Abstract
Mycobacterium tuberculosis (Mtb) is the most common co-infection in HIV patients and a serious co-epidemic. Apart from increasing the risk of reactivation of latent tuberculosis (TB), HIV infection also permits opportunistic infection of environmental non-pathogenic mycobacteria. To gain insights into mycobacterial survival inside host macrophages and identify mycobacterial proteins or processes that influence HIV propagation during co-infection, we employed proteomics approach to identify differentially expressed intracellular mycobacterial proteins during mono- and HIV co-infection of human THP-1 derived macrophage cell lines. Of the 92 proteins identified, 30 proteins were upregulated during mycobacterial mono-infection and 40 proteins during HIV-mycobacteria co-infection. We observed down-regulation of toxin-antitoxin (TA) modules, up-regulation of cation transporters, Type VII (Esx) secretion systems, proteins involved in cell wall lipid or protein metabolism, glyoxalate pathway and branched chain amino-acid synthesis during co-infection. The bearings of these mycobacterial factors or processes on HIV propagation during co-infection, as inferred from the proteomics data, were validated using deletion mutants of mycobacteria. The analyses revealed mycobacterial factors that possibly via modulating the host environment, increased viral titers during co-infection. The study provides new leads for investigations towards hitherto unknown molecular mechanisms explaining HIV-mycobacteria synergism, helping address diagnostics and treatment challenges for effective co-epidemic management.
Resurgence of tuberculosis (TB), a curable infectious disease, with the pandemic of Human Immunodeficiency Virus (HIV), the virus that causes Acquired Immunodeficiency Syndrome (AIDS) is a major health issue worldwide1. The current statistics by WHO divulges that a third of HIV infected people worldwide are simultaneously infected with Mycobacterium species, mainly Mycobacterium tuberculosis (Mtb), the TB causing bacteria (WHO report, 2014). Disease management of mycobacterial infections in HIV patients is highly challenging because of drug to drug interactions, drug toxicity, increased incidence of TB related Immune reconstitution inflammatory syndrome (IRIS)2,3,4 and emerging threat from opportunistic infections by environmental mycobacteria. These mycobacteria are ubiquitously present in the environment including soil, water, air and food5. Some of the common opportunistic mycobacteria include Mycobacterium avium complex (MAC), M. kansasii, M. fortuitum, M. gordonae, M. phlei, M. xenopi and attenuated strains like M. bovis BCG (BCG)5,6. HIV patients with mycobacterial co-infections have been reported to progress to AIDS faster than those without co-infection7, at the same time, mycobacteria that can otherwise be cleared establish infection in HIV patients, pointing to a strong mutualism between these two pathogens8.
Mycobacteria reside inside the phagosomes of infected host macrophages. They adapt to the hostile intracellular conditions by concerted modulation of their protein expression and host signaling9. The adaptation of pathogenic mycobacterial proteome to the hostile intra-phagosomal environment has been studied using both transcriptomics and proteomics approaches10,11,12,13,14. These studies have been successful to a large extent in understanding the pathogenesis of TB bacteria. However, being restricted to mono-infection, these studies do not explain how a mycobacterium adapts to macrophages, where the cells have had a prior exposure to HIV. A population of monocytes/macrophages is colonized and used as reservoir by HIV during infection, but a large fraction of monocytes are also stimulated owing to immune environment generated by infected macrophages that makes them more aggressive phagocytes than usual15,16,17,18. Mycobacteria, during co-infection, encounters altered, quite possibly, activated macrophages as the host is already infected with HIV-1, whereas during mono-infection, mycobacteria faces the challenges within a naïve monocyte19. This prompts one to look at the changes in the early adaptive responses of mycobacteria to survive inside the phagosomes of infected macrophages during HIV-mycobacteria co-infections. Further, mycobacterial infection exacerbates HIV disease progression20 and it is not clear if mycobacterial factors or processes are directly or indirectly involved in increasing HIV titers during co-infection. In order to understand these two aspects of HIV-mycobacteria co-infection biology, we undertook a proteomics approach to understand the early adaptive changes in intraphagosomal mycobacterial protein in the phagosome-enriched fractions of HIV-mycobacteria co-infected macrophages.
The proteomics studies were performed using human derived macrophages THP-1 infected either with Bacille-Calmette-Guérin (BCG), a live attenuated strain of Mycobacterium bovis alone or in the background of M-tropic strain ADA-8 of HIV-1. This cell culture based set up simulated a condition when an HIV patient acquired secondary mycobacterial infection. In compliance with the clinical observations on HIV-mycobacterial co-infections, the co-infection of THP-1 with HIV and mycobacteria resulted in higher propagation of both the virus and the bacteria21.
In this paper, we discuss the differentially expressed intra-phagosomal mycobacterial proteins in response to mycobacterial mono-infection and HIV co-infection and correlate the same with mycobacterial survival and viral titers. The intra-phagosomal mycobacterial proteome suggested key alterations in the toxin-antitoxin modules, lipid metabolism, cation transporters and Type VII Esx secretory systems during co-infection. Some of the leads from the proteomics analyses were validated through deletion mutants or over-expressing mycobacterial strains that revealed involvement of factors from mycobacteria that possibly via modulating the host environment, promote HIV propagation during co-infection. The study is the first attempt to catalogue the early changes in the mycobacterial proteomes in the phagosomes of host cell during co-infection to understand the synergism between HIV and mycobacteria. Identification of mycobacterial proteins differentially expressed during mono- and co-infection will help us decipher the mycobacterial strategies employed to survive inside phagosomes of macrophages during co-infection and identify factors that have impact on viral propagation during co-infection.
Results and Discussion
Comparative proteomes of intra-phagosomal mycobacteria from mono- and HIV co-infected THP-1 macrophages
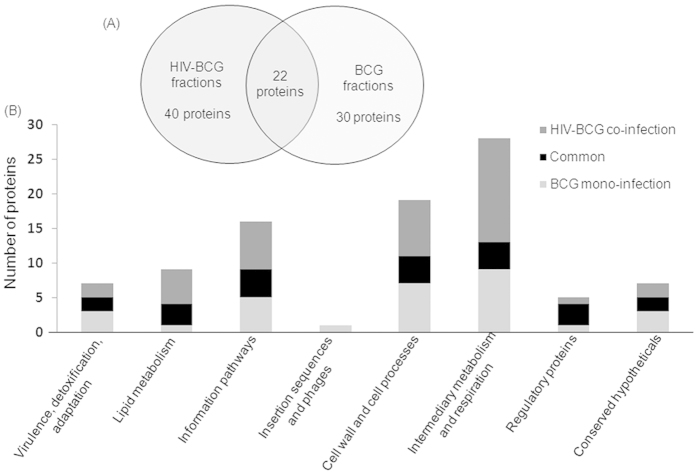
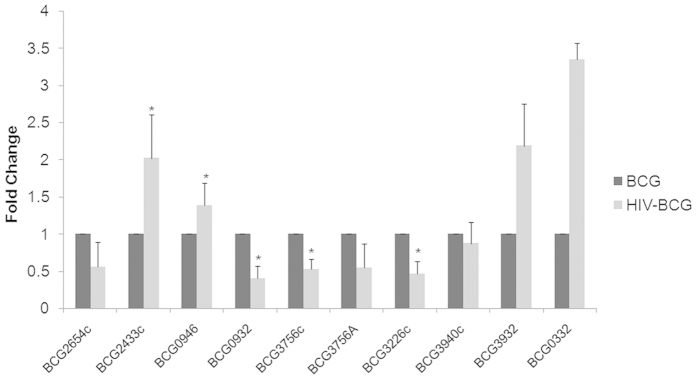
We studied the early adaptive changes in the proteome of the intraphagosomal mycobacteria in the background of HIV infection. We used THP-1 derived macrophages which were either infected with M. bovis BCG (Multiplicity of infection (MOI) = 100) alone or co-infected with ADA-8 HIV-1 (at 30 ng/mL p24 equivalents) and BCG (refer methods). High MOI of 100 was used to enrich mycobacteria-laden phagosomes to identify the intra-phagosomally expressed mycobacterial proteins. To capture the early events, after 24 hr post-infection, we isolated phagosome-enriched fractions, to minimize the host background, from BCG mono- and HIV-BCG co-infected cells and performed LC-MALDI-MS/MS of the fractions. The results of proteomic data were from the three experiments of BCG mono-infected fractions and four experiments of HIV-BCG co-infected fractions. Proteins identified (95% confidence) from these experiments of a condition (mono- and co-infection) were pooled together into their respective categories. Then, the proteins identified in at the least two experiments per category were considered for further analyses. Overall, we have identified 92 differentially expressed proteins between BCG mono- and HIV-BCG co-infected fractions (Table 1, S1 and S2). Of these proteins, 22 proteins overlapped between both the fractions, 30 and 40 proteins were exclusively present in BCG mono-infected and HIV-BCG co-infected fractions, respectively (Table 1, S1, S2 and Fig. 1A). Proteins exclusively present in one condition but absent in the other condition were considered for further analyses. The proteins were categorized into different functional categories as described in Tuberculist22 database (Fig. 1B). The proteins belonging to two major groups, that is, lipid metabolism and intermediary metabolism and respiration, were upregulated during co-infection. The other group that was categorically enriched during co-infection was information pathways (Fig. 1B).The proteomic data were further validated by the Real-Time PCR of randomly picked 10 mycobacterial genes using the RNA isolated from the intraphagosomal BCG during BCG mono- and HIV-BCG co-infection (Fig. 2). The Real-Time PCR results reiterated the proteomic data, where BCG0946 and BCG3940c were common for both mono- and co-infections, BCG2654c, BCG0932, BCG3756c and BCG3226c were observed upregulated in mono-infection and BCG3932, BCG0332 and BCG2433c were observed upregulated in co-infection. Some of the important observations are discussed below. These observations are supported by co-infection studies with the knock-out/over-expression mutants of M.smegmatis to understand the synergistic impact of factors from mycobacteria on HIV propagation during co-infection.
Table 1. Mycobacterial proteins identified by LC-MALDI-MS/MS from the phagosomal fractions of BCG mono- and HIV-BCG co-infected cells.
| S# | BCG gene# | pI | MW (kDa) | Functional category | Product/Function (Known/probable/annotated) | Ortholog in M.tuberculosis H37Rv (Rv#) |
|---|---|---|---|---|---|---|
| Proteins Observed in fractions from BCG mono-infected cells | ||||||
| 1 | BCG1194c | 4.9 | 81.5 | intermediary metabolism and respiration | Involved de novo biosynthesis of Methionine | Rv1133c |
| 2 | BCG3477 | 10.7 | 11 | virulence, detoxification, adaptation | Unknown; Possible Anti-toxin VapB47 | Rv3407 |
| 3 | BCG3226c | 6.1 | 116.6 | information pathways | Probable ATP-dependent DNA helicase/Has both ATPase and helicase activities | Rv3201c |
| 4 | BCG0136c | 7.3 | 77.4 | cell wall and cell processes | Probable cation-transporter P-type ATPase B CtpB/Cation-transporting ATPase | Rv0103c (ctpB) |
| 5 | BCG0422c | 4.9 | 92.5 | virulence, detoxification, adaptation | Probable endopeptidase ATP binding protein (chain B) ClpB (ClpB protein) (heat shock protein F84.1) | Rv0384c |
| 6 | BCG2367c | 6.1 | 46.8 | intermediary metabolism and respiration | Probable dGTP triphosphohydrolase | Rv2344c |
| 7 | BCG0677c | 6.3 | 118.7 | information pathways | Probable exonuclease V (beta chain) RecB/Involved in homologous recombination. | Rv0630c |
| 8 | BCG2238c | 7.5 | 109.1 | intermediary metabolism and respiration | Glutamate-ammonia-ligase adenylyltransferaseGlnE/Regulatory protein involved in the regulation of glutamine synthetase activity | Rv2221c |
| 9 | BCG2457c | 5.4 | 74.6 | intermediary metabolism and respiration | Glutamine-dependent NAD(+) synthetaseNadE (NAD(+) synthase/Involved in biosynthesis of NAD | RV2438c |
| 10 | BCG1867 | 5.3 | 99.4 | intermediary metabolism and respiration | Probable glycine dehydrogenase GcvB/The glycine cleavage system catalyses the degradation of glycine. | Rv1832 |
| 11 | BCG1419c | 5.9 | 33.9 | conserved hypotheticals | Conserved hypothetical protein/Unknown | Rv1357c |
| 12 | BCG2119 | 6.5 | 58.9 | insertion seqs and phages | Conserved hypothetical protein/Unknown | Rv2100 |
| 13 | BCG0079c | 7.5 | 30.8 | cell wall and cell processes | Possible membrane protein/Unknown | Rv0048c |
| 14 | Mb2002c | 4.5 | 24.8 | cell wall and cell processes | Immunogenic protein Mpt64/Unknown. | Rv1980c |
| 15 | BCG3662c | 10.69 | 12 | information pathways | Iron-regulated H-NS-like protein Lsr2/Has DNA-bridging activity. | Rv3597c |
| 16 | BCG3174 | 5.2 | 85.3 | intermediary metabolism and respiration | Probable NADH dehydrogenase I (chain G) NuoG/Involved in aerobic|anaerobic respiration | Rv3151 |
| 17 | BCG1088 | 6.3 | 74.6 | cell wall and cell processes | Probable potassium-transporting P-type ATPase B chain KdpB | Rv1030 (kdpB) |
| 18 | BCG1391c | 6.5 | 70.1 | information pathways | Probable ATP-dependent helicase DinG/Probable helicase involved in DNA repair and perhaps also replication. | Rv1329c |
| 19 | BCG1182 | 4.99 | 52.1 | intermediary metabolism and respiration | Probable glucose-6-phosphate 1-dehydrogenase Zwf1 (G6PD)/Involved in pentose phosphate pathway (first step) | Rv1121 |
| 20 | BCG3756c | 9.6 | 16.3 | virulence, detoxification, adaptation | Possible toxin VapC48/Unknown | Rv3697c |
| 21 | BCG0453c | 6.1 | 23.2 | intermediary metabolism and respiration | Thiamine-phosphate pyrophosphorylaseThiE/Involved in thiamine biosynthesis | Rv0414c |
| 22 | BCG3326; BCG3362 | 8.7 | 28.5 | information pathways | Probable endonuclease VIII Nei/Involved in damage reversal of DNA | Rv3297 |
| 23 | BCG1573 | 5 | 63.1 | lipid metabolism | Probable fatty-acid-AMP ligase FadD25/Function unknown, but involvement in lipid degradation | Rv1521 |
| 24 | BCG0932 | 10.9 | 15.5 | Regulatory proteins | Possible transcriptional regulatory protein (possibly MarR-family)/Thought to be involved in transcriptional mechanism. | Rv0880 |
| 25 | BCG1357 | 5.4 | 65.1 | information pathways | Probable transcription termination factor Rho homolog/Facilitates transcription termination by a mechanism that involves rho binding to the nascent RNA, activation of rho’S RNA-dependent ATPase activity, and release of the mRNA from the DNA template | Rv1297 |
| 26 | BCG0937 | 8.7 | 39.7 | conserved hypotheticals | Conserved hypothetical protein/Unknown | Rv0885 |
| 27 | BCG0788 | 5.2 | 19.1 | conserved hypotheticals | Conserved protein/Unknown | Rv0738 |
| 28 | BCG2654c | 10 | 46.2 | conserved hypotheticals | Conserved protein/Unknown | Rv2627c |
| 29 | BCG0489c* | 7.11 | 105.2 | cell wall and cell processes | Probable conserved transmembrane transport protein MmpL4/Unknown. Thought to be involved in fatty acid transport. | Rv0450c |
| 30 | BCG2361; BCG2362 | 6.5 | 89.9 | cell wall and cell processes | Probable conserved transmembrane transport protein MmpL9/Unknown. Thought to be involved in fatty acid transport. | Rv2339 |
| Proteins Observed in fractions from HIV-BCG co-infected cells | ||||||
| 1 | BCG3770 | 4.78 | 70 | intermediary metabolism and respiration | 2-isopropylmalate synthase LeuA/Involved in leucine biosynthesis (at the first step) | Rv3710 |
| 2 | BCG3025c | 6.5 | 66 | intermediary metabolism and respiration | Acetolactate synthase (large subunit) IlvB1/Involved in valine and isoleucine biosynthesis (at the first step) | Rv3003c |
| 3 | BCG0626 | 10.5 | 7.5 | virulence, detoxification, adaptation | Possible antitoxin VapB26/Unknown | Rv0581 |
| 4 | BCG0391 | 8.2 | 41.3 | virulence, detoxification, adaptation | Probable chaperone protein DnaJ1/Acts as a co-chaperone. | Rv0352 |
| 5 | BCG2216c | 7.97 | 40.4 | intermediary metabolism and respiration | Probable transmembrane cytochrome C oxidase (subunit II) CtaC/Involved in aerobic respiration. | Rv2200c |
| 6 | BCG3007c | 12.4 | 22.1 | Information pathways | DNA-binding protein HU homolog HupB (histone-like protein)/This protein belongs to the histone like family of prokaryotic DNA-binding proteins which are capable of wrapping DNA to stabilize it, and prevent its denaturation under extreme environmental conditions. | Rv2986c |
| 7 | BCG0006; BCG0036 | 5.2 | 92.2 | Information pathways | DNA gyrase (subunit A) GyrA | Rv0006c |
| 8 | BCG3932 | 7.7 | 51 | cell wall and cell processes | ESX conserved component EccB1/Unknown. | Rv3869 |
| 9 | BCG0332* | 10.5 | 35.9 | cell wall and cell processes | ESX conserved component EccE3. ESX-3 type VII secretion system protein/Unknown. | Rv0292 |
| 10 | BCG3331c | 6.7 | 62 | intermediary metabolism and respiration | Probable glycerol-3-phosphate dehydrogenase GlpD2/Involved in aerobic respiration and oxidation of glycerol. | Rv3302c |
| 11 | BCG3470 | 5.6 | 87 | intermediary metabolism and respiration | Conserved protein/Function unknown; probably enzyme involved in cellular metabolism. | Rv3401 |
| 12 | BCG2921c | 7.7 | 84.5 | intermediary metabolism and respiration | Possible formate dehydrogenase H FdhF/Decomposes formic acid to hydrogen and carbon dioxide under anaerobic conditions in the absence of exogenous electron acceptors | Rv2900c |
| 13 | BCG0535c | 7.3 | 35.5 | conserved hypotheticals | Conserved protein/Function Unknown. | Rv0493c |
| 14 | BCG2433c | 6.5 | 28.4 | conserved hypotheticals | Conserved protein/Function Unknown. | Rv2417c |
| 15 | BCG1633* | 6.7 | 53.7 | intermediary metabolism and respiration | Probable L-aspartate oxidase NadB/Quinolinate biosynthesis. | Rv1595 |
| 16 | BCG1872c | 4.8 | 80.4 | intermediary metabolism and respiration | Malate synthase G GlcB/Involved in glyoxylate bypass (second step), an alternative to the tricarboxylic acid cycle | Rv1837c |
| 17 | BCG1689 | 4.9 | 88 | information pathways | Probable phenylalanyl-tRNAsynthetase, beta chain PheT/Charging PHE-tRNA | Rv1650 |
| 18 | BCG0447 | 5.04 | 72.9 | intermediary metabolism and respiration | Probable phosphate acetyltransferasePta/Involved at the last step (of two) in the conversion of acetate to acetyl-CoA | Rv0408 |
| 19 | BCG2957* | 5.5 | 158 | lipid metabolism | Phenolpthiocerol synthesis type-I polyketide synthase PpsE/Involved in phenolpthiocerol and phthioceroldimycocerosate (dim) biosynthesis: | Rv2935 |
| 20 | BCG1855 | 5.7 | 57.2 | intermediary metabolism and respiration | Probable acetolactate synthase IlvG/Valine and isoleucine biosynthesis (first step). | Rv1820 |
| 21 | BCG3661c | 5.6 | 93.5 | intermediary metabolism and respiration | Probable ATP-dependent protease ATP-binding subunit ClpC1/Hydrolyses proteins in presence of ATP. | Rv3596c |
| 22 | BCG3299 | 7.4 | 76.5 | cell wall and cell processes | Probable metal cation-transporting P-type ATPase C CtpC/Metal cation-transporting ATPase | Rv3270 |
| 23 | BCG0957 | 6.3 | 26 | lipid metabolism | Possible enoyl-CoA hydratase EchA6/Could possibly oxidize fatty acids using specific components | Rv0905 |
| 24 | BCG0976c | 10.7 | 44.9 | cell wall and cell processes | Divalent cation-transport integral membrane protein MntH (BRAMP)/H(+)-stimulated, highly selective, divalent cation uptake system. | Rv0924c |
| 25 | BCG2596 | 6.5 | 28.1 | conserved hypotheticals | Conserved protein/Function Unknown. | Rv2573 |
| 26 | BCG3443 | 9.7 | 48.8 | lipid metabolism | Possible triacylglycerol synthase/May be involved in synthesis of triacylglycerol | Rv3371 |
| 27 | BCG0992 | 7.8 | 83.5 | Information pathways | ATP dependent DNA ligase LigD/Involved in DNA double-strand break repair, by nonhomologous end joining (NHEJ). | Rv0938 |
| 28 | BCG1479 | 8.4 | 24.2 | cell wall and cell processes | Probable lipoprotein LprH/Function Unknown | Rv1418 |
| 29 | BCG1655 | 5.3 | 50.5 | intermediary metabolism and respiration | Probable pyruvate kinase PykA/Produces phosphoenol pyruvate in glycolysis | Rv1617 |
| 30 | BCG2858c | 5.3 | 18.9 | information pathways | Probable ribosome-binding factor aRbfA/Associates with free 30S ribosomal subunits (but not with 30S subunits that are part of 70S ribosomes or polysomes). | Rv2838c |
| 31 | BCG2870c | 8.5 | 66.9 | intermediary metabolism and respiration | Possible magnesium chelatase/Function unknown; possibly introduces a magnesium ion into specific substrate/compound. | Rv2850c |
| 32 | BCG1671 | 4.7 | 78 | information pathways | Probable excinuclease ABC (subunit B - helicase) UvrB/Involved in nucleotide excision repair. | Rv1633 |
| 33 | BCG1667 | 4.78 | 98.4 | information pathways | Probable DNA polymerase I PolA/Involved in post-incision events. | Rv1629 |
| 34 | BCG3633c | 5.96 | 33.58 | intermediary metabolism and respiration | 3,4-DHSA dioxygenase/Catalyzes the extradiol cleavage of 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (3,4-DHSA) | Rv3568c |
| 35 | BCG1816 | 5.1 | 152.8 | cell wall and cell processes | ESX conserved component EccC5. ESX-5 type VII secretion system protein/Function Unknown | Rv1783 |
| 36 | BCG2636c* | 8.96 | 35.16 | lipid metabolism | Probable acyltransferase/Catalyzes the acylation of the 6-position of the mannose residue linked to position 2 of the myo-inositol in phosphatylinositol mono- and DI-mannosides. | Rv2611c |
| 37 | BCG3639 | 9.4 | 21.9 | regulatory proteins | Transcriptional regulatory protein KstR (probably TetR-family)/Involved in transcriptional mechanism. Predicted to control regulon involved in lipid metabolism. | Rv3574 |
| 38 | BCG2891c | 8.69 | 42.9 | cell wall and cell processes | Membrane bound metalloprotease/Controls membrane composition | Rv2869c |
| 39 | BCG0602 | 10.63 | 41.24 | lipid metabolism | MannosyltransferaseMgtA/Involved in lipomannan (LM) biosynthesis | Rv0557 |
| 40 | BCG0733 | 4.69 | 77.20 | Information pathways | Probable elongation factor G FusA1 (EF-G)/This protein promotes the GTP-dependent translocation of the nascent protein chain from the A-site to the P-site of the ribosome. | Rv0684 |
| Proteins Observed in fractions from both mono- and co-infected cells | ||||||
| 1 | BCG1389c | 5.3 | 78.6 | intermediary metabolism and respiration | Unknown; probably involved in polysaccharide degradation | Rv1327c |
| 2 | BCG1368 | 4.7 | 59.2 | intermediary metabolism and respiration | Probable ATP synthase alpha chain AtpA | Rv1308 |
| 3 | BCG1370 | 4.5 | 53.09 | intermediary metabolism and respiration | Probable ATP synthase beta chain AtpD | Rv1310 |
| 4 | BCG1627 | 4.4 | 37.5 | intermediary metabolism and respiration | Probable biotin synthetase BioB/ Involved in biotin synthesis. | Rv1589 |
| 5 | BCG0389 | 4.5 | 66.8 | virulence, detoxification, adaptation | Probable chaperone protein DnaK (heat shock protein 70)/Acts as chaperone | Rv0350 |
| 6 | BCG2562c | 6.5 | 41.7 | intermediary metabolism and respiration | Probable chorismate synthase AroF/Involved in the synthesis of chorismate | Rv2540c |
| 7 | BCG2944c | 4.9 | 130.6 | cell wall and cell processes | Probable chromosome partition protein Smc/Plays an important role in chromosome structure and partitioning | Rv2922c |
| 8 | BCG0717 | 6 | 146.7 | information pathways | DNA-directed RNA polymerase (beta’ chain) RpoC | Rv0668 |
| 9 | BCG0716 | 4.67 | 129.2 | information pathways | DNA-directed RNA polymerase (beta chain) RpoB | Rv0667 |
| 10 | BCG3940c | 10.6 | 57.6 | cell wall and cell processes | ESX conserved component EccE2. ESX-2 type VII secretion system protein/Function unknown | Rv3885 |
| 11 | BCG0946 | 7.2 | 42.7 | Regulatory proteins | Possible transcriptional regulatory protein (possibly LuxR-family)/Thought to be involved in transcriptional mechanism. | Rv0894 |
| 12 | BCG2301c; BCG2302c | 6.3 | 25.4 | conserved hypotheticals | Possible membrane protein/Unknown | Rv2286c |
| 13 | BCG2962c* | 4.8 | 224 | lipid metabolism | Probable multifunctional mycocerosic acid synthase membrane-associated Mas/Catalyzes the elongation of N-fatty acyl-CoA with methylamalonyl-CoA (not malonyl-CoA) as the elongating agent to form mycocerosyl lipids | Rv2940c |
| 14 | BCG1539 | 9.3 | 49.8 | virulence, detoxification, adaptation | Peptidoglycan hydrolase/Unknown | Rv1477 |
| 15 | BCG2953 | 5 | 198.8 | lipid metabolism | Phenolpthiocerol synthesis type-I polyketide synthase PpsA/Involved in phenolpthiocerol and phthioceroldimycocerosate (dim) biosynthesis | Rv2931 |
| 16 | BCG2112c | 7.4 | 99.57 | information pathways | ATP-dependent DNA helicase HelY/DNA helicase activity | Rv2092c |
| 17 | Mb2007c | 9.3 | 32.8 | Regulatory proteins | Probable transcriptional regulatory protein (probably LysR-family)/Involved in transcriptional mechanism. | Rv1985c |
| 18 | BCG1085c | 5.8 | 92.7 | Regulatory proteins | Probable sensor protein KdpD/Member of the two-component regulatory system KDPD/KDPE involved in the regulation of the KDP operon. | Rv1028 |
| 19 | BCG0123 | 10.6 | 27.8 | cell wall and cell processes | Possible membrane protein/Unknown | Rv0090 |
| 20 | BCG1550 | 6.1 | 41.2 | cell wall and cell processes | Possible exported conserved protein/Unknown | Rv1488 |
| 21 | BCG3215c | 9.2 | 107.4 | cell wall and cell processes | Probable conserved transmembrane protein/Unknown | Rv3193c |
| 22 | BCG2500c | 8.7 | 88.2 | lipid metabolism | Probable glycerol-3-phosphate acyltransferase PlsB2/Involved in phospholipid biosynthesis | Rv2482c |
Note: *Corresponding protein belongs to IdeR regulon24.
Figure 1. Functional categorization of mycobacterial proteins identified from the phagosome-enriched fractions of BCG mono- and HIV-BCG co-infected cells.
(A) Venn diagram of mycobacterial proteins distributed between mono- and co-infections. (B) Graphical representation of functional categories of the proteins from mono- and co-infections.
Figure 2. Real-time PCR of RNA from intracellular BCG reiterates the proteomic data.
Statistical analyses were done with Student’s t-test. *represents p < 0.05.
Reduced expression of Toxin-Antitoxin systems in mycobacteria during co-infection
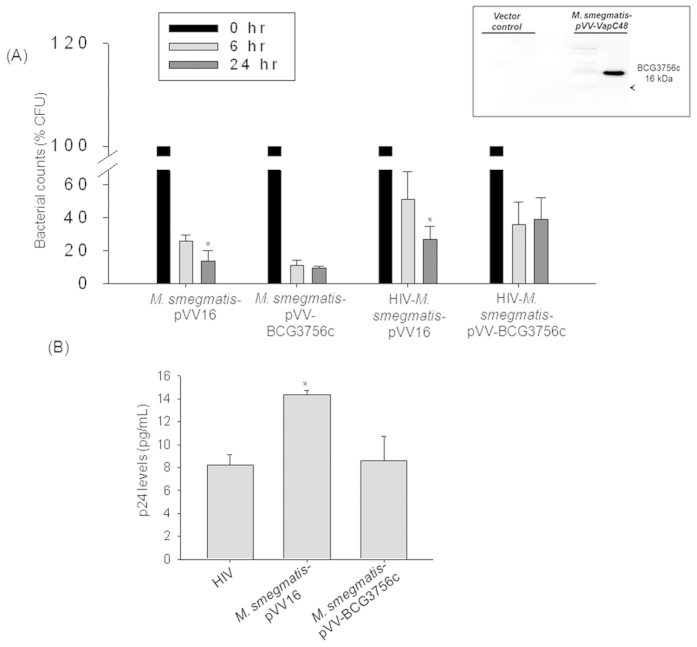
During mono-infection, expression of proteins belonging to Virulence-detoxification-adaptation category such as BCG3756c (VapC48) and BCG3477 (VapB47) were upregulated. BCG3756c (toxin) and BCG3477 (anti-toxin) belong to Toxin-Antitoxin (TA) family of proteins22. Toxin-Antitoxin systems are expressed during stress and help in the viability of the bacteria by allowing the expression of stress responsive genes23. The up regulation of BCG3756c during mono-infection was validated by qRT-PCR (Fold change in transcript levels - BCG mono-infection: 1 ± 0.001 fold; HIV-BCG co-infection: 0.53 ± 0.135 fold; Fig. 2). As BCG3756c was over-expressed during BCG mono-infection, we expected that it may play a role in intracellular survival of mycobacteria. We expressed BCG3756c gene with C-terminus 6× His-tag in M.smegmatis, that does not have an ortholog of BCG3756c, using a mycobacterial shuttle vector, pVV16 (Fig. 3A inset). The recombinant M.smegmatis-pVV-BCG3756c was used for mono- and HIV co-infections and scored for CFU by plating on 7H10 agar plates after 0, 6 and 24 hours post-infection. Accordingly, the recombinant strain survived better over time, 6 hr to 24 hr, inside macrophages as compared to M.smegmatis-pVV16 both during mono- and co-infection. The clearance of recombinant strain M.smegmatis-pVV-BCG3756c (6 hr: 10.92 ± 3.21%CFU; 24 hr: 9.42 ± 1.04%CFU) was not significant from 6 hr to 24 hr (Fig. 3A) compared to the vector control, M.smegmatis-pVV16 (6 hr: 25.98 ± 3.39%CFU; 24 hr: 13.75 ± 6.25%CFU; p < 0.05) which was cleared by 45% from 6 hr to 24 hr time point during mono-infections (Fig. 3A). During co-infection, M.smegmatis-pVV-BCG3756c (6 hr: 35.64 ± 14.19%CFU; 24 hr: 38.95 ± 13.47%CFU) was not cleared efficiently from 6 hr to 24 hr (Fig. 3A) whereas M.smegmatis-pVV16 (6 hr: 51.31 ± 16.74%CFU; 24 hr: 26.78 ± 8.07%CFU; p < 0.05) was cleared by 48% from 6 hr to 24 hr time point. The percentage CFU was normalized to that of 0 hr. Evident from these results, over-expression of BCG3756c, irrespective of mono- or co-infections, helped in the survival of mycobacteria.
Figure 3. BCG3756c (VapC48) expression in M.smegmatis helps its survival without affecting HIV titers.
(A) Plot representing M.smegmatis-pVV16 and M.smegmatis-pVV-BCG3756c survival inside macrophages upon mono- and co-infections in percentage CFU (%CFU) at 0, 6 and 24 hrs post-infection. Inset: Western blot with anti-His antibody confirming the expression His-tagged BCG3756c in M.smegmatis. (B) Bar graph represents HIV p24 titers in the supernatants of HIV mono-, HIV-M.smegmatis-pVV16 and HIV-M.smegmatis-pVV-BCG3756c co-infected macrophages at 24 hr post-infection. The HIV titers were not detectable in the culture supernatants at 6 hr post-infection.
Since, BCG3756c was down-regulated during co-infection as per the proteomics data we scored the impact of over-expression of this protein on HIV titers during co-infection. It was observed that the co-infection with the Vector control increased the p24 equivalents of viral titers (HIV-M.smegmatis-pVV16: 14.36 ± 0.36 pg/mL; p < 0.05) compared to HIV infection (HIV: 8.21 ± 0.91 pg/mL) alone (Fig. 3B), but co-infection with the recombinant strain failed to provide such support to viral propagation (HIV-M.smegmatis-pVV-BCG3756c: 8.63 ± 2.12 pg/mL) as compared to HIV infection (HIV: 8.21 ± 0.91 pg/mL) (Fig. 3B). VapBC TA systems are considered as stress responsive elements which help the bacteria persist during stress probably by decreasing the metabolic activity and activating the required pathways. This function is probably carried out by its annotated ribonuclease activity on mRNAs and thus inhibiting the translation of avoidable proteins during stress23. During this process probably, toxin also inhibits the factors which can influence the viral titers. Therefore, expression of proteins belonging to TA systems helps in the persistence of mycobacteria upon infection but probably has no impact on HIV propagation during co-infection.
This explained the down-regulation of this factor in the proteomics data during co-infection.
Mycobacterial iron dependent transcription regulator (IdeR) promoted viral titers during co-infection
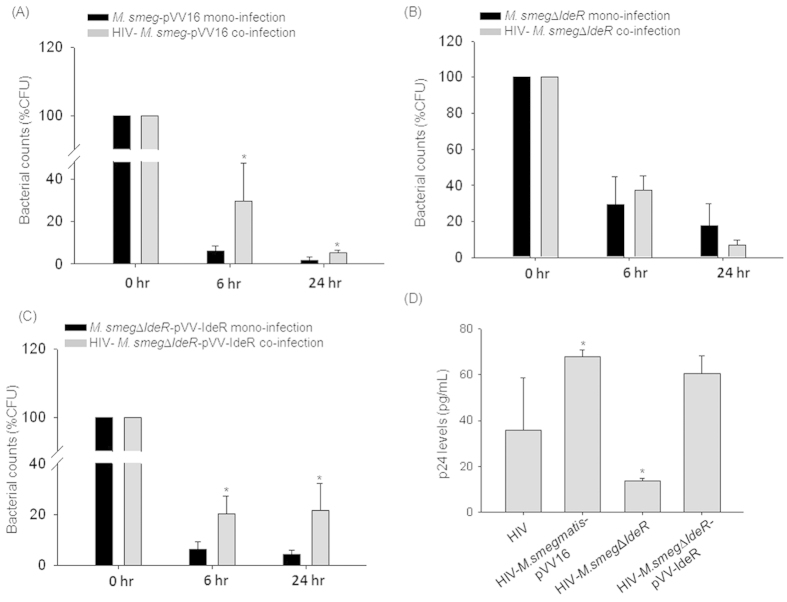
Amongst the identified mycobacterial proteins upregulated during co-infection, BCG0332 (52% similar to MSMEG_0626), BCG1633 (76.5% similar to MSMEG_3200), BCG2957 (52% similar to MSMEG_0408), BCG2636c (78.5% similar to MSMEG_3200), BCG2962c (75% similar to MSMEG_4727) belonged to IdeR regulon (Table 1)24. IdeR (91% similar to MSMEG_2750) is a global iron dependent transcriptional factor that regulates the expression of several critical proteins in mycobacteria24. Hence, using the mutant, M.smegmatis (M.smegΔideR) and complemented strain (M.smegΔideR-pVV-IdeR)25, we checked the effect on mycobacterial survival and HIV titers during co-infection. IdeR complemented strain was prepared by expressing the IdeR gene cloned into pVV16 shuttle vector in the mutant, M.smegΔideR (Figure S1).
HIV co-infection helped in the survival of the wildtype mycobacteria (M.smegmatis-pVV16) as observed in terms of percentage CFU (M.smegmatis-pVV16 mono-infection: 1.72 ± 1.59%CFU; co-infection: 5.14 ± 1.29%CFU at 24 hr) (Fig. 4A); however HIV co-infection could not support the survival of the IdeR mutant M.smegΔideR (mono-infection: 17.88 ± 11.88%CFU; co-infection: 7.13 ± 2.36%CFU at 24 hr; Fig. 4B), which was rescued in the complement strain M.smegΔideR-pVV-IdeR (mono-infection: 4.28 ± 1.87%CFU; co-infection: 21.56 ± 10.68%CFU at 24 hr; Fig. 4C), suggesting that IdeR is essential for HIV-supported survival of mycobacteria. The percentage CFU (%CFU) was normalized to 0 hr.
Figure 4. IdeR or IdeR dependent proteins support both mycobacteria and HIV titers during co-infection.
(A) Plot representing (A) M.smeg-pVV16, (B) M.smeg∆IdeR and (C) M.smeg∆IdeR-pVV-IdeR survival inside macrophages upon mono- and co-infections in percentage CFU (%CFU) at 0, 6 and 24 hr post-infection. (D) Bar graph represents HIV p24 titers in the supernatants of HIV mono-, HIV-M.smeg-pVV16, HIV-M. smeg∆IdeR and HIV-M. smeg∆IdeR-pVV-IdeR co-infected macrophages at 24 hr post-infection. The HIV titers were not detectable in the culture supernatants at 6 hr post-infection.
We then compared the p24 equivalent of viral titers during co-infection using all the three strains, viz; with the vector control, HIV-M.smegmatis-pVV16 (HIV: 35.97 ± 22.55 pg/mL; HIV-M.smegmatis-pVV16: 67.80 ± 2.94 pg/mL; p < 0.05), with the mutant, HIV-M.smegΔideR (HIV-M.smegΔideR: 13.76 ± 1.10 pg/mL; p < 0.05) and with the complemented strain, HIV-M.smegΔideR-pVV-IdeR (HIV-M.smegΔideR-pVV-IdeR: 60.37 ± 7.99 pg/mL) (Fig. 4D). We observed that the co-infection with the mutant strain decreased virus production as compared to the wild type. This decreased viral titers were restored upon complementation in the mutant (HIV: 35.97 ± 22.55 pg/mL; HIV-M.smegΔideR-pVV-IdeR: 60.37 ± 7.99 pg/mL). The defects in the survival of the IdeR mutant and the drastic decrease in the HIV p24 titers during co-infection compared to HIV mono-infection or HIV-M.smeg-pVV16 co-infection suggested that IdeR is essential for mycobacterial survival and also involved in the increase of viral titers during co-infection. This could be a direct effect of IdeR or because of the proteins belonging to IdeR regulon. BCG2957 and BCG2636c are involved in the synthesis of cell wall lipids, phosphatidylinositol mannoside and lipoarabinomannans, respectively26,27,28. The alterations in the composition of mycobacterial cell wall lipids could alter the stimulation of the macrophages and thereby macrophage responses, which in turn can influence HIV propagation in a host29.
Alterations in the mycobacterial cell wall and lipid metabolism influence viral production and mycobacterial survival during co-infection
Mycobacterium relies on fatty acids as a source of energy and probably synthesizes triacylglycerols as energy reserves during infections for long-term sustenance inside the host30,31. BCG proteome indicated expression of proteins that are involved in synthesis of Phthiocerol Dimycocerosates (PDIMs) during both mono- and co-infection conditions. PDIMs are key components of cell wall determining the virulence of mycobacteria32. These proteins included Long-chain-fatty-acid-CoA ligase FadD15/BCG2202, Mycocerosic acid synthase (BCG2962c), Phenolpthiocerol synthesis polyketide synthase PpsA/BCG2953 and probable enoyl-CoA hydratase echA6/BCG0957. However, Phenolpthiocerol synthesis polyketide synthase type I PpsE/BCG2957 (involved in the synthesis and translocation of PDIM to cell wall surface), Phosphatidylinositol mannoside acyltransferase/BCG2636c (involved in the synthesis of lipomannans and lipoarabinomannans) and Putative diacyglycerol O-acyltransferase/BCG3443 (involved in the synthesis of triacylglycerols)26,27,28 were upregulated during co-infection. A putative zinc metalloprotease (Rip1/BCG2891c), a protein known to regulate the cell envelope lipid composition important for the virulence of mycobacterium33 was over-expressed during HIV-BCG co-infection. Rip1 was also shown to positively regulate transcription of other proteins BCG2962c and BCG295334, which concurrently appeared in our proteomics data (Table 1). Together the over-expression of BCG2891c, BCG2962c and BCG2953 can regulate the composition of cell wall affecting the virulence of the mycobacterium.
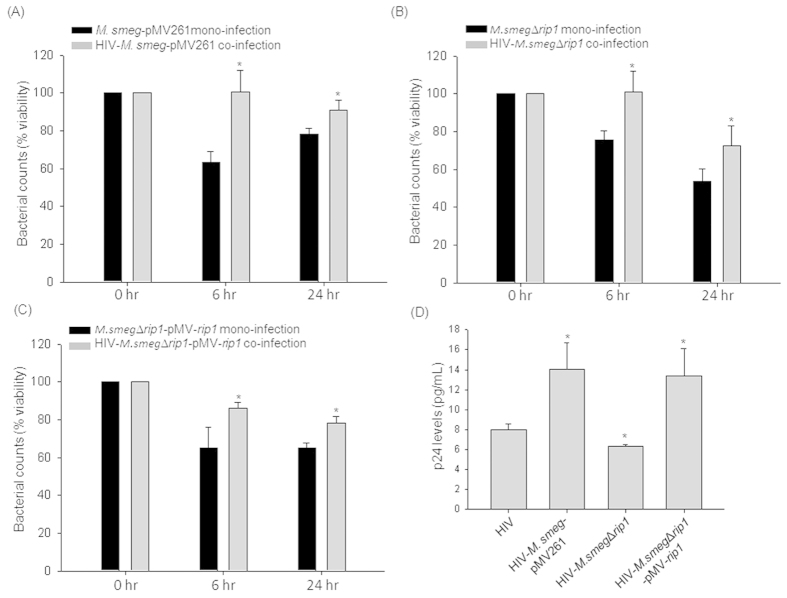
The significance of the over-expression of mycobacterial proteins involved in the cell wall and lipid metabolism in the context of co-infection was studied next using knock-out (KO) mutants of M. smegmatis for the proteins, Rip1/MSMEG_2579 (81% similar to BCG2891c) and Polyketide synthase (Pks) (52% similar to BCG2957). As these proteins were over-expressed during co-infection, it was speculated that they may influence both mycobacterial survival and HIV titers during co-infection. The Pks was selected as it is reported to be involved in the synthesis of surface glycopeptidolipid (GPL) of mycobacteria35. THP-1 cells were either infected with mutants M.smegmatis::Δrip1 (Δrip1)/ M.smegmatis::Δpks (M.smegΔpks)35,36 alone or in the presence of HIV. As compared to mono-infection by vector control (M.smeg-pMV261), rip1 mutant (M.smegΔrip1) or the complemented strain (M.smegΔrip1-pMV-rip1), all the strains demonstrated increased survival upon co-infection with HIV (Fig. 5A–C). The comparative % viability upon mono- and co-infections at 24 hrs post mycobacterial infection are: M.smeg-pMV261: 78.10 ± 3.24% while HIV-M.smeg-pMV261: 91.14 ± 4.96% (Fig. 5A); M.smegΔrip1: 53.77 ± 6.69% while HIV-M.smegΔrip1: 72.48 ± 10.80% (Fig. 5B) and M.smegΔrip1-pMV-rip1: 64.98 ± 2.95% while HIV- M.smegΔrip1-pMV-rip1: 78.47 ± 3.20% (Fig. 5C). With this one can infer, that HIV background helped the survival of all the strains of mycobacteria, while deletion of rip1 had no impact on survival. However, HIV-M.smegΔrip1 co-infection showed significantly (p < 0.05) lesser p24 levels (6.35 ± 0.18 pg/mL) compared to HIV mono-infection (8.01 ± 0.57 pg/mL) and HIV-M.smeg-pMV261co-infection (14.07 ± 2.65 pg/mL) (Fig. 5D). The expression of Rip1 in trans in the mutant (M.smegΔrip1-pMV-rip1) restored the HIV titers equivalent to the co-infection with the vector control (13.38 ± 2.74 pg/mL) (Fig. 5D). These experiments indicated that directly or indirectly, mycobacterial factor Rip1 definitely augmented HIV production during co-infection.
Figure 5. Mycobacterial Rip1 protein impacts the viral titers during co-infection.
Plot representing (A) M.smeg-pMV261, (B) M.smeg∆rip1 and (C) M.smeg∆rip1-pMV-rip1 persistence inside macrophages upon mono- and co-infections in % viability (refer methods) at 0, 6 and 24 hr post-infection; (D) Bar graph represents HIV p24 titers in the supernatants of HIV mono-, HIV-M.smeg-pMV261, HIV-M.smeg∆rip1 and HIV-M.smeg∆rip1-pMV-rip1 co-infected macrophages at 24 hr post-infection. The HIV titers were not detectable in the culture supernatants at 6 hr post-infection.
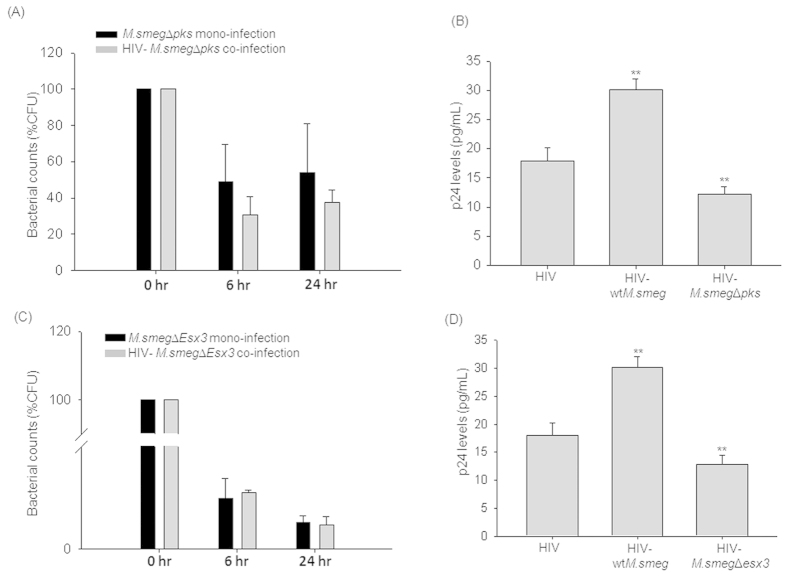
The Pks protein, involved in the synthesis of cell wall glycopeptidolipid, is a large multi-functional protein corresponding to 390 kDa with 11 active functional domains. The M.smegmatis pks mutant (M.smegΔpks) was used to score for the CFU and the impact on the viral titers. Percentage CFU of intracellular M.smegΔpks showed defect in survival during co-infection (37.68 ± 6.58%CFU at 24 hr) and mono-infection (54.21 ± 26.66%CFU at 24 hr) suggesting that it is required for the intracellular survival during co-infection (Fig. 6A). Similarly, HIV-Δpks (12.24 ± 1.25 pg/mL) co-infection showed significantly (p < 0.001) lesser p24 levels compared to both HIV mono-infection (17.94 ± 2.22 pg/mL) and HIV-wtM.smeg co-infection (30.11 ± 1.87 pg/mL) (Fig. 6B). Due to the large size (390 kDa) of the Pks protein making a complement of the mutant was difficult. But, however, owing to the multi-functional domain structure of the Pks protein, it would be interesting to investigate the above impact on the viral titers by M.smegΔpks is due to the intact multi-functional Pks protein or due to individual domains, which is part of future investigations in the laboratory. One can thus infer that mycobacterial protein Pks supported both the survival of mycobacteria and viral titers during co-infection. Taken together, one may hypothesize that the differences in the cell envelope proteins or lipid composition of mycobacteria modulate host cell response, increasing viral propagation during co-infection. This corroborates the earlier findings that the HIV propagation is differentially regulated by mycobacteria in a strain-dependent manner which was attributed to the differences in the cell wall composition29.
Figure 6. Validation of the inferences from the proteomics data using M.smegmatis knock-out (KO) mutants.
The cells were infected with M.smegmatis KO mutants for mono-infections and along with HIV for co-infection to score for mycobacterial survival and viral titers. Percentage CFU (%CFU) of (A) M.smegΔpks and (C) M.smegΔesx3 upon mono- or co-infection at 0, 6 and 24 hr post-infection, represented as bar plots. HIV p24 titers from the culture supernatants were measured at 24 hr post-infection and represented as bar graphs accordingly (B) M.smegΔpks and (D) M.smegΔesx3. The HIV titers were not detectable in the culture supernatants at 6 hr post-infection.
Increased expression of Esx system and cation transporter proteins in mycobacteria during co-infection and impact on viral titers
We observed increased expression of components of Esx system (EccE3/BCG0332 and EccC5/BCG1816) along with cation transporter proteins (MntH/BCG0976c and BCG3299) during co-infection as compared to mycobacterial mono-infection. Esx systems are Type VII secretion systems of mycobacteria and are implicated in both in-vitro and intracellular survival37. BCG0332 (EccE3) (52% similar to MSMEG_0626), a component of Esx-3 system, confirmed by qRT-PCR (Fig. 2) was upregulated by 3.35 ± 0.221 folds during co-infection. Esx-3 system has been reported to be essential for in-vitro growth38 and sensed iron and zinc availability39 in pathogenic mycobacteria. One may speculate that the Esx-3 secretion system may indirectly be involved in iron and zinc acquisition for survival inside macrophages where these ions are limiting. This speculation was further strengthened by the concomitant increased expression of cation transporters, MntH (orthologue of eukaryotic Nramp)40,41 and BCG3299 (probable cation transporter P type ATPase C)42 during co-infection. Similarly, EccC5/ BCG1816, an Esx-5 system protein was present during co-infection. Esx-5 system, in pathogenic mycobacteria, has been implicated in the secretion of PPE proteins, maintenance of cell wall integrity and modulation of macrophage response43,44 and helps the survival of intracellular mycobacteria.
To understand the significance of these observations in the context of HIV-mycobacteria co-infection, we used Esx-3 as representative system and studied the M.smegmatis esx3 deletion mutant (M.smegΔesx3) where the whole cassette of esx-3 system was disrupted45. We expected that Esx-3 system, whose component protein (EccE3/BCG0332) was over-expressed selectively during HIV-mycobacteria co-infections may play a role not only in the intracellular survival of the attenuated mycobacteria, but also in inducing HIV production. THP-1 cells were either infected with Δesx3 alone or co-infected with HIV. The cells were lysed after 0, 6 and 24 hrs of M.smegΔesx3 infection and plated onto 7H10 agar plates and CFU were counted. The percentage CFU was observed to be nearly similar for both M.smegΔesx3 mono-infection (3.86 ± 1.00%CFU at 24 hr) and HIV-M.smegΔesx3 co-infection (3.50 ± 1.23%CFU at 24 hr) (Fig. 6C). With these observations, one can say that Esx-3 system may provide additional survival advantage to mycobacteria during co-infection. When HIV titers in the culture supernatants were compared between HIV-wtM.smeg co-infection and HIV-M.smegΔesx3 co-infection, it was observed that deletion of esx-3 reduced the mycobacterial induced increase in HIV titers by more than 50%. The viral titers in the culture supernatant for HIV mono-infection was 17.94 ± 2.22 pg/mL, which significantly increased in HIV-wtM.smeg co-infection to 30.11 ± 1.87 pg/mL, but HIV-M.smegΔesx3 co-infection showed only 12.79 ± 1.69 pg/mL of viral titers (Fig. 6D). This provided an experimental evidence of factors (here, Esx-3 system) from mycobacteria involved in promotion of HIV propagation, explaining the alliance between HIV and mycobacteria.
A challenging but interesting quest would be to characterize the secretome of opportunistic mycobacteria either alone or in tandem with HIV co-infection, to identify the mycobacterial factors that make the macrophage environment more conducive, making these factors attractive for anti-mycobacterial drug targeting.
Heightened Intermediary metabolism and respiration in BCG during co-infection to support intracellular survival
Proteins belonging to intermediary metabolism and respiration category such as Malate synthase G (BCG1872c), Isopropylmalate synthase (BCG3770), Acetolactate synthase (BCG3025c and BCG1855), L-Aspartate oxidase (BCG1633), Glycerol-3-phosphate dehydrogenase (BCG3331c), pyruvate kinase (BCG1655) and probable L-lysine-epsilon-aminotransferase (BCG3319c and BCG3354c) were upregulated in the phagosomal fraction of co-infected cells. Malate synthase along with isocitrate lyase are the unique enzymes of glyoxylate shunt pathway. Earlier reports suggested that the glyoxylate shunt pathway is upregulated in pathogenic mycobacteria during the persistent phase in vivo and is important for the virulence46,47,48. Glyoxylate pathway utilizes the C2 substrates from fatty acids, which are abundantly found in mammalian cells, without generating carbon dioxide and help in the persistence of mycobacteria during nutrient stress46. This was consistent with our observation of increased lipid body accumulation in host during co-infection21 and mycobacteria adaptation to the host niche by concomitant expression of malate synthase. Isopropylmalate synthase and Acetolactate synthase, which were upregulated during co-infection, are involved in the synthesis of branched chain amino acids, Leucine, Isoleucine and Valine which may provide survival advantage during co-infection49,50. Glycerol-3-phosphate dehydrogenase and pyruvate kinase help bacteria utilize glycerol as carbon source51. L-Aspartate oxidase (involved in the biosynthesis of NAD+)52 was expressed more during co-infection. NAD+ plays an essential role in cellular metabolism, given its involvement in almost all metabolic pathways; hence, biosynthesis of NAD+ can be used as a potential target against pathogenic bacteria53. Over-expression of these enzymes during co-infection may be supporting the survival of mycobacteria under co-infection conditions.
The above results clearly demonstrated that intra-phagosomal mycobacteria indeed adapted differently during mono- and co-infection and while adapting to the HIV induced-conducive intracellular niche of co-infected cells for persistence, the mycobacterial factors, possibly through influencing host responses, also influence the viral propagation.
Conclusions
Proteomics approach to catalog changes in the pathogen proteome helped us understand possible molecular mechanisms that explain how mycobacterial factors may help HIV propagation during co-infection, mutually benefiting both the pathogens. The study, to our knowledge, is the first attempt to catalog the intracellular mycobacterial differential proteins during co-infection. Considering the challenges of proteome identification of intracellular mycobacteria, which allows preferential identification of abundant proteins, getting a high coverage has been observed to be problematic, as also evident from other studies10,54,55. Hence, though coverage of 92 proteins with 70 differentially expressed proteins was fairly informative, we substantiated the analyses of our data with mycobacterial KO and over-expression mutant strains. The key mechanisms like toxin-antitoxin systems, Type VII (Esx) secretory systems, cell wall and lipid metabolism pathways, intermediary metabolism and respiration were modulated during HIV co-infection. We also observed that mycobacteria tend to adapt to the intracellular niche provided by HIV during co-infection by altering its protein complement. For instance, concurrent to the increased host lipid accumulation inside macrophages during co-infection21, it increases the expression of lipid utilizing mycobacterial enzymes or pathways during co-infection. Another significant observation from the data is the increased expression of proteins involved in altering the lipid and protein composition of the cell wall. The differences in the cell wall alter the macrophage signaling pathways probably influencing the viral replication and propagation. This corroborates the observation that different clinical strains affect viral replication in a strain-dependent manner during co-infection which was attributed to the differences in the cell wall composition29. All the proteins identified in the current study have homologues in the virulent Mtb H37Rv strain (Table 1). Upon comparisons with the intracellular mycobacterial proteome from granuloma of guinea pigs infected with Mtb reported by Kruh et al. group56, our proteomic data has shown an overlap of only 22 proteins out of 92 (23.91%). Elucidating their role would help understand the pathogenesis of Mtb during co-infection. Thus, the leads from the study can be pursued to understand co-infection biology, helping decipher new intervention strategies and biomarker to overcome the synergism between HIV and mycobacteria.
Materials and Methods
The cell lines used were HEK293T (Human embryonic kidney) cell line (NCCS, Pune) and THP-1 monocyte leukemia cell line (Cat#TIB-202, ATCC). Bacterial strains used were Mycobacterium bovis BCG, M.smegmatis mc2155, M.smegmatis::Δrip1 and M.smegmatis::Δrip1-complemented (gift by Prof. Michael S. Glickman), M.smegmatisΔEsx3 (gift by Prof. Eric J. Rubin), M.smegmatisΔpks (gift by Dr. Rajesh S. Gokhale) and M.smegmatisΔIdeR (gift by Dr. Marcela Rodriguez)25,35,36,45. Macrophage tropic HIV (NL-ADA8 was gifted by Dr. Jayant Bhattacharya) was used. The THP-1 cell line was maintained in RPMI 1640 media (Invitrogen, USA) supplemented with 10% FBS (South American origin, Gibco, USA) and incubated at 37 °C with 5% CO2. The media was changed when the cells were 90% confluent. The 293T HEK cell line was maintained in DMEM media (Invitrogen, USA) supplemented with 10% FBS and incubated at 37 °C with 5% CO2. When the cells were 90% confluent, the cells were trypsinized with the Trypsin-EDTA (Sigma-Aldrich, USA) and washed with Phosphate buffered saline pH 7.4 (PBS) and fresh complete media was added.
The protocols of handling mycobacterial and HIV strains for co-infection in the laboratory of SB were approved by the Institutional Biosafety Committee No. UH/SLS/IBSC/Review/SB-R-11 and SB-R-14. All experiments were performed in the facilities (F-60 and F-70) approved for Mycobacterial and HIV cultures by University of Hyderabad Institutional Biosafety Committee under Department of Biotechnology, Govt. of India.
Note: The detailed protocol for Growth conditions of mycobacteria, Preparation and quantification of infectious HIV-1 (ADA8) particles, Infection of macrophages and phagosome isolation, Sample preparation and LC-MALDI-MS/MS Analyses, MALDI-TOF/TOF Analyses, Peptide Identification and Statistical Analyses have been described earlier21 and provided the same as supplementary text along with this manuscript. Kindly refer supplementary material.
CFU enumeration and intracellular bacilli viability measurement
As previously described21, for the CFU assay, 24-well tissue culture plates carrying 0.2 million THP-1 macrophages per well was used. The mono- and co-infected cells after the incubation were washed thrice with PBS to remove any extracellular bacilli followed by lysis of macrophages with sterile water at 37 °C for 10 min. The lysates were diluted and were plated on 7H10 agar plates. The plates were incubated at 37 °C with 5% CO2 and 95% humidity for 2–3 weeks (for BCG) and 2–3 days (for M.smegmatis) and the colonies were enumerated and represented as percentage CFU which was normalized to CFU at 0 hr as 100%. The intracellular viability of Mycobacterium was measured using the Alamar blue assay57,58 in a 24-well plate. The fluorescence was read in a fluorescence plate reader with excitation at 530 nm and emission at 590 nm. % viability was calculated by normalizing the fluorescence intensity at 0 hr as 100%.
Intracellular Mycobacterial RNA isolation
The monolayers of the infected cells after 24 h were washed thrice with PBS. The cells were lysed in sterile water for 10 min at 37 °C. The lysate was centrifuged at 1500 rpm for 5 min to remove cell debris. The bacterium from the supernatant was pelleted at high speed (10000 g) for 10 min and then the bacterial pellet was subjected to RNA isolation. The bacterial pellet was resuspended in Trizol and added to pre-chilled, acid washed 0.1 mm glass beads and lysed by bead beating - pulse on: 1 min and pulse off: 2 min on ice. Added glycogen to a final concentration of 200 μg/mL and kept at RT for 10 min. 1/10th volume of chloroform was added and vortexed vigorously for 15 sec and kept at RT for 10 min. Centrifuged at 12000 rpm for 15 min at 4 °C and collected the upper aqueous layer. The RNA was precipitated using isopropanol in presence of glycogen (200 μg/mL) and pellet was washed with 75% ethanol. Air dried and resuspended in RNase free water. Prior to reverse transcription the RNA was subjected to DNase treatment to remove any residual DNA contamination. The DNase treated RNA was reverse transcribed using Superscript III Reverse Transcriptase (Invitrogen) with random hexamers as primer. The reverse transcribed RNA was used for Real-time PCR.
Cloning and Expression of BCG3756c (VapC48) and IdeR gene into pVV16 shuttle vector
The BCG3756c gene was PCR amplified from BCG genomic DNA and IdeR gene was PCR amplified from Mtb H37Rv genomic DNA (Table S3). The mycobacterial shuttle vector, pVV1659,60, was amplified in Escherichia coli DH5α strain in Luria Bertani broth media with kanamycin at 50μg/mL. The vector, BCG3756c and IdeR inserts were double digested with NdeI and HindIII (Fermentas, Thermo Fisher Scientific Inc., USA). The digested insert and vector were ligated using T4 DNA ligase (Fermentas, Thermo Fisher Scientific Inc., USA) downstream of heat shock protein 60 (hsp60) promoter and transformed into Escherichia coli DH5α. The genes were cloned with C-terminus 6X Histidine tag. The recombinant plasmids carrying BCG3756c and IdeR were electroporated into M.smegmatis mc2155 and M.smegmatis::ΔIdeR, respectively. The transformants were picked and grown in 7H9 broth media at 37 °C and culture was harvested and lysed. Supernatant and cell pellet were separated and fractionated on SDS-PAGE followed by immunoblotting using anti-His antibody to confirm the expression of BCG3756c and IdeR. The growth curve for all the transformants was performed (Supplementary Figure 1B and 1C).
Immunoblot
The Mycobacterial cell lysates and supernatants were fractionated on 10% SDS-PAGE, transferred to Nitrocellulose membrane using GE-Amersham Western wet-transfer apparatus. The membrane was blocked with 5% non-fat milk powder in PBS with 0.1% Tween-20 at RT for 2 hrs. The primary antibody, Anti-His tag mouse antibody (SantaCruz Biotechnology Inc., USA) was prepared in PBS-T (PBS + 0.1% Tween-20) with 2% BSA 1:1000 dilutions, was added and incubated at 4 °C on platform rocker overnight. After 3 PBS-T washes, added the Anti-mouse HRP-conjugated secondary antibody (Santa cruz Biotechnology Inc., USA), diluted 1:2000 in PBS-T with 2% BSA, incubated at RT for 2 hrs. After 3 washes with PBS, ECL substrate (Femtolucent Plus-HRP, chemiluminiscent reagent from G-biosciences, USA) was added to the membranes and was scanned for chemiluminiscence using Versadoc Imaging system (Biorad).
Real-time PCR
Equal amount of RNA was considered for equal loading and the corresponding cDNA was used for RT-PCR. As endogenous controls, 16S rRNA gene was used (Table S3). The 2X SyBr green mix (Takara Bio Inc., Japan) was used. The real-time PCR data analyses were done using the 2−ΔΔCt method with endogenous 16S rRNA gene as control. The fold change in the transcript levels were calculated with respect to the BCG mono-infection. All the experiments were performed more than three times. For genes, BCG3932, BCG0332 and BCG3940c data is representative of two experiments.
Additional Information
How to cite this article: Ganji, R. et al. Understanding HIV-Mycobacteria synergism through comparative proteomics of intra-phagosomal mycobacteria during mono- and HIV co-infection. Sci. Rep. 6, 22060; doi: 10.1038/srep22060 (2016).
Supplementary Material
Acknowledgments
This study was supported by grants from Department of Biotechnology (BT/PR3260/BRB/10/967/2011 and BT/05/IYBA/2011) to SB. RG and AR are funded by research fellowship from CSIR. Infrastructure support by DBT-CREB, DST-FIST and DST-PURSE to the Department of Biochemistry and School of Life Sciences, University of Hyderabad is acknowledged. We thank Prof. Michael S. Glickman (M.smegΔrip1 and M.smegΔrip1-pMV-rip1), Prof. Eric J. Rubin (M.smegΔesx3), Dr. Rajesh S. Gokhale (M.smegΔpks) and Dr. Marcela Rodriguez (M.smegΔIdeR) for providing the M.smegmatis mutant strains. We thank Dr. Jayant Bhattacharya for pNL-ADA8 and Prof. Jaya S Tyagi for mycobacterial RNA isolation protocol. We thank Dr. Shekhar C. Mande and Dr. Gaurang Mahajan for the critical inputs and scientific discussions. We thank Mr. Kannan Balakrishnan for cloning of ideR gene into pVV16 vector. We also thank Ms. Swetha Sankati and Mrs. Mani Harika Vemula for critical inputs and proof reading of the manuscript.
Footnotes
Author Contributions S.B. and R.G. conceptualized the research idea and designed the experiments. S.D. and S.R. collected LC-MALDI-MS/MS data. S.R. and S.B. contributed the reagents. R.G., S.D. and A.R. performed the experiments. R.G., S.D., A.R., S.R. and S.B. analyzed the data. R.G., S.R. and S.B. wrote the manuscript.
References
- Pawlowski A., Jansson M., Skold M., Rottenberg M. E. & Kallenius G. Tuberculosis and HIV co-infection. PLoS Pathog 8, e1002464, 10.1371/journal.ppat.1002464PPATHOGENS-D-11-01902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo K. et al. The immune reconstitution inflammatory syndrome after antiretroviral therapy initiation in patients with tuberculosis: findings from the SAPiT trial. Ann Intern Med 157, 313–324, 1355683 10.7326/0003-4819-157-5-201209040-00004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. K. & Soneja M. HIV & immune reconstitution inflammatory syndrome (IRIS). Indian J Med Res 134, 866–877, IndianJMedRes_2011_134_6_866_92632 10.4103/0971-5916.92632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo K., Padayatchi N. & Abdool Karim Q. HIV-Associated Tuberculosis. Clin Dev Immunol 2011, 10.1155/2011/585919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primm T. P., Lucero C. A. & Falkinham J. O. 3rd. Health impacts of environmental mycobacteria. Clin Microbiol Rev 17, 98–106 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane P. J. & Glassroth J. Pulmonary Disease Due to Nontuberculous Mycobacteria: Current State and New Insights. Chest, 2422757 10.1378/chest.15-0458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goletti D. et al. Effect of Mycobacterium tuberculosis on HIV replication. Role of immune activation. J Immunol 157, 1271–1278 (1996). [PubMed] [Google Scholar]
- Pathak S., Wentzel-Larsen T. & Asjo B. Effects of in vitro HIV-1 infection on mycobacterial growth in peripheral blood monocyte-derived macrophages. Infect Immun 78, 4022–4032, 10.1128/IAI.00106-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann S. H. & McMichael A. J. Annulling a dangerous liaison: vaccination strategies against AIDS and tuberculosis. Nat Med 11, S33–44, 10.1038/nm1221 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattow J. et al. Proteins unique to intraphagosomally grown Mycobacterium tuberculosis. Proteomics 6, 2485–2494 (2006). [DOI] [PubMed] [Google Scholar]
- Schnappinger D. et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med 198, 693–704, 10.1084/jem.20030846 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan I. M., Betts J., Banerjee D. K. & Butcher P. D. Differential expression of mycobacterial proteins following phagocytosis by macrophages. Microbiology 147, 459–471 (2001). [DOI] [PubMed] [Google Scholar]
- Li M. S. et al. cDNA-RNA subtractive hybridization reveals increased expression of mycocerosic acid synthase in intracellular Mycobacterium bovis BCG. Microbiology 147, 2293–2305 (2001). [DOI] [PubMed] [Google Scholar]
- Rienksma R. A. et al. Comprehensive insights into transcriptional adaptation of intracellular mycobacteria by microbe-enriched dual RNA sequencing. BMC Genomics 16, 34, 10.1186/s12864-014-1197-2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. A. & Ehrlich L. S. Cell biology of HIV-1 infection of macrophages. Annu Rev Microbiol 62, 425–443, 10.1146/annurev.micro.62.081307.162758 (2008). [DOI] [PubMed] [Google Scholar]
- Guirado E., Schlesinger L. S. & Kaplan G. Macrophages in tuberculosis: friend or foe. Semin Immunopathol 35, 563–583, 10.1007/s00281-013-0388-2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson G. S. et al. HIV-1 infection of macrophages dysregulates innate immune responses to Mycobacterium tuberculosis by inhibition of interleukin-10. J Infect Dis 209, 1055–1065, 10.1093/infdis/jit621 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser D. M. The many faces of macrophage activation. J Leukoc Biol 73, 209–212 (2003). [DOI] [PubMed] [Google Scholar]
- Lawn S. D., Butera S. T. & Shinnick T. M. Tuberculosis unleashed: the impact of human immunodeficiency virus infection on the host granulomatous response to Mycobacterium tuberculosis. Microbes Infect 4, 635–646, S1286457902015824 (2002). [DOI] [PubMed] [Google Scholar]
- Diedrich C. R. & Flynn J. L. HIV-1/mycobacterium tuberculosis coinfection immunology: how does HIV-1 exacerbate tuberculosis ? Infect Immun 79, 1407–1417, 10.1128/IAI.01126-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji R. et al. Proteomics approach to understand reduced clearance of mycobacteria and high viral titers during HIV-mycobacteria co-infection. Cell Microbiol, 10.1111/cmi.12516 (2015). [DOI] [PubMed] [Google Scholar]
- Lew J. M., Kapopoulou A., Jones L. M. & Cole S. T. TubercuList--10 years after. Tuberculosis (Edinb) 91, 1–7, 10.1016/j.tube.2010.09.008 (2011). [DOI] [PubMed] [Google Scholar]
- Ramage H. R., Connolly L. E. & Cox J. S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5, e1000767, 10.1371/journal.pgen.1000767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez G. M., Voskuil M. I., Gold B., Schoolnik G. K. & Smith I. ideR, An essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun 70, 3371–3381 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussurget O., Rodriguez M. & Smith I. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol Microbiol 22, 535–544 (1996). [DOI] [PubMed] [Google Scholar]
- Kordulakova J. et al. Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of mycobacterium species. J Biol Chem 278, 36285–36295, 10.1074/jbc.M303639200 (2003). [DOI] [PubMed] [Google Scholar]
- Daniel J. et al. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol 186, 5017–5030, 10.1128/JB.186.15.5017-5030.2004186/15/5017 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M. & Cox J. S. Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathog 1, e2, 10.1371/journal.ppat.0010002 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar S. et al. HIV-1 replication is differentially regulated by distinct clinical strains of Mycobacterium tuberculosis. PLoS One 4, e6116, 10.1371/journal.pone.0006116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. G. Phagosomes, fatty acids and tuberculosis. Nat Cell Biol 5, 776–778, 10.1038/ncb0903-776 (2003). [DOI] [PubMed] [Google Scholar]
- Sirakova T. D. et al. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology 152, 2717–2725, 152/9/2717 10.1099/mic.0.28993-0 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren M. B., Brokl O. & Schaefer W. B. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: phthiocerol dimycocerosate and the attenuation indicator lipid. Infect Immun 9, 150–158 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinoshima H. & Glickman M. S. Regulation of Mycobacterium tuberculosis cell envelope composition and virulence by intramembrane proteolysis. Nature 436, 406–409, 10.1038/nature03713 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar J. G., Makinoshima H., Schneider J. S. & Glickman M. S. M.. tuberculosis intramembrane protease Rip1 controls transcription through three anti-sigma factor substrates. Mol Microbiol 77, 605–617, MMI7232 10.1111/j.1365-2958.2010.07232.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats A. et al. Retrobiosynthetic approach delineates the biosynthetic pathway and the structure of the acyl chain of mycobacterial glycopeptidolipids. J Biol Chem 287, 30677–30687, 10.1074/jbc.M112.384966 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. S., Sklar J. G. & Glickman M. S. The Rip1 protease of Mycobacterium tuberculosis controls the SigD regulon. J Bacteriol 196, 2638–2645, 10.1128/JB.01537-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltcher M. E., Gibbons H. S., Ligon L. S. & Braunstein M. Protein export by the mycobacterial SecA2 system is determined by the preprotein mature domain. J Bacteriol 195, 672–681, 10.1128/JB.02032-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H. & Rubin E. J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48, 77–84, 10.1046/j.1365-2958.2003.03425 (2003). [DOI] [PubMed] [Google Scholar]
- Serafini A., Pisu D., Palu G., Rodriguez G. M. & Manganelli R. The ESX-3 secretion system is necessary for iron and zinc homeostasis in Mycobacterium tuberculosis. PLoS One 8, e78351, 10.1371/journal.pone.0078351PONE-D-13-29715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranoff D., Monahan I. M., Mangan J. A., Butcher P. D. & Krishna S. Mycobacterium tuberculosis expresses a novel pH-dependent divalent cation transporter belonging to the Nramp family. J Exp Med 190, 717–724 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace K. M. & Maguire M. E. Manganese transport and the role of manganese in virulence. Annu Rev Microbiol 60, 187–209, 10.1146/annurev.micro.60.080805.142149 (2006). [DOI] [PubMed] [Google Scholar]
- Novoa-Aponte L. et al. In silico identification and characterization of the ion transport specificity for P-type ATPases in the Mycobacterium tuberculosis complex. BMC Struct Biol 12, 25, 10.1186/1472-6807-12-25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah A. M. et al. The ESX-5 secretion system of Mycobacterium marinum modulates the macrophage response. J Immunol 181, 7166–7175, 181/10/7166 (2008). [DOI] [PubMed] [Google Scholar]
- Bottai D. et al. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol 83, 1195–1209, 10.1111/j.1365-2958.2012.08001.x (2012). [DOI] [PubMed] [Google Scholar]
- Siegrist M. S. et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci USA 106, 18792–18797, 10.1073/pnas.0900589106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney J. D. et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738, 10.1038/35021074 (2000). [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S., Haddix P. L. & Russell D. G. The interaction between Mycobacterium and the macrophage analyzed by two-dimensional polyacrylamide gel electrophoresis. Electrophoresis 18, 2558–2565, 10.1002/elps.1150181411 (1997). [DOI] [PubMed] [Google Scholar]
- Hou J. Y., Graham J. E. & Clark-Curtiss J. E. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS). Infect Immun 70, 3714–3726 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange F. C., Brown A. M. & Jacobs W. R. Jr. Leucine auxotrophy restricts growth of Mycobacterium bovis BCG in macrophages. Infect Immun 64, 1794–1799 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam R. A. et al. In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect Immun 63, 1004–1012 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan T., Murthy P. S. & Gopinathan K. P. Intermediary metabolism of mycobacteria. Bacteriol Rev 36, 65–108 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W. & Moat A. G. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev 44, 83–105 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. et al. Functional characterization of the Mycobacterium tuberculosis serine/threonine kinase PknJ. Microbiology 156, 1619–1631, 10.1099/mic.0.038133-0 (2010). [DOI] [PubMed] [Google Scholar]
- Lee B. Y. et al. The Mycobacterium bovis bacille Calmette-Guerin phagosome proteome. Mol Cell Proteomics 9, 32–53, 10.1074/mcp.M900396-MCP200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N., Sharma P., Kumar M., Joshi B. & Bisht D. Analysis of intracellular expressed proteins of Mycobacterium tuberculosis clinical isolates. Proteome Sci 10, 14, 10.1186/1477-5956-10-14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruh N. A., Troudt J., Izzo A., Prenni J. & Dobos K. M. Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo. PLoS One 5, e13938, 10.1371/journal.pone.0013938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajko D. M. et al. Colorimetric method for determining MICs of antimicrobial agents for Mycobacterium tuberculosis. J Clin Microbiol 33, 2324–2327 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M. et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe 17, 345–356, 10.1016/j.chom.2015.01.007 (2015). [DOI] [PubMed] [Google Scholar]
- Stover C. K. et al. New use of BCG for recombinant vaccines. Nature 351, 456–460, 10.1038/351456a0 (1991). [DOI] [PubMed] [Google Scholar]
- Chatrath S., Gupta V. K., Dixit A. & Garg L. C. The Rv1651c-encoded PE-PGRS30 protein expressed in Mycobacterium smegmatis exhibits polar localization and modulates its growth profile. FEMS Microbiol Lett 322, 194–199, 10.1111/j.1574-6968.2011.02354.x (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.