin Microbiol Infect 18:591–597. doi: 10.1111/j.1469-0691.2011.03605.x. [DOI] [PubMed] [Google Scholar]
- 23.Fujisawa T, Suda T, Matsuda H, Inui N, Nakamura Y, Sato J, Toyoshima M, Nakano Y, Yasuda K, Gemma H, Hayakawa H, Chida K. 2009. Real-time PCR is more specific than conventional PCR for induced sputum diagnosis of Pneumocystis pneumonia in immunocompromised patients without HIV infection. Respirology 14:203–209. doi: 10.1111/j.1440-1843.2008.01457.x. [DOI] [PubMed] [Google Scholar]
- 24.Flori P, Bellete B, Durand F, Raberin H, Cazorla C, Hafid J, Lucht F, Sung RTM. 2004. Comparison between real-time PCR, conventional PCR and different staining techniques for diagnosing Pneumocystis jirovecii pneumonia from bronchoalveolar lavage specimens. J Med Microbiol 53:603–607. doi: 10.1099/jmm.0.45528-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.