Abstract
Listeriosis is a serious foodborne infection that disproportionately affects elderly adults, pregnant women, newborns, and immunocompromised individuals. Diagnosis is made by culturing Listeria monocytogenes from sterile body fluids or from products of conception. This report describes the investigations of two listeriosis pseudo-outbreaks caused by contaminated laboratory media made from sheep blood.
TEXT
Listeria monocytogenes is a bacteria that can cause serious illnesses in humans, including meningitis, sepsis, miscarriage, stillbirth, and preterm labor. Transmission is most often foodborne. Older adults, pregnant women, newborns, and immunocompromised individuals are at the highest risk of developing severe disease. L. monocytogenes is also a well-recognized cause of disease in livestock, having first been described in sheep in the 1930s (1).
Invasive listeriosis is diagnosed in ill patients by culturing L. monocytogenes primarily from blood or cerebrospinal fluid (CSF) but also from other specimen sites, and it is a nationally notifiable disease (http://www.cdc.gov/listeria/surveillance.html). Sheep blood agar is routinely used in clinical laboratories to cultivate fastidious bacteria, including L. monocytogenes. Blood cannot be sterilized during media production. Therefore, bacteria present in sheep blood are a possible source of contamination of media (2). An apparent cluster of infections that is found to be an artifact is referred to as a pseudo-outbreak (3).
Pseudo-outbreak—2013.
In February 2013, Ohio Department of Health investigators identified seven L. monocytogenes isolates from patient urine samples. Urine is an unusual source of L. monocytogenes, and none of the patients had symptoms of listeriosis. Clinical laboratories noticed L. monocytogenes subsurface growth, growth outside streak lines, and growth on uninoculated culture medium. Shortly after, other state health departments reported L. monocytogenes isolates from uncommon specimen sites. In response, the Centers for Disease Control and Prevention (CDC) sent notices to state health departments and clinical laboratories regarding possible laboratory media contamination. By May 2013, eight states had reported a total of 23 isolates, 22 (96%) of which were from atypical sites (14 urine, 6 wound, 1 gastric aspirate, and 1 gastrostomy tube) (Table 1). The remaining isolate was cultured from the CSF of an infant who died suddenly. Two pulsed-field gel electrophoresis (PFGE) patterns were observed among the 23 isolates. The two PFGE patterns were identical at six of seven loci according to multilocus sequence typing (MLST); whole-genome sequencing (WGS) was not available.
TABLE 1.
Description of Listeria monocytogenes clinical isolates from 2013 and 2014 pseudo-outbreaks in the United States
| Parameter | 2013 Pseudo-outbreak | 2014 Pseudo-outbreak |
|---|---|---|
| No. of isolates | 23 | 27 |
| Isolation dates | 13 January to 27 March 2013 | 16 April to 28 May 2014 |
| Median age of patients (range), yr | 57 (1–81) | 62 (2–89) |
| Specimen source, no. (%) | ||
| Urine | 14 (61) | 10 (37) |
| Wound/abscess | 6 (26) | 14 (52) |
| Othera | 3 (13) | 3 (11) |
| States from which clinical isolates were identified | California, Florida, Michigan, Ohio, Tennessee, Washington | California, Georgia, Michigan, Mississippi, New Mexico, New York, Oklahoma, Washington |
Other specimen sources in 2013 include 1 gastric aspirate, 1 percutaneous gastrostomy tube, and 1 cerebrospinal fluid. Other specimen sources in 2014 include 1 sputum, 1 blood, and 1 unknown sample.
Media production information was available for 20 of the culture plates used for patient samples. All 20 were made with sheep blood, and 18 (90%) were produced by Manufacturer A. This manufacturer reviewed production records for the identified lots and determined that the sheep blood units used to produce these culture plates came from a single supplier who collected blood from sheep at a single feedlot. Other manufacturers also used the same blood supplier. Sheep deaths on the feedlot had increased in the month before the pseudo-outbreak was detected. Twenty of 55,000 lambs died on 7 January 2013, representing a peak well above the baseline rate of 4 to 5 deaths per day. Cultures of necropsy specimens from three lambs that died in January yielded L. monocytogenes isolates indistinguishable from pseudo-outbreak isolates by PFGE.
On 11 March 2013, Manufacturer A issued a field correction notice to purchasers, recalling 12 lots among four products (varieties of Trypticase soy agar, MacConkey agar, and anaerobe agar made with sheep blood), noting that contamination was present on a small number of media plates and was found at low levels. In response to this incident, Manufacturer A added an additional incubation step when testing blood used in culture media, began requiring notification of events, such as an increase in deaths among blood-supplying sheep, and implemented a testing program of sheep feed.
Pseudo-outbreak—2014.
In May 2014, the New Mexico Department of Health notified the CDC about two L. monocytogenes isolates from specimen sites rarely associated with Listeria infection, one surgical wound and one abscess. Four days later, the Washington Department of Health reported two wound isolates and one urine isolate. The CDC sent notices of possible laboratory media contamination to state health departments and clinical laboratories. By August 2014, nine states identified 25 L. monocytogenes isolates from atypical specimen sites (14 wound specimens, 10 urine specimens, and 1 sputum specimen) and 5 isolates from uninoculated blood agar plates. Laboratories again reported subsurface growth, typically with just a few CFU. Three PFGE patterns were identified among these isolates, and a review of the national PFGE database (part of PulseNet, the national molecular subtyping network for foodborne disease surveillance) subsequently identified eight other patient isolates (seven from blood and one from an unknown site). Based on patient symptoms, six of the seven blood isolates were thought to represent true cases of listeriosis and were excluded from the pseudo-outbreak. The seventh patient had no signs of listeriosis. PFGE patterns were not similar between the 2014 and 2013 pseudo-outbreaks.
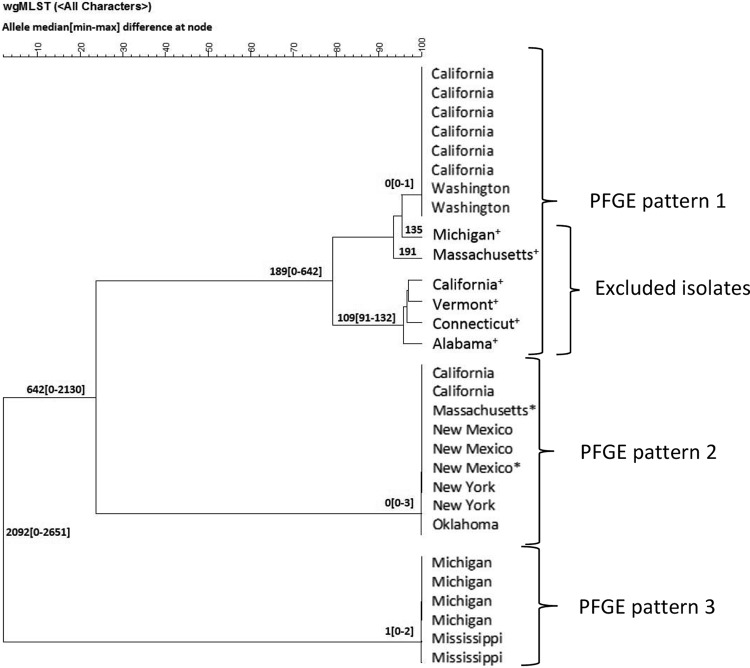
In 2014, we performed whole-genome MLST (wgMLST) using BioNumerics 7.5, which compares allele differences between isolates at >4,000 loci (compared with 7 loci examined by traditional MLST). Alleles were defined using an allele database generated to reflect the diversity of L. monocytogenes (Listeria database v.3) (4). The 21 pseudo-outbreak isolates, the 6 excluded blood isolates described above, and 2 isolates from uninoculated plates were tested by wgMLST (Fig. 1). Pseudo-outbreak isolates within each PFGE pattern designation were nearly identical (≤3 allele differences), supporting that they originated from a common source like contaminated media. The six excluded blood isolates were not closely related (≥135 alleles) to pseudo-outbreak isolates with indistinguishable PFGE patterns, which provided additional evidence that these isolates were not part of the pseudo-outbreak.
FIG 1.
Relationships by whole-genome multilocus sequencing type (wgMLST) of Listeria monocytogenes isolates with three pulsed-field gel electrophoresis (PFGE) patterns from 11 U.S. states associated with the 2014 pseudo-outbreak. Median (minimum to maximum) allele differences at node by wgMLST analysis performed using BioNumerics 7.5 and alleles were defined using an allele database generated to reflect the diversity of L. monocytogenes (Listeria database v.3). *, Isolates from uninoculated laboratory media plates; +, Isolates excluded from the pseudo-outbreak because of a clinical history compatible with listeriosis.
Information about the culture media manufacturer was available for 21 of the 27 pseudo-outbreak isolates from 2014, and 20 (95%) of these were made by Manufacturer A. An investigation by Manufacturer A again traced the source of contamination to the sheep blood used to manufacture the media. No information was available about the condition of the sheep or feedlot. Manufacturer A again issued a field correction notice.
Although nearly all isolates in these investigations involved specimen sites uncommonly associated with Listeria infection, one isolate from each pseudo-outbreak was cultured from a more typical site. WGS results suggest that the blood isolate resulted from contaminated media, but it remains unknown if the CSF isolate represented a true infection.
Because of the importance of laboratory culture in clinical diagnosis and in public health surveillance of listeriosis, microbiologists, clinicians, and public health officials should be aware of the possibility of false-positive results from media contamination. Data from these investigations demonstrate that listeriosis in livestock used to supply blood for culture media can result in false-positive clinical cultures for L. monocytogenes. In the two pseudo-outbreaks, alert epidemiologists and microbiologists rapidly detected contamination, which allowed for wider outreach through microbiology electronic mailing lists, direct communication with clinical laboratories, and prevention of inappropriate medical diagnosis and treatment. This points to the importance of monitoring culture results to ensure that they are clinically relevant. In several instances, laboratory workers visualized media contamination before inoculation, which is an important quality control point that may be missed during automated laboratory workflows. The media plates in these two outbreaks may have passed manufacturing quality control because of the low level and nonuniform contamination. However, effective product tracking by Manufacturer A allowed for a traceback investigation.
Several interventions might reduce the risk of blood agar contamination. L. monocytogenes infection prevention in sheep includes monitoring of silage pH and moisture content and keeping sheep in clean, low-stress environments (http://www.cdc.gov/onehealth/) (5). Measures that can prevent L. monocytogenes from entering the supply chain for blood agar include monitoring illness and death rates among sheep so that harvesting can be suspended when a significant increase is detected, using sterile harvesting practices, and avoiding harvesting blood from younger animals who are more likely to have L. monocytogenes sepsis. Additionally, increasing postharvest blood testing and finished laboratory media testing with optimized incubation times and temperatures might increase the chance of detecting contamination before distribution. Further investigation is needed to determine which interventions are most feasible and effective in this industry.
ACKNOWLEDGMENTS
We acknowledge the clinicians, laboratories, and health departments involved in these investigations, including the Wadsworth Center Bacteriology Laboratory and the Applied Genomic Technologies Core Facility.
REFERENCES
- 1.Low JC, Donachie W. 1997. A review of Listeria monocytogenes and listeriosis. Vet J 153:9–29. doi: 10.1016/S1090-0233(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 2.Bemis DA, Kania SA. 2007. Isolation of Bartonella sp. from sheep blood. Emerg Infect Dis 13:1565–1567. doi: 10.3201/eid1310.070570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shears P. 1996. Pseudo-outbreaks. Lancet 347:138. doi: 10.1016/S0140-6736(96)90337-2. [DOI] [PubMed] [Google Scholar]
- 4.Carleton HA, Katz LS, Stroika S, Sabol A, Roache K, Kucerova Z, Ribot EM, Evans P, Holt K, Kubota K, Pouseele H, Besser J, Tarr C, Trees E, Gerner-Smidt P. 2015. The application of whole genome multi-locus sequence typing to characterize Listeria Monocytogenes, p 57 In International Conference on Emerging Infectious Diseases Program and Abstracts Book. American Society for Microbiology, Washington, DC. [Google Scholar]
- 5.Wesley IV. 2007. Listeriosis in animals, p 56–60. In Ryser ET, Marth EH (ed), Listeria, listeriosis, and food safety. Taylor and Francis Group, Boca Raton, FL. [Google Scholar]