Abstract
Molecular diagnostics that rapidly and accurately predict resistance to fluoroquinolone drugs and especially later-generation agents promise to improve treatment outcomes for patients with multidrug-resistant tuberculosis and prevent the spread of disease. Mutations in the gyr genes are known to confer most fluoroquinolone resistance, but knowledge about the effects of gyr mutations on susceptibility to early- versus later-generation fluoroquinolones and about the role of mutation-mutation interactions is limited. Here, we sequenced the full gyrA and gyrB open reading frames in 240 multidrug-resistant and extensively drug-resistant tuberculosis strains and quantified their ofloxacin and moxifloxacin MIC by testing growth at six concentrations for each drug. We constructed a multivariate regression model to assess both the individual mutation effects and interactions on the drug MICs. We found that gyrB mutations contribute to fluoroquinolone resistance both individually and through interactions with gyrA mutations. These effects were statistically significant. In these clinical isolates, several gyrA and gyrB mutations conferred different levels of resistance to ofloxacin and moxifloxacin. Consideration of gyr mutation combinations during the interpretation of molecular test results may improve the accuracy of predicting the fluoroquinolone resistance phenotype. Further, the differential effects of gyr mutations on the activity of early- and later-generation fluoroquinolones requires further investigation and could inform the selection of a fluoroquinolone for treatment.
INTRODUCTION
Global surveillance for drug-resistant tuberculosis (TB) suggests that at least 3.5% of the 9 million incident TB cases are multidrug resistant (MDR), i.e., resistant to isoniazid and rifampin. Fluoroquinolones are among the most effective drugs available for the treatment of MDR TB (1, 2). The importance of the fluoroquinolone drug class is implied in how extensively drug-resistant (XDR) TB is defined, i.e., as MDR TB with additional in vitro resistance to second-line injectables and any member of the fluoroquinolone drug class. The different fluoroquinolone agents vary in their potency against Mycobacterium tuberculosis, with third- and fourth-generation agents (e.g., levofloxacin, gatifloxacin, and moxifloxacin) having higher activity than second-generation agents (e.g., ciprofloxacin or ofloxacin) (3, 4). These differences have led the World Health Organization (WHO) to recommend against extrapolating in vitro culture-based resistance results from second- to later-generation fluoroquinolones and to revise the in vitro testing drug concentration upward to 2 mg/liter for moxifloxacin (4, 5). Revisions of drug testing concentrations also highlight the need for quantitative resistance testing with MIC measurements when new diagnostic technologies are being compared to culture-based drug susceptibility tests.
Fluoroquinolones exert their antibacterial activity by inhibiting DNA gyrase and topoisomerase IV, limiting the cell's capacity for DNA replication and transcription. M. tuberculosis lacks topoisomerase IV, and its gyrase consists of two subunits encoded by the genes gyrA and gyrB (6, 7). Mutations in these genes, particularly in the gyrA quinolone resistance-determining region (QRDR), are the main cause of fluoroquinolone resistance in M. tuberculosis (8). In addition to mutations that confer resistance, the gyrA QRDR also harbors neutral mutations that do not confer resistance (6–9).
Molecular diagnostics for isoniazid and rifampin resistance are now available, with advantages over conventional culture-based drug susceptibility (7, 10, 11). As these tests can be performed directly on sputum, their use does not require the biosafety facilities needed for conventional culture. They can be performed by relatively unskilled workers, and results can be available in 90 min (12). Molecular technique-based diagnostics that detect gyrA QRDR mutations have been developed, but the sensitivity for predicting phenotypic fluoroquinolone resistance is highly variable, ranging from 69% to 99% (13). In addition, studies often report aggregate test performance, pooling sensitivity and specificity across early- and later-generation fluoroquinolone agents, and infrequently report on MIC measurements (14). To improve the accuracy of molecular diagnostics, detailed analyses assessing differential effects of the gyr mutations on resistance to early- versus late-generation fluoroquinolones is important, as resistance to the latter more active agents is the increasingly clinically relevant readout. In addition, the identification of more than one mutation in the gyr genes in a given isolate is not uncommon, and although there is compelling evidence that these can affect the MIC in laboratory strains (15) and in other bacterial systems (16), the role of mutation additive effects and mutation-mutation interactions (17, 18) in clinical isolates has been rarely reported on. Because the incorporation of any additive and interaction effects can improve the prediction of fluoroquinolone resistance from the gyr genotype, we sought to systematically examine these associations in 240 MDR and XDR clinical M. tuberculosis isolates. Here, we performed quantitative resistance phenotyping with MIC measurements for both early- and late-generation fluoroquinolones (specifically, ofloxacin and moxifloxacin) and performed targeted sequencing of the entire open reading frames and promoter regions of the gyr genes.
MATERIALS AND METHODS
Ethics review.
This study was deemed not to constitute human subject research by the Partners Health Care Human Research Committee.
Strain selection and MIC testing.
Using an archive of M. tuberculosis isolates from patients referred for individualized M/XDR TB treatment in Lima, Peru, between 1 February 1997 and 31 July 2003 (1), we selected isolates that had undergone ciprofloxacin drug susceptibility testing (DST) using the indirect agar proportion method on 7H10 medium (19). In 2001, the testing laboratory altered its standard ciprofloxacin critical concentration, reducing it from 2 mg/liter to 1 mg/liter. We randomly selected 175 ciprofloxacin-resistant and 100 ciprofloxacin-sensitive isolates from this archive for ofloxacin and moxifloxacin MIC testing using the indirect proportion method on 7H10 agar. The concentrations tested were 1.0, 2.0, 4.0, 6.0, 8.0, and 10.0 mg/liter ofloxacin and 0.125, 0.25, 0.5, 1.0, 4.0, and 8.0 mg/liter moxifloxacin for each isolate. At the time these concentrations were chosen, the WHO's revised critical concentration of 2.0 mg/liter for moxifloxacin had not yet been adopted. Colony counts were recorded for the control and for dilutions of 10−2 and 10−4 at each of the 6 drug concentrations. For quality control, we selected a subset of isolates for repeat MIC testing. The repeat MIC results were retained for all data analysis.
Sequencing.
In brief the gyrA and gyrB gene sequences were captured using molecular inversion probes (MIPs) (20) designed to cover both DNA strands of the open reading frames, promoter regions, and 100 flanking bases on either side of the selected genes (primers are listed in Table S1 in the supplemental material). DNA was extracted from cultures using the cetyltrimethylammonium bromide (CTAB) method (21) without colony selection. Barcodes and Illumina adapters were attached to the captured sequences during the amplification phase to allow for parallel sequencing. We sequenced the amplified targets on an Illumina GAIIx device, producing raw reads of 75 nucleotides. We repeated this process on strains for which fewer than 95% of the targeted nucleotide positions were covered by at least 20 reads, and we retained in the analysis only those resequenced strains that met these criteria. We called mutations using Bowtie (22) 0.12.7/SAMtools (23) 0.1.18 using the thresholds listed in Table S2 in the supplemental material. We excluded any synonymous mutations and the polymorphisms E21Q, T80A, S95T, G247S, and G668D in gyrA, as they are either recognized to be neutral variants or are known markers of genetic lineage (13, 24, 25). Mutations in gyrB were numbered based on the http://tuberculist.epfl.ch/quicksearch.php?gene+name=gyrB&submit=Search, similar to what was reported by Malik et al. (15).
Statistical analyses.
We treated the MIC as a continuous variable, as has been done previously (26). For MICs measured at the extremes of the concentrations tested, we used the agar colony counts to predict the MIC using linear extrapolation (“approximately” function in R v 3.1.3). The extrapolated concentrations were highly similar across different strains. For an ofloxacin MIC of ≤1 mg/liter, we used an extrapolated concentration of 0.99 mg/liter; for an ofloxacin MIC of >10 mg/liter, we used 10.025 mg/liter; for a moxifloxacin MIC of ≤0.125 mg/liter, we used 0.12375 mg/liter; and for a moxifloxacin MIC of >8 mg/liter, we used 8.5 mg/liter. We performed a sensitivity analysis by repeating the univariate and multivariate model construction described below and altering the value assigned to the extreme MICs.
We used the Wilcoxon rank sum test using R version 3.1.3 for univariate association testing of the gyr mutation with the MIC. As MIC distributions are recognized to be Gaussian on a 2-fold log scale (27), for the multivariate analysis, we transformed the MIC measures to the log2 scale and performed linear regression. All nonsynonymous gyr mutations excluding gyrA E21Q, T80A, S95T, G247S, and G668D were included in the multivariate model. A multiplicative interaction term was added for each pair of mutations only if two or more strains carried the mutation pair (4 interaction pairs). We applied forward, backward, and bidirectional stepwise selection using the Akaike information criterion (AIC) to arrive at the final model. We performed a generalized linear F test to assess whether coefficients of gyrA D94G, A90V, and D94A were equal in the moxifloxacin model. Throughout, we used a P value cutoff of 0.01 to assess statistical significance.
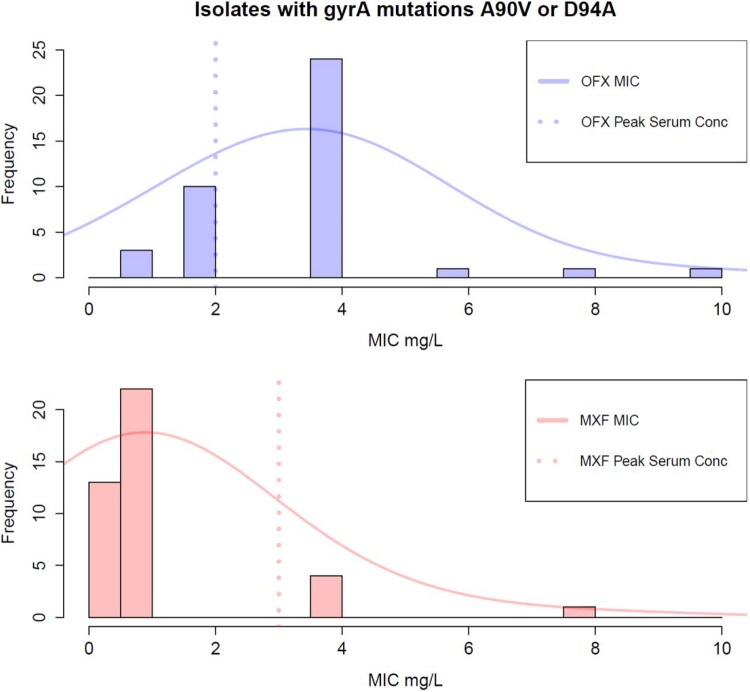
We estimated the MIC distribution curves as follows. Using the observed MICs for the pooled isolates harboring either gyrA A90V or D94A, we computed a Gaussian kernel density with a standard deviation (bandwidth) of two using the “density” function in R. We computed this separately for each drug.
Fingerprinting and phylogeny construction.
Molecular fingerprinting by either spoligotyping or IS6110 restriction fragment length polymorphism (RFLP) analysis was performed for 44 of the isolates using standard methods (28, 29), and lineages were identified by comparison with those from publically available databases (30). For all isolates, data from 28 drug resistance genetic loci, 44,997 kbp, was available from a previous study (M. R. Farhat, S. Razvan, S. Boseman, O. Iartchouk, J. Galagan, P. Sisk, H. Nebenzahl-Guimaraes, K. Jocobson, A. Sloutsky, D. Kaur, J. Posey, B. N. Kreiswirth, N. Kurpina, L. Rigouts, E. M. Streicher, T. C. Victor, R. M. Warren, D. van Soolingen, and M. Murray, submitted for publication). These data, after exclusion of variants presumed to be associated with drug resistance (31), were used to construct and annotate a neighbor-joining tree with the Phylip (32) Neighbor program and Figtree v1.4.0. We also determined the presence or absence of 23 lineage-defining SNPs (33, 34) in these loci (see Table S3 in the supplemental material).
RESULTS
Of the 275 MDR TB isolates, 35 duplicate samples from the same patient were excluded. Among the remaining 240, 76 were previously determined to be ciprofloxacin sensitive and 165 were ciprofloxacin resistant. For quality control, we repeated MIC testing on a randomly selected subset of 20 isolates. Sixteen isolates were reconfirmed to have the exact moxifloxacin and ofloxacin MIC. For two isolates, the measured moxifloxacin MIC increased from 0.250 mg/liter to 0.5 mg/liter. For one, the moxifloxacin MIC changed from ≤0.125 to 0.250 mg/liter, and for the last, the moxifloxacin MIC decreased from 8 to 4 and the ofloxacin MIC changed from 8 to 6 mg/liter.
Across the 240 isolates, 96% of the gyr gene bases were covered with 20 or more reads, and 88% were covered with more than 100 reads. The isolates were genetically diverse, and both the Euro-African American lineages (LAM, T, H, and X) and the East Asian lineage (Beijing) were represented (see Fig. S1 and Table S3 in the supplemental material).
We found 37 distinct mutations in the gyr genes, all single nucleotide substitutions, 29 (76%) of which were nonsynonymous changes; 15 were located in gyrA and 14 were in gyrB. We found no variants in the gyr promoter region. The mutations most strongly associated with ofloxacin and moxifloxacin resistance on univariate analyses were the gyrA D94G, A90V, and D94Y mutations and the gyrB V340L and N538T mutations (Table 1). The gyrA D94A and A90V mutations were associated with median MICs of 2 and 4 mg/liter, respectively, for ofloxacin and with a median moxifloxacin MIC of 1 mg/liter. We found 17 mutations outside the QRDRs (Table 1).
TABLE 1.
Mutations in gyr genes by drug and their univariate association P value using the Wiloxon rank sum testa
| Gene | Mutation | Amino acid change | Mutation frequency (of 240) | Ofloxacinb |
Moxifloxacinb |
||||
|---|---|---|---|---|---|---|---|---|---|
| Median MIC (IQR) | MIC (min, max) | P | Median MIC (IQR) | MIC (min, max) | P | ||||
| gyrA | 7582AG | D94G | 32 | 6.0 (4.0, 6.0) | <1.0, >10.0 | <0.001 | 4.0 (1.0, 4.0) | 0.250, >8.0 | <0.001 |
| gyrA | 7570CT | A90V | 26 | 4.0 (4.0, 4.0) | <1.0, 10.0 | <0.001 | 1.0 (0.6, 1.0) | <0.125, 8.0 | <0.001 |
| gyrB | 6052TG | I310M | 16 | <1.0 (<1.0, 2.5) | <1.0, >10.0 | 0.7 | 0.25 (<0.125, 1.0) | <0.125, 8.0 | 0.7 |
| gyrB | 7221GC | S700T | 16 | <1.0 (<1.0, 2.5) | <1.0, >10.0 | 0.7 | 0.25 (<0.125, 1.0) | <0.125, 8.0 | 0.7 |
| gyrA | 7582AC | D94A | 14 | 2.0 (2.0, 4.0) | <1.0, 8.0 | 0.006 | 1.0 (0.50, 1.0) | 0.250, 4.0 | 0.02 |
| gyrB | 6140GT | V340L | 11 | 4.0 (2.0, 4.0) | <1.0, >10.0 | 0.01 | 1.0 (0.8, 4.0) | 0.250, 4.0 | 0.003 |
| gyrA | 7581GT | D94Y | 5 | 6.0(4.0, 8.0) | 4.0, >10.0 | 0.001 | 4.0 (1.0, 8.0) | 1.0, 8.0 | 0.003 |
| gyrB | 6735AC | N538T | 4 | >10 (7.8, >10) | <1.0, >10.0 | 0.01 | 8.0 (6.1, 8.0) | 0.25, >8.0 | 0.04 |
| gyrA | 7572TC | S91P | 3 | 4.0 (3.0, 4.0) | 2.0, 4.0 | 0.13 | 4.0 (2.5, 4.0) | 1.0,4.0 | 0.03 |
| gyrA | 7566GA | D89N | 3 | <1.0 (<1.0, 2.5) | <1.0, 4.0 | 0.7 | 4.0 (2.5, 4.0) | 1.0, 4.0 | 0.03 |
| gyrA | 7581GA | D94N | 2 | 8.0 | 6.0, >10.0 | 0.01 | 6.1 | 4.0, >8.0 | 0.02 |
| gyrA | 8101CT | T267I | 2 | 4.0 | 4.0,4.0 | 0.1 | 1.0 | 1.0, 1.0 | 0.2 |
| gyrA | 8475CT | R392C | 2 | 3.5 | <1.0, 6.0 | 0.6 | 2.1 | 0.250, 4.0 | 0.4 |
| gyrA | 7521GT | A74S | 1 | <1.0 | 0.4 | 1.0 | 0.4 | ||
| gyrA | 7605CT | P102S | 1 | 4.0 | 0.3 | 1.0 | 0.4 | ||
| gyrA | 7684GA | R128K | 1 | 2.0 | 0.6 | 0.5 | 0.8 | ||
| gyrA | 8164CA | A288D | 1 | 4.0 | 0.3 | 1.0 | 0.4 | ||
| gyrA | 9111CT | P604S | 1 | <1.0 | 0.4 | <0.125 | 0.2 | ||
| gyrB | 5354GA | E78K | 1 | 2.0 | 0.6 | <0.125 | 0.2 | ||
| gyrB | 6479GT | A453S | 1 | <1.0 | 0.4 | 0.250 | 0.9 | ||
| gyrB | 6575CT | R485C | 1 | 2.0 | 0.6 | <0.125 | 0.2 | ||
| gyrB | 6576GA | R485H | 1 | <1.0 | 0.4 | 1.0 | 0.4 | ||
| gyrB | 6579CA | S486Y | 1 | <1.0 | 0.4 | 1.0 | 0.4 | ||
| gyrB | 6734AG | N538D | 1 | 4.0 | 0.3 | 4.0 | 0.2 | ||
| gyrB | 6737AG | T539A | 1 | 4.0 | 0.3 | 0.250 | 0.9 | ||
| gyrB | 6742AC | E540D | 1 | <1.0 | 0.4 | 4.0 | 0.2 | ||
| gyrB | 7158AC | D679A | 1 | <1.0 | 0.4 | <0.125 | 0.2 | ||
| gyrB | 7163GC | A681P | 1 | 6.0 | 0.1 | 4.0 | 0.2 | ||
Mutations are ordered by their observed frequency. In bold are QRDR mutations as defined by Tagliani et al. (35). MICs are in milligrams per liter. IQR, interquartile range.
Values are for all isolates with the indicated mutation.
We estimated the association between MIC and gyr mutations and their pairwise interactions through multivariate regression (Table 2). As 31% (35/112) of the isolates with gyr mutations harbored more than one mutation (see Table S4 in the supplemental material), the multivariate models allowed us to isolate and compare the effects of each mutation on the moxifloxacin and ofloxacin MICs. Several gyrA mutations were associated with a higher MIC of both ofloxacin and moxifloxacin. The gyrA D89N mutation, however, was significantly associated with an increase in the moxifloxacin MIC but was not significantly associated with a change in the ofloxacin MIC in both univariate and multivariate analyses.
TABLE 2.
Linear regression multivariate model results for log2 transformed MICsa
| Drug, gene, and mutation | Log2 MIC change (95% CI) | P |
|---|---|---|
| Ofloxacin | ||
| gyrA D94G | 1.8 (1.5, 2.1) | <0.0001* |
| gyrA A90V | 1.6 (1.3, 1.9) | <0.0001* |
| gyrA D94Y | 2.4 (1.8, 3.0) | <0.0001* |
| gyrA D94A | 1.1 (0.7, 1.5) | <0.0001* |
| gyrA D94N | 2.7 (1.6, 3.8) | <0.0001* |
| gyrB A681P | 2.4 (1.1, 3.8) | 0.0006* |
| gyrA A288D | 1.8 (0.5, 3.2) | 0.009* |
| gyrB T539A | 1.8 (0.5, 3.2) | 0.009* |
| gyrB N538D | 1.8 (0.5, 3.2) | 0.009* |
| gyrA S91P | 0.9 (0.1, 1.7) | 0.03 |
| gyrA D94G:gyrB N538T | 1.2 (−0.2, 2.7) | 0.10 |
| gyrB N538T | 0.1 (−1.0, 1.2) | 0.83 |
| Moxifloxacin | ||
| gyrA D94G | 3.1 (2.7, 3.5) | <0.0001* |
| gyrA A90V | 2.2 (1.7, 2.7) | <0.0001* |
| gyrA D94Y | 4.3 (3.2, 5.4) | <0.0001* |
| gyrA D89N | 3.7 (2.4, 4.9) | <0.0001* |
| gyrA D94A | 1.7 (1.1, 2.3) | <0.0001* |
| gyrA D94N | 4.6 (2.8, 6.3) | <0.0001* |
| gyrA S91P | 2.6 (1.4, 3.9) | 0.0001* |
| gyrB E540D | 4.3 (2.2, 6.5) | 0.0001* |
| gyrB A681P | 4.3 (2.2, 6.5) | 0.0001* |
| gyrB N538D | 4.3 (2.2, 6.5) | 0.0001* |
| gyrB E78K | −2.9 (−5.1, −0.7) | 0.01 |
| gyrB V340L | 0.8 (0.1, 1.5) | 0.04 |
| gyrB S486Y | 2.3 (0.1, 4.5) | 0.04 |
| gyrA A74S | 2.3 (0.1, 4.5) | 0.04 |
| gyrB R485H | 2.3 (0.1, 4.5) | 0.04 |
| gyrA A288D | 2.3 (0.1, 4.5) | 0.04 |
| gyrA P102S | −2.0 (−4.4, 0.4) | 0.11 |
| gyrA D94G gyrB N538T | 1.7 (−0.7, 4.1) | 0.16 |
| gyrB N538T | 0.6 (−1.2, 2.3) | 0.54 |
Mutations or their interactions are ordered by their P value for each drug. An asterisk indicates significance at the 0.01 threshold. The values represents the increase in log2 MIC (in milligrams per liter) when the mutation is present versus when it is absent, controlling for the other mutations and interactions listed.
The gyrB mutations N538T, N538D, T539A, E540D, and A681P, although infrequent, were associated with significant effects on the MIC in either the univariate or multivariate analysis (Table 2). The gyrB mutation N538T was the most frequently observed of these mutations, occurring in 4 isolates. Although statistically significant in the univariate analysis, it was no longer significant after adjustment for the presence of other gyr mutations; specifically, 3 of the 4 isolates with gyrB N538T carried a mutation in codon 94 of gyrA. The fourth isolate carried only gyrB N538T and had moxifloxacin and ofloxacin MICs of 0.250 and <1 mg/liter, respectively. Notably, all three isolates that carried gyrB N538T and gyrA codon 94 mutations (1 with D94N and 2 with D94G) had moxifloxacin MICs of ≥8 mg/liter and ofloxacin MICs of ≥10 mg/liter, whereas the remaining 31 isolates that carried D94N or D94G alone had moxifloxacin MICs of ≤4 mg/liter (median, 4 mg/liter) and ofloxacin MICs of ≤10 mg/liter (median, 6 mg/liter). Although the interaction term between gyrB N538T and gyrA D94G was not significant in the multivariate model, including the interaction term improved the models' MIC prediction for both moxifloxacin and ofloxacin. In the multivariate model, the gyrB mutations N538D and A681P were associated with significant elevations in MICs of both drugs; gyrB E540D was significantly associated with an increase in the MIC of moxifloxacin but not to ofloxacin, and gyrB T539A was associated with an increase in ofloxacin MIC but not moxifloxacin MIC. Although the gyrB mutation V340L was individually associated with a higher MIC of both drugs, it was not significantly associated with a higher MIC in the multivariate analysis; we observed that 9 of the 11 isolates that carried this mutation also had a mutation in gyrA (3 with D94G, 5 with A90V, and 1 with D94A). Forward, backward, and bidirectional stepwise selection all yielded an identical final multivariate model for each drug. In the sensitivity analysis, we confirmed that altering the value assigned to the extreme MICs did not change the final MIC models' coefficients by >10% and did not alter their significance.
Of the gyrA QRDR mutations, the mutations A90V and D94A had the smallest effect on the moxifloxacin MIC (log2 MIC change of 2.2 [95% confidence interval {CI}, 1.7 to 2.7] and 1.7 [95% CI, 1.1 to 2.3], respectively). These effects were statistically different from the effect of gyrA D94G, with a 3.1 log2 MIC change (95% CI, 2.7 to 3.5) (F test P values of 0.003 and 0.0005, respectively). For ofloxacin, the mutations A90V and D94A had a comparable effect on the MIC as D94G (F test P values of 0.4 and 0.02, respectively). We estimated the ofloxacin and moxifloxacin MIC distribution for isolates with these gyrA mutations. The MIC distribution's mode was 0.9 mg/liter for moxifloxacin (Fig. 1) and 3.5 mg/liter for ofloxacin. Only 5/40 (13%) of these isolates would be classified as resistant to moxifloxacin (MIC > 2 mg/liter), and 27/40 (68%) would be classified as resistant to ofloxacin (MIC > 2 mg/liter).
FIG 1.
Estimated ofloxacin (OFX) and moxifloxacin (MXF) MIC distributions in isolates with the gyrA mutation A90V or D94A (there were no isolates with both mutations). MIC distributions are superimposed on a histogram of the observed MICs. The MXF MIC peaks lower than the lower limit of the peak MXF serum concentrations (dotted pink line; 3 mg/liter) (41); however, the OFX MIC peaks higher than the peak serum concentration for OFX (dotted blue line; 2 mg/liter) (42).
We determined the percentage of isolates with high moxifloxacin MICs that harbored only gyrB mutations. Of 39 isolates with a moxifloxacin MIC of >2 mg/liter, 13 carried a gyrB mutation. Only four isolates carried a gyrB mutation without a cooccurring QRDR mutation in gyrA. Three of the 39 isolates (8%) carried a gyrB mutation that was associated with resistance in the multivariate analysis (N538D, A681P, and E540D); the fourth isolate carried the gyrB S700T mutation. The gyrB mutations N538D, A681P, and E540D did not occur in any isolate with a moxifloxacin MIC of <2 mg/liter.
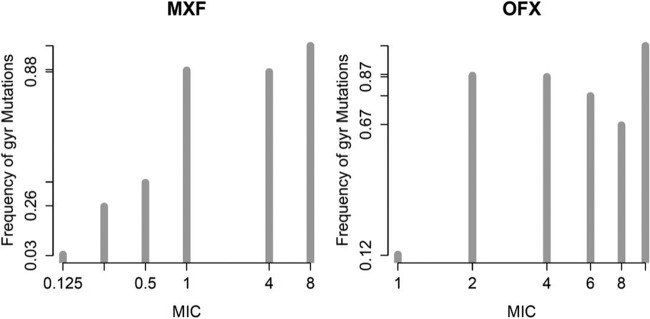
Isolates with higher MICs were more likely to carry a gyr mutation (Fig. 2) and all the isolates within the highest MIC categories (ofloxacin MIC of ≥10 mg/liter, 6 isolates; moxifloxacin MIC of ≥8 mg/liter, 7 isolates) harbored gyr mutations. In isolates with intermediate MICs, there were no gyr mutations found in 17% (12/71) of isolates for ofloxacin (MIC, 4 to 8 mg/liter) and in 12% (9/77) of isolates for moxifloxacin (MIC, 1 to 4 mg/liter). gyrA and gyrB individual mutations, excluding gyrB I310M, V340L, and S700T, which were not significant by multivariate analysis and interaction effects, have sensitivities of 75% and 90% for predicting moxifloxacin MICs of >0.25 and >2 mg/liter, respectively. The specificity of this set of mutations at the respective moxifloxacin MICs is 90% and 70%. Restricting to the gyr QRDR regions tested in version 2.0 of the GenoType MTBDRsl assay (35), the sensitivities are 70% and 87% and the specificities are 93% and 73% for moxifloxacin MICs of >0.25 and >2 mg/liter, respectively.
FIG 2.
Histogram of gyr mutation frequency as a function of MIC by drug. MICs are plotted using a log2 scale. The two extreme MIC bins were collapsed into the adjacent MIC bin. All nonsynonymous mutations were counted except for the lineage gyrA mutations E21Q, T80A, S95T, G247S, and G668D and the gyrB mutations I310M, V340L, and S700T, as the latter 3 were not found to be associated with drug resistance in the multivariate model.
DISCUSSION
Our data support the conclusion that gyr mutations can have differential effects on resistance to early- versus late-generation fluoroquinolones. We found the gyrA mutations A90V and D94A to have a smaller effect on the moxifloxacin MIC than the gyrA D94G mutation, but we did not observe a similar difference for ofloxacin. Previous studies have also found gyrA A90V to have a weaker effect on moxifloxacin resistance, including allelic exchange experiments, where the ofloxacin MIC was found to be in the 2- to 4-mg/liter range and the moxifloxacin MIC was consistently ≤1 mg/liter and sensitive by current WHO standards (36–38). We could not find any experimental or allelic exchange data on the effect of D94A on fluoroquinolone resistance, but this mutation is regarded as a canonical cause for any fluoroquinolone resistance in MTB (38–40). The estimated moxifloxacin MIC distribution for strains carrying either D94A or A90V peaks lower than the expected peak serum concentration (41, 42), suggesting that it may be possible to nevertheless “treat through” this resistance with moxifloxacin but not with ofloxacin (Fig. 1). Studies of dose increases and patient treatment outcome are needed to confirm this.
We also found that gyrA D89N and gyrB E540D cause differential resistance to moxifloxacin in clinical strains; this is supported by previous allelic exchange experiments for gyrB E540D, where resistance to ofloxacin, ciprofloxacin, and levofloxacin was also tested (15). Our data also suggest that interactions between gyrA and gyrB mutations influence the fluoroquinolone MIC. In particular, isolates harboring both gyrB N538T and either gyrA D94G or gyrA D94N may have higher MICs than isolates carrying the gyrA mutations alone. However, we observed these mutation combinations in a small proportion of isolates, and further analysis in larger data sets and perhaps with allelic exchange is needed to confirm this interaction.
In addition, we found that gyrB mutations alone explained 8% of moxifloxacin resistance, and although all the gyrA mutations significantly associated with moxifloxacin resistance were in the QRDR, we found several gyrA mutations outside the QRDR in fluoroquinolone-resistant isolates. We found that isolates with intermediate MICs were less likely to carry gyr mutations than those in the highest-MIC categories. Potential etiologies of the unexplained phenotypic resistance in these isolates include the presence of rarer subpopulations of bacilli that carry gyr mutations that were not detected despite the high depth of sequencing. Alternatively, other genetic mechanisms, including mutation-mutation interactions outside the gyr regions may explain fluoroquinolone resistance in these strains. Alterations in both efflux pump function and the DNA gyrase-associated protein MfpA have been proposed, but neither has been confirmed to be a relevant cause of fluoroquinolone resistance in clinical isolates (13).
Overall, our study highlights the complexity of phenotypic resistance prediction from the gyr genotype. Capturing the differential effect of the mutations on early- and later-generation fluoroquinolone agents will require an improved interpretation of the molecular test results, one that is drug specific and perhaps classifies mutations into three or more risk categories for resistance. If future research, especially studies that involve the whole-genome sequences, reveals and confirms the role of mutation-mutation interactions, more complex statistical models of predicting resistance may be required to help improve the genotype sensitivity for predicting the fluoroquinolone resistance phenotype.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Peruvian team for their patient care and for providing the clinical isolates that made this study possible.
M.R.F and K.R.J. designed the study with key contributions from C.D.M. and M.M. A.S. and D.K. maintained the strain bank and performed the MIC measurements. M.R.F. performed the data analysis with key input from M.F.F. and wrote the first draft of the manuscript. M.M., C.D.M., M.F.F., and K.R.J. provided key edits of the manuscript.
This work was supported by the Harvard Center for AIDS Research (CFAR), a NIH-funded program supported by the following NIH Co-Funding and Participating Institutes and Centers: NIAID, NCI, NICHD, NHLBI, NIDA, NIMH, NIA, NIDDK, NIGMS, FIC, and OAR.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02775-15.
REFERENCES
- 1.Mitnick CD, Franke MF, Rich ML, Alcantara Viru FA, Appleton SC, Atwood SS, Bayona JN, Bonilla CA, Chalco K, Fraser HSF, Furin JJ, Guerra D, Hurtado RM, Joseph K, Llaro K, Mestanza L, Mukherjee JS, Muñoz M, Palacios E, Sanchez E, Seung KJ, Shin SS, Sloutsky A, Tolman AW, Becerra MC. 2013. Aggressive regimens for multidrug-resistant tuberculosis decrease all-cause mortality. PLoS One 8:e58664. doi: 10.1371/journal.pone.0058664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moadebi S, Harder CK, Fitzgerald MJ, Elwood KR, Marra F. 2007. Fluoroquinolones for the treatment of pulmonary tuberculosis. Drugs 67:2077–2099. doi: 10.2165/00003495-200767140-00007. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson KR, Tierney DB, Jeon CY, Mitnick CD, Murray MB. 2010. Treatment outcomes among patients with extensively drug-resistant tuberculosis: systematic review and meta-analysis. Clin Infect Dis 51:6–14. doi: 10.1086/653115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farhat MR, Mitnick CD, Franke MF, Kaur D, Sloutsky A, Murray M, Jacobson KR. 2015. Concordance of Mycobacterium tuberculosis fluoroquinolone resistance testing: implications for treatment. Int J Tuberc Lung Dis 19:339–341. doi: 10.5588/ijtld.14.0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. 2014. Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 6.Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E, Aubry A. 2012. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother 67:819–831. doi: 10.1093/jac/dkr566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillemann D, Rüsch-Gerdes S, Richter E. 2009. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 47:1767–1772. doi: 10.1128/JCM.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aubry A, Veziris N, Cambau E, Truffot-Pernot C, Jarlier V, Fisher LM. 2006. Novel gyrase mutations in quinolone-resistant and -hypersusceptible clinical isolates of Mycobacterium tuberculosis: functional analysis of mutant enzymes. Antimicrob Agents Chemother 50:104–112. doi: 10.1128/AAC.50.1.104-112.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feuerriegel S, Cox HS, Zarkua N, Karimovich HA, Braker K, Rüsch-Gerdes S, Niemann S. 2009. Sequence analyses of just four genes to detect extensively drug-resistant Mycobacterium tuberculosis strains in multidrug-resistant tuberculosis patients undergoing treatment. Antimicrob Agents Chemother 53:3353–3356. doi: 10.1128/AAC.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brossier F, Veziris N, Aubry A, Jarlier V, Sougakoff W. 2010. Detection by GenoType MTBDRsl test of complex mechanisms of resistance to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J Clin Microbiol 48:1683–1689. doi: 10.1128/JCM.01947-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small PM, Pai M. 2010. Tuberculosis diagnosis—time for a game change. N Engl J Med 363:1070–1071. doi: 10.1056/NEJMe1008496. [DOI] [PubMed] [Google Scholar]
- 13.Miotto P, Cirillo DM, Migliori GB. 2015. Drug resistance in Mycobacterium tuberculosis: molecular mechanisms challenging fluoroquinolones and pyrazinamide effectiveness. Chest 147:1135–1143. doi: 10.1378/chest.14-1286. [DOI] [PubMed] [Google Scholar]
- 14.Molina-Moya B, Lacoma A, Prat C, Pimkina E, Diaz J, García-Sierra N, Haba L, Maldonado J, Samper S, Ruiz-Manzano J, Ausina V, Dominguez J. 2015. Diagnostic accuracy study of multiplex PCR for detecting tuberculosis drug resistance. J Infect 71:220–230. doi: 10.1016/j.jinf.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Malik S, Willby M, Sikes D, Tsodikov OV, Posey JE. 2012. New insights into fluoroquinolone resistance in Mycobacterium tuberculosis: functional genetic analysis of gyrA and gyrB mutations. PLoS One 7:e39754. doi: 10.1371/journal.pone.0039754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344:1519–1522. doi: 10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips PC. 2008. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woods RJ, Barrick JE, Cooper TF, Shrestha U, Kauth MR, Lenski RE. 2011. Second-order selection for evolvability in a large Escherichia coli population. Science 331:1433–1436. doi: 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CLSI document M24-A. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. Approved standard. CLSI, Wayne, PA. [PubMed] [Google Scholar]
- 20.Hardenbol P, Banér J, Jain M, Nilsson M, Namsaraev EA, Karlin-Neumann GA, Fakhrai-Rad H, Ronaghi M, Willis TD, Landegren U, Davis RW. 2003. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol 21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 21.Parish T, Brown AC. 2009. Mycobacteria protocols, 2nd ed Humana Press, Totowa, NJ. [Google Scholar]
- 22.Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11:Unit 11.7. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau RWT, Ho P-L, Kao RYT, Yew W-W, Lau TCK, Cheng VCC, Yuen K-Y, Tsui SKW, Chen X, Yam W-C. 2011. Molecular characterization of fluoroquinolone resistance in Mycobacterium tuberculosis: functional analysis of gyrA mutation at position 74. Antimicrob Agents Chemother 55:608–614. doi: 10.1128/AAC.00920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker TM, Kohl TA, Omar SV, Hedge J, Del Ojo Elias C, Bradley P, Iqbal Z, Feuerriegel S, Niehaus KE, Wilson DJ, Clifton DA, Kapatai G, Ip CLC, Bowden R, Drobniewski FA, Allix-Béguec C, Gaudin C, Parkhill J, Diel R, Supply P, Crook DW, Smith EG, Walker AS, Ismail N, Niemann S, Peto TEA, Modernizing Medical Microbiology (MMM) Informatics Group . 2015. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis 15:1193–1202. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherrard LJ, Schaible B, Graham KA, McGrath SJ, McIlreavey L, Hatch J, Wolfgang MC, Muhlebach MS, Gilpin DF, Schneiders T, Elborn JS, Tunney MM. 2014. Mechanisms of reduced susceptibility and genotypic prediction of antibiotic resistance in Prevotella isolated from cystic fibrosis (CF) and non-CF patients. J Antimicrob Chemother 69:2690–2698. doi: 10.1093/jac/dku192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infect 12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- 28.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol 29:2578–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, de Viedma DG, Garzelli C, Gazzola L, Gomes HM, Guttierez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ly HM, Martin C, Martin C, Mokrousov I, Narvskaïa O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim Z, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Rüsch-Gerdes S, Sajduda A, Samper S, Shemyakin IG, Singh UB, Somoskovi A, Skuce RA, van Soolingen D, Streicher EM, Suffys PN, Tortoli E, Tracevska T, Vincent V, Victor TC, Warren RM, Yap SF, Zaman K, Portaels F, Rastogi N, Sola C. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol 6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. 2009. Tuberculosis drug resistance mutation database. PLoS Med 6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felsenstein J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- 33.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, Portugal I, Pain A, Martin N, Clark TG. 2014. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feuerriegel S, Köser CU, Niemann S. 2014. Phylogenetic polymorphisms in antibiotic resistance genes of the Mycobacterium tuberculosis complex. J Antimicrob Chemother 69:1205–1210. doi: 10.1093/jac/dkt535. [DOI] [PubMed] [Google Scholar]
- 35.Tagliani E, Cabibbe AM, Miotto P, Borroni E, Toro JC, Mansjö M, Hoffner S, Hillemann D, Zalutskaya A, Skrahina A, Cirillo DM. 2015. Diagnostic performance of the new version (v2.0) of GenoType MTBDRsl assay for detection of resistance to fluoroquinolones and second-line injectable drugs: a multicenter study. J Clin Microbiol 53:2961–2969. doi: 10.1128/JCM.01257-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nebenzahl-Guimaraes H, Jacobson KR, Farhat MR, Murray MB. 2014. Systematic review of allelic exchange experiments aimed at identifying mutations that confer drug resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 69:331–342. doi: 10.1093/jac/dkt358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirgel FA, Warren RM, Streicher EM, Victor TC, van Helden PD, Böttger EC. 2012. gyrA mutations and phenotypic susceptibility levels to ofloxacin and moxifloxacin in clinical isolates of Mycobacterium tuberculosis. J Antimicrob Chemother 67:1088–1093. doi: 10.1093/jac/dks033. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Gao X, Luo T, Wu J, Sun G, Liu Q, Jiang Y, Zhang Y, Mei J, Gao Q. 2014. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect 3:e19. doi: 10.1038/emi.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakravorty S, Aladegbami B, Thoms K, Lee JS, Lee EG, Rajan V, Cho E-J, Kim H, Kwak H, Kurepina N, Cho S-N, Kreiswirth B, Via LE, Barry CE, Alland D. 2011. Rapid detection of fluoroquinolone-resistant and heteroresistant Mycobacterium tuberculosis by use of sloppy molecular beacons and dual melting-temperature codes in a real-time PCR assay. J Clin Microbiol 49:932–940. doi: 10.1128/JCM.02271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Chen Z, Li Y, Xia W, Chen X, Chen T, Zhou L, Xu B, Xu S. 2012. Characterization of gyrA and gyrB mutations and fluoroquinolone resistance in Mycobacterium tuberculosis clinical isolates from Hubei Province, China. Braz J Infect Dis 16:136–141. doi: 10.1016/S1413-8670(12)70294-5. [DOI] [PubMed] [Google Scholar]
- 41.Peloquin CA, Hadad DJ, Molino LPD, Palaci M, Boom WH, Dietze R, Johnson JL. 2008. Population pharmacokinetics of levofloxacin, gatifloxacin, and moxifloxacin in adults with pulmonary tuberculosis. Antimicrob Agents Chemother 52:852–857. doi: 10.1128/AAC.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Böttger EC. 2011. The ins and outs of Mycobacterium tuberculosis drug susceptibility testing. Clin Microbiol Infect 17:1128–1134. doi: 10.1111/j.1469-0691.2011.03551.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.