Abstract
The therapeutic monoclonal antibody (mAb) TGN1412 (anti-CD28 superagonist) caused near-fatal cytokine release syndrome (CRS) in all six volunteers during a phase-I clinical trial. Several cytokine release assays (CRAs) with reported predictivity for TGN1412-induced CRS have since been developed for the preclinical safety testing of new therapeutic mAbs. The whole blood (WB) CRA is the most widely used, but its sensitivity for TGN1412-like cytokine release was recently criticized. In a comparative study, using group size required for 90% power with 5% significance as a measure of sensitivity, we found that WB and 10% (v/v) WB CRAs were the least sensitive for TGN1412 as these required the largest group sizes (n = 52 and 79, respectively). In contrast, the peripheral blood mononuclear cell (PBMC) solid phase (SP) CRA was the most sensitive for TGN1412 as it required the smallest group size (n = 4). Similarly, the PBMC SP CRA was more sensitive than the WB CRA for muromonab-CD3 (anti-CD3) which stimulates TGN1412-like cytokine release (n = 4 and 4519, respectively). Conversely, the WB CRA was far more sensitive than the PBMC SP CRA for alemtuzumab (anti-CD52) which stimulates FcγRI-mediated cytokine release (n = 8 and 180, respectively). Investigation of potential factors contributing to the different sensitivities revealed that removal of red blood cells (RBCs) from WB permitted PBMC-like TGN1412 responses in a SP CRA, which in turn could be inhibited by the addition of the RBC membrane protein glycophorin A (GYPA); this observation likely underlies, at least in part, the poor sensitivity of WB CRA for TGN1412. The use of PBMC SP CRA for the detection of TGN1412-like cytokine release is recommended in conjunction with adequately powered group sizes for dependable preclinical safety testing of new therapeutic mAbs.
Abbreviations: CRA, cytokine release assay; WB, whole blood; PBMCs, peripheral blood mononuclear cells; WBC, white blood cells; SP, Solid Phase; AQ, Aqueous Phase; RBCs, Red Blood Cells; mAb, monoclonal antibody; GYPA, glycophorin A; CRS, cytokine release syndrome; PBS, phosphate buffered saline; HDC, high density culture; IL-2, interleukin-2; IL-8, interleukin-8; IL-13, interleukin-13; IL-17, interleukin-17; TNFα, tumor necrosis factor alpha; IFNγ, interferon gamma; IL-2R, interleukin-2 receptor; TCR, T cell receptor
Keywords: Cytokine release assay, Group size, Whole blood, PBMC, Therapeutic mAb, TGN1412
Highlights
-
•
Sensitivity of different CRA formats determined by power analysis of group sizes.
-
•
PBMC solid phase CRAs were highly predictive for TGN1412 but not alemtuzumab.
-
•
Whole blood CRAs were predictive for alemtuzumab but poorly predictive for TGN1412.
-
•
TGN1412 cytokine release was inhibited by glycophorin A and red blood cells.
1. Introduction
One of the most dangerous side effects caused by the administration of therapeutic monoclonal antibodies is CRS or “cytokine storm”. The most dramatic example to date occurred in March 2006, during the first-in-man phase-I clinical trial of the therapeutic mAb TGN1412. TGN1412 caused severe infusion reactions in all six healthy volunteers characterized by the rapid systemic release of high levels of pro-inflammatory cytokines later termed a “cytokine storm” (Suntharalingam et al., 2006). Pre-clinical safety testing, which included in vitro incubation of TGN1412 in aqueous-phase (AQ) with human PBMC and intravenous administration of TGN1412 to cynomolgus macaques, had both failed to predict an adverse response in man (TeGenero, 2005, Stebbings et al., 2007). This failure to predict cytokine release in man demonstrated an urgent need for a new generation of CRAs that would be both sensitive and predictive of clinical outcome in man (Vidal et al., 2010). Several groups subsequently developed CRA methods with reported predictivity for TGN1412, although the in vivo mechanism resulting in TGN1412-induced cytokine storm in man is not fully understood. These CRA methods include: PBMC incubated with therapeutic mAbs either wet or dry coated onto tissue culture plates for SP presentation (Stebbings et al., 2007, Eastwood et al., 2010, Eastwood et al., 2013), WB incubated with therapeutic mAbs bound to protein A capture beads for SP presentation (Walker et al., 2011, Xiong et al., 2014); high density pre-culture (HDC) of PBMC followed by incubation with AQ therapeutic mAbs (Römer et al., 2011, Bartholomaeus et al., 2014); WB or diluted WB incubated with AQ therapeutic mAbs (Wolf et al., 2012, Bailey et al., 2013); and co-culturing PBMC with AQ therapeutic mAbs over human umbilical vein endothelial cells (Stebbings et al., 2007, Findlay et al., 2011, Dhir et al., 2012), although this latter approach is not widely used. However, for the safety evaluation of new therapeutic mAbs there is no consensus regarding which CRA methods are the most relevant, which controls are the most appropriate, which cytokine measurements are the most appropriate, what group size should be employed and how that data is best interpreted (European Medicines Agency, 2007, Finco et al., 2014).
A recent survey found that the WB CRA was the most widely used method, even though the WB CRA has been criticized for being poorly predictive for TGN1412 activity in vivo due to the low levels of cytokine release obtained and the low frequency of responding donors obtained, when compared to the PBMC SP CRA and in vivo responses (Finco et al., 2014, Thorpe et al., 2013). Conversely, it has been argued that the WB CRA more closely mimics the in vivo environment and contains factors at physiological concentration that may influence mechanisms of cytokine release whereas PBMC isolation results in the removal of RBCs, neutrophils, platelets, serum components and other factors (Wolf et al., 2012, Wolf et al., 2013, Finco et al., 2014). Moreover, WB CRA can be performed more rapidly and conveniently than PBMC CRA in non-specialized settings using smaller volumes of blood. Since the consequence of failing to predict an adverse response in man would be very serious, as occurred in 2006, a sensitive CRA with a high statistical power of 90% (probability of detecting a response if present) with a significance level of 5% (probability of obtaining a false positive or negative result) is required for the best chance of detecting a true effect if present. An insensitive CRA is more likely to be underpowered and have a greater chance of false positive and negative results due to the requirement for very large group sizes in excess of numbers typically or that can be practically employed (Finco et al., 2014). In addition, an insensitive CRA would lack the dynamic range required to distinguish between mild, moderate and severe CRS, so it couldn't be claimed to be predictive of clinical outcome (Thorpe et al., 2013, Stebbings et al., 2013). To better understand the differences between WB and PBMC CRAs, we compared the most widely used of these methods with a range of therapeutic mAbs associated with severe (TGN1412; Suntharalingam et al., 2006, Stebbings et al., 2007), moderate (muronomab-CD3; Suthanthiran et al., 1989, Chatenoud et al., 1990) and mild-moderate (alemtuzumab and rituximab; Wing et al., 1996, Moreau et al., 1996, Winkler et al., 1999, Lim et al., 1999) CRS, using the group size required for 90% power and 5% significance as a measure of sensitivity.
2. Materials and methods
2.1. Therapeutic mAbs, controls and CRA methods
CRAs were performed using the following therapeutic mAbs: TGN1412 (humanized IgG4k, anti-CD28 superagonist; TeGenero AG), rituximab (chimeric mouse/human IgG1κ, anti-CD20, mabthera; Hoffmann–La Roche Ltd), alemtuzumab (humanized IgG1, anti-CD52, Campath-1H; Genzyme Corp.), muromonab-CD3 (murine IgG2a, anti-CD3, Orthoclone-OKT3; Janssen-Cilag Ltd). Purified human IgG4κ and IgG1 were used as isotype controls (AMS Biotechnology Ltd, UK) and anti-CD28 agonist (Biolegend, UK) was used as a control to assess specificity for a CD28 superagonist. Sodium azide was removed from control mAbs using Amicon® Ultra-4 centrifugal filter units (Millipore Ltd, UK) and confirmed endotoxin-free using the limulus amebocyte lysate gel clot test. Since human FcγRI and FcγRIIIa bind murine IgG2a as per human IgG1, muromonab-CD3 responses were compared to human IgG1 isotype control (Bruhns, 2012). Phytohaemagglutinin (PHA; Sigma-Aldrich Ltd, UK) at 10 μg ml− 1 was used as a positive control. PBMC SP CRAs were performed by wet coating wells of microtitre plates at a mAb concentration of 1 μg well− 1 for 1 h, followed by washing to remove unbound mAb and then the addition of PBMC for 48 h, as previously described (Eastwood et al., 2010, Eastwood et al., 2013). PBMC HDC CRAs were performed by addition of mAb at 1 μg ml− 1, to PBMC pre-incubated at high density for 48 h, and 24 h stimulation, as previously described (Römer et al., 2011, Bartholomaeus et al., 2014). WB and 10% (v/v) WB CRAs were performed at a mAb concentration of 5 μg ml− 1 and 48 h stimulation, as previously described (Wolf et al., 2012, Bailey et al., 2013). All concentrations were chosen for comparability with previously published findings on cytokine release using these methods. Although not a published method, WB SP and 10% (v/v) WB SP CRAs were performed as per the PBMC SP CRA with a mAb coating concentration of 1 μg well− 1, as controls. All assays were carried out in 96-well round bottom microtitre plates (Sigma Aldrich Ltd), PBMC SP and HDC CRA utilized 2 × 105 PBMC in 200 μl of complete media per well, the WB CRA utilized 200 μl of WB per well containing 0.7–2.0 × 106 WBCs and the 10% (v/v) WB CRA utilized a tenth of the latter in complete media at 200 μl well− 1. CRA comparisons were performed using the same set of donors, except for the PBMC HDC CRA which utilized a different set of 8 donors. The effect of selective depletion of RBCs from heparinized WB was assessed, using EasySep™ glycophorin A positive cell depletion cocktail (Stemcell Technologies, UK) according to the manufacturer's instructions. Briefly, buffy coats were prepared from whole blood by centrifugation and incubated with anti-glycophorin A reagent for 15 min at room temperature before addition of magnetic nanoparticles added and a further incubation for 10 min. RBCs were then removed by immuno-magnetic separation. Depletion of 95–99% of RBCs was confirmed by visual assessment. The resultant white blood cell (WBC) suspension consisting almost entirely of plasma depleted polymorphonuclear leukocytes, lymphocytes and monocytes was adjusted to 1 × 106 ml− 1 in complete media and used in a WBC SP CRA at 200 μl well− 1 and a mAb coating concentration of 1 μg well− 1 TGN1412. To investigate inhibition of TGN1412-associated cytokine release, WBCs were resuspended in autologous WB or 10–200 μg well− 1 of GYPA (Sigma-Aldrich Ltd) was added. To assess the effect of IL-2 on TGN1412-associated cytokine release, daclizumab (IL-2R antagonist) at a concentration of 5 μg ml− 1 was added to cells and pre-incubated for 10 min prior to plating in a PBMC SP CRA as described above. Concentrations of IFNγ, IL-2, IL-13 and IL-8 in culture supernatants were measured using custom made MSD plates, according to the manufacturer's instructions (Meso Scale Discovery, USA). TNFα and IL-17 concentrations were quantified by ELISA as previously described (Eastwood et al., 2010, Eastwood et al., 2013).
2.2. Statistical analysis
Statistical analysis was performed using Minitab 16 software (Minitab). A general linear model was employed for repeated measures analysis of log10 transformed data using Tukey's test with 95% confidence to counteract for multiple comparisons. Group size requirements were calculated for paired t-tests on log transformed data to compare each mAb treatment against the IgG1 control, except for TGN1412 which was compared to the IgG4 and CD28 agonist controls. All mAb and cytokine response combinations were assumed to have equally variable responses for each CRA method and the standard deviations used for the calculations were pooled.
3. Results
3.1. The PBMC SP CRA is highly predictive for TGN1412-induced cytokine release
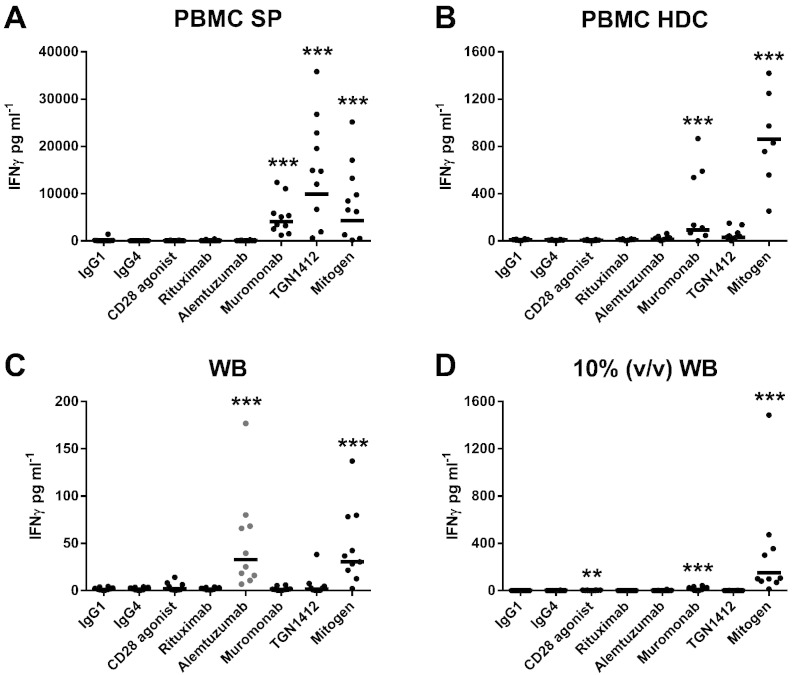
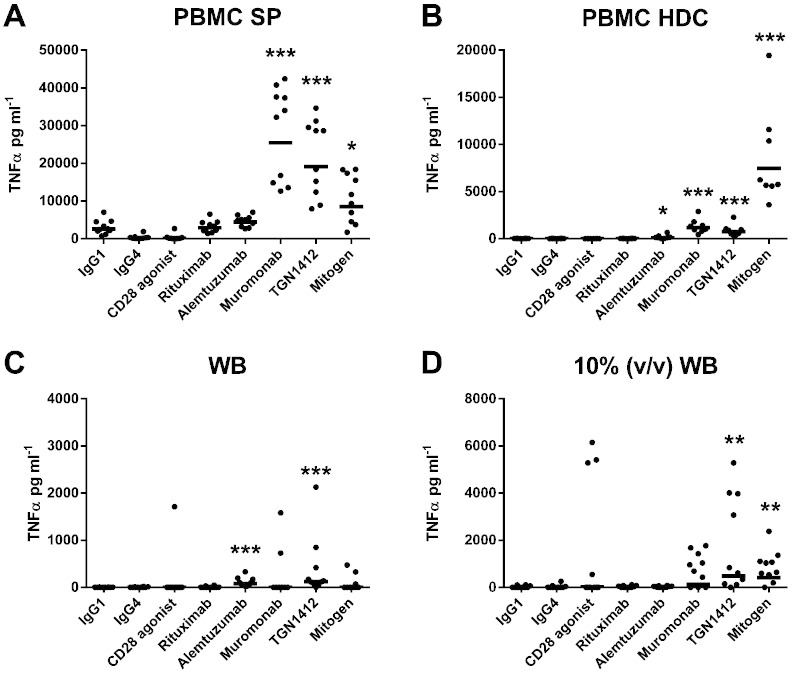
The levels of IFNγ, IL-2 and TNFα release following stimulation with a range of therapeutic mAbs using different CRA methods are shown in Fig. 1, Fig. 2, Fig. 3, respectively. The levels of IL-8, IL-13 and IL-17 release are shown in Table 1. Only the PBMC SP CRA produced significant IFNγ (p < 0.0001 and 9908 pg ml− 1), IL-8 (p < 0.0001 and 136,048 pg ml− 1), IL-13 (p < 0.0001 and 2451 pg ml− 1) and IL-17 (p < 0.0001 and 346 pg ml− 1) release with TGN1412 (Fig. 1, Table 1). Both the PBMC SP and PBMC HDC CRAs produced significant IL-2 release (both p < 0.0001, 5469 pg ml− 1 and 158 pg ml− 1, respectively) with TGN1412 (Fig. 2). In order of magnitude, the PBMC SP, PBMC HDC, 10% (v/v) WB and WB CRAs all produced significant TNFα release (p < 0.0001 and 19,090 pg ml− 1, p < 0.0001 and 748 pg ml− 1, p = 0.0046 and 488 pg ml− 1, p = 0.0003 and 125 pg ml− 1, respectively), with TGN1412 (Fig. 3). Low but significant amounts of IFNγ (p = 0.0015 and 2.4 pg ml− 1), IL-2 (p = 0.0019 and 15 pg ml− 1) and IL-13 (p < 0.0001 and 41 pg ml− 1) release was observed with the 10% (v/v) WB CRA and CD28 agonist control (Fig. 1, Fig. 2, Table 1). Significant IL-13 (p = 0.0044 and 300 pg ml− 1) release was also noted with the PBMC SP CRA and CD28 agonist control, but the level obtained with TGN1412 was in comparison significantly (p < 0.0001) greater (Table 1).
Fig. 1.
Analysis of IFNγ release obtained with different CRA methods: IFNγ release from human (A) PBMC SP (1 μg well− 1 coating concentration), (B) PBMC HDC (1 μg ml− 1), (C) WB and (D) 10% (v/v) WB CRA (5 μg ml− 1) stimulated with the therapeutic mAbs Rituximab, Alemtuzumab, Muromonab-CD3 and TGN1412. PBMC SP and HDC CRA utilized 2 × 105 PBMC in 200 μl of complete media per well, the WB CRA utilized 200 μl of WB per well containing 0.7–2.0 × 106 WBCs of which 2–8 × 105 were lymphocytes and the 10% (v/v) WB CRA utilized a tenth of the latter in 200 μl well− 1. IFNγ release expressed in pg ml− 1 was assessed after 48 h stimulation, with the exception of the PBMC HDC CRA which utilizes a 48 h HDC followed by 24 h stimulation with mAbs. PHA at 10 μg ml− 1 was used as a mitogen control. All responses were compared against IgG1 isotype control with the exception of TGN1412, which was compared against IgG4κ isotype control. Significant results are denoted by asterisks (repeated measures analysis of log10 transformed data using Tukey's method with 95.0% confidence — *p < 0.05, **p < 0.001, ***p < 0.0001). The geometric mean of group responses is denoted by a horizontal bar. Donor responses shown are n = 10 from 3 independent experiments, except for the PBMC HDC CRA for which n = 8 from 2 independent experiments. CRA comparisons were performed using the same set of donors, except for the PBMC HDC CRA which utilized a different set of 8 donors.
Fig. 2.
Analysis of IL-2 release obtained with different CRA methods: IL-2 release from human (A) PBMC SP (1 μg well− 1 coating concentration), (B) PBMC HDC (1 μg ml− 1), (C) WB and (D) 10% (v/v) WB CRA (5 μg ml− 1) stimulated with the therapeutic mAbs rituximab, alemtuzumab, muromonab-CD3 and TGN1412. PBMC SP and HDC CRA utilized 2 × 105 PBMC in 200 μl of complete media per well, the WB CRA utilized 200 μl of WB per well containing 0.7–2.0 × 106 WBCs of which 2–8 × 105 were lymphocytes and the 10% (v/v) WB CRA utilized a tenth of the latter in 200 μl well− 1. IFNγ release expressed in pg ml− 1 was assessed after 48 h stimulation, with the exception of the PBMC HDC CRA which utilizes a 48 h HDC followed by 24 h stimulation with mAbs. PHA at 10 μg ml− 1 was used as a mitogen control. All responses were compared against IgG1 isotype control with the exception of TGN1412, which was compared against IgG4κ isotype control. Significant results are denoted by asterisks (repeated measures analysis of log10 transformed data using Tukey's method with 95.0% confidence — *p < 0.05, **p < 0.001, ***p < 0.0001). The geometric mean of group responses is denoted by a horizontal bar. Donor responses shown are n = 10 from 3 independent experiments, except for the PBMC HDC CRA for which n = 8 from 2 independent experiments. CRA comparisons were performed using the same set of donors, except for the PBMC HDC CRA which utilized a different set of 8 donors.
Fig. 3.
Analysis of TNFα release obtained with different CRA methods: TNFα release from human (A) PBMC SP (1 μg well− 1 coating concentration), (B) PBMC HDC (1 μg ml− 1), (C) WB and (D) 10% (v/v) WB CRA (5 μg ml− 1) stimulated with the therapeutic mAbs rituximab, alemtuzumab, muromonab-CD3 and TGN1412. PBMC SP and HDC CRAs utilized 2 × 105 PBMC in 200 μl of complete media per well, the WB CRA utilized 200 μl of WB per well containing 0.7–2.0 × 106 WBCs of which 2–8 × 105 were lymphocytes and the 10% (v/v) WB CRA utilized a tenth of the latter in 200 μl well− 1. IFNγ release expressed in pg ml− 1 was assessed after 48 h stimulation, with the exception of the PBMC HDC CRA which utilizes a 48 h HDC followed by 24 h stimulation with mAbs. PHA at 10 μg ml− 1 was used as a mitogen control. All responses were compared against IgG1 isotype control with the exception of TGN1412, which was compared against IgG4κ isotype control. Significant results are denoted by asterisks (repeated measures analysis of log10 transformed data using Tukey's method with 95.0% confidence — *p < 0.05, **p < 0.001, ***p < 0.0001). The geometric mean of group responses is denoted by a horizontal bar. Donor responses shown are n = 10 from 3 independent experiments, except for the PBMC HDC CRA for which n = 8 from 2 independent experiments. CRA comparisons were performed using the same set of donors, except for the PBMC HDC CRA which utilized a different set of 8 donors.
Table 1.
Comparative analysis of IL-8, IL-13 and IL-17 release with different CRA methods: Cytokine release expressed in pg ml− 1 from human PBMC SP (1 μg well− 1 coating concentration), PBMC HDC (1 μg ml− 1), WB and 10% (v/v) WB CRA (5 μg ml− 1) stimulated with the indicated therapeutic mAbs after 48 h stimulation, with the exception of the PBMC HDC CRA which utilizes a 48 h HDC followed by 24 h stimulation with mAbs. PHA at 10 μg ml− 1 was used as a mitogen control. All responses were compared against IgG1 isotype control with the exception of TGN1412, which was compared against IgG4κ isotype control. Group responses shown are the geometric means followed by the 95% confidence intervals in brackets. Significant results (p < 0.05) are shown in bold (repeated measures analysis of log10 transformed data using Tukey's method with 95.0% confidence). Donor responses shown are n = 10 from 3 independent experiments, except for the PBMC HDC CRA for which n = 8 from 2 independent experiments. CRA comparisons were performed using the same set of donors, except for the PBMC HDC CRA which utilized a different set of 8 donors.
| Cytokine | CRA method | IgG1 control | IgG4 control | Rituximab | Alemtuzumab | Muromonab-CD3 | TGN1412 | CD28 agonist | Mitogen control |
|---|---|---|---|---|---|---|---|---|---|
| IL-8 | PBMC SP | 9202 (8696, 9738) | 7066 (6206, 8045) | 7520 (6803, 8313) | 7648 (7088, 8254) | 193,381 (150,646, 248,239) | 136,048 (91,305, 202,716) | 9291 (2934, 29,415) | 210,755 (156,467, 283,879) |
| PBMC HDC | 3566 (2715, 4683) | 3156 (2098, 4746) | 1117 (53, 23383) | 1137 (54, 23921) | 1148(54, 24,265) | 1137 (54, 23,946) | 2510 (1298, 4854) | 4112 (3279, 5155) | |
| WB | 2102 (879, 5028) | 2538 (1038, 6205) | 1815 (896, 3676) | 7162 (5033, 10192) | 1256 (814, 1938) | 2474 (842, 7274) | 1504 (882, 2566) | 19,529 (6838, 55,771) | |
| 10% (v/v) WB | 188 (88, 400) | 252 (79, 806) | 82 (41, 163) | 302 (224, 408) | 2855 (1156, 7048) | 83 (41, 170) | 130 (55, 304) | 41,750 (20,714, 84,151) | |
| IL-13 | PBMC SP | 140 (91, 213) | 67 (45, 101) | 136 (112, 166) | 192 (113, 326) | 1625 (1289, 2049) | 2451 (1719, 3494) | 300 (172, 521) | 1932 (1402, 2662) |
| PBMC HDC | 119 (80, 176) | 101 (66, 155) | 94 (24, 364) | 96 (24, 383) | 184 (38, 899) | 293 (55, 1567) | 67 (31, 147) | 779 (553, 1098) | |
| WB | 13 (6.6, 24) | 15 (7.6, 28) | 16 (11, 24) | 39 (27, 56) | 9.7 (3.7, 25) | 19 (5.4, 68) | 31 (14, 68) | 224 (102, 493) | |
| 10% (v/v) WB | 2.9 (1.8, 4.9) | 3.6 (1.8, 7.3) | 2 (1.5, 2.5) | 3 (2.3, 3.9) | 26 (6.8, 100) | 6.4 (2.3, 18) | 41 (22, 77) | 372 (218, 635) | |
| IL-17 | PBMC SP | 1.3 (0.75, 2.1) | 2.3 (0.66, 7.7) | 1.6 (0.79, 3.1) | 1.9 (0.72, 5) | 629 (365, 1085) | 346 (123, 974) | 1.7 (0.75, 3.9) | 71 (36, 138) |
| PBMC HDC | 1.5 (0.58, 3.8) | 2.7 (0.83, 9) | 3.5 (0.8, 16) | 3.5 (0.77, 16) | 1.6 (0.55, 4.4) | 3 (0.54, 17) | 2.7 (0.84, 8.5) | 6.4 (1.1, 37) | |
| WB | 1.6 (0.78, 3.3) | 1.5 (0.8, 2.8) | 1.4 (0.64, 3.2) | 1.4 (0.65, 3) | 1.4 (0.63, 3.3) | 1 (1, 1) | 2 (0.87, 4.7) | 1.2 (0.77, 2) | |
| 10% (v/v) WB | 1.3 (0.71, 2.4) | 4.1 (1.3, 13) | 1.8 (0.73, 4.4) | 1.7 (0.73, 4.2) | 3.5 (1.1, 11) | 2.2 (0.86, 5.9) | 2.5 (0.84, 7.3) | 152 (42, 552) |
In order of magnitude, the PBMC SP, PBMC HDC and 10% (v/v) WB CRAs all produced significant IFNγ (p < 0.0001 and 4035 pg ml− 1, p = 0.0006 and 92 pg ml− 1, p < 0.0001 and 9.7 pg ml− 1, respectively) and IL-2 (p < 0.0001 and 547 pg ml− 1, p = 0.0185 and 54 pg ml− 1, p < 0.0001 and 25 pg ml− 1, respectively) release following stimulation with muromonab-CD3 (Fig. 1, Fig. 2). Only the PBMC SP and PBMC HDC CRAs produced significant TNFα release (p < 0.0001 and 25,437 pg ml− 1, p < 0.0001 and 1173 pg ml− 1, respectively) with muromonab-CD3 (Fig. 3), whereas, only the PBMC SP and 10% (v/v) WB CRAs produced significant IL-8 (p < 0.0001 and 193,381 pg ml− 1, p < 0.0001 and 2855 pg ml− 1, respectively) and IL-13 (p < 0.0001 and 1625 pg ml− 1, p = 0.0001 and 26 pg ml− 1, respectively) release with muromonab-CD3 (Table 1). Only the PBMC SP CRA produced significant IL-17 (p < 0.0001 and 629 pg ml− 1) release with muromonab-CD3 (Table 1). No significant cytokine release was measured using the WB CRA with muromonab-CD3.
3.2. The WB CRA is predictive for alemtuzumab-induced cytokine release
Only the WB CRA produced significant IFNγ (p < 0.0001 and p = 33 pg ml− 1) and IL-8 (p = 0.0089 and 7162 pg ml− 1) release with alemtuzumab (Fig. 1, Table 1). Both the PBMC HDC and WB CRAs induced significant TNFα release (p = 0.0005 and 81 pg ml− 1, p = 0.0105 and 83 pg ml− 1, respectively) with alemtuzumab (Fig. 3). No significant cytokine release with alemtuzumab was observed using the PBMC SP and 10% (v/v) WB CRAs (Fig. 1, Fig. 2, Fig. 3, Table 1). Interestingly, if presentation of alemtuzumab in a WB CRA was changed from AQ to SP then no significant cytokine release was detected (Supplementary Table 1). However, using the WB or 10% (v/v) WB SP CRA, low but significant IL-2 (p = 0.0097 and 24 pg ml− 1, p = 0.0025 and 7.6 pg ml− 1, respectively) and TNFα (p = 0.0043 and 135 pg ml− 1, p = 0.0414 and 11 pg ml− 1) release was achieved with TGN1412 (Supplementary Table 1). No significant cytokine release was observed with rituximab using any CRA method tested.
3.3. The WB CRA is not predictive for TGN1412-induced cytokine release
Shown in Table 2 are estimated group sizes required by each CRA method to detect significant cytokine release at the 5% significance level with 90% power for the therapeutic mAbs alemtuzumab, muromonab-CD3 and TGN1412 compared to isotype control. For the detection of TGN1412 responses the least sensitive method was the 10% (v/v) WB CRA followed by the WB CRA which required group sizes of n = 79 and 52, respectively. The WB CRA was essentially insensitive to muromonab-CD3 as it required a group size of n = 4519. Paradoxically, the 10% (v/v) WB and 10% (v/v) WB SP CRAs were the second most sensitive methods for muromonab-CD3 as they only required group sizes of n = 9 and n = 8, respectively (Table 2). The PBMC SP CRA was the most sensitive method for TGN1412 and muromonab-CD3, requiring a group size of just n = 4 for both mAbs, whereas the PBMC HDC CRA required group sizes of n = 27 and 31 for TGN1412 and muromonab-CD3, respectively. Compared to the PBMC HDC CRA, the WB SP CRA required a smaller group size (n = 13) for TGN1412, but a larger group size (n = 43) for muromonab-CD3 (Table 2). While the WB CRA was the most sensitive method for alemtuzumab requiring a group size of n = 8, the 10% (v/v) WB SP, 10% (v/v) WB, PBMC SP, PBMC HDC and SP WB CRA were not sensitive to alemtuzumab as these required group sizes of n = 66, 114, 180, 341 and 5069, respectively (Table 2). Group sizes required for 80% power are shown in Supplementary Table 2.
Table 2.
Estimated group sizes required to detect cytokine release with different CRA methods with a power of 90%: Group size requirements for human PBMC SP, PBMC HDC, WB and 10% (v/v) WB CRA, WB SP and 10% (v/v) WB SP were calculated for paired t-tests on log transformed data to compare each therapeutic mAb against IgG1 isotype control, with the exception of TGN1412 which was compared to IgG4κ isotype control and a CD28 agonist*. All therapeutic mAb and cytokine response combinations were assumed to have equally variable responses for each CRA method and the standard deviations used for the calculations were pooled standard deviations (all 6 cytokine responses for the different therapeutic mAbs). The group sizes given for each therapeutic mAb would detect the difference observed in cytokine release for each CRA method with a power of 90% at the 5% significance level.
| CRA method | Power analysis | Alemtuzumab | Muromonab-CD3 | TGN1412 | TGN1412* |
|---|---|---|---|---|---|
| PBMC SP | Relative mean | 1.2 | 32.9 | 110.9 | 71.8 |
| Group size required | 180 | 4 | 4 | 5 | |
| PBMC HDC | relative mean | 1.5 | 4.0 | 4.4 | 6.7 |
| Group size required | 341 | 31 | 27 | 19 | |
| WB | Relative mean | 5.5 | 0.9 | 2.2 | 1.7 |
| Group size required | 8 | 4519 | 52 | 117 | |
| 10% (v/v) WB | Relative mean | 1.5 | 13.7 | 2.1 | 0.7 |
| Group size required | 114 | 9 | 79 | 284 | |
| WB SP | Relative mean | 0.9 | 3.4 | 3.4 | 4.3 |
| Group size required | 5069 | 43 | 13 | 13 | |
| 10% (v/v) WB SP | Relative mean | 1.8 | 24.3 | 2.4 | 1.4 |
| Group size required | 66 | 8 | 46 | 313 |
Although individual donors responded to the CD28 agonist control with IFNγ, IL-2 and TNFα release in the WB CRA, group responses were not significant (Fig. 1, Fig. 2, Fig. 3, respectively). In the 10% (v/v) WB CRA significant but very low IFNγ (2.4 pg ml− 1, p = 0.0015) and IL-2 release and (27 pg ml− 1, p < 0.0001), but not TNFα release, was obtained with the CD28 agonist control (Fig. 1, Fig. 2, respectively). To determine the specificity of different CRA for a CD28 superagonist, group sizes were estimated compared to CD28 agonist controls (Table 2). For the WB, 10% (v/v) WB and 10% (v/v) SP WB CRAs the group sizes required to distinguish between TGN1412 and the CD28 agonist control were more than doubled, but for the PBMC SP, PBMC HDC and WB SP CRAs the group size requirements barely changed (Table 2). By calculating the group sizes required for 90% significance with the omission of TNFα release, the robustness of these results were further tested by excluded reliance upon a single cytokine (Supplementary Table 3). For the PBMC SP CRA the group size required to detect a TGN1412 response was unaffected (n = 4) whereas for the WB and 10% (v/v) WB CRA the group sizes required were dramatically increased (n = 1135 and n = 18,843, respectively).
3.4. RBCs, GYPA and daclizumab inhibit TGN1412-induced cytokine release
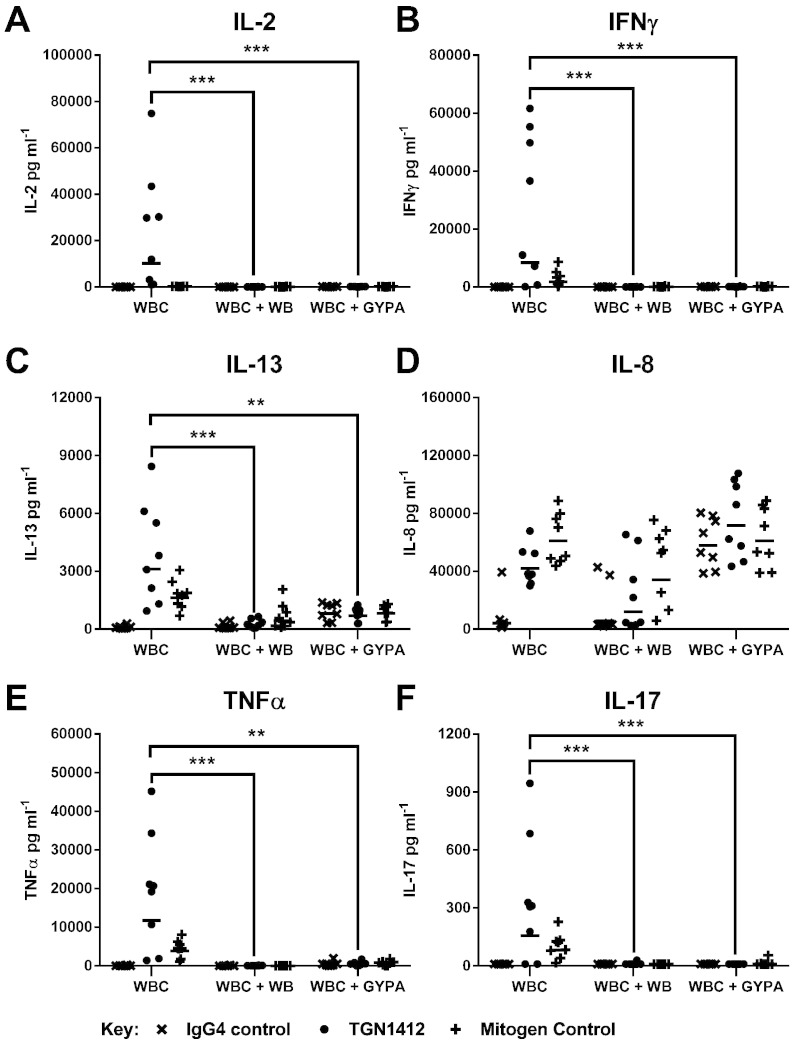
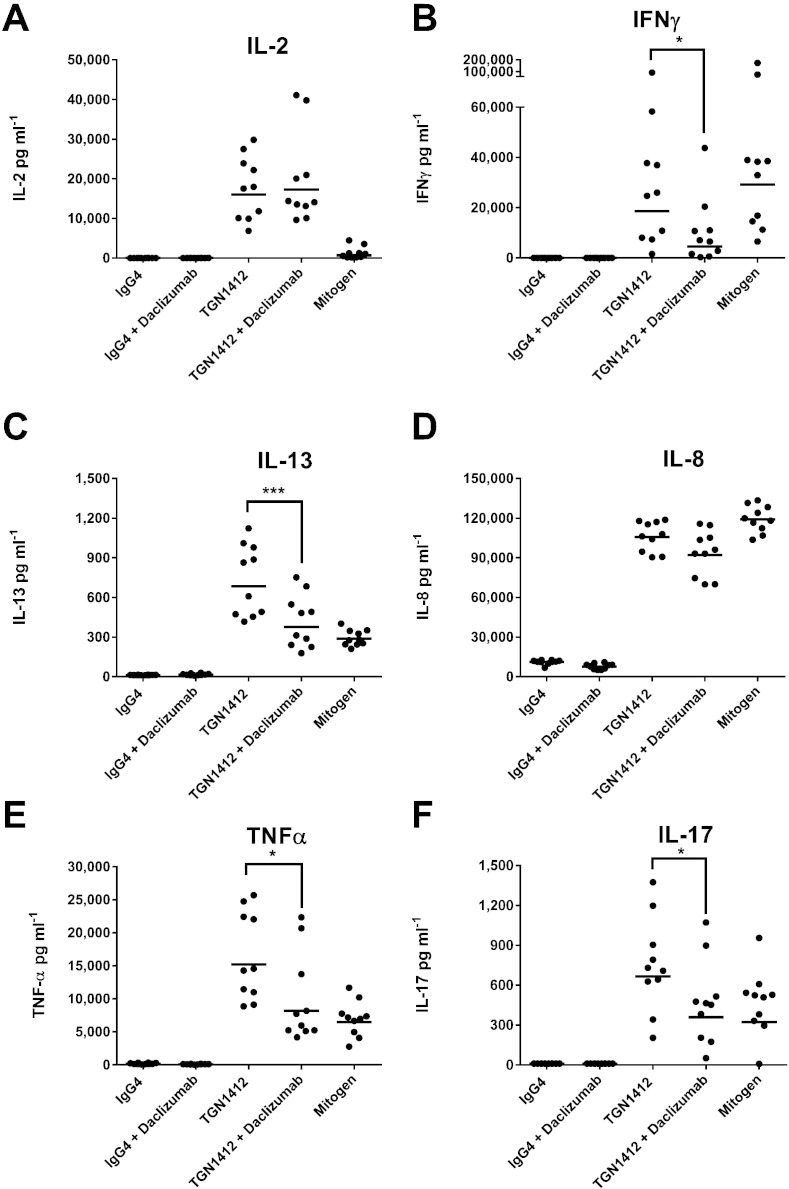
Depletion of RBCs from WB allowed significant (all p < 0.0001) IL-2 (10,220 pg ml− 1), IFNγ (8366 pg ml− 1), IL-13 (3113 pg ml− 1), IL-8 (41,913 pg ml− 1), TNFα (11,750 pg ml− 1) and IL-17 (156 pg m1− 1) release to occur with TGN1412 in a WBC SP CRA (Fig. 4). Re-suspension of WBC in WB prior to stimulation with TGN1412 in a SP CRA significantly (all p < 0.0001) inhibited IL-2, IFNγ, IL-13, TNFα and IL-17, but not IL-8 release (Fig. 4). The addition of GYPA to WBC SP CRA also significantly inhibited TGN1412 mediated IL-2 (p < 0.0001), IFNγ (p < 0.0001), IL-13 (p = 0.0017), TNFα (p = 0.0026) and IL-17 (p < 0.0001) release, but not IL-8 release (Fig. 4). A second batch of GYPA used in the range 10–200 μg well− 1 gave similar results that were dose dependent (Supplementary Fig. 1). A significant (p < 0.0001) increase in IL-8 release with the addition of GYPA to WB SP CRA control wells was retrospectively ascribed to contamination with 6.0 IU mg− 1 of endotoxin and the second batch of GYPA with 2.4 IU mg− 1 endotoxin (data not shown). Contamination of GYPA with these levels of endotoxin was not associated with any reduction in cell viability when used in the range 10–200 μg well− 1 (Supplementary Fig. 2). Addition of daclizumab to PBMC SP CRA significantly reduced IFN-γ (p = 0.011), IL-13 (p = 0.0001), TNF-α (p = 0.031) and IL-17 (p = 0.031) release with TGN1412, but did not significantly reduce IL-8 release (Fig. 5). No significant reduction in IL-2 release was noted with TGN1412 and daclizumab (Fig. 5).
Fig. 4.
RBCs and GYPA inhibit TGN1412-associated cytokine release: (A) IL-2, (B) IFNγ, (C) IL-13, (D) IL-8, (E) TNFα and (F) IL-17 release expressed in pg ml− 1 was assessed using a human WBC SP CRA stimulated for 48 h with a coating concentration of 1 μg well− 1 of TGN1412 and IgG4κ isotype control. PHA at 10 μg ml− 1 was used as a mitogen control. WBCs consisted of RBCs and plasma depleted polymorphonuclear leukocytes, lymphocytes and monocytes resuspended in complete media and adjusted to 1 × 106 ml. WBCs + WB consisted of WBCs resuspended in autologous WB. WBCs + GYPA consisted of WBCs in complete media to which 200 μg well− 1 of GYPA was added. Significant results are denoted by asterisks (repeated measures analysis of log10 transformed data using Tukey's method with 95.0% confidence — *p < 0.05, **p < 0.001, ***p < 0.0001). The geometric mean of group responses is denoted by a horizontal bar. Donor responses shown are n = 8 from 2 independent experiments.
Fig. 5.
Daclizumab inhibits TGN1412-associated cytokine release: (A) IL-2, (B) IFNγ, (C) IL-13, (D) IL-8, (E) TNFα and (F) IL-17 release expressed in pg ml− 1 was assessed using a human PBMC SP CRA stimulated for 48 h with a coating concentration of 1 μg well− 1 of TGN1412 and IgG4κ isotype control. PHA at 10 μg ml− 1 was used as a mitogen control. The influence of pre-incubation of PBMC for 10 min with 1 μg ml− 1 of daclizumab (anti-CD25) on cytokine release with TGN1412 was compared. Significant results are denoted by asterisks (repeated measures analysis of log10 transformed data using Tukey's method with 95.0% confidence — *p < 0.05, **p < 0.001, ***p < 0.0001). The geometric mean of group responses is denoted by a horizontal bar. Donor responses shown are n = 10 from 3 independent experiments.
4. Discussion
This comparative study demonstrates that in terms of the group size required to detect a real effect, the PBMC SP CRA is by far the most sensitive method for the detection of TGN1412-like cytokine release (n = 4), whereas WB CRAs are the least sensitive (n = 52–79). Of concern is a survey which reports that the majority of laboratories and contract research organizations use WB CRAs with between n = 6 and 14 donors to detect a TGN1412-like response, which our results show is statistically underpowered and therefore poorly predictive of clinical outcome in man (Finco et al., 2014). Despite recent criticism, it is argued that the WB CRA is a valid approach to detect TGN1412-like cytokine release (Thorpe et al., 2013, Wolf et al., 2013). Yet, even for candidate screening of therapeutic mAbs, where only relatively large effects may be of interest and a power of 80% may be sufficient since false positive or negative results would not have serious consequences, WB CRAs still require very large group sizes (n = 39–59).
Signaling through the co-stimulatory molecule CD28, either by its natural ligands CD80/CD86 or an agonist mAb cannot induce T cell activation and cytokine release without simultaneous engagement of the T cell receptor, which restricts responses to antigen-specific cells which typically comprise only 0.01–1.0% of the total T cell population (Acuto and Michel, 2003, Rudd and Schneider, 2003, Harari et al., 2004, Calarota and Baldanti, 2013). In contrast, a CD28 superagonist such as TGN1412 is able to dispense with primary signaling through the TCR and is capable of polyclonal activation of all antigen-specific T cells (Lühder et al., 2003, Stebbings et al., 2007, Waibler et al., 2008). Therefore, greater levels of cytokine release would be expected in a CRA stimulated with a CD28 superagonist compared to a CD28 agonist, used here as a control. Yet here the responses of WB CRAs to a CD28 agonist control resembled or in certain cases exceeded that of TGN1412, indicating poor specificity and very weak responses to TGN1412. Some cytokine release at very low levels would be expected within a subset of donors as a CD28 agonist would be capable of the re-stimulation of recently primed T-cells (e.g. recent vaccination or infection). It is possible that AQ TGN1412 is behaving like a CD28 agonist in the WB CRAs and that the low levels of cytokine release observed within a subset of donors may have been misinterpreted as a superagonist response in the absence of appropriate controls. For example, the use of natalizumab (anti-α4 integrin) as an isotype control for TGN1412 is not recommended as it suppresses background TNFα and IL-8 release, which may lead to over interpretation of weak positive or background cytokine release (Eastwood et al., 2013). When TGN1412 responses were compared with a CD28 agonist control the group size requirement for 90% power more than doubled for WB CRAs (n = 117–284), further illustrating poor specificity and low responses to TGN1412. Moreover, greater cytokine release overall was obtained with a CD28 agonist control than TGN1412 using the 10% (v/v) WB CRA method. In contrast, the group sizes required for the PBMC SP to detect a TGN1412 response compared with a CD28 agonist control hardly changed (n = 5).
The PBMC HDC CRA was the third most sensitive method for the detection of TGN1412 induced cytokine release, in terms of the group size required (n = 27). However, the 31 pg ml− 1 of IFNγ release obtained here is considerably lower than the > 1000 pg ml− 1 reported by Rӧmer et al. (2011). One difference between these assays is that we only used freshly isolated PBMC, whereas Rӧmer et al. (2011) used PBMC recovered from leukoreduction filters which are discarded after the preparation of therapeutic red blood cell concentrates from individual blood donations. We have found that cells recovered from leukoreduction filters gave higher background cytokine release implying non-specific priming occurs during processing, which may have enhanced IFNγ release with TGN1412. A longer stimulation of 48 h with the PBMC HDC CRA may have resulted in higher levels of IFNγ release, as is seen with the PBMC SP CRA. Alternatively, the majority of IFNγ release reported by Rӧmer et al. (2011) may not in fact have come from CD4 + T cells. As the level of TGN1412-associated IL-2 release with the PBMC HDC was comparable with Rӧmer et al. (2011), both methods likely stimulate similar levels of CD4 + T cells since this cytokine is restricted to production by this cell type, whereas IFNγ release is not restricted so it could have come from a different cell type e.g. monocytes or neutrophils. Intriguingly, the WB SP CRA included as a control, but which is akin to the protein A bead WB CRA method (Walker et al., 2011, Xiong et al., 2014), proved more sensitive for TGN1412 than the PBMC HDC CRA (n = 13), likely due to significant IL-13 release in addition to TNFα and IL-2, a situation not observed with muromonab-CD3. Differences in the pattern of TGN1412-associated cytokines detected with different CRA may reflect the strength of the signal; with TNFα being the easiest to detect, followed by IL-2 and IFNγ, then IL-13 and IL-17 being the hardest to detect. Conversely, different patterns of cytokine release may reflect the involvement of different subsets or mechanisms or in the case of WB CRA the inability to detect certain cytokines due to with the inhibitory effects of GYPA.
Criticism of SP presentation of therapeutic mAbs as artificial or the use of PBMC compared to WB as unrepresentative of the in vivo environment is not valid as only the combination of both these elements produces the most sensitive CRA to detect TGN1412-like cytokine responses with a power of 90% and a significance level of 5% (Stebbings et al., 2007, Römer et al., 2011, Wolf et al., 2012). It is reported that binding of TGN1412 to inhibitory FcγRIIb is necessary for in vitro superagonistic activity with the PBMC HDC CRA and in vivo with agonistic anti-TNFR antibodies (Bartholomaeus et al., 2014, Hussain et al., 2015, Li and Ravetch, 2012, White et al., 2014). If FcγRIIIb cross-linking was responsible for the TGN1412 cytokine storm in vivo, then SP presentation of mAbs in vitro at least partially emulates it. A qualitative factor to consider is that TGN1412 caused the release of high levels of multiple pro-inflammatory cytokines in trial volunteers and that the PBMC SP CRA similarly produced high levels of all 6 cytokines measured here, with the caveat that one is an in vivo response and the other an in vitro assay. In contrast, the WB CRA produced significant release of just one out of six cytokines: TNFα at levels 153 fold lower than the PBMC SP CRA. The next best CRA with TGN1412 was the PBMC HDC CRA which produced significant release of TNFα and IL-2, but at levels that were still 25.5 and 34.6 times lower, respectively, than with the PBMC SP CRA. Thus, in terms of the range and magnitude of cytokine release obtained here we have concluded that compared to other CRA the PBMC SP method more closely reflects the observed biological response to TGN1412 in vivo (Suntharalingam et al., 2006, Stebbings et al., 2007, Eastwood et al., 2013).
Selective depletion of the RBC component in WB demonstrated its central role in inhibiting TGN1412-associated cytokine release in the WB CRA. However, it should be noted that this result was achieved in the context of SP presentation and that depletion of RBCs alone would be unlikely to fully restore responses to TGN1412 as it would be similar to the insensitive PBMC CRA which failed to predict the severity of the in vivo outcome (TeGenero, 2005, Stebbings et al., 2007). Conversely, the absence of a RBC component in PBMC explains why the PBMC SP CRA proved to be a much more sensitive method for the detection of TGN1412-like cytokine release than WB CRAs. Since TGN1412 predominantly stimulates T cells, then this finding is also consistent with the reported underestimation of antigen-specific T cell cytokine responses using WB assays (Stebbings et al., 2007, Hoffmeister et al., 2003). An examination of the literature provided a potential mechanism to account for the ability of RBCs to block the response to TGN1412: GYPA, a major membrane glycoprotein of RBCs, can interact with IL-2 and inhibit IL-2-dependent T cell proliferation (Chu and Sharom, 1992). This potential mechanism was confirmed when purified GYPA was shown to block TGN1412-associated cytokine release in a dose dependent manner when added to a WBC SP CRA. Therefore, the presence of RBCs likely at least partially explains the poor sensitivity of WB CRAs for TGN1412-associated cytokine release, mediated through the abundant RBC membrane protein GYPA (106 copies per cell).
TGN1412 and CD28 stimulation in vitro are both characterized by IL-2 release (Stebbings et al., 2007, Van Berkel and Oosterwegel, 2006). Moreover, TGN1412-associated IL-2 release has been ascribed to the stimulation of CD4 + effector memory T cells and shown to be dysregulated (Eastwood et al., 2010, Eastwood et al., 2013). Although daclizumab-mediated inhibition of IL-2 signaling significantly inhibited TGN1412-associated TNFα, IFNγ, IL-13 and IL-17 release, it did not inhibit IL-2 release, implying that this cytokine amplifies the release of other cytokines. Since CD4 + effector memory T cells are more responsive to IL-7 and IL-15 than IL-2, then they would be less sensitive to daclizumab, whereas CD4 + naïve and central memory T cells are more responsive to IL-2 so would be sensitive to daclizumab (Geginat et al., 2001). However, the inhibition of cytokine release by daclizumab was not as profound as that observed with RBCs or GYPA. Some IL-2 signaling may have occurred via the intermediate-affinity IL-2R (βγc chain complex) present on resting T cells and CD56bright NK cells, since daclizumab only blocks the high-affinity IL-2R (αβγc chain complex) present on activated T cells (Sheridan et al., 2011, Liao et al., 2013). Alternatively, GYPA may also be interacting with other cytokines to suppress TGN1412-associated cytokine release or the measurement of those cytokines. Alemtuzumab does not stimulate T cell mediated cytokine release but rather activating FcγR (FcγRI, FcγRIIa, FcγRIIc, FcγRIIIa) mediated cytokine release from NK cells, monocytes and neutrophils (Wing et al., 1996, Hu et al., 2009, Siders et al., 2010). Neutrophils account for 40–75% of total WBC but are depleted during density gradient isolation of PBMC, which may explain the poor sensitivity of PBMC SP CRA for alemtuzumab-associated cytokine release. Conversely, the presence of neutrophils in WB may be responsible for the greater sensitivity of WB CRA for alemtuzumab, compared to the PBMC SP CRA. Neutrophils can also inhibit T cell responses which may explain the opposing sensitivities of WB and 10% (v/v) WB CRA for muromonab-CD3 and alemtuzumab if related to the dilution of this component (McKenna et al., 2009, Afonso et al., 2010).
While high levels of alemtuzumab-associated cytokine release were observed with the PBMC SP CRA, only levels of TNFα release markedly exceeded the levels of other cytokines compared to the isotype control due to the high background. This background was likely caused by activating FcγR-mediated cytokine release due to cross-linking caused by SP presentation, as it is observed with therapeutic mAbs of the IgG1 subclass that have activating FcγR mediated effector function e.g. alemtuzumab (Eastwood et al., 2013). No background was observed with TGN1412 since agonistic and antagonistic therapeutic mAbs tend to be of the IgG4 subclass due to its very low FcγRIIa, FcγRIIc and FcγRIIIa binding affinity or have been engineered to silence unwanted activating FcγR binding (Ascierto et al., 2010, Alegre et al., 1992). Conversely, the small amount of cytokines produced in WB CRA assays raises the biological significance of results even when they are statistically significant when compared to isotype controls e.g. 33 pg ml− 1 of IFNγ with alemtuzumab in the WB CRA. However, even lower levels than this are considered significant by some proponents of WB CRA, which questions the significance and validity of very low levels of cytokine release. The validity of statistically significant but low level cytokine release may be enhanced if the patterns of cytokines released are consistent with a particular mechanism or cellular repertoire. For example, in the case of IFNγ release with alemtuzumab in the WB CRA it was accompanied by significant TNFα and IL-8 release but no T cell-associated IL-2 release, which is consistent with an activating FcγRs, mediated mechanism. However, for TGN1412 only significant TNFα release was observed with the WB CRA, which alone cannot be ascribed to a particular mechanism and when excluded from the analysis dramatically increases the group size requirement (n = 1135) to the point of insensitivity. In contrast, exclusion of TNFα release has no effect on the predictive power of the PBMC SP CRA (n = 4) because the response is much broader and not reliant upon a single cytokine response.
The presence of RBCs does not appear to inhibit alemtuzumab associated cytokine release, presumably because this does not involve a T cell mediated mechanism driven by IL-2 and GYPA is not known to affect other pro-inflammatory pathways e.g. IL-1 (Chu and Sharom, 1992). NK cells have been implicated in alemtuzumab mediated cytokine release but as these are not depleted by PBMC isolation they are unlikely to account for observed differences between PBMC SP and WB CRA sensitivity (Wing et al., 1996). Rituximab is associated with adverse events in patients with B-cell chronic lymphocytic leukemia (B-CLL), ascribed to activating FcγRs mediated cytokine release, but we observed no significant rituximab mediated cytokine release with any CRA (Winkler et al., 1999, Lim et al., 1999). We have previously detected NK cell cytokine production with rituximab in a PBMC SP CRA when using intracellular cytokine staining as the readout, but here there was insufficient released cytokine for quantification (Stebbings et al., 2013). Since we used normal healthy donors for PBMC then it is possible that when compared to B-CLL patients there were insufficient B-cell targets for rituximab to stimulate significant cytokine release. Enriching the B-cell fraction of normal donor WB to emulate B-CLL patients may have resulted in more significant cytokine release. Rituximab is not negative in all CRA (Walker et al., 2011), in particular IL-6 release and B-cell activation is prominent in vitro and in vivo but this cytokine was not included in our panel (Agarwal et al., 2004, Jones et al., 2014).
The levels of cytokine release detected across the board with the PBMC SP CRA and TGN1412 are vastly higher than those generated using the WB CRA. In terms of group size required for 90% power and 5% significance, the PBMC SP CRA is highly predictive for TGN1412 and WB CRA are poorly predictive. Inhibition of TGN1412-associated cytokine release by RBCs/GYPA provides a clear and convincing explanation for the observed differences in cytokine levels release between the PBMC and WB CRA, at least when using a SP format. It is clear that a “one size fits all” approach for the preclinical safety testing of therapeutic mAbs does not work because different mechanisms of cytokine release require different CRA for dependable sensitivity in conjunction with appropriately powered groups.
Disclosure of conflicts of interest
The authors declare no competing financial interests or conflict of interests to declare.
Authorship contributions
Contribution: RS and SV conceived and designed the experiments, SV, DE, BF and SS performed experiments; SV, DE, BF, RS and SS analyzed results; TD performed statistical analysis; SV, JS, SJT, RT and RS wrote the manuscript.
Acknowledgements
The authors thank the staff of NIBSC who kindly donated their blood for this study.
This work was funded by the UK Department of Health project “NIBSC — Regulatory science research unit”.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jim.2015.04.020.
Contributor Information
S. Vessillier, Email: Sandrine.vessillier@nibsc.org.
R. Stebbings, Email: stebbingsr@medimmune.com.
Appendix A. Supplementary data
Additional analyses of cytokine release and inhibitory effect of GYPA.
References
- Acuto O., Michel F. CD28-mediated co-stimulation: a quantitative support for TCR signalling. Nat. Rev. Immunol. 2003;3:939. doi: 10.1038/nri1248. [DOI] [PubMed] [Google Scholar]
- Alegre M.-L., Collins A.M., Pulito V.L., Brosius R.A., Olson W.C., Zivin R.A., Knowles R., Thistlethwaite J.R., Jolliffe L.K., Bluestone J.A. Effect of a single amino acid mutation on the activating and immunosuppressive properties of a “humanized” OKT3 monoclonal antibody. J. Immunol. 1992;148:3461. [PubMed] [Google Scholar]
- Afonso G., Scotto M., Renand A., Arvastsson J., Vassilieff D., Cilio C.M., Mallone R. Critical parameters in blood processing for T-cell assays: validation on ELISpot and tetramer platforms. J. Immunol. Methods. 2010;359:28. doi: 10.1016/j.jim.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Agarwal A., Vieira C.A., Book B.K., Sidner R.A., Fineberg N.S., Pescovitz M.D. Rituximab, anti-CD20, induces in vivo cytokine release but does not impair ex vivo T-cell responses. Am. J. Transplant. 2004;4:1357. doi: 10.1111/j.1600-6143.2004.00502.x. [DOI] [PubMed] [Google Scholar]
- Ascierto P.A., Simeone E., Sznol M., Fu Y.X., Melero I. Clinical experiences with anti-CD137 and anti-PD1 therapeutic antibodies. Semin. Oncol. 2010;37:508. doi: 10.1053/j.seminoncol.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Bailey L., Moreno L., Manigold T., Krasniqi S., Kropshofer H., Hinton H., Singer T., Suter L., Hansel T.T., Mitchell J.A. A simple whole blood bioassay detects cytokine responses to anti-CD28SA and anti-CD52 antibodies. J. Pharmacol. Toxicol. Methods. 2013;68:231. doi: 10.1016/j.vascn.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Bartholomaeus P., Semmler L.Y., Bukur T., Boisguerin V., Römer P.S., Tabares P., Chuvpilo S., Tyrsin D.Y., Matskevich A., Hengel H., Castle J., Hünig T., Kalinke U. Cell contact-dependent priming and Fc interaction with CD32 + immune cells contribute to the TGN1412-triggered cytokine response. J. Immunol. 2014;192:2091. doi: 10.4049/jimmunol.1302461. [DOI] [PubMed] [Google Scholar]
- Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. 2012;119:5640. doi: 10.1182/blood-2012-01-380121. [DOI] [PubMed] [Google Scholar]
- Calarota S.A., Baldanti F. Enumeration and characterization of human memory T cells by enzyme-linked immunospot assays. Clin. Dev. Immunol. 2013;2013:637649. doi: 10.1155/2013/637649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatenoud L., Ferran C., Legendre C., Thouard I., Merite S., Reuter A., Gevaert Y., Kreis H., Franchimont P., Bach J.F. In vivo cell activation following OKT3 administration. Systemic cytokine release and modulation by corticosteroids. Transplantation. 1990;49:697. doi: 10.1097/00007890-199004000-00009. [DOI] [PubMed] [Google Scholar]
- Chu J.W., Sharom F.J. Glycophorin A interacts with interleukin-2 and inhibits interleukin-2-dependent T-lymphocyte proliferation. Cell. Immunol. 1992;145:223. doi: 10.1016/0008-8749(92)90327-l. [DOI] [PubMed] [Google Scholar]
- Dhir V., Fort M., Mahmood A., Higbee R., Warren W., Narayanan P., Wittman V. A predictive biomimetic model of cytokine release induced by TGN1412 and other therapeutic monoclonal antibodies. J. Immunotoxicol. 2012;9:34. doi: 10.3109/1547691X.2011.613419. [DOI] [PubMed] [Google Scholar]
- Eastwood D., Findlay L., Poole S., Bird C., Wadhwa M., Moore M., Burns C., Thorpe R., Stebbings R. Monoclonal antibody TGN1412 trial failure explained by species differences in CD28 expression on CD4 + effector memory T-cells. Br. J. Pharmacol. 2010;161:512. doi: 10.1111/j.1476-5381.2010.00922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood D., Bird C., Dilger P., Hockley J., Findlay L., Poole S., Thorpe S.J., Wadhwa M., Thorpe R., Stebbings R. Severity of the TGN1412 trial disaster cytokine storm correlated with IL-2 release. Br. J. Clin. Pharmacol. 2013;76:299. doi: 10.1111/bcp.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Medicines Agency . CHMP/SWP/28367/07. 2007. Guideline on strategies to identify and mitigate risks for first-in-human clinical trials with investigational medicinal products. (Available at: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/09/WC500002988.pdf (last accessed 20/3/2015)) [Google Scholar]
- Finco D., Grimaldi C., Fort M., Kiessling A., Wolf B., Salcedo T., Faggioni R., Schneider A., Ibraghimov A., Scesney S., Serna D., Prell R., Stebbings R., Narayanan P.K. Cytokine release assays: current practices and future directions. Cytokine. 2014;66:143. doi: 10.1016/j.cyto.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Findlay L., Sharp G., Fox B., Ball C., Robinson C.J., Bird C., Stebbings R., Eastwood D., Wadhwa M., Poole S., Thorpe R., Thorpe S.J. Endothelial cells co-stimulate peripheral blood mononuclear cell responses to monoclonal antibody TGN1412 in culture. Cytokine. 2011;55:141. doi: 10.1016/j.cyto.2011.03.019. [DOI] [PubMed] [Google Scholar]
- Geginat J., Sallusto F., Lanzavecchia A. Cytokine-driven proliferation and differentiation of human naïve, central memory, and effector memory CD4 + T cells. J. Exp. Med. 2001;194:1711. doi: 10.1084/jem.194.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari A., Zimmerli S.C., Pantaleo G. Cytomegalovirus (CMV)-specific cellular immune responses. Hum. Immunol. 2004;65:500. doi: 10.1016/j.humimm.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Hoffmeister B., Bunde T., Rudawsky I.M., Volk H.D., Kern F. Detection of antigen-specific T cells by cytokine flow cytometry: the use of whole blood may underestimate frequencies. Eur. J. Immunol. 2003;33:3484. doi: 10.1002/eji.200324223. [DOI] [PubMed] [Google Scholar]
- Hu Y., Turner M.J., Shields J., Gale M.S., Hutto E., Roberts B.L., Siders W.M., Kaplan J.M. Investigation of the mechanism of action of alemtuzumab in a human CD52 transgenic mouse model. Immunology. 2009;128:260. doi: 10.1111/j.1365-2567.2009.03115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain K., Hargreaves C.E., Roghanian A., Oldham R.J., Chan H.T., Mockridge C.I., Chowdhury F., Frendéus B., Harper K.S., Strefford J.C., Cragg M.S., Glennie M.J., Williams A.P., French R.R. Upregulation of FcγRIIb on monocytes is necessary to promote the superagonist activity of TGN1412. Blood. 2015;125:102. doi: 10.1182/blood-2014-08-593061. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Hamilton B.J., Skopelja S., Rigby W.F. Induction of interleukin-6 production by rituximab in human B cells. Arthritis Rheum. 2014;66:2938. doi: 10.1002/art.38798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ravetch J.V. A general requirement for FcγRIIB co-engagement of agonistic anti-TNFR antibodies. Cell Cycle. 2012;11:3343. doi: 10.4161/cc.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Lin J.X., Leonard W.L. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.C., Koh L.P., Tan P. Fatal cytokine release syndrome with chimeric anti-CD20 monoclonal antibody rituximab in a 71-year-old patient with chronic lymphocytic leukemia. J. Clin. Oncol. 1999;17:1962. doi: 10.1200/jco.1999.17.6.1962. [DOI] [PubMed] [Google Scholar]
- Lühder F., Huang Y., Dennehy K.M., Guntermann C., Müller I., Winkler E., Kerkau T., Ikemizu S., Davis S.J., Hanke T., Hünig T. Topological requirements and signaling properties of T cell-activating, anti-CD28 antibody superagonists. J. Exp. Med. 2003;197:955. doi: 10.1084/jem.20021024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna K.C., Beatty K.M., Vicetti Miguel R., Bilonick R.A. Delayed processing of blood increases the frequency of activated CD11b + CD15 + granulocytes which inhibit T cell function. J. Immunol. Methods. 2009;341:68. doi: 10.1016/j.jim.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Moreau T., Coles A., Wing M., Isaacs J., Hale G., Waldmann H., Compston A. Transient increase in symptoms associated with cytokine release in patients with multiple sclerosis. Brain. 1996;119:225. doi: 10.1093/brain/119.1.225. [DOI] [PubMed] [Google Scholar]
- Römer P.S., Berr S., Avota E., Na S.Y., Battaglia M., ten Berge I., Einsele H., Hünig T. Preculture of PBMCs at high cell density increases sensitivity of T-cell responses, revealing cytokine release by CD28 superagonist TGN1412. Blood. 2011;118:6772. doi: 10.1182/blood-2010-12-319780. [DOI] [PubMed] [Google Scholar]
- Rudd C.E., Schneider H. Unifying concepts in CD28, ICOS and CTLA4 co-receptor signalling. Nat. Rev. Immunol. 2003;3:544. doi: 10.1038/nri1131. [DOI] [PubMed] [Google Scholar]
- Sheridan J.P., Zhang Y., Riester K., Tang M.T., Efros L., Shi J., Harris J., Vexler V., Elkins J.S. Intermediate-affinity interleukin-2 receptor expression predicts CD56 (bright) natural killer cell expansion after daclizumab treatment in the CHOICE study of patients with multiple sclerosis. Mult. Scler. 2011;17:1441. doi: 10.1177/1352458511414755. [DOI] [PubMed] [Google Scholar]
- Siders W.M., Shields J., Garron C., Hu Y., Boutin P., Shankara S., Weber W., Roberts B., Kaplan J.M. Involvement of neutrophils and natural killer cells in the anti-tumor activity of alemtuzumab in xenograft tumor models. Leuk. Lymphoma. 2010;51:1293. doi: 10.3109/10428191003777963. [DOI] [PubMed] [Google Scholar]
- Stebbings R., Findlay L., Edwards C., Eastwood D., Bird C., North D., Mistry Y., Dilger P., Liefooghe E., Cludts I., Fox B., Tarrant G., Robinson J., Meager T., Dolman C., Thorpe S.J., Bristow A., Wadhwa M., Thorpe R., Poole S. “Cytokine storm” in the phase I trial of monoclonal antibody TGN1412: better understanding the causes to improve preclinical testing of immunotherapeutics. J. Immunol. 2007;179:3325. doi: 10.4049/jimmunol.179.5.3325. [DOI] [PubMed] [Google Scholar]
- Stebbings R., Eastwood D., Poole S., Thorpe R. After TGN1412: recent developments in cytokine release assays. J. Immunotoxicol. 2013;10:75. doi: 10.3109/1547691X.2012.711783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe S.J., Stebbings R., Findlay L., Eastwood D., Poole S., Thorpe R. How predictive are in vitro assays for cytokine release syndrome in vivo? A comparison of methods reveals worrying differences in sensitivity and frequency of response. Cytokine. 2013;64:471. doi: 10.1016/j.cyto.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Suthanthiran M., Fotino M., Riggio R.R., Cheigh J.S., Stenzel K.H. OKT3-associated adverse reactions: mechanistic basis and therapeutic options. Am. J. Kidney Dis. 1989;14:39. [PubMed] [Google Scholar]
- Suntharalingam G., Perry M.R., Ward S., Brett S.J., Castello-Cortes A., Brunner M.D., Panoskaltsis N. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 2006;355:1018. doi: 10.1056/NEJMoa063842. [DOI] [PubMed] [Google Scholar]
- TeGenero A.G. TGN1412 Investigational Medicinal Product Dossier. 2005. www.circare.org/foia5/tgn1412dossier.pdf Available at. (last accessed 20/3/2015)
- Van Berkel M.E.A.T., Oosterwegel M.A. CD28 and ICOS: similar or separate costimulators of T cells? Immunol. Lett. 2006;105:115. doi: 10.1016/j.imlet.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Vidal J.M., Kawabata T.T., Thorpe R., Silva-Lima B., Cederbrant K., Poole S., Mueller-Berghaus J., Pallardy M., Van der Laan J.W. In vitro cytokine release assays for predicting cytokine release syndrome: the current state-of-the-science. Report of a European Medicines Agency Workshop. Cytokine. 2010;51:213. doi: 10.1016/j.cyto.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Waibler Z., Sender L.Y., Merten C., Hartig R., Kliche S., Gunzer M., Reichardt P., Kalinke U., Schraven B. Signaling signatures and functional properties of anti-human CD28 superagonistic antibodies. PLoS One. 2008;3:e1708. doi: 10.1371/journal.pone.0001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M.R., Makropoulos D.A., Achuthanandam R., Van Arsdell S., Bugelski P.J. Development of a human whole blood assay for prediction of cytokine release similar to anti-CD28 superagonists using multiplex cytokine and hierarchical cluster analysis. Int. Immunopharmacol. 2011;11:1697. doi: 10.1016/j.intimp.2011.06.001. [DOI] [PubMed] [Google Scholar]
- White A.L., Dou L., Chan H.T., Field V.L., Mockridge C.I., Moss K., Williams E.L., Booth S.G., French R.R., Potter E.A., Butts C., Al-Shamkhani A., Cragg M.S., Verbeek J.S., Johnson P.W., Glennie M.J., Beers S.A. Fcγ receptor dependency of agonistic CD40 antibody in lymphoma therapy can be overcome through antibody multimerization. J. Immuno. 2014;193:1828. doi: 10.4049/jimmunol.1303204. [DOI] [PubMed] [Google Scholar]
- Wing M.G., Moreau T., Greenwood J., Smith R.M., Hale G., Isaacs J., Waldmann H., Lachmann P.J., Compston A. Mechanism of first-dose cytokine-release syndrome by CAMPATH 1-H: involvement of CD16 (FcgammaRIII) and CD11a/CD18 (LFA-1) on NK cells. J. Clin. Invest. 1996;98:2819. doi: 10.1172/JCI119110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler U., Jensen M., Manzke O., Schulz H., Diehl V., Engert A. Cytokine-release syndrome in patients with B-cell chronic lymphocytic leukemia and high lymphocyte counts after treatment with an anti-CD20 monoclonal antibody (rituximab, IDEC-C2B8) Blood. 1999;94:2217. [PubMed] [Google Scholar]
- Wolf B., Morgan H., Krieg J., Gani Z., Milicov A., Warncke M., Brennan F., Jones S., Sims J., Kiessling A. A whole blood in vitro cytokine release assay with aqueous monoclonal antibody presentation for the prediction of therapeutic protein induced cytokine release syndrome in humans. Cytokine. 2012;60:828. doi: 10.1016/j.cyto.2012.08.018. [DOI] [PubMed] [Google Scholar]
- Wolf B., Morgan H., Brennan F., Krieg J., Gani Z., Jones S., Kiessling A. Response to the letter to the editor by Susan Thorpe et al.: how predictive are in vitro assays for cytokine release syndrome in vivo? A comparison of methods reveals worrying differences in sensitivity and frequency of response. Cytokine. 2013;64:473. doi: 10.1016/j.cyto.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Xiong F., Janko M., Walker M., Makropoulos D., Weinstock D., Kam M., Hrebien L. Analysis of cytokine release assay data using machine learning approaches. Int. Immunopharmacol. 2014;22:465. doi: 10.1016/j.intimp.2014.07.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional analyses of cytokine release and inhibitory effect of GYPA.