Abstract
Pseudomonas aeruginosa has emerged as an important opportunistic human pathogen that is often highly resistant to eradication strategies, mediated in part by the formation of multicellular aggregates. Cellular aggregates may occur attached to a surface (biofilm), at the air-liquid interface (pellicle), or as suspended aggregates. Compared to surface attached communities, knowledge about the regulatory processes involved in the formation of suspended cell aggregates is still limited. We have recently described the SiaA/D signal transduction module that regulates macroscopic cell aggregation during growth with, or in the presence of the surfactant SDS. Targets for SiaA/D mediated regulation include the Psl polysaccharide, the CdrAB two-partner secretion system and the CupA fimbriae. While the global regulators c-di-GMP and RsmA are known to inversely coordinate cell aggregation and regulate the expression of several adhesins, their potential impact on the expression of the cupA operon remains unknown. Here, we investigated the function of SiaA (a putative ser/thr phosphatase) and SiaD (a di-guanylate cyclase) in cupA1 expression using transcriptional reporter fusions and qRT-PCR. These studies revealed a novel interaction between the RsmA posttranscriptional regulatory system and SiaA/D mediated macroscopic aggregation. The RsmA/rsmY/Z system was found to affect macroscopic aggregate formation in the presence of surfactant by impacting the stability of the cupA1 mRNA transcript and we reveal that RsmA directly binds to the cupA1 leader sequence in vitro. We further identified that transcription of the RsmA antagonist rsmZ is controlled in a SiaA/D dependent manner during growth with SDS. Finally, we found that the siaD transcript is also under regulatory control of RsmA and that overproduction of RsmA or the deletion of siaD results in decreased cellular cyclic di-guanosine monophosphate (c-di-GMP) levels quantified by a transcriptional reporter, demonstrating that SiaA/D connects c-di-GMP and RsmA/rsmY/Z signaling to reciprocally regulate cell aggregation in response to environmental conditions.
Keywords: c-di-GMP signaling, RsmA, Pseudomonas aeruginosa, cellular aggregation, biofilm, SiaA, SiaD, CupA fimbriae
Introduction
It is now well established that most bacteria can form multicellular aggregates and that this is believed to represent the dominant mode of microbial life (Costerton et al., 1995; Hall-Stoodley et al., 2004). Cellular aggregates can occur attached to a surface as biofilms, as a pellicle at the air-liquid interface, or as suspended aggregates of cells. The evolutionary and ecological success of the multicellular lifestyle has often been attributed to the increased resistance of the resident cells compared to their planktonic free living counterparts (Davey and O’Toole, 2000; Hall-Stoodley et al., 2004). The increased resistance of cellular aggregates can impact human health, for example by allowing the persistence of bacteria in health-care settings despite the use of hygiene regimens, which provides a potential reservoir for the transmission of infections (Kerr and Snelling, 2009; Asghari et al., 2013; Williams et al., 2013). Pseudomonas aeruginosa has emerged as an opportunistic pathogen for patients with compromised immunity, such as those with the genetic disorder cystic fibrosis (Bjarnsholt, 2013). In the context of chronic lung infections of cystic fibrosis patients, P. aeruginosa is thought to survive predominantly as cell aggregates growing in mucus that are not associated with any surface (Bjarnsholt et al., 2009). In addition, recent studies indicate that aggregation is commonly observed for P. aeruginosa growing in liquid environments (Schleheck et al., 2009).
The reciprocal regulation of cellular aggregation mediated by the RsmA and c-di-GMP global regulatory systems has been well documented, with high levels of c-di-GMP leading to a sessile lifestyle, in contrast to RsmA, which promotes planktonic physiology (Hengge, 2009; Timmermans and Van Melderen, 2010; Romling et al., 2013; Vakulskas et al., 2015). Both regulators can function at multiple levels of gene expression either directly or by influencing additional regulators. C-di-GMP has been found to specifically bind to mRNA riboswitches and a range of proteins to exert its regulatory potential (Amikam and Galperin, 2006; Hickman and Harwood, 2008; Sudarsan et al., 2008; Boehm et al., 2010; Chambers et al., 2014; Trampari et al., 2015). The RsmA protein acts directly by binding to the mRNA of target genes, typically by binding to a consensus sequence (Dubey et al., 2005) overlapping the Shine-Dalgarno sequence (SD), which prevents ribosome assembly, reducing translation, and most often resulting in more rapid degradation of the transcript by RNAses (Timmermans and Van Melderen, 2010; Vakulskas et al., 2015). The GacA/S two-component system is the best characterized regulatory system that influences the activity of RsmA, where GacA/S controlled rsmY/Z sRNA expression leads to their binding and functional sequestration of the RsmA protein (Goodman et al., 2004; Kay et al., 2006; Sorger-Domenigg et al., 2007; Brencic et al., 2009).
The RsmA/rsmY/Z and c-di-GMP pathways can affect the expression of a large number of target genes in P. aeruginosa, and are predicted to display significant cross-talk (Coggan and Wolfgang, 2012). Interestingly, recent evidence indicates that rsmY or rsmZ regulation is not limited to the GacA/S two-component system, suggesting that additional mechanisms influence the regulation of genes targeted by RsmA in response to certain environmental conditions. This system has been shown to be affected by oxygen concentration (via the Anr responsive NarL transcription factor; O’Callaghan et al., 2011), changes in biofilm growth stage (via BfiR; Petrova and Sauer, 2010), envelope stress resulting from loss of the OprF porin (possibly via SigX; Bouffartigues et al., 2015) as well as controlling virulence factor production (through RetS and the hybrid sensor PA1611; Kong et al., 2013). Thus, the RsmA/rsmY/Z regulatory system impacts many aspects of growth and development of P. aeruginosa.
In the context of cellular aggregation, c-di-GMP and RsmA are well known to be involved in the expression of several cell surface adhesins including the Psl polysaccharide and the CdrAB two-partner secretion system (Burrowes et al., 2006; Brencic and Lory, 2009; Borlee et al., 2010; Irie et al., 2010). Accordingly, either mutation of rsmA or artificially increasing c-di-GMP levels leads to Psl and CdrAB dependent autoaggregation irrespective of the environmental conditions (Starkey et al., 2009; Borlee et al., 2010; Irie et al., 2010, 2012). Interestingly, the CupA fimbriae are also known to be involved in cellular aggregation at surfaces (Meissner et al., 2007), although the genes of the cupA operon have not previously been shown to be influenced by RsmA, and the evidence that c-di-GMP regulates the cupA operon is conflicting (Meissner et al., 2007; Starkey et al., 2009).
We have recently described that macroscopic cellular aggregation of P. aeruginosa can be induced by exposure to sub-inhibitory concentrations of the surfactant sodium dodecyl sulfate (SDS) and that these aggregated cells display an increased resistance to additional stresses (Klebensberger et al., 2006, 2007). By mutational analysis and phenotypic characterisation, SiaA/D was shown to play a central role in this process in a c-di-GMP dependent manner (Klebensberger et al., 2009). The functions of both the putative ser/thr dependent phosphatase SiaA and the di-guanylate cyclase (DGC) SiaD were shown to be required for macroscopic aggregate formation during growth with SDS by regulating the expression of a core set of genes including those encoding the CupA fimbriae (Klebensberger et al., 2009). In the present study, we show that RsmA signaling is a negative regulator of macroscopic aggregation during growth with SDS, and that cupA1 represents a novel target for RsmA binding. Further, we provide strong evidence that this is achieved by SiaA/D dependent transcriptional regulation of rsmZ and that the siaABCD operon itself is under negative feedback regulation of RsmA. From these data we conclude that SiaA/D connects the c-di-GMP and RsmA networks to reciprocally coordinate the aggregation of cells in response to environmental conditions.
Materials and Methods
Bacterial Strains and Growth Conditions
Pseudomonas aeruginosa and Escherichia coli strains (Supplementary Table S1) were maintained on Lysogeny Broth (LB) (Bertani, 1951) solid medium (1.5% agar w/v) and routinely cultured in 10 mL LB medium in 50 mL Falcon tubes (Greiner Bio-one) with shaking at 200 rpm and incubation at 37°C (Ratek Orbital Mixer Incubator). Where appropriate, antibiotics were added at the following concentrations for maintenance or selection respectively: 50 or 100 μg mL-1 carbenicillin, 15 or 30 μg mL-1 gentamycin, 10 or 20 μg mL-1 tetracycline for E. coli, and 50 or 200 μg mL-1 carbenicillin, 30 or 120 μg mL-1 gentamycin, 20 or 200 μg mL-1 tetracycline for P. aeruginosa. For cupA::lacZ experiments, 10 μg mL-1, gentamicin and 10 μg mL-1 tetracycline was added to the growth medium for plasmid maintenance.
Strain and Plasmid Construction
The E79tv2 phage was used to construct mutant strains by transferring mutant alleles into the wild-type PAO1 background used in this study as previously described (Malone et al., 2010). The PW8792 strain was obtained from the University of Washington Genome Centre and used as the source of the tetracycline resistant cdrB mutation. The resulting ΔcdrB strain was subsequently cured of the antibiotic resistance cassette by Cre/loxP recombination using the pCre1 plasmid (Jacobs et al., 2003). The MPAO1_KOcupA3 and MPAO1_KOpslBCD strains (Kirisits et al., 2005; Starkey et al., 2009) were kindly provided by the Parsek lab (University of Washington, USA) and used as the sources of gentamycin resistant cupA3 and psl mutations respectively. The resulting ΔcupA3 and Δpsl strains were subsequently cured of the antibiotic resistance cassettes by FLP/FRT recombination using the pFLP2 plasmid (Hoang et al., 1998). The arabinose inducible rsmA allele was inserted at the neutral attTn7 site of the P. aeruginosa chromosome by transposition using miniTn7T[PBAD::rsmA] (kindly provided by Tina Jäger from the Biozentrum Basel) and the pTNS2 plasmid (Choi et al., 2005). A detailed description for the construction of the pBBR[siaD], pUC18[cupA], pUC19[lolB], pUC19[rsmZ], pBAD33[RsmA-His6] plasmids and the primers used (Supplementary Table S2) is provided in the Supplementary Material.
Growth and Cell Suspension Experiments
Growth experiments were performed in a modified M9 minimal medium (Klebensberger et al., 2006). Overnight cultures of P. aeruginosa strains grown in LB medium were inoculated into 2 mL M9 medium at an OD600 = 0.01 in 12 well microtitre plates (Corning) supplemented with either 10 mM succinate or 3.5 mM SDS and incubated at 30°C with shaking at 200 rpm for 18 h (Ratek orbital mixer incubator).
For cell suspension experiments, LB overnight cultures were used to inoculate 100 mL of M9 medium with an OD600 = 0.01 in a 500 mL Erlenmeyer flask. The medium was supplemented with either 10 mM succinate or 3.5 mM SDS as a growth substrate and incubated at 30°C with shaking at 200 rpm to the late-logarithmic phase of growth. Subsequently, the cells were washed twice with 10 mL M9 medium with centrifugation for 2 min at 10,000 × g at room temperature (Beckman Avanti J25-I), re-suspended in 1 mL M9 and used to generate cell suspensions with an OD600 = 1. Cell suspensions were supplemented with 10 mM succinate or 3.5 mM SDS, transferred to 12 well microtitre plates (Corning) in 2 mL samples and incubated as indicated below. Where appropriate, L-arabinose (2% final concentration) was included in both the 100 mL pre-culture and the cell suspension experiment. After incubation, images were acquired from growth and cell suspension experiments using an Umax Powerlook 1000 flatbed scanner with V4.71 software in a dark room. Images were normalized to the first image scanned using this experimental format by using Adobe Photoshop CS5 Software by using the ‘match color’ image adjustment. Experiments were performed multiple times (n ≥ 3) and a representative image is shown.
Quantification of Macroscopic Aggregation
Scanned images of cultures grown in 12 well plates were analyzed using ImageJ (Schneider et al., 2012). Images were preprocessed by converting to 8-bit grayscale images and applying a rolling ball background subtraction to correct for the uneven background present within each well. A radius of 11 pixels was observed to retain aggregate structures yet flatten out the background suitable for thresholding. To remove bias, thresholds were determined automatically using an implementation of Huang’s thresholding (Huang and Wang, 1995) already available in ImageJ. Each well was then measured using ‘Analyze Particles’ with measurements restricted to circular regions of interest of consistent size, each positioned in the center to avoid any artifacts resulting from the edges of the wells. Individual data (circles) are shown together with the mean value of the three biological replicates (bars).
cupA1::lacZ Transcriptional Reporter Studies
Cell suspensions for P. aeruginosa strains harboring the cupA1::lacZ transcriptional promoter fusion (Vallet et al., 2004) were prepared as described above. Following 1 h incubation at 30°C with shaking at 200 rpm, strains were harvested by centrifugation for 30 s at 16,060 × g (Heraues Biofuge Pico), washed twice in 1 mL fresh M9 medium, resuspended in 1 mL M9 medium and lysed with ∼65 mg of Cellytic express (Sigma) at 37°C in a water bath for 15 min with occasional mixing. Intact cells and cell debris were removed from the crude cell extracts by centrifugation for 1 min at 16,060 × g (Heraues Biofuge Pico) and 800 μL of the supernatant was transferred to a fresh Eppendorf tube and stored at -20°C for further analysis.
Beta-galactosidase (β-galactosidase) activity was measured based on the hydrolysis of ortho-nitrophenyl-β-galactoside (ONPG). For activity measurements, 20 μL of cell extract was incubated with 80 μL of β-galactosidase buffer (60 mM Na2HPO4, 40 mM NaH2PO4 and 30 mM β-mercaptoethanol, pH 7) in a 96 well plate (Sarstedt, flat bottom) at 30°C for 10 min. Immediately following this incubation, the reaction was started by the addition of 20 μL ONPG from a freshly prepared 4 μg mL-1 stock and incubated at 30°C. Reactions were stopped at specific time points by the addition of 50 μL of 1 M Na2CO3 solution and the absorbance measured at 420 nm in a plate reader (Perkin Elmer; Wallac Victor2). The concentration of ortho-nitrophenol (ONP), the enzymatic product formed by the hydrolysis of ONPG due to β-galactosidase activity, was calculated from a freshly prepared calibration curve. Specific β-galactosidase activity was determined by normalizing the values of β-galactosidase activity to the total protein content of the corresponding cell extract measured by the Bicinchoninic Acid (BCA) assay (Pierce), and is expressed as nmol ONPG min-1 mg-1 total protein. Individual data (circles) are shown together with the mean value of the three biological replicates (bars).
Reporter Assays Using cdrA, rsmY and rsmZ Transcriptional gfp Fusions
To quantify intracellular c-di-GMP levels or the transcriptional activity of the sRNAs rsmZ/Y, P. aeruginosa strains harboring the CdrA::gfpc, rsmZ::gfp or rsmY::gfp fusion constructs (Rybtke et al., 2012; Chua et al., 2014) were grown in 10 mL LB at 30°C with shaking at 200 rpm for 10 h (Ratek orbital mixer incubator). After incubation, the cells were washed twice in 1 mL M9 and used to inoculate either 10 mL M9 medium containing either 10 mM succinate or 3.5 mM SDS with an OD600 = 0.01. Succinate cultures were grown for 9 h, while SDS cultures were grown for 18 h with incubation at 30°C and shaking at 200 rpm. These cultures were then washed twice in 10 mL M9 medium, resuspended in 1 mL M9 without carbon source and adjusted to an OD600 = 0.5. These cell suspensions were inoculated in 2 mL aliquots in 12 well plates and grown at 30°C shaking at 200 rpm for 30 min.
To quantify the CdrA::gfpc reporter activity, fluorescence was measured in a plate reader using an excitation/emission of 485/535 nm with constant voltage for 1 s and the use of a large emission aperture (Perkin Elmer, Wallac victor2). The samples were normalized by the initial optical density and experiments consisted of six biological samples that were repeated as two technical replicates to ensure reproducibility of the data. Individual data (circles) are shown together with the mean value of the six biological replicates (bars).
To quantify the rsmY and rsmZ transcriptional reporter activity, each sample was passed through a 25 gauge syringe three times before the samples were lysed with Cellytic B reagent according to the manufacturer’s instructions (Sigma). The total protein content and the GFP signal of each sample was then measured with a Nanodrop (ND1000) using the protein determination (absorbance at 280 nm with baseline correction) and Alexaflour 488 (absorbance at 488 nm with dye slope correction) label quantification functions, respectively (Nanodrop v3.12 software). Finally, the relative GFP signal for each sample was determined by dividing the absorbance value at 488 nm by the corresponding total protein content. Experiments were performed in three biological replicates, each performed as two technical replicates. Individual data (circles) are shown together with the mean value of the three biological replicates (bars).
mRNA Structural Prediction, mRNA Decay, and mRNA Abundance Experiments
mRNA structural prediction was performed using version 2.3 of the mFOLD web server (Zuker, 2003). Initially, sequences were scanned for the presence of the SELEX derived RsmA binding consensus A/UCANGGANGU/A (Dubey et al., 2005), and also for the presence of the primary and secondary RsmA binding sites used in the algorithm for predicting novel RsmA targets (Kulkarni et al., 2014). For the analysis of the cupA1 leader sequence, the 200 nucleotides upstream and including the AUG start codon was used for analysis. For the analysis of the siaA leader sequence a 93 nt sequence from the previously described transcriptional start site from the AmrZ promoter (Jones et al., 2014) up to and including the AUG start codon and was used. mRNA secondary structures were predicted at 30°C using the mFOLD web server (version 2.3 energies) with standard settings, and the most stable structure according to the free energy (ΔG) predictions is presented.
The mRNA decay experiments were performed by generating cell suspensions of an OD600 = 0.5 as described above. These cell suspensions were incubated for 1 h at 30°C with shaking at 200 rpm (Ratek orbital mixer incubator), and then 300 μg mL-1 rifampicin was added to prevent de novo mRNA synthesis. Samples were taken immediately, and following 10, 20, and 30 min incubation at 30°C with shaking at 200 rpm. Samples were harvested by centrifugation for 30 s at 16,060 × g (Heraues Biofuge Pico) and washed once in 1 mL ice cold M9 before being treated with 1 mL RNAlater bacterial mRNA stabilization reagent according to the manufacturer’s instructions (Qiagen). Total RNA was extracted from the samples by using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions, including the use of the optional on-column DNAse treatment with RNAse free DNAse (Qiagen). Purified RNA was subjected to a second DNAse treatment with a Turbo DNAse kit (Ambion) to remove residual traces of genomic DNA. RNA samples were quantified with a NanoDrop (ND-1000) and random RNA samples were selected for Bioanalyser analysis to validate RNA integrity and accuracy of NanoDrop measurements (Agilent 2100 Electrophoresis Bioanalyser; Ramaciotti centre, UNSW) (available on request). cDNA was synthesized from 500 ng total RNA with an iScript cDNA Synthesis Kit using random hexamers according to the manufacturer’s instructions (Bio-rad). qPCR was performed in a black hard-shell 96-well plate with white wells (Bio-rad) in a CFX-1000 machine (Bio-rad) using Evagreen supermix (Bio-rad), 0.5 μL cDNA, 300 nM each primer in a final reaction volume of 25 μL. Each sample was measured in triplicate, and a cDNA synthesis reaction performed without the addition of reverse transcriptase enzyme was included as a control for the detection of gDNA. The reaction conditions were as follows; 3 min at 98°C for 1 cycle, followed by 60°C for 5 s, 72°C for 10 s and data acquisition, 98°C for 10 s for 40 cycles, and a melt curve performed from 60°C to 98°C with 0.5°C intervals. CFX1000 software was used to calculate cycle threshold (Ct) values, and data was analyzed according to the 2-ΔCt method for mRNA decay analysis (Livak and Schmittgen, 2001). Individual data (circles) are shown together with the mean value of the three biological replicates (bars).
The same experimental format described for the mRNA decay experiments was used to quantify the relative abundance of mRNA, with the exception that following the 1 h incubation of cell suspensions total RNA was harvested immediately for analysis. Samples were processed as described above, and data was analyzed by the 2-ΔΔCt method (Livak and Schmittgen, 2001) with the proC housekeeping gene used for normalization. Individual data (circles) are shown together with the mean value of the three biological replicates (bars).
Surface Plasmon Resonance
RsmA binding studies were performed with purified RsmA-His6 protein and in vitro generated RNAs of rsmZ, lolB, cupA1 and cupA1[AAA] using surface plasmon resonance measurements on a BIACore 3000 system equipped with a NTA sensor chip (GE Healthcare). Detailed information about the experimental procedure for protein purification, in vitro transcription, BIACore measurements and the data analysis can be found in the Supplementary Material.
Statistical Methods
Bar graphs represent the mean value of all available biological replicates and are accompanied with the individual data points. Statistical significance was determined using a two-tailed, unpaired student t-test, with a p-value ≤0.05 considered significant.
Results
Essential Components of Aggregate Formation
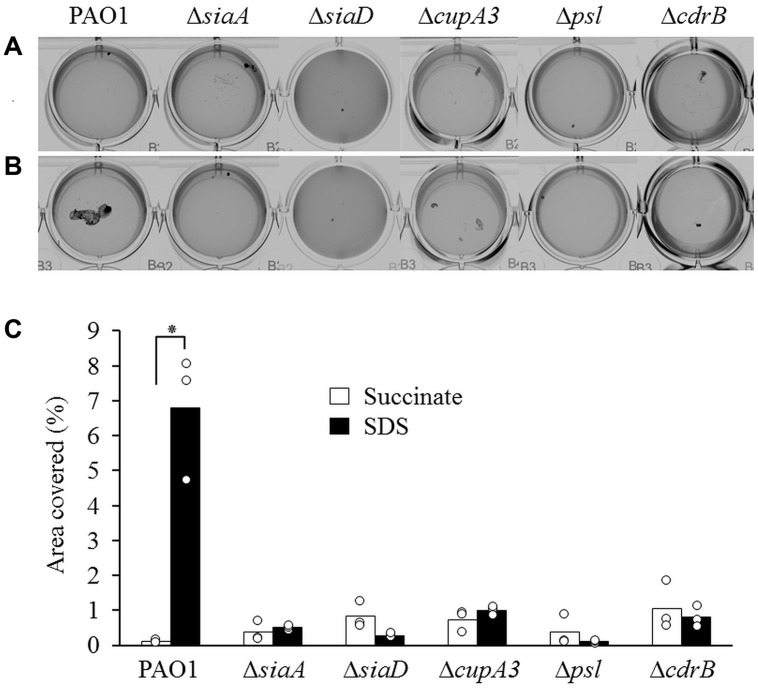
Previous studies have demonstrated that the putative ser/thr phosphatase SiaA and the DGC SiaD are essential for macroscopic aggregate formation during growth with, or in the presence of the surfactant SDS (Klebensberger et al., 2006, 2009). SiaA harbors a HAMP domain characteristic of sensory proteins that functions to link sensory perception with an intracellular response, such as the predicted phosphatase activity of its N-terminal PP2C ser/thr phosphatase domain. Further genetic analysis identified the CupA fimbriae, the CdrAB two-partner secretion system, and the Psl polysaccharide as regulatory targets for SiaA/D (Klebensberger et al., 2009). In this study, we used the ΔsiaA and ΔsiaD mutants to validate our experimental conditions and constructed defined mutations of cupA3, the psl operon, and cdrB to confirm their role in macroscopic aggregation during growth with SDS (Figures 1A,B). The wild-type strain PAO1 produced macroscopic aggregates during growth with SDS as previously described (Klebensberger et al., 2006). In contrast, mutation of siaA, siaD, cupA3, psl, or cdrB eliminated this phenotype, confirming their essential role in macroscopic aggregation. Quantification analysis revealed that growth with SDS increased the area of coverage by macroscopic aggregates 58 fold for the wild-type strain compared to growth with succinate (Figure 1C). In contrast, none of the, ΔsiaA, ΔsiaD, ΔcupA3, psl, or ΔcdrB mutant strains showed an increase in aggregation during growth with SDS.
FIGURE 1.
Phenotypic characterisation of Pseudomonas aeruginosa and isogenic mutant strains during growth with 10 mM succinate (A) or 3.5 mM SDS (B). (C) Quantification of the percentage of area covered by macroscopic aggregates of cells during growth with succinate (white bars) or SDS (black bars) from different strains. Bars represent the mean value from three individual experiments and are accompanied by the individual data points. Statistical significance is indicated (∗p = 0.003).
The GGEEF Active Site Motif of SiaD is Required for Aggregate Formation
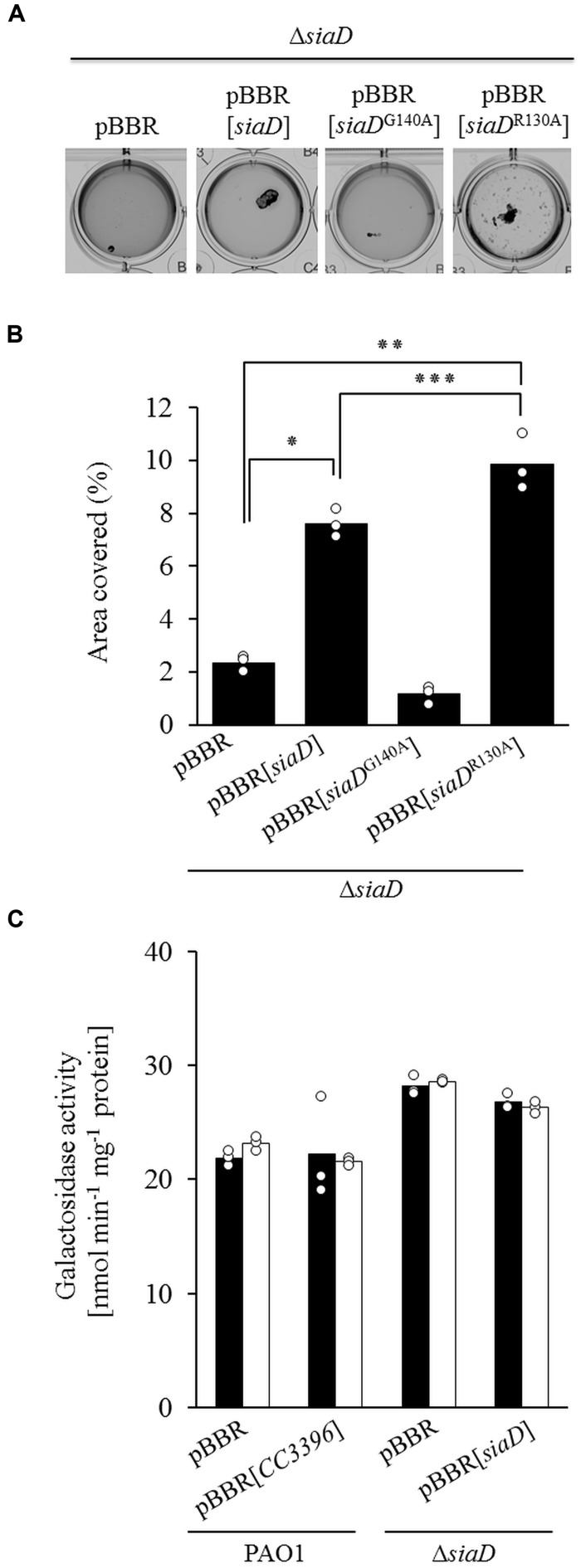
Macroscopic aggregate formation involves c-di-GMP and the DGC SiaD (Klebensberger et al., 2009). This protein harbors both a GGEEF active-site motif (A-site) characteristic of c-di-GMP biosynthesis and a conserved RXXD motif (I-site) characteristic of c-di-GMP binding and allosteric regulation. Site specific mutagenesis was used to produce two siaD alleles with either a non-functional A-site (siaDG140A) or I-site (siaDR130A), and the resulting constructs were tested for their ability to complement the ΔsiaD mutant (Figure 2A). Macroscopic aggregation assays and their subsequent quantitative analysis indicated that growth with SDS increased the area covered by macroscopic aggregates of the ΔsiaD mutant expressing the native siaD gene or the mutant I-site allele siaDR130A, but not for the non-functional A-site allele siaDG140A (Figure 2B). In addition, the observed increase was greater for cultures of the ΔsiaD mutant expressing the defective I-site allele siaDR130A compared to the strain complemented with the native siaD allele (9.85 vs. 7.60%).
FIGURE 2.
(A) Phenotypic analysis of the ability for the pBBR empty vector, the native siaD gene, and the mutated A-site siaDG140A or I-site siaDR130A alleles to restore aggregation to the ΔsiaD mutant during growth with 3.5 mM SDS. (B) Quantitative analysis of the percentage of area covered by macroscopic aggregates of different strains. (C) Transcription of cupA1 assessed with a cupA::lacZ reporter for the PAO1 pBBR plasmid control, the PAO1 pBBR[CC3396] strain encoding a functional c-di-GMP specific phosphodiesterase from Caulobacter crescentus, the ΔsiaD mutant, and ΔsiaD pBBR[siaD] complemented strain during growth in 10 mM succinate (black bars) or 3.5 mM SDS (white bars). Bars represent the mean value from three individual experiments and are accompanied by the individual data points. Statistical significance is indicated (∗p < 0.001, ∗∗p < 0.001, ∗∗∗p = 0.03).
Regulation of cupA Fimbriae
Our previous transcriptomics based approach indicated that cupA1 transcripts were 19 fold more abundant in wild-type cells during SDS exposure compared to cells grown with only succinate, and also when compared to the ΔsiaD mutant grown with SDS (12 fold difference) (Klebensberger et al., 2009). To investigate a potential SiaA/D dependent transcriptional regulation of cupA1, we used a previously described functional cupA1::lacZ reporter (Figure 2C) (Vallet et al., 2004).
Surprisingly, no differences in cupA1 transcription could be observed between the different strains and growth conditions tested. The fact that the increased cupA1 mRNA levels detected in earlier studies did not correlate with increased activation of the transcriptional cupA1 reporter under identical conditions could be indicative of a posttranscriptional regulatory mechanism. The RsmA/rsmY/Z system has been shown to play an important role in the posttranscriptional regulation of many genes in P. aeruginosa, including those involved in biofilm formation (Burrowes et al., 2006; Brencic and Lory, 2009). Analysis of the cupA1 leader sequence revealed a potential primary RsmA binding site that shared 90% sequence identity with the known SELEX binding consensus (ACAAGGAAcA; sequence divergence is indicated by lowercase), including the GGA core motif (bold and underlined) located 6–15 nucleotides upstream from the cupA1 start codon (Figure 3A; positions 186–195). In addition, one primary (29–33) and one secondary binding site (76–80) were identified as defined by a recent computational approach (Kulkarni et al., 2014). This sequence was then further analyzed by structural prediction using the mFold web server (Zuker, 2003). The structural prediction with the highest stability was found to harbor multiple stem–loop structures, and notably the SELEX derived RsmA consensus sequence is predicted to be located at the end of a stem–loop (nucleotides 186–195) which overlaps with the predicted SD sequence for cupA1 (nucleotides 188–193), which is predicted to be the ideal RsmA binding conditions.
FIGURE 3.
(A) Structural prediction of the cupA1 mRNA leader sequence using mFold (Zuker, 2003) with predicted primary (P) and secondary (S) RsmA binding sites. (B) Phenotypic analysis of strains PAO1 and PAO1::rsmA and the impact of RsmA induction (+) on macroscopic cell aggregation during growth with SDS. (C) Quantitative analysis of the percentage covered by macroscopic aggregates of different strains. Bars represent the mean value from three individual experiments and are accompanied by the individual data points. Statistical significance is indicated (∗p = 0.001).
To explore the potential influence of RsmA on macroscopic cell aggregation during growth with SDS, we used a strain harboring an arabinose inducible copy of rsmA at a neutral site of the P. aeruginosa chromosome. The induction of RsmA completely abolished macroscopic aggregate formation during growth with SDS compared to the non-induced control (Figure 3B). Quantitative analysis of these strains revealed that the induction of rsmA caused a fourfold reduction in the area covered by macroscopic aggregates when compared to the non-induced control during growth with SDS (Figure 3C).
RsmA Overexpression Reduces cupA1 mRNA Stability
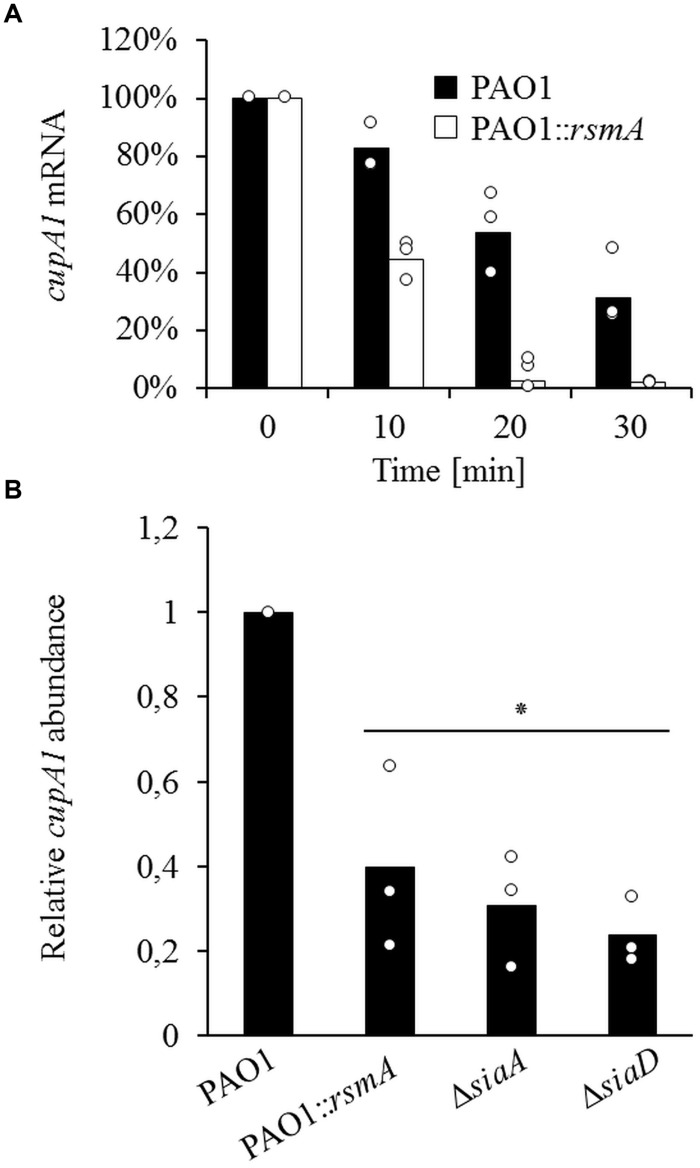
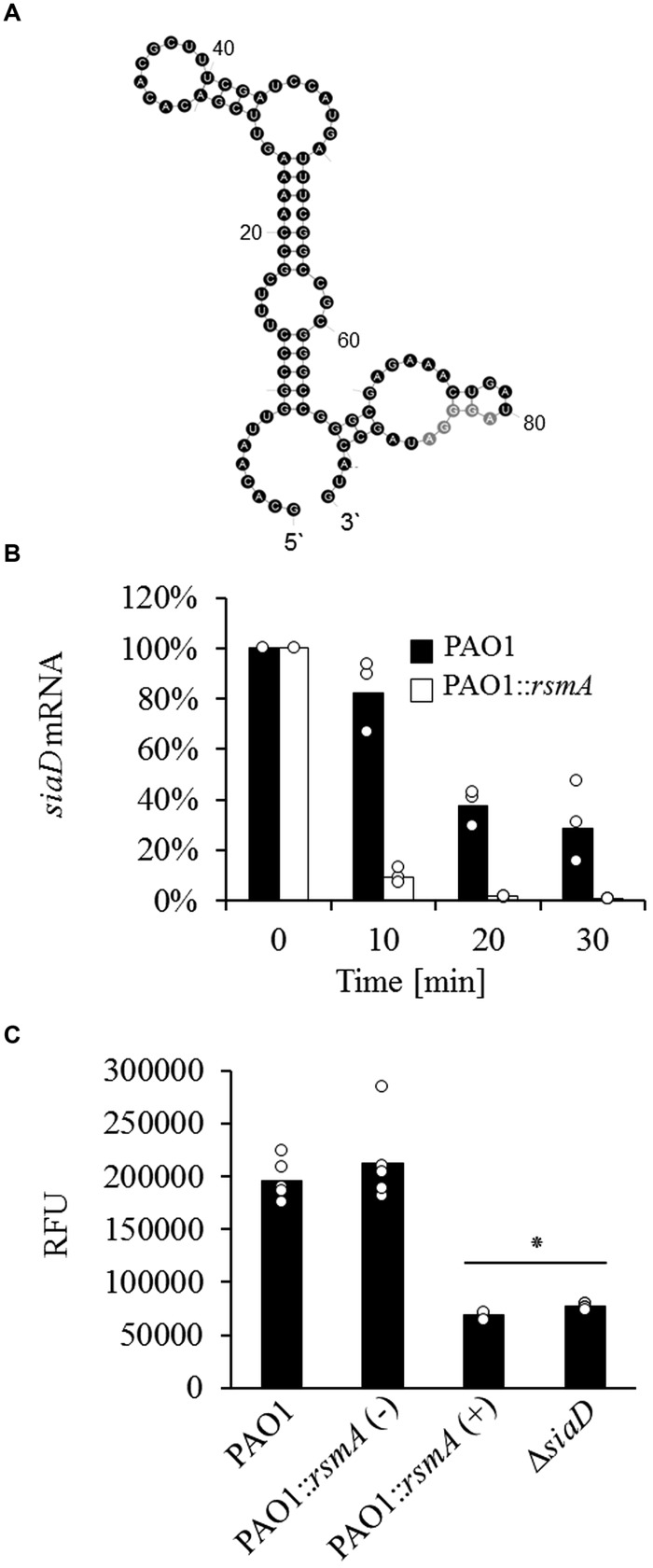
To investigate whether RsmA influences cupA1 mRNA stability, we analyzed the mRNA decay of cupA1 by qRT-PCR and found that the overproduction of RsmA strongly reduced the stability of the cupA1 transcript for all time points tested (Figure 4A). Compared to cells of the wild-type strain, which retained 83, 54, and 31% of the initial cupA1 transcript levels after 10, 20, and 30 min respectively, cells containing increased levels of RsmA showed a much faster rate of decay with only 43, 2, and 2% of the initially detected transcript levels present at the same time points. In addition, quantification of the relative abundance of the cupA1 transcript revealed that cupA1 levels were reduced during growth with SDS when RsmA was overproduced compared to the parental strain (0.35 fold) (Figure 4B). Further, the non-aggregative ΔsiaA and ΔsiaD mutant strains also displayed reduced cupA1 mRNA levels (0.46 fold and 0.23 fold, respectively) when compared to wild-type cells.
FIGURE 4.
(A) The effect of RsmA overexpression on the stability of cupA mRNA as determined by qRT-PCR. Data were normalized by the use of equivalent mRNA and analyzed by the 2-ΔCt method. (B) Relative abundance of cupA1 mRNA during growth in SDS for strains PAO1, PAO1::rsmA and the mutant strains ΔsiaA and ΔsiaD. Data were normalized to the proC housekeeping gene by the 2-ΔΔCt method. Bars represent the mean value from three individual experiments and are accompanied by the individual data points. Statistical significance compared to PAO1 is indicated (∗p ≤ 0.01).
RsmA Directly Binds to cupA1 mRNA
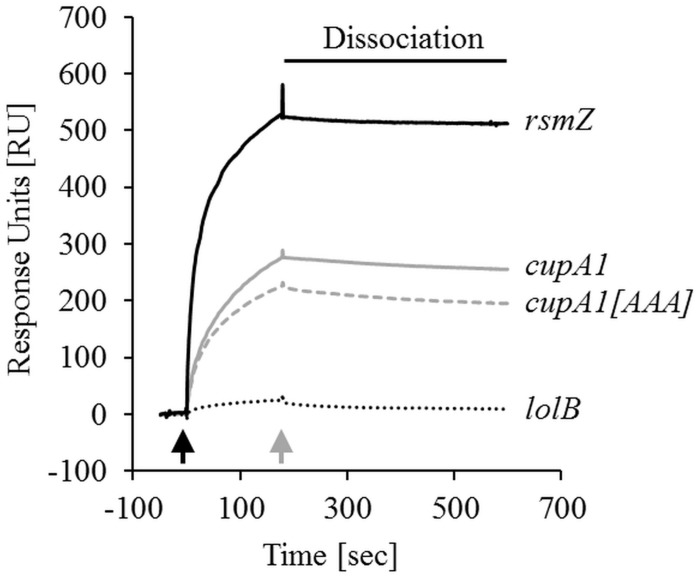
In order to test if RsmA regulates cupA1 stability by directly binding to its mRNA, we performed surface plasmon resonance measurements. For these experiments, 961 (±18.5) response units (RU) of purified RsmA-His6 protein was immobilized on a Ni-NTA sensor chip, which reliably allowed the binding analysis of different in vivo generated RNA molecules in independent runs. These experiments revealed that samples with 50, 100, and 200 nM of RNA comprised of the 200 nt upstream of the cupA1 starting codon resulted in a specific increase in RU over time (Figure 5; Supplementary Figures S1A,B). In contrast, the negative control RNA lolB showed no specific increase in RU at 200 nM, demonstrating a specific interaction of the cupA1 RNA with the immobilized RsmA protein. Compared to RNA of the known RsmA antagonist rsmZ, the cupA1 RNA displayed 67, 59, and 50% lower RU at concentrations of 50, 100, and 200 nM (determined at the end of the dissociation) when normalized to molecular weight (Figure 5; Supplementary Figures S1A,B). In addition, mutation of the core motif of the potential RsmA binding site overlapping with the SD sequence of cupA1 (GGA to AAA mutation; cupA1[AAA]) resulted in 28, 31, and 24% lower RU compared to the same concentrations of the native cupA1 RNA. Thus, RsmA appears to bind the cupA1 mRNA leader sequence and the GGA motif at the SD sequence is involved in this binding, which would be consistent with the reduced mRNA stability observed when RsmA was overexpressed in the presence of SDS.
FIGURE 5.
Protein-RNA binding studies. Surface plasmon resonance with purified RsmA-His6 protein (961 RU ± 18.5 RU) immobilized on an NTA sensor chip (GE Healthcare) and 200 nM of the in vitro generated RNAs as interaction partners, namely the cupA1 RNA, the cupA1[AAA] RNA harboring a GGA to AAA mutation of the RsmA binding site overlapping with the SD sequence, the positive control RNA rsmZ and the negative control RNA lolB. The response units (RU), which indicate direct binding to RsmA, were normalized to the molecular weight of the rsmZ RNA. The start of sample injection (black arrow) and the end of sample injection (gray arrow) are indicated. Detailed information about the experimental setup and data analysis can be found in the Supplementary Material.
SiaA/D are Required for rsmZ Expression and Increased Intracellular c-di-GMP Levels During Growth with SDS
The ability of RsmA to negatively influence macroscopic aggregate formation during growth with SDS and its impact on the stability of the cupA1 mRNA transcript raised the question of whether SiaA/D could influence RsmA activity. As such, we investigated the transcriptional activation of the known RsmA antagonists rsmY/Z during growth with SDS. Experiments with previously described rsmY and rsmZ transcriptional reporters (Chua et al., 2014) found that the transcription of rsmZ was increased threefold for wild-type cells grown with SDS when compared to those grown with succinate, whereas the transcription of rsmY was not altered (Figure 6). This increase in rsmZ transcription was dependent on both SiaA and SiaD, as no increase was observed for either of the ΔsiaA or ΔsiaD mutant strains.
FIGURE 6.
(A) Phenotypic analysis of rsmY and rsmZ overexpression in the non-aggregative ΔsiaA and ΔsiaD mutants during growth with 3.5 mM SDS. (B) Transcriptional reporter analysis of rsmZ and rsmY for strain PAO1 and the ΔsiaA and ΔsiaD mutant strains. Data is expressed as the signal intensity at 488 nm normalized to the total protein of the sample (Abs. 488 nm mg-1 protein). Bars represent the mean value from three individual experiments and are accompanied by the individual data points. Statistical significance is indicated (∗p = 0.004).
It has been previously shown that the RsmA/rsmY/Z system influences DGC expression in P. aeruginosa (Chua et al., 2014; Moscoso et al., 2014; Bouffartigues et al., 2015). Given that RsmA and SiaD inversely regulate macroscopic aggregate formation during growth with SDS, we speculated that this may involve a regulatory feedback where RsmA negatively affects the expression of siaD. The siaD gene is transcribed in an operon together with siaA, siaB (PA0171) and siaC (PA0170) (Klebensberger et al., 2009), so the 90 nt leader sequence of siaA from the previously reported transcriptional start site (Jones et al., 2014) to the translational start codon was analyzed for RsmA binding sites. Indeed, a potential primary RsmA binding site with 70% similarity to the SELEX sequence (AuAGGGAUag, sequence divergence is indicated in lowercase), including the core GGA motif (bold and underlined), was found 3–12 nucleotides upstream of the start codon of siaA (Figure 7A). Structural prediction of this sequence using mFold placed the potential binding site as part of a stem–loop structure (nucleotides 67–90) overlapping with the SD sequence of siaA, suggesting that RsmA could be involved in the regulation of siaABCD mRNA stability. In order to test this hypothesis, we quantified the mRNA decay of siaD. While the wild-type strain retained 82, 32, and 28% of the initial siaD transcript levels after 10, 20, and 30 min, the RsmA overproducing strain showed a much faster rate of decay with only 9, 2, and 1% of the initially detected transcript levels being detected at these same time points (Figure 7B).
FIGURE 7.
(A) Structural prediction of the siaABCD leader sequence using mFold (Zuker, 2003) with the predicted primary RsmA binding site (gray) (B) Impact of RsmA overexpression on the stability of siaD mRNA as determined by qRT-PCR. Data were normalized by the use of equivalent mRNA and analyzed by the 2-ΔCt method. (C) c-di-GMP levels of cells quantified as relative fluorescence units (RFU with excitation/emission of 485/535 nm) normalized to optical density by using the pCdrA::gfpc reporter for the wild-type, PAO1::rsmA [with (+) and without (-) RsmA induction (2% arabinose)], and the ΔsiaD strains. Bars represent the mean value from three (B) or six (C) individual experiments and are accompanied by the individual data points. Statistical significance compared to strain PAO1 and PAO1::rsmA is indicated (∗p = 0.001).
The ability for RsmA overproduction to reduce the stability of the mRNA encoding the DGC SiaD suggested that it could negatively impact the levels of c-di-GMP within the cell during growth with SDS. As such, we quantified the intracellular levels of c-di-GMP with the previously described c-di-GMP responsive CdrA::gfpC reporter construct for the wild-type strain, the RsmA overproducing strain and the ΔsiaD mutant during growth with SDS (Rybtke et al., 2012). While the non-induced control retained wild-type levels of c-di-GMP, the overproduction of RsmA reduced the intracellular levels of c-di-GMP to just 35.3% of those present for cells of the wild-type strain during growth with SDS (Figure 7C). Notably, a similar reduction in c-di-GMP levels was found for the ΔsiaD strain, further supporting the link between SiaA/D dependent c-di-GMP biosynthesis, the connection with RsmA and macroscopic cell aggregation.
Discussion
We have previously described that macroscopic aggregate formation of P. aeruginosa during growth with, or in the presence of the surfactant SDS (Klebensberger et al., 2006). We proposed that activation of the sensor protein and putative ser/thr phosphatase SiaA and the regulation of c-di-GMP levels by SiaD modulate this phenotype (Klebensberger et al., 2007, 2009). The functional role of SiaD in c-di-GMP biosynthesis is based on several lines of evidence. Firstly, loss of siaD or the depletion of c-di-GMP eliminates macroscopic aggregate formation (Klebensberger et al., 2009). Secondly, SiaD was found to be essential for purified Psl polysaccharide to stimulate increased c-di-GMP levels, in addition to the cyclase SadC (Irie et al., 2012). Thirdly, mutation of the active site responsible for c-di-GMP catalysis produces a siaD allele that is unable to complement the ΔsiaD mutant (this study). Fourthly, mutation of siaD strongly reduced the activity of the pCdrA::gfpC c-di-GMP responsive reporter (this study). Lastly, growth in the presence of stress-inducing tellurite was found to increase c-di-GMP levels in a SiaD dependent manner (Chua et al., 2015). The lack of c-di-GMP catalysis previously reported for purified SiaD protein (Kulesekara et al., 2006) together with the fact that the overexpression of SiaD does not complement a ΔsiaA mutant (Klebensberger et al., 2009) suggests that functional activation of SiaD is dependent on SiaA during cell stress (e.g., growth with SDS or exposure to tellurite). In addition to the predicted biosynthesis of c-di-GMP, our data further indicate a functional regulatory role for the I-site in SiaD. This is based on the observation that mutation of the I-site in SiaD produced an allele that increased macroscopic aggregation greater than the native siaD allele. This motif is well known for binding c-di-GMP and thereby limiting further activity of the DGCs in a negative feedback loop (Christen et al., 2006). From our data, we suggest that this is also a plausible explanation for its function in SiaD.
Regulation of cupA
At least three cell surface adhesins are essential for macroscopic cell aggregation during growth with, or in the presence of the surfactant SDS, and are targets for the SiaA/D pathway; the CdrB proteinaceous adhesin, the Psl polysaccharide and the CupA fimbriae (Klebensberger et al., 2009). Each of these adhesins has also been shown to play an important role in promoting surface attached biofilm formation (Vallet et al., 2001; Jackson et al., 2004; Borlee et al., 2010). In contrast to Psl and CdrAB (Burrowes et al., 2006; Brencic and Lory, 2009; Borlee et al., 2010; Irie et al., 2012), the evidence that c-di-GMP regulates CupA fimbriae is conflicting (Meissner et al., 2007; Starkey et al., 2009). Further, no role for RsmA has been suggested. In this study, we present compelling evidence that the cupA1 leader is a novel, direct target for RsmA, and show that RsmA overproduction results in loss of macroscopic aggregation during growth with SDS. Sequence analysis and structural prediction first identified several potential RsmA binding sites in the cupA1 leader. Most strikingly, one of the sites is predicted to be exposed in single stranded RNA at the end of a stem loop structure overlapping the SD sequence of cupA1, which represents the most common feature of mRNA targets directly regulated by RsmA (Timmermans and Van Melderen, 2010; Vakulskas et al., 2015). Subsequent surface plasmon resonance analysis demonstrated that in vitro generated cupA1 RNA specifically binds to purified RsmA protein. The comparison of relative binding affinities for different RsmA-RNA interactions demonstrated that rsmZ exhibits ∼2 fold stronger affinity to RsmA than that of the cupA1 RNA. Interestingly, mutation of the GGA core motif of the predicted primary binding site overlapping the SD sequence of cupA1 reduced RsmA binding, although an interaction with lower affinity was still detected that displayed stable binding over time. This could be explained by the presence of additional RsmA binding sites within the cupA1 RNA molecule (Babitzke and Romeo, 2007; Timmermans and Van Melderen, 2010; Romeo et al., 2013). The RsmA homolog CsrA functions as a homodimer that is able to bind to two locations of target mRNA that are separated by 10–63 nt (Mercante et al., 2009). The predicted additional primary RsmA binding site and the experimentally defined RsmA target in the cupA1 leader are separated by 14 nt, a distance similar to the 18 nt separation that has been described as the optimal bridging distance (Mercante et al., 2009). It has also been hypothesized that RsmA could function by a sequential binding mechanism, where an initial binding event enables the second binding surface present in the homodimer to access the mRNA (Mercante et al., 2009; Lapouge et al., 2013). The binding of multiple sites within target mRNA is known to be physiologically relevant for regulating target gene expression for CsrA (Mercante et al., 2009). While further experiments would be required to prove that the additional binding sites in the cupA1 leader are indeed functional, this study clearly shows that RsmA specifically binds the cupA1 transcript in a manner that is partially dependent on the primary binding site overlapping its SD sequence. Given that RsmA has been well characterized for its ability to impede ribosome access when binding to mRNA overlapping a SD sequence, resulting in translational repression and increased mRNA turnover (Romeo et al., 2013), our results suggest a similar mechanism for the regulation of cupA1 mRNA.
Notably, a recent sequence based informatics approach did not identify the cupA1 gene as a potential target for RsmA regulation (Kulkarni et al., 2014). However, this can be explained by the high levels of stringency required to minimize the number of false positive sequences, which is problematic because of the similarity between the RsmA consensus and the widespread SD sequence (Kulkarni et al., 2014). While the 200 nt upstream of cupA1 contains two primary and one secondary potential binding sites for RsmA, the informatics algorithm used in the aforementioned study required either three primary, or two primary and two secondary sequence sites for an mRNA leader to be subjected to further analysis (Kulkarni et al., 2014). In addition, earlier experimental approaches using ΔrsmA mutants also did not identify genes of the cupA operon as part of the RsmA regulon (Burrowes et al., 2006; Brencic and Lory, 2009). In this regard, it is important to note that these experiments were performed in rich LB medium, a growth condition in which the transcription of cupA1 is known to be strongly repressed by the global repressor MvaT, and to a lesser degree by MvaU (Vallet et al., 2004; Vallet-Gely et al., 2005, 2007). In addition, we show that the CupA fimbriae are essential for macroscopic aggregation during growth with SDS. As such, it seems possible that transcriptomic studies comparing wild-type cells with those of a ΔrsmA mutant would fail to identify the influence of RsmA on cupA1 regulation due to the use of rich medium for growth, and the low levels of cupA1 transcription in the absence of the signal generated by SDS.
The global regulator RsmA and the second messenger c-di-GMP have been shown to inversely regulate various phenotypic traits in P. aeruginosa (Frangipani et al., 2014; Moscoso et al., 2014; Chen et al., 2015; Petrova et al., 2015). Accordingly, each of these regulators is influenced by a large range of different sensory inputs. Here, we show that the SiaA/D signal transduction module regulates phenotype expression in a c-di-GMP- and RsmA dependent manner. These results are in accordance with the recent finding that SiaD influences the production of RsmA regulated Psl polysaccharide at the posttranscriptional level (Irie et al., 2012). Our present study demonstrates that the connection between SiaD and RsmA involves the presence of a functional SiaA protein and the transcriptional regulation of rsmZ. This is in line with the observation that SiaD dependent pyoverdine expression in cells with artificially increased c-di-GMP levels involves the expression of rsmY/Z (Chen et al., 2015). While the specific mechanism by which SiaA/D impacts the transcription of rsmZ remains unknown, our data and the study of Chen et al. (2015) indicate that it involves the functional activation of SiaD and as a consequent the modulation of c-di-GMP levels. Finally, we provide strong evidence that the siaABCD operon is also under regulatory control of the RsmA/rsmY/Z system. The siaABCD operon contains a potential RsmA binding site overlapping with the SD sequence, and the decay of siaD was strongly dependent on RsmA induction. This is supported by earlier microarray studies comparing a ΔrsmA and a ΔretS mutant (a hybrid sensor kinase and response regulator that regulates rsmZ to inhibit RsmA) with their respective parental controls, which identified a significant up-regulation of siaC and the complete siaABCD operon, respectively (Goodman et al., 2004; Brencic and Lory, 2009). As further support, we demonstrate that overexpression of RsmA results in a similar decrease in intracellular c-di-GMP levels during growth with SDS as the deletion of siaD, providing strong evidence of a novel interconnection between these two global regulatory networks.
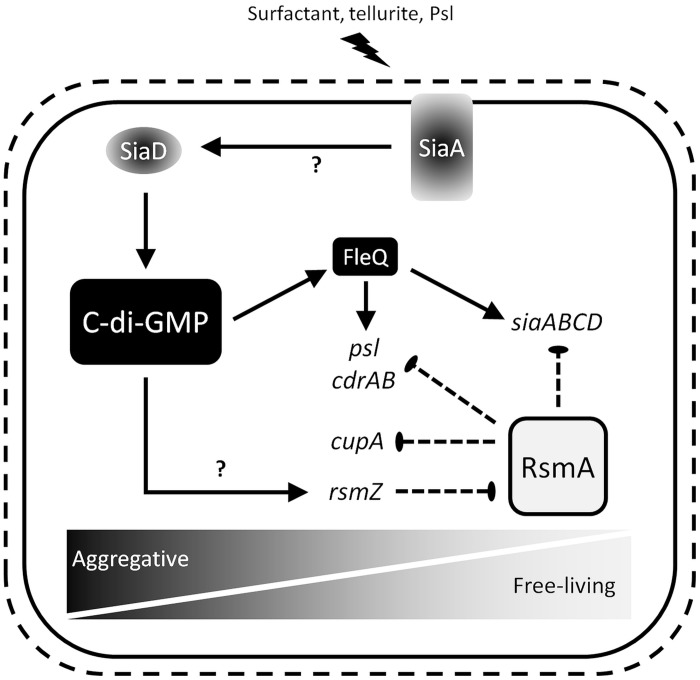
From these data, we propose a working hypothesis (Figure 8) in which SiaA/D coordinates the antagonistic activities of c-di-GMP and RsmA to control cellular aggregation in response to multiple environmental conditions, including the presence of SDS and tellurite during growth, and the signaling properties of the matrix component Psl (Irie et al., 2012; Chua et al., 2015). In the absence of surfactant stress, the DGC SiaD is inactive and thus RsmA is not repressed by the c-di-GMP dependent transcription of rsmZ, resulting in a shift toward a planktonic, non-aggregative phenotype. In the presence of surfactant stress, the activation of SiaD by SiaA influences the expression of target genes by c-di-GMP biosynthesis and the transcriptional activation of rsmZ. The exact mechanisms of SiaD and rsmZ activation remain incompletely understood, and are under current investigation in our laboratory. Notably, both RsmA, and c-di-GMP via the transcriptional regulator FleQ (Baraquet and Harwood, 2015), are also proposed to exhibit a regulatory feedback on the siaABCD operon. This complex and unique regulatory feedback circuit by which SiaA/D seems to be interconnected to c-di-GMP and RsmA signaling likely reflects the importance of appropriately regulating cellular aggregation, and could allow for a rapid and energy-efficient adaptation in response to external stimuli. This is of particular interest as both RsmA and c-di-GMP regulate phenotypic traits which are of clinical relevance, such as stress tolerance, acute virulence, and chronic persistence.
FIGURE 8.
Working hypothesis for SiaA/D function. Detailed information is given in the text.
Author Contributions
BC, SR, SK, and JK designed research. BC, VD, MC, and JK performed experiments. All authors analyzed data. BC and JK wrote the paper with contributions of VD, SR, and SK during revision.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer MS and handling Editor declared a current collaboration and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
The authors would like to thank Prof. Alain Filloux, Dr. Joana Moscoso, Prof. Urs Jenal, Dr. Tina Jaeger, Prof. Matthew Parsek, Dr. Melissa Starkey, Prof. Yang Liang, and Mr. Philipp Scheller for valuable discussions and providing different strains and plasmids. Prof. Bernhard Hauer is acknowledged for his continuous support. We further gratefully acknowledge the financial support of Dr. JK by the Deutsche Forschungsgemeinschaft (DFG, KL 2340/2-1) and support from the Centre for Marine Bio-Innovation.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00179
References
- Amikam D., Galperin M. Y. (2006). PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22 3–6. 10.1093/bioinformatics/bti739 [DOI] [PubMed] [Google Scholar]
- Asghari F. B., Nikaeen M., Mirhendi H. (2013). Rapid monitoring of Pseudomonas aeruginosa in hospital water systems: a key priority in prevention of nosocomial infection. FEMS Microbiol. Lett. 343 77–81. 10.1111/1574-6968.12132 [DOI] [PubMed] [Google Scholar]
- Babitzke P., Romeo T. (2007). CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10 156–163. 10.1016/j.mib.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Baraquet C., Harwood C. S. (2015). A FleQ DNA binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa. J. Bacteriol. 198 178–186. 10.1128/JB.00539-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. (1951). Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T. (2013). The role of bacterial biofilms in chronic infections. APMIS 121 1–58. 10.1111/apm.12099 [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T., Jensen P. O., Fiandaca M. J., Pedersen J., Hansen C. R., Andersen C. B., et al. (2009). Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44 547–558. 10.1002/ppul.21011 [DOI] [PubMed] [Google Scholar]
- Boehm A., Kaiser M., Li H., Spangler C., Kasper C. A., Ackermann M., et al. (2010). Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141 107–116. 10.1016/j.cell.2010.01.018 [DOI] [PubMed] [Google Scholar]
- Borlee B. R., Goldman A. D., Murakami K., Samudrala R., Wozniak D. J., Parsek M. R. (2010). Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75 827–842. 10.1111/j.1365-2958.2009.06991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffartigues E., Moscoso J. A., Duchesne R., Rosay T., Fito-Boncompte L., Gicquel G., et al. (2015). The absence of the Pseudomonas aeruginosa OprF protein leads to increased biofilm formation through variation in c-di-GMP level. Front. Microbiol. 6:630 10.3389/fmicb.2015.00630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A., Lory S. (2009). Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72 612–632. 10.1111/j.1365-2958.2009.06670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A., McFarland K. A., McManus H. R., Castang S., Mogno I., Dove S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73 434–445. 10.1111/j.1365-2958.2009.06782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrowes E., Baysse C., Adams C., O’Gara F. (2006). Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152 405–418. 10.1099/mic.0.28324-0 [DOI] [PubMed] [Google Scholar]
- Chambers J. R., Liao J., Schurr M. J., Sauer K. (2014). BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol. Microbiol. 92 471–487. 10.1111/mmi.12562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yuan M., Mohanty A., Yam J. K., Liu Y., Chua S. L., et al. (2015). Multiple diguanylate cyclase-coordinated regulation of pyoverdine synthesis in Pseudomonas aeruginosa. Environ Microbiol. Rep. 7 498–507. 10.1111/1758-2229.12278 [DOI] [PubMed] [Google Scholar]
- Choi K. H., Gaynor J. B., White K. G., Lopez C., Bosio C. M., Karkhoff-Schweizer R. R., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2 443–448. 10.1038/nmeth765 [DOI] [PubMed] [Google Scholar]
- Christen B., Christen M., Paul R., Schmid F., Folcher M., Jenoe P., et al. (2006). Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281 32015–32024. 10.1074/jbc.M603589200 [DOI] [PubMed] [Google Scholar]
- Chua S. L., Liu Y., Yam J. K., Chen Y., Vejborg R. M., Tan B. G., et al. (2014). Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 5:4462 10.1038/ncomms5462 [DOI] [PubMed] [Google Scholar]
- Chua S. L., Sivakumar K., Rybtke M., Yuan M., Andersen J. B., Nielsen T. E., et al. (2015). C-di-GMP regulates Pseudomonas aeruginosa stress response to tellurite during both planktonic and biofilm modes of growth. Sci. Rep. 5:10052 10.1038/srep10052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggan K. A., Wolfgang M. C. (2012). Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 14 47–70. [PubMed] [Google Scholar]
- Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49 711–745. 10.1146/annurev.mi.49.100195.003431 [DOI] [PubMed] [Google Scholar]
- Davey M. E., O’Toole G. A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64 847–867. 10.1128/MMBR.64.4.847-867.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey A. K., Baker C. S., Romeo T., Babitzke P. (2005). RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11 1579–1587. 10.1261/rna.2990205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangipani E., Visaggio D., Heeb S., Kaever V., Camara M., Visca P., et al. (2014). The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ. Microbiol. 16 676–688. 10.1111/1462-2920.12164 [DOI] [PubMed] [Google Scholar]
- Goodman A. L., Kulasekara B., Rietsch A., Boyd D., Smith R. S., Lory S. (2004). A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7 745–754. 10.1016/j.devcel.2004.08.020 [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W., Stoodley P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2 95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Hickman J. W., Harwood C. S. (2008). Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69 376–389. 10.1111/j.1365-2958.2008.06281.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212 77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Huang L.-K., Wang M.-J. J. (1995). Image thresholding by minimizing the measures of fuzziness. Pattern Recognit. 28 41–51. 10.1016/0031-3203(94)E0043-K [DOI] [Google Scholar]
- Irie Y., Borlee B. R., O’Connor J. R., Hill P. J., Harwood C. S., Wozniak D. J., et al. (2012). Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 109 20632–20636. 10.1073/pnas.1217993109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y., Starkey M., Edwards A. N., Wozniak D. J., Romeo T., Parsek M. R. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78 158–172. 10.1111/j.1365-2958.2010.07320.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson K. D., Starkey M., Kremer S., Parsek M. R., Wozniak D. J. (2004). Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186 4466–4475. 10.1128/JB.186.14.4466-4475.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., et al. (2003). Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100 14339–14344. 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Newsom D., Kelly B., Irie Y., Jennings L. K., Xu B., et al. (2014). ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10:e1003984 10.1371/journal.ppat.1003984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E., Humair B., Denervaud V., Riedel K., Spahr S., Eberl L., et al. (2006). Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 188 6026–6033. 10.1128/JB.00409-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr K. G., Snelling A. M. (2009). Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 73 338–344. 10.1016/j.jhin.2009.04.020 [DOI] [PubMed] [Google Scholar]
- Kirisits M. J., Prost L., Starkey M., Parsek M. R. (2005). Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71 4809–4821. 10.1128/AEM.71.8.4809-4821.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebensberger J., Birkenmaier A., Geffers R., Kjelleberg S., Philipp B. (2009). SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa. Environ. Microbiol. 11 3073–3086. 10.1111/j.1462-2920.2009.02012.x [DOI] [PubMed] [Google Scholar]
- Klebensberger J., Lautenschlager K., Bressler D., Wingender J., Philipp B. (2007). Detergent-induced cell aggregation in subpopulations of Pseudomonas aeruginosa as a preadaptive survival strategy. Environ. Microbiol. 9 2247–2259. 10.1111/j.1462-2920.2007.01339.x [DOI] [PubMed] [Google Scholar]
- Klebensberger J., Rui O., Fritz E., Schink B., Philipp B. (2006). Cell aggregation of Pseudomonas aeruginosa strain PAO1 as an energy-dependent stress response during growth with sodium dodecyl sulfate. Arch. Microbiol. 185 417–427. 10.1007/s00203-006-0111-y [DOI] [PubMed] [Google Scholar]
- Kong W., Chen L., Zhao J., Shen T., Surette M. G., Shen L., et al. (2013). Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 88 784–797. 10.1111/mmi.12223 [DOI] [PubMed] [Google Scholar]
- Kulesekara H., Lee V., Brencic A., Liberati N., Urbach J., Miyata S., et al. (2006). Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3’-5’)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U.S.A. 103 2839–2844. 10.1073/pnas.0511090103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni P. R., Jia T., Kuehne S. A., Kerkering T. M., Morris E. R., Searle M. S., et al. (2014). A sequence-based approach for prediction of CsrA/RsmA targets in bacteria with experimental validation in Pseudomonas aeruginosa. Nucleic Acids Res. 42 6811–6825. 10.1093/nar/gku309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K., Perozzo R., Iwaszkiewicz J., Bertelli C., Zoete V., Michielin O., et al. (2013). RNA pentaloop structures as effective targets of regulators belonging to the RsmA/CsrA protein family. RNA Biol. 10 1031–1041. 10.4161/rna.24771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Malone J. G., Jaeger T., Spangler C., Ritz D., Spang A., Arrieumerlou C., et al. (2010). YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 6:e1000804 10.1371/journal.ppat.1000804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A., Wild V., Simm R., Rohde M., Erck C., Bredenbruch F., et al. (2007). Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ. Microbiol. 9 2475–2485. 10.1111/j.1462-2920.2007.01366.x [DOI] [PubMed] [Google Scholar]
- Mercante J., Edwards A. N., Dubey A. K., Babitzke P., Romeo T. (2009). Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J. Mol. Biol. 392 511–528. 10.1016/j.jmb.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso J. A., Jaeger T., Valentini M., Hui K., Jenal U., Filloux A. (2014). The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 196 4081–4088. 10.1128/JB.01850-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan J., Reen F. J., Adams C., O’Gara F. (2011). Low oxygen induces the type III secretion system in Pseudomonas aeruginosa via modulation of the small RNAs rsmZ and rsmY. Microbiology 157 3417–3428. 10.1099/mic.0.052050-0 [DOI] [PubMed] [Google Scholar]
- Petrova O. E., Cherny K. E., Sauer K. (2015). The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J. Bacteriol 197 174–187. 10.1128/JB.02244-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova O. E., Sauer K. (2010). The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 192 5275–5288. 10.1128/JB.00387-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T., Vakulskas C. A., Babitzke P. (2013). Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15 313–324. 10.1111/j.1462-2920.2012.02794.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U., Galperin M. Y., Gomelsky M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77 1–52. 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybtke M. T., Borlee B. R., Murakami K., Irie Y., Hentzer M., Nielsen T. E., et al. (2012). Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78 5060–5069. 10.1128/AEM.00414-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleheck D., Barraud N., Klebensberger J., Webb J. S., McDougald D., Rice S. A., et al. (2009). Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS ONE 4:e5513 10.1371/journal.pone.0005513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger-Domenigg T., Sonnleitner E., Kaberdin V. R., Blasi U. (2007). Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 352 769–773. 10.1016/j.bbrc.2006.11.084 [DOI] [PubMed] [Google Scholar]
- Starkey M., Hickman J. H., Ma L., Zhang N., De Long S., Hinz A., et al. (2009). Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 191 3492–3503. 10.1128/JB.00119-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N., Lee E. R., Weinberg Z., Moy R. H., Kim J. N., Link K. H., et al. (2008). Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321 411–413. 10.1126/science.1159519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans J., Van Melderen L. (2010). Post-transcriptional global regulation by CsrA in bacteria. Cell. Mol. Life Sci. 67 2897–2908. 10.1007/s00018-010-0381-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trampari E., Stevenson C. E., Little R. H., Wilhelm T., Lawson D. M., Malone J. G. (2015). Bacterial rotary export ATPases are allosterically regulated by the nucleotide second messenger Cyclic-di-GMP. J. Biol. Chem. 290 24470–24483. 10.1074/jbc.M115.661439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulskas C. A., Potts A. H., Babitzke P., Ahmer B. M., Romeo T. (2015). Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 79 193–224. 10.1128/MMBR.00052-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet I., Diggle S. P., Stacey R. E., Camara M., Ventre I., Lory S., et al. (2004). Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol. 186 2880–2890. 10.1128/JB.186.9.2880-2890.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet I., Olson J. W., Lory S., Lazdunski A., Filloux A. (2001). The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 98 6911–6916. 10.1073/pnas.111551898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I., Donovan K. E., Fang R., Joung J. K., Dove S. L. (2005). Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 102 11082–11087. 10.1073/pnas.0502663102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I., Sharp J. S., Dove S. L. (2007). Local and global regulators linking anaerobiosis to cupA fimbrial gene expression in Pseudomonas aeruginosa. J. Bacteriol. 189 8667–8676. 10.1128/JB.01344-1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. M., Armbruster C. R., Arduino M. J. (2013). Plumbing of hospital premises is a reservoir for opportunistically pathogenic microorganisms: a review. Biofouling 29 147–162. 10.1080/08927014.2012.757308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.