Abstract
RNA-binding proteins (RBPs) are key regulators of gene expression. There are several distinct families of RBPs and they are involved in the cellular response to environmental changes, cell differentiation and cell death. The RBPs can differentially combine with RNA molecules and form ribonucleoprotein (RNP) complexes, defining the function and fate of RNA molecules in the cell. RBPs display diverse domains that allow them to be categorized into distinct families. They play important roles in the cellular response to physiological stress, in cell differentiation, and, it is believed, in the cellular localization of certain mRNAs. In several protozoa, a physiological stress (nutritional, temperature or pH) triggers differentiation to a distinct developmental stage. Most of the RBPs characterized in protozoa arise from trypanosomatids. In these protozoa gene expression regulation is mostly post-transcriptional, which suggests that some RBPs might display regulatory functions distinct from those described for other eukaryotes. mRNA stability can be altered as a response to stress. Transcripts are sequestered to RNA granules that ultimately modulate their availability to the translation machinery, storage or degradation, depending on the associated proteins. These aggregates of mRNPs containing mRNAs that are not being translated colocalize in cytoplasmic foci, and their numbers and size vary according to cell conditions such as oxidative stress, nutritional status and treatment with drugs that inhibit translation.
Keywords: Gene expression regulation, RNA-binding proteins, RNA-protein complexes, RNA granules, Protozoa, Stress and cell differentiation
Core tip: RNA-binding proteins (RBPs) are numerous and widely distributed in nature. In addition to having different domains, these proteins are key modulators of gene expression and are involved in the cellular response to environmental changes, cell differentiation and cell death. In protozoa RBPs are crucial for the rapid gene expression remodeling that occurs in the course of cell differentiation or the stress response.
INTRODUCTION
RNA-binding proteins (RBPs) are numerous and widely distributed in nature and are involved in gene expression control at all levels. There are several distinct families of RBPs. They are key modulators of gene expression and are involved in the cellular response to environmental changes, cell differentiation and cell death. The RBPs can differentially combine with RNA molecules and form ribonucleoprotein (RNP) complexes, thereby defining the function and fate of the RNA molecules within the cell.
RBPs and the RNA-binding domains
RBPs have multiple and diverse domains; many of them have a combinatorial set of RNA-binding domains (RBDs) that allow various modes of recognition to exert their function in the cell. The RBD recognizes and interacts with the RNA molecule based on sequence and/or structure. In addition to the RBD domain, many of the RBPs also have protein-protein domains that form complexes that are also subjected to intense post-translational modifications, such as methylation and phosphorylation, further increasing the complexity of the study and characterization of RBPs.
One of the most common domains found in RBPs is the RNA-recognition motif (RRM); it shows affinity with single-stranded RNA, DNA and proteins. Approximately 90 amino acid residues are involved in the recognition and binding of a target RNA sequence of 2 to 6 nucleotides. The proteins that contain this domain are involved in all pathways of the RNA cycle, from splicing to mRNA turnover. The RRM is usually present in multiple copies within a protein, and this multiplicity enhances the specificity of the ligand[1].
The zinc finger domain was first described as a DNA-binding domain (with the CCHH amino acid motif) and was subsequently described in proteins that interact with RNA (with the CCCH motif), such as Tis11 and Tristetraprolin (TTP)[2,3]. TTP is involved in transcript degradation through its interaction with the AU-rich elements in the 3’-untranslated regions (3’-UTRs) of some mRNAs, as described for tumor necrosis factor alpha and granulocyte-macrophage colony-stimulating factor mRNAs[4,5].
The Pumilio (PUF) protein takes part in regulating different processes in the cell, such as embryogenesis, development, and differentiation; however, the best-characterized function of PUF is as a post-transcriptional repressor[6,7]. The domain is characterized by a highly conserved C-terminal RBD composed of eight tandem repeats[6-10].
The hnRNP K-homology domain recognizes a specific sequence of single-stranded RNA and is found in both eukaryotes and prokaryotes. The domain is approximately 70 amino acids long, and the proteins are associated with a diversity of gene expression regulation mechanisms, such as splicing, transcriptional regulation, and translational control[6].
The Arg-Gly-Gly repeat (RGG) domain can interact with either RNAs or proteins, and this interaction can be based on the positive charge of the arginine amino acids[7]. Proteins containing RGG motifs are associated with several cellular processes, including transcription, splicing and mRNA export. One example of an RGG domain protein is the yeast Scd6 protein. It modulates translation by preventing the formation of the 48S pre-initiation complex by binding eIF4G through its RGG motif[8,9].
RBPs in protozoa
Most of the RBPs characterized in protozoa arise from trypanosomatids. Proteins such as TcDHH1[10] are involved in the formation of mRNA granules in Trypanosoma cruzi and Trypanosoma brucei, but it is believed that the function and composition of these granules can be different from those described for higher eukaryotes[11]. TcDHH1 binds to RNAs encoding stage-specific surface antigens from the infective trypomastigote forms. However, these mRNAs are also found at low levels in the non-infective epimastigote forms, leading to the hypothesis that TcDHH1 might be involved in the degradation of the targets in these forms[10,12].
Trypanosome gene expression regulation is mostly post-transcriptional, which suggests that some RBPs might display regulatory functions distinct from those described for other eukaryotes. One example is the poly-A binding proteins PABP1 and PABP2: Only PABP2 accumulates in the nucleus, indicating that PABP1 and PABP2 might be part of distinct mRNPs that associate with distinct populations of mRNAs[13]. Some proteins, such as TbDRBD3, TbPTB2, TbZPF3, TbPUF9, TbPUF1 and TbPUF2, can function as mRNA destabilization factors. However, the modulation mechanisms for most RBPs in trypanosomatids remain unknown[14].
In T. cruzi, the zinc finger protein TcZFP1 binds preferentially to C-rich regions[15]. However, TcZFP2 has a higher affinity for A-rich regions, and the mRNAs associated with this protein appear to be upregulated in metacyclic trypomastigotes, suggesting a possible role in parasite differentiation[16].
Some RBPs, such as TcPUF6[17] and TcUBP1[18], are involved in the downregulation of their target transcripts, while others, such as RBP19, can negatively regulate their own mRNAs[19]. Some RBPs, such as RBP42[20] and the Alba proteins[21], associate with the translation machinery. The RBP42 protein from T. brucei binds to the coding region of mRNAs of the insect form that encode metabolic proteins. RBP42 is an essential protein and associates with mRNAs bound to polysomes in the cytoplasm[20]. However, some RBPs, such as TbRBP10, a cytoplasmic protein essential to the T. brucei bloodstream forms, appear to affect the expression of their targets indirectly through other proteins that form the mRNP complex[22].
TcRBP40 is found dispersed within the cytoplasm and is also concentrated in reservosomes, a storage organelle present in T. cruzi and some bat trypanosomes during a specific life cycle stage[23,24], suggesting that this organelle can also be involved in RNA processing[25]. There are also some RBPs with assigned nuclear functions, such as TbPTB2, which is involved in trans-splicing processing of some mRNAs[26]. Similarly, TbRRM1 is a nuclear RBP that associates with mRNAs from T. brucei and with the auxiliary splicing factor polypyrimidine tract-binding protein (PTB) 2, but not with components of the core spliceosome. A knockdown of TbRRM1 caused extensive alterations in mRNA abundance and regions enriched for the downregulated mRNAs were identified. In addition, in cells subjected to heat shock, TbRRM1 shifted from the nucleus to the cytoplasm, which led to the compaction of chromatin[27].
The T. brucei nucleolar RBPs PUF7 and PUF10 are involved in ribosomal RNA maturation and transcriptional control of procyclic stage-specific genes transcribed by RNA polymerase I[28]. TcSUB2 is involved in the transcription/export pathway and is mainly localized to the nucleus[29].
In Plasmodium, DDX-6 class DEAD box RNA helicase (DOZI) is essential to zygote development because of its role in controlling mRNA stability[30,31]. In P. falciparum, both PfPuf1 (PFE0935c) and PfPuf2 (PFD0825c) are upregulated in gametocytes[32,33]. Microarray analysis detected increasing amounts of PfPuf1 and PfPuf2 mRNAs during gametocyte maturation[34]. Interestingly, the highest level of PfPuf2 expression was observed in sporozoites[35].
In Toxoplasma gondii, TgPuf1 displays different expression levels in tachyzoites and bradyzoites, indicating that it might function in regulating processes such as proliferation and/or differentiation that would enable the parasites to respond rapidly to changes in environmental conditions[36]. Another protein from T. gondii is the RGG single-strand-binding protein (TgSsossB), which interacts with the TgAlba proteins that are involved in translation regulation[37]. When the RGG domain was deleted from the protein, the mutant strain produced fewer plaques in stress conditions, a defect associated to a slow growth phenotype due to the exposure of extracellular parasites to stress. Moreover, the mutant lost its capacity to interact with the TgAlba complex[38].
RBPs and post-transcriptional control in stress
In unicellular and multicellular organisms, there is a very subtle balance between the cell’s capacity to respond to stress and cell death. For the former, this balance needs to be even more strictly regulated, and there are many variables involved that take into account the strength and duration of the stimuli[39]. The stress response needs to be fast to allow cell survival, if we consider the cell’s time scale, at the moment the stress is triggered, the response starts immediately (within nanoseconds), and the cell metabolism adapts dynamically to the environment. Accordingly, there is modulation of gene expression, and trans-acting factors such as RBPs are key to this prompt response.
Post-transcriptional gene regulation occurs at different levels: Splicing, mRNA stability and translation regulation. With regard to splicing, it has been shown that when a cell is subjected to a low dose of ultraviolet radiation irradiation, the C-terminal domain of RNA polymerase II is phosphorylated, and the splicing dynamics are altered. This alteration allows, via alternative splicing, the inclusion of exons with weaker splice sites[40,41]. One example of alternative splicing occurs with the MDM2 RNA during genotoxic stress. Under normal conditions, the MDM2 protein targets the p53 protein for degradation via ubiquitination; however, during stress, an exon is skipped in the MDM2 RNA, which ultimately results in the maintenance of p53 in the cell. This effect is only reversed upon restoration of the normal condition[42]. Splicing factors are also targets of regulation during stress. For example, hnRNP1 shuttles to the cytoplasm during osmotic stress, while the other splicing factors remain in the nucleus[43].
Extensive studies have been performed and highlight the importance of translational control during stress. For the cell, it is vital to control and arrest translation during stress, as 50% of the cell’s energy is consumed during translation[44].
For most mRNAs, translation initiation occurs when the trimeric eIF4F complex binds to the transcript. This complex is formed by the protein eIF4E (the cap-binding protein), eIF4A (RNA helicase) and eIF4G (which forms the scaffold protein complex via its binding sites for eIF4E and eIF4A). The trimeric complex binds to the cap, followed by ribosome scanning from the RNA 5’-end to the first AUG site. Then, eukaryotic initiation factor 2 (eIF2) associates with tRNAimet, part of eIF2-GTP ternary complex that is required to bring the initiator tRNA to the ribosome[45,46]. When a stress signal is generated, two basic mechanisms are activated: The availability of eIF4E, which can be sequestered by eIF4EBPs (eIF4E-binding proteins), and the phosphorylation of eIF2α. Global translational arrest occurs, and only a few proteins are translated to help the cell overcome the stress. The most studied genes upregulated during stress are those encoding heat shock proteins (HSPs), whose transcription is stimulated by the heat shock factor that senses misfolded proteins in the cytoplasm and shuttles to the nucleus. HSPs are chaperones that help properly fold proteins[47]. Aside from HSPs, some transcripts that contain upstream open reading frames can be translated even under conditions of eIF2α phosphorylation[48,49]. The transcripts that contain internal ribosome entry sites (IRESs) on their 5’-UTRs can also bypass the inhibition of cap-dependent control (eIF4F). Interestingly, some RBPs function as IRES-trans acting factors and interact with these IRES elements, resulting in the coordination of the subset of transcripts that will be translated[50]. For example, during stress, the PTB recognizes the IRES element in p53 transcripts and in other mRNAs associated with apoptosis and starvation and triggers their translation to respond to the stress conditions[51].
mRNA stability can be altered as a response to stress. Transcripts are sequestered to RNA granules that ultimately modulate their availability to the translation machinery, storage or degradation, depending on the associated proteins. Among the RNA granules, the processing bodies (P-bodies) are multiprotein aggregates of mRNPs containing mRNAs that are not being translated, associated with proteins involved in RNA degradation pathways and translation inhibition factors. These aggregates colocalize in cytoplasmic foci, and their numbers and size vary according to cell conditions such as oxidative stress, nutritional status and treatment with drugs that inhibit translation[52]. The proteins that have been identified as being involved in mRNA decapping [DCP1, DCP2, DHH1 and their activators, RCK/p54, pat1 (as part of the Lsm1-7 complex), the XRN1 exonuclease complex, and CCR4/POP2/NOT] are also found in P-bodies of mammalian and yeast cells[53-55]. Some proteins are essential for the formation of P-bodies, such as GW182 (which is involved in the route of degradation by micro RNA), RCK/p54 and Lsm4, as the absence of these proteins prevents the formation of these granules[54,56-58]. Some of the proteins characteristic of P-bodies are depicted.
There is a spatial separation between the mRNAs that are being translated from those that are associated with P-bodies. The mRNA translation status depends on the proteins associated with the mRNP complexes, which direct the mRNA molecules either to the polysomes for translation or to RNA granules for storage and/or degradation[55,59]. Observations indicate that several mRNAs present in the P-bodies undergo cap removal and are consequently prone to degradation[60,61]. However, the P-bodies may also act as mRNA storage sites, allowing mRNAs to return to the translation machinery. Experiments have shown that specific mRNAs accumulate in P-bodies in growing cells or cells under stress and that they emerge from the granules and migrate to the polysomes to be translated when normal conditions are restored[60,61].
Another type of mRNP granule found in eukaryotes is the stress granule. Unlike P-bodies, the stress granules are composed mainly of translation initiation factors, the 40 s ribosomal subunit and several RBPs[62]. Studies indicate that physical interaction occurs between the P-bodies and stress granules and that the mRNAs can switch between them[63] (Figure 1). The translation initiation complexes bound to mRNAs, which include eIF3, the eIF4F complex, eIF4B, the 40S ribosomal subunit and PABP-1, form the core of the stress granules and are considered markers of these structures[64]. Other proteins characteristic of stress granules may affect mRNA translation, such as T cell antigen internal-1 (TIA-1) and TIA-1-related, Fragile X mental retardation protein (FMRP) and Fragile X mental retardation-related protein-1, Argonaute, cytoplasmic polyadenylation element-binding protein, Pumilio and RNA-associated protein 55. Still others are involved in mRNA stability, such as RNA helicase RCK/p54, 5’-exonuclease 3’ XRN1, proteins that bind to the ARE element, and Hur and TTP, which respectively stabilize and destabilize mRNA[65]. In mammalian cells, stress granules are formed as a result of phosphorylation of eIF2α by stress-activated kinases. The granules assemble when RBPs such as TIA-1, TTP, FMRP or G3BP bind to specific transcripts that are linked to the stalled pre-initiation 48S complex. After this initial nucleation stage, the polyA-binding protein (PABP) promotes the aggregation of these small granules by making them microscopically visible. Some proteins that do not have a canonical RBP domain can also associate with the granules through protein-protein interactions, which suggests that stress granules can interact with other signaling pathways in the cell[62,66].
Figure 1.
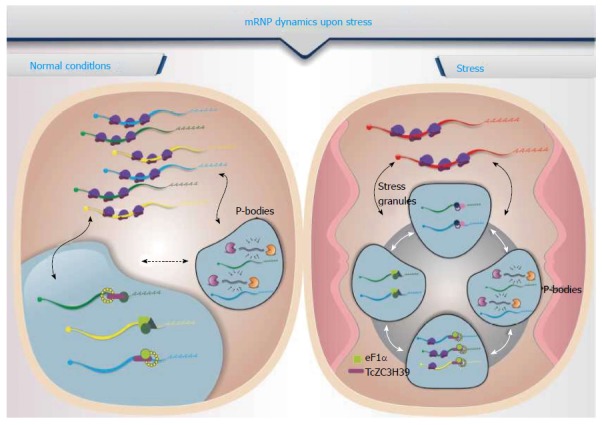
Messenger ribonucleoprotein compex dynamics during stress. Under normal conditions (left panel), several mRNAs exist in the cytoplasm and are normally translated. Some of these mRNAs can be degraded depending on the several translation rounds, and translation is repressed for some of them. Upon stress (right panel), most mRNAs leave the polysomes and are directed to either the degradation machinery or specific mRNP complexes (RNA granules), where they remain silent until normal conditions are restored. Few mRNAs are translated under conditions of stress. mRNP: Messenger ribonucleoprotein compex.
Sheth et al[67] proposed an RNA cycle model describing the dynamics of mRNAs in the cytoplasm, from polysomes to P-bodies and stress granules. In this model, mRNAs are able to associate with polysomes being effectively translated into proteins; however, when a stress signal is triggered, mRNAs interact with components of the degradation/storage pathway, translation is stopped, and the mRNP that is formed is held in P-bodies. This mRNA can then be stored or degraded in the P-bodies or returned to the translation apparatus[55,66]. Stress granules represent a translation arrest site that increases in abundance as mRNAs are dissociated from polysomes; the association of mRNA with initiation factors may facilitate their re-entry for translation when the stress ceases[55]. The molecular mechanisms encompassed in the transition between polysomes, P-bodies and stress granules need to be clearly elucidated.
In several unicellular eukaryotes, stress is involved in triggering differentiation. This association is exemplified in Figure 2, which shows that nutritional, pH and temperature stresses are involved in the life cycle of the human pathogen Trypanosoma cruzi. Accordingly, it has been shown that formation of RNA granules increases during the nutritional stress preceding metacyclogenesis[10,12] (Figure 1). This process is dynamic because there is an important switch in the mRNAs sequestered to RNA granules in the course of the differentiation process[68].
Figure 2.
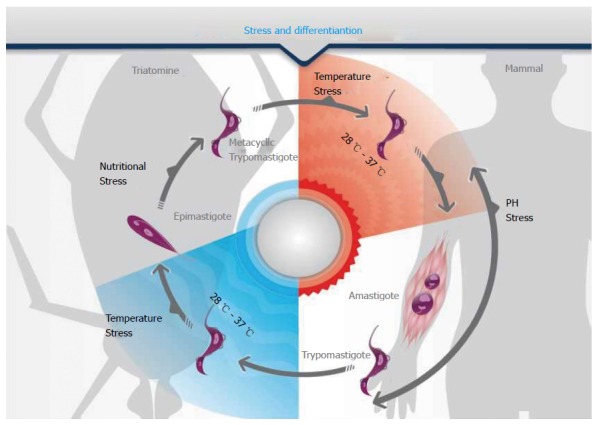
Physiological stresses and differentiation. The schematic representation shows the life cycle of Trypanosoma cruzi. This parasite alternates between two distinct intermediary hosts. The change to each host involves a temperature stress, either a heat shock (28 °C to 37 °C) during the passage from the invertebrate to the vertebrate or a cold shock (37 °C to 28 °C) when the parasite switches from the mammalian to the triatomine vector. Inside the hosts, nutritional and pH stresses are determinants for the differentiation from the non-replicative to the replicative forms (either epimastigotes in the insect or amastigotes in the mammal host) and from those to the infective trypomastigote forms.
RBPs in stress in protozoa
RBPs are crucial for the rapid gene expression remodeling that occurs in the course of cell differentiation or the stress response. In some case, the changes in gene expression are accompanied by changes in mRNA localization within the cell. Accordingly, the localization of specific mRNAs is a meaningful tool to optimize protein synthesis and to maintain cell polarity[69]. Fluorescence in situ hybridization analysis using poly-T showed the increase of fluorescent granules during stress conditions in trypanosomatids, suggesting that the mRNAs were mobilized to form RNA granules. It was also observed that the cytoplasmic localization of specific transcripts changed during differentiation[70].
Proteomic analysis evidenced a global change in the protein composition of T. cruzi mRNPs isolated from polysomal and post-polysomal fractions from epimastigotes in exponential growth and epimastigotes under nutritional stress. More than 500 proteins were identified, and among them were several RBPs. Interestingly, a dynamic shift in protein composition was also observed in response to nutritional stress, especially for those proteins identified in the polysomal-free fraction of stressed parasites[71].
Some specific RBPs play important roles during the stress response in parasites. One example is TcZC3H39, a zinc finger protein from T. cruzi that exhibits a shift in the composition of the bound mRNA targets depending on the physiological conditions of the cell. Under stress conditions, the TcZC3H39-mRNP complex sequesters highly expressed mRNAs, thereby slowing translation activity. Accordingly, the number of bound mRNA targets was higher in stressed parasites than in non-stressed parasites. Under stress conditions, the TcZC3H39-mRNP complex was enriched in targets that are highly expressed under non-stress conditions, such as ribosomal and oxidative phosphorylation pathway proteins[68]. The protein content of TcZC3H39-mRNP consists of ribosomes, translation factors, RNA helicases and other RBPs, suggesting that this zinc finger protein might be part of complexes similar to stress granules, sequestering highly expressed mRNAs and the associated ribosomes, possibly slowing down translation in response to stress conditions[68].
The zinc finger protein ZC3H11 plays an important role during T. brucei heat shock. This protein is essential for the bloodstream form of the parasite and stabilizes HSP70 mRNA and other transcripts that encode chaperones necessary for proper protein folding or refolding during heat stress[72]. ZC3H11 interacts with the MKT1 and PBP1 proteins, forming a complex that stabilizes the associated transcripts[73].
TbDRBD3 is a cytoplasmic RRM protein from T. brucei that relocalizes to the nucleus and forms stress granules upon oxidative stress. It has been suggested that DRBD3 participates in the transport of target mRNAs that are remodeled in response to stress[74]. Accordingly, RNA-seq showed that the mRNAs associated with this protein encode ribosomal proteins, translation factors and enzymes, suggesting a role in protein synthesis in T. brucei[75].
TcSR62 is a nuclear RBP from T. cruzi that relocalizes to the nucleolus during actinomycin-D treatment[76]. Other RBPs, such as DRBD4 (PTB2 in T. brucei, a PTB homolog) and PABP, have also been observed to show this behavior during treatment with actinomycin-D or during severe heat shock[76]. It has been hypothesized that the nucleolus plays a role as a sensor or regulator of cell metabolism during the stress response[77,78].
T. brucei Alba 3 and 4 are found in stress granule-like structures during nutritional deprivation. They partially co-migrate with polysomes and also associate with the helicase DHH1, translation initiation factors and PABP1, indicating a role in translation modulation[21].
The heat shock response in T. brucei involves a reduction of the polysomes and a corresponding increase in the number of RNA granules, such as P-bodies and stress granules. The P-bodies from T. brucei are composed of XRNA (mRNA decay machinery), Scd6 and DHH1. These RNA granules could act as sites for RNA storage or degradation during stress and differentiation[79].
In T. cruzi, the DHH1 protein is also localized in cytoplasmic foci that resemble P-bodies and/or stress granules. The protein interacts with PABP, and its localization changes in response to nutritional stress; the granules increase in number and also become bigger, likely due to the shift of mRNAs from the polysomes to the RNA granules[12].
It is important to mention that proteins lacking canonical RBDs but having other functions in the cell can also associate with RNAs and become structural components of RNP complexes[80]. The moonlighting theory proposes that some multifunctional enzymes normally associated with metabolic pathways can also play a role in regulating gene expression in prokaryotes and eukaryotes. This “moonlighting” role was previously described for glyceraldehyde-3-phosphate dehydrogenase, which can be found within the nuclei of mammalian cells acting in transcription, DNA repair, RNA binding and transport, and telomere binding[73,81,82]. Other examples are inosine monophosphate dehydrogenase[83], acetyl-CoA carboxylase[84], lipin-1[85], aminoacyl-tRNA synthetase[86], Llv5p, which is required for the synthesis of branched chain amino acids[87], phosphofructokinase[88] and the mitochondrial enzyme Arg5,6, that participates in the transcription of target genes in addition to its role in arginine biosynthesis[89].
Another example of a moonlighting protein in T. cruzi is EF-1α, which associates with a specific subset of mRNAs during stress conditions. This protein can associate with large protein complexes independent of the translation machinery. A specific subset of mRNAs associates with EF-1α-mRNPs in unstressed or stressed epimastigotes. Some mRNAs are common to both physiological conditions, whereas others are specific to a given physiological condition. Gene ontology analysis identified enrichment of gene sets involved in single-organism metabolic processes, amino acid metabolic processes, ATP and metal ion binding, glycolysis, glutamine metabolic processes, and cobalt and iron ion binding[90].
RBPs and eukaryotic cell differentiation
In parasites, several RBPs involved in differentiation have been described. The RRM-containing domain proteins RBP10 and RBP6 from T. brucei are implicated in life-cycle differentiation[22,91]. Accordingly, RBP10 knockdown led to a great alteration in the mRNA levels of transcripts normally found in high levels in the bloodstream forms[22]. The CCCH zinc finger proteins ZFP1, ZFP2 and ZFP3 have also been described as having roles in differentiation in trypanosomes[92,93]. Knockdown of ZFP1 disturbed the bloodstream differentiation by repositioning the kinetoplast in T. brucei[94]. A similar result was obtained with the RNAi knockdown for ZFP2: The parasite was unable to differentiate[92]. ZFP3 overexpression enhanced the differentiation levels to procyclic forms, and it is believed that these three proteins are part of the same differentiation pathway[93,95].
CONCLUSION
Despite evidence of the role for RBPs in gene expression regulation, particularly in protozoa as a means to respond promptly to environmental changes, the detailed mechanisms remain to be clearly elucidated. We anticipate that the complexity of these mechanisms should increase, as we do not yet know how different combinations of RBPs in a given complex affect the fate of the bound mRNAs. Furthermore, the modulation of the interactions within a complex could also be affected by post-translational modifications of the RBPs, creating an RBP code analogous to the histone code.
ACKNOWLEDGMENTS
We wish to thank Wagner Nagib Birbeire for the illustrations. Samuel Goldenberg is a research fellow from Conselho Nacional do Desenvolvimento Científico e Tecnologico (CNPq, Brazil)
Footnotes
Conflict-of-interest statement: Authors declare no conflict of interests for this article.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: May 29, 2015
First decision: August 19, 2015
Article in press: November 17, 2015
P- Reviewer: Gervois P, Jia JH, Kan L, Lawen A S- Editor: Qiu S L- Editor: A E- Editor: Jiao XK
References
- 1.Cléry A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda M, Sawa H, Tobiume M, Tokunaga K, Hasegawa H, Ichinohe T, Sata T, Moriyama M, Hall WW, Kurata T, et al. Tristetraprolin inhibits HIV-1 production by binding to genomic RNA. Microbes Infect. 2006;8:2647–2656. doi: 10.1016/j.micinf.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 5.Carballo E, Blackshear PJ. Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood. 2001;98:2389–2395. doi: 10.1182/blood.v98.8.2389. [DOI] [PubMed] [Google Scholar]
- 6.Nicastro G, Taylor IA, Ramos A. KH-RNA interactions: back in the groove. Curr Opin Struct Biol. 2015;30:63–70. doi: 10.1016/j.sbi.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Godin KS, Varani G. How arginine-rich domains coordinate mRNA maturation events. RNA Biol. 2007;4:69–75. doi: 10.4161/rna.4.2.4869. [DOI] [PubMed] [Google Scholar]
- 8.Rajyaguru P, Parker R. RGG motif proteins: modulators of mRNA functional states. Cell Cycle. 2012;11:2594–2599. doi: 10.4161/cc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajyaguru P, She M, Parker R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol Cell. 2012;45:244–254. doi: 10.1016/j.molcel.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holetz FB, Correa A, Avila AR, Nakamura CV, Krieger MA, Goldenberg S. Evidence of P-body-like structures in Trypanosoma cruzi. Biochem Biophys Res Commun. 2007;356:1062–1067. doi: 10.1016/j.bbrc.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 11.Kramer S. RNA in development: how ribonucleoprotein granules regulate the life cycles of pathogenic protozoa. Wiley Interdiscip Rev RNA. 2014;5:263–284. doi: 10.1002/wrna.1207. [DOI] [PubMed] [Google Scholar]
- 12.Holetz FB, Alves LR, Probst CM, Dallagiovanna B, Marchini FK, Manque P, Buck G, Krieger MA, Correa A, Goldenberg S. Protein and mRNA content of TcDHH1-containing mRNPs in Trypanosoma cruzi. FEBS J. 2010;277:3415–3426. doi: 10.1111/j.1742-4658.2010.07747.x. [DOI] [PubMed] [Google Scholar]
- 13.Kramer S, Bannerman-Chukualim B, Ellis L, Boulden EA, Kelly S, Field MC, Carrington M. Differential localization of the two T. brucei poly(A) binding proteins to the nucleus and RNP granules suggests binding to distinct mRNA pools. PLoS One. 2013;8:e54004. doi: 10.1371/journal.pone.0054004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández-Moya SM, Estévez AM. Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. Wiley Interdiscip Rev RNA. 2010;1:34–46. doi: 10.1002/wrna.6. [DOI] [PubMed] [Google Scholar]
- 15.Mörking PA, Dallagiovanna BM, Foti L, Garat B, Picchi GF, Umaki AC, Probst CM, Krieger MA, Goldenberg S, Fragoso SP. TcZFP1: a CCCH zinc finger protein of Trypanosoma cruzi that binds poly-C oligoribonucleotides in vitro. Biochem Biophys Res Commun. 2004;319:169–177. doi: 10.1016/j.bbrc.2004.04.162. [DOI] [PubMed] [Google Scholar]
- 16.Mörking PA, Rampazzo Rde C, Walrad P, Probst CM, Soares MJ, Gradia DF, Pavoni DP, Krieger MA, Matthews K, Goldenberg S, et al. The zinc finger protein TcZFP2 binds target mRNAs enriched during Trypanosoma cruzi metacyclogenesis. Mem Inst Oswaldo Cruz. 2012;107:790–799. doi: 10.1590/s0074-02762012000600014. [DOI] [PubMed] [Google Scholar]
- 17.Dallagiovanna B, Correa A, Probst CM, Holetz F, Smircich P, de Aguiar AM, Mansur F, da Silva CV, Mortara RA, Garat B, et al. Functional genomic characterization of mRNAs associated with TcPUF6, a pumilio-like protein from Trypanosoma cruzi. J Biol Chem. 2008;283:8266–8273. doi: 10.1074/jbc.M703097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volpon L, D’Orso I, Young CR, Frasch AC, Gehring K. NMR structural study of TcUBP1, a single RRM domain protein from Trypanosoma cruzi: contribution of a beta hairpin to RNA binding. Biochemistry. 2005;44:3708–3717. doi: 10.1021/bi047450e. [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Díaz L, Pastro L, Smircich P, Dallagiovanna B, Garat B. Evidence for a negative feedback control mediated by the 3’ untranslated region assuring the low expression level of the RNA binding protein TcRBP19 in T. cruzi epimastigotes. Biochem Biophys Res Commun. 2013;436:295–299. doi: 10.1016/j.bbrc.2013.05.096. [DOI] [PubMed] [Google Scholar]
- 20.Das A, Morales R, Banday M, Garcia S, Hao L, Cross GA, Estevez AM, Bellofatto V. The essential polysome-associated RNA-binding protein RBP42 targets mRNAs involved in Trypanosoma brucei energy metabolism. RNA. 2012;18:1968–1983. doi: 10.1261/rna.033829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mani J, Güttinger A, Schimanski B, Heller M, Acosta-Serrano A, Pescher P, Späth G, Roditi I. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS One. 2011;6:e22463. doi: 10.1371/journal.pone.0022463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wurst M, Seliger B, Jha BA, Klein C, Queiroz R, Clayton C. Expression of the RNA recognition motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei. Mol Microbiol. 2012;83:1048–1063. doi: 10.1111/j.1365-2958.2012.07988.x. [DOI] [PubMed] [Google Scholar]
- 23.Cunha-e-Silva N, Sant’Anna C, Pereira MG, Porto-Carreiro I, Jeovanio AL, de Souza W. Reservosomes: multipurpose organelles? Parasitol Res. 2006;99:325–327. doi: 10.1007/s00436-006-0190-3. [DOI] [PubMed] [Google Scholar]
- 24.Sant’Anna C, Parussini F, Lourenço D, de Souza W, Cazzulo JJ, Cunha-e-Silva NL. All Trypanosoma cruzi developmental forms present lysosome-related organelles. Histochem Cell Biol. 2008;130:1187–1198. doi: 10.1007/s00418-008-0486-8. [DOI] [PubMed] [Google Scholar]
- 25.Guerra-Slompo EP, Probst CM, Pavoni DP, Goldenberg S, Krieger MA, Dallagiovanna B. Molecular characterization of the Trypanosoma cruzi specific RNA binding protein TcRBP40 and its associated mRNAs. Biochem Biophys Res Commun. 2012;420:302–307. doi: 10.1016/j.bbrc.2012.02.154. [DOI] [PubMed] [Google Scholar]
- 26.Stern MZ, Gupta SK, Salmon-Divon M, Haham T, Barda O, Levi S, Wachtel C, Nilsen TW, Michaeli S. Multiple roles for polypyrimidine tract binding (PTB) proteins in trypanosome RNA metabolism. RNA. 2009;15:648–665. doi: 10.1261/rna.1230209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naguleswaran A, Gunasekera K, Schimanski B, Heller M, Hemphill A, Ochsenreiter T, Roditi I. Trypanosoma brucei RRM1 is a nuclear RNA-binding protein and modulator of chromatin structure. MBio. 2015;6:e00114. doi: 10.1128/mBio.00114-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schumann Burkard G, Käser S, de Araújo PR, Schimanski B, Naguleswaran A, Knüsel S, Heller M, Roditi I. Nucleolar proteins regulate stage-specific gene expression and ribosomal RNA maturation in Trypanosoma brucei. Mol Microbiol. 2013;88:827–840. doi: 10.1111/mmi.12227. [DOI] [PubMed] [Google Scholar]
- 29.Serpeloni M, Moraes CB, Muniz JR, Motta MC, Ramos AS, Kessler RL, Inoue AH, daRocha WD, Yamada-Ogatta SF, Fragoso SP, et al. An essential nuclear protein in trypanosomes is a component of mRNA transcription/export pathway. PLoS One. 2011;6:e20730. doi: 10.1371/journal.pone.0020730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braks JA, Mair GR, Franke-Fayard B, Janse CJ, Waters AP. A conserved U-rich RNA region implicated in regulation of translation in Plasmodium female gametocytes. Nucleic Acids Res. 2008;36:1176–1186. doi: 10.1093/nar/gkm1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mair GR, Braks JA, Garver LS, Wiegant JC, Hall N, Dirks RW, Khan SM, Dimopoulos G, Janse CJ, Waters AP. Regulation of sexual development of Plasmodium by translational repression. Science. 2006;313:667–669. doi: 10.1126/science.1125129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui L, Fan Q, Li J. The malaria parasite Plasmodium falciparum encodes members of the Puf RNA-binding protein family with conserved RNA binding activity. Nucleic Acids Res. 2002;30:4607–4617. doi: 10.1093/nar/gkf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan Q, Li J, Kariuki M, Cui L. Characterization of PfPuf2, member of the Puf family RNA-binding proteins from the malaria parasite Plasmodium falciparum. DNA Cell Biol. 2004;23:753–760. doi: 10.1089/dna.2004.23.753. [DOI] [PubMed] [Google Scholar]
- 34.Young JA, Fivelman QL, Blair PL, de la Vega P, Le Roch KG, Zhou Y, Carucci DJ, Baker DA, Winzeler EA. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol Biochem Parasitol. 2005;143:67–79. doi: 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Lindner SE, Mikolajczak SA, Vaughan AM, Moon W, Joyce BR, Sullivan WJ, Kappe SH. Perturbations of Plasmodium Puf2 expression and RNA-seq of Puf2-deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell Microbiol. 2013;15:1266–1283. doi: 10.1111/cmi.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, Miao J, Liu T, Sullivan WJ, Cui L, Chen X. Characterization of TgPuf1, a member of the Puf family RNA-binding proteins from Toxoplasma gondii. Parasit Vectors. 2014;7:141. doi: 10.1186/1756-3305-7-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gissot M, Walker R, Delhaye S, Alayi TD, Huot L, Hot D, Callebaut I, Schaeffer-Reiss C, Dorsselaer AV, Tomavo S. Toxoplasma gondii Alba proteins are involved in translational control of gene expression. J Mol Biol. 2013;425:1287–1301. doi: 10.1016/j.jmb.2013.01.039. [DOI] [PubMed] [Google Scholar]
- 38.Boulila Y, Tomavo S, Gissot M. A RGG motif protein is involved in Toxoplasma gondii stress-mediated response. Mol Biochem Parasitol. 2014;196:1–8. doi: 10.1016/j.molbiopara.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Thomas MP, Lieberman J. Live or let die: posttranscriptional gene regulation in cell stress and cell death. Immunol Rev. 2013;253:237–252. doi: 10.1111/imr.12052. [DOI] [PubMed] [Google Scholar]
- 40.Muñoz MJ, Pérez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, Dujardin G, Le Jossic-Corcos C, Corcos L, Auboeuf D. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol. 2010;17:1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- 43.van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Cáceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 45.Pain VM. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 46.Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 47.Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Wouters BG, van den Beucken T, Magagnin MG, Koritzinsky M, Fels D, Koumenis C. Control of the hypoxic response through regulation of mRNA translation. Semin Cell Dev Biol. 2005;16:487–501. doi: 10.1016/j.semcdb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 50.Martínez-Salas E, Lozano G, Fernandez-Chamorro J, Francisco-Velilla R, Galan A, Diaz R. RNA-binding proteins impacting on internal initiation of translation. Int J Mol Sci. 2013;14:21705–21726. doi: 10.3390/ijms141121705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol. 2008;28:40–49. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain S, Parker R. The discovery and analysis of P Bodies. Adv Exp Med Biol. 2013;768:23–43. doi: 10.1007/978-1-4614-5107-5_3. [DOI] [PubMed] [Google Scholar]
- 56.Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 57.Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 60.Beelman CA, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 61.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 65.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 66.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alves LR, Oliveira C, Mörking PA, Kessler RL, Martins ST, Romagnoli BA, Marchini FK, Goldenberg S. The mRNAs associated to a zinc finger protein from Trypanosoma cruzi shift during stress conditions. RNA Biol. 2014;11:921–933. doi: 10.4161/rna.29622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alves LR, Guerra-Slompo EP, de Oliveira AV, Malgarin JS, Goldenberg S, Dallagiovanna B. mRNA localization mechanisms in Trypanosoma cruzi. PLoS One. 2013;8:e81375. doi: 10.1371/journal.pone.0081375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alves LR, Avila AR, Correa A, Holetz FB, Mansur FC, Manque PA, de Menezes JP, Buck GA, Krieger MA, Goldenberg S. Proteomic analysis reveals the dynamic association of proteins with translated mRNAs in Trypanosoma cruzi. Gene. 2010;452:72–78. doi: 10.1016/j.gene.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 72.Droll D, Minia I, Fadda A, Singh A, Stewart M, Queiroz R, Clayton C. Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathog. 2013;9:e1003286. doi: 10.1371/journal.ppat.1003286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh R, Green MR. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- 74.Fernández-Moya SM, García-Pérez A, Kramer S, Carrington M, Estévez AM. Alterations in DRBD3 ribonucleoprotein complexes in response to stress in Trypanosoma brucei. PLoS One. 2012;7:e48870. doi: 10.1371/journal.pone.0048870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Das A, Bellofatto V, Rosenfeld J, Carrington M, Romero-Zaliz R, del Val C, Estévez AM. High throughput sequencing analysis of Trypanosoma brucei DRBD3/PTB1-bound mRNAs. Mol Biochem Parasitol. 2015;199:1–4. doi: 10.1016/j.molbiopara.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 76.Názer E, Verdún RE, Sánchez DO. Nucleolar localization of RNA binding proteins induced by actinomycin D and heat shock in Trypanosoma cruzi. PLoS One. 2011;6:e19920. doi: 10.1371/journal.pone.0019920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olson MO. Sensing cellular stress: another new function for the nucleolus? Sci STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 79.Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J Cell Sci. 2008;121:3002–3014. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Müller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 81.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 82.Demarse NA, Ponnusamy S, Spicer EK, Apohan E, Baatz JE, Ogretmen B, Davies C. Direct binding of glyceraldehyde 3-phosphate dehydrogenase to telomeric DNA protects telomeres against chemotherapy-induced rapid degradation. J Mol Biol. 2009;394:789–803. doi: 10.1016/j.jmb.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McLean JE, Hamaguchi N, Belenky P, Mortimer SE, Stanton M, Hedstrom L. Inosine 5’-monophosphate dehydrogenase binds nucleic acids in vitro and in vivo. Biochem J. 2004;379:243–251. doi: 10.1042/BJ20031585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benson BK, Meades G, Grove A, Waldrop GL. DNA inhibits catalysis by the carboxyltransferase subunit of acetyl-CoA carboxylase: implications for active site communication. Protein Sci. 2008;17:34–42. doi: 10.1110/ps.073186408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reue K, Zhang P. The lipin protein family: dual roles in lipid biosynthesis and gene expression. FEBS Lett. 2008;582:90–96. doi: 10.1016/j.febslet.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martinis SA, Plateau P, Cavarelli J, Florentz C. Aminoacyl-tRNA synthetases: a new image for a classical family. Biochimie. 1999;81:683–700. doi: 10.1016/s0300-9084(99)80126-6. [DOI] [PubMed] [Google Scholar]
- 87.Bateman JM, Perlman PS, Butow RA. Mutational bisection of the mitochondrial DNA stability and amino acid biosynthetic functions of ilv5p of budding yeast. Genetics. 2002;161:1043–1052. doi: 10.1093/genetics/161.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rabinovitz M. Evidence for a role of phosphofructokinase and tRNA in the polyribosome disaggregation of amino acid deficiency. FEBS Lett. 1991;283:270–272. doi: 10.1016/0014-5793(91)80605-3. [DOI] [PubMed] [Google Scholar]
- 89.Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- 90.Alves LR, Oliveira C, Goldenberg S. Eukaryotic translation elongation factor-1 alpha is associated with a specific subset of mRNAs in Trypanosoma cruzi. BMC Microbiol. 2015;15:104. doi: 10.1186/s12866-015-0436-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hendriks EF, Robinson DR, Hinkins M, Matthews KR. A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J. 2001;20:6700–6711. doi: 10.1093/emboj/20.23.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Walrad PB, Capewell P, Fenn K, Matthews KR. The post-transcriptional trans-acting regulator, TbZFP3, co-ordinates transmission-stage enriched mRNAs in Trypanosoma brucei. Nucleic Acids Res. 2012;40:2869–2883. doi: 10.1093/nar/gkr1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hendriks EF, Matthews KR. Disruption of the developmental programme of Trypanosoma brucei by genetic ablation of TbZFP1, a differentiation-enriched CCCH protein. Mol Microbiol. 2005;57:706–716. doi: 10.1111/j.1365-2958.2005.04679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kolev NG, Ullu E, Tschudi C. The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei. Cell Microbiol. 2014;16:482–489. doi: 10.1111/cmi.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]


