Abstract
In mammals, haem degradation to biliverdin (BV) through the action of haem oxygenase (HO) is a critical step in haem metabolism. The malaria parasite converts haem into the chemically inert haemozoin to avoid toxicity. We discovered that the knock-out of HO in P. berghei is lethal; therefore, we investigated the function of biliverdin (BV) and haem in the parasite. Addition of external BV and haem to P. falciparum-infected red blood cell (RBC) cultures delays the progression of parasite development. The search for a BV molecular target within the parasites identified P. falciparum enolase (Pf enolase) as the strongest candidate. Isothermal titration calorimetry using recombinant full-length Plasmodium enolase suggested one binding site for BV. Kinetic assays revealed that BV is a non-competitive inhibitor. We employed molecular modelling studies to predict the new binding site as well as the binding mode of BV to P. falciparum enolase. Furthermore, addition of BV and haem targets the phosphorylation of Plasmodium falciparum eIF2α factor, an eukaryotic initiation factor phosphorylated by eIF2α kinases under stress conditions. We propose that BV targets enolase to reduce parasite glycolysis rates and changes the eIF2α phosphorylation pattern as a molecular mechanism for its action.
The expansion of malaria intervention has had a tremendous impact on malaria incidence and mortality worldwide; however, the lack of a commercial vaccine and the increase in drug-resistant strains highlight the importance of identifying new mechanisms to combat the disease. Malaria symptoms appear when red blood cell (RBC) infection occurs and follows a series of developmental stages known as ring (R), trophozoite (T) and schizont (S). After cell lysis, the invasive form merozoites are released and subsequently invade other RBCs to reinitiate the cycle1,2. During the intraerythrocytic phase, the host haemoglobin is degraded and used as a source of amino acids3. This process generates haem, a toxic molecule that is scavenged by the malaria parasite through the formation of haemozoin polymer4.
Most mammalian cells use a different strategy to nullify haem toxicity by converting it to biliverdin (BV), a step catalysed by haem oxygenase (HO). BV is subsequently converted to bilirubin (BR) by biliverdin reductase (BVR)5,6. In the past, BV and its by-product BR have been exclusively viewed as waste products of haem catabolism7. However, there are indications that haem, BV and BR may play important roles in gene expression, oxidative response, and cellular signal transduction pathways in several biological systems8,9,10. Haem is a prosthetic group indispensable for the growth of malaria parasites11,12,13. Recently, it has been demonstrated that P. falciparum possess a de novo haem biosynthetic pathway6. The existence of a haem oxygenase enzyme in P. falciparum (PfHO) was demonstrated14,15, however, its activity is controversial16,17. Therefore, the role of Plasmodium HO in the haem metabolism and the impact of host BV on host-parasite interaction remain unclear.
Phosphorylation is among the most important post-translational modifications for eukaryotic cells as well for Plasmodium18. In general, the kinases that regulate the eukaryote translation of proteins in response to stress are called eukaryotic initiation factor 2α kinases (eIF2α kinases), and the mechanism of action involves changes in the phosphorylation of eIF2α under stress conditions, which inhibits the translation of most proteins. In Plasmodium, three eIF2α kinases were identified: PfeIK1, PfeIK2 and PfPK418. PfeIK1 and PfPK4 are more abundant in asexual stages, whereas PfeIK2 is more abundant in sporozoites19. A.P. Han et al. 200120 identified the presence of an eIF2α kinase in reticulocytes whose action is modulated by haem. The three-dimensional structure has two haem-binding sites that suppress the phosphorylation activity of eIF2α. Thus, eIF2α kinase HRI (haem-regulated inhibitor) prevents the development of reticulocytes in the absence of haem through the phosphorylation of eIF2α21. Here, we investigated changes in the phosphorylation pattern of Plasmodium eIF2α factor upon the addition of BV and haem into P. falciparum-infected RBCs.
To the best of our knowledge, this work is the first to investigate the effect of BV on intraerythrocytic development and its potential targets in P. falciparum. We demonstrate that BV is present in infected RBC cultures. The higher BV concentration affects the P. falciparum cell cycle by increasing the abundance of early stages and reducing late stages. We identify enolase as a target for BV binding in P. falciparum and evaluate the stoichiometry and energetic components underlying BV binding. Moreover, our data indicate BV as a non-competitive inhibitor at the low micromolar range, and molecular modelling suggests a BV-binding mode for P. falciparum enolase.
Results
Detection of BV in P. falciparum–infected red blood cell supernatant culture
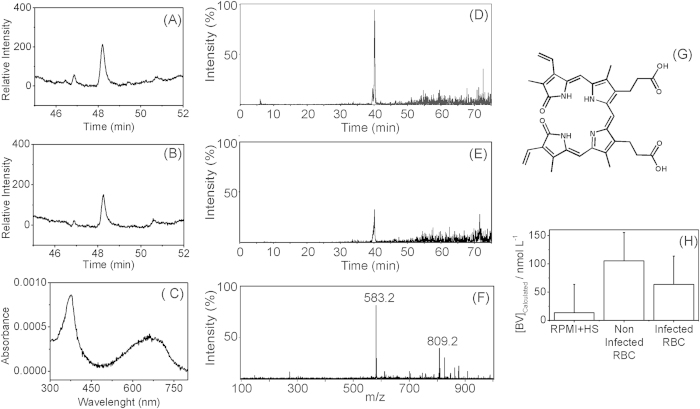
To investigate the host BV in Plasmodium culture, the amounts of BV in the supernatant of uninfected and P. falciparum-infected RBC cultures were measured by high-performance liquid chromatography (HPLC-UV-Vis and HPLC-MS; Fig. 1). Both uninfected (Fig. A) and infected (Fig. 1B) RBC culture supernatant chromatograms exhibited a UV-Vis peak at 48.3 min due to BV. ESI-MS analysis of the eluate exhibited a strong peak at m/z 583.2 (Fig. 1D,E), corresponding to the molecular ion of HBV+ (protonated BV). BV quantification, which was performed by the addition of external standards, revealed no significant difference between BV concentration in supernatant samples obtained from uninfected and infected RBC cultures (105 ± 30 and 63 ± 30 nM, respectively; Fig. 1H).
Figure 1. BV characterization in P. falciparum culture and non-infected supernatant.
(A,B) are chromatographic analysis at 660 nm of uninfected and infected culture supernatants, respectively. (C) is the absorption spectra observed for (A) at 48.3 min retention time. (D,E) are ESI + MS chromatographic analysis (EIC m/z 583) of non-infected and infected culture supernatants, respectively. (F) is the mass spectrum observed for (D) at 40 min retention time. Different RTs are due to changes in the mobile phase necessary for LC-MS. BV structure (G) presents a calculated monoisotopic mass of 582.2 Da. Quantitative analysis (H) reveals similar concentrations of BV in infected and non-infected culture supernatants.
Effect of BV in P. falciparum cell cycle and P. berghei haem oxygenase (PbOH) knockout
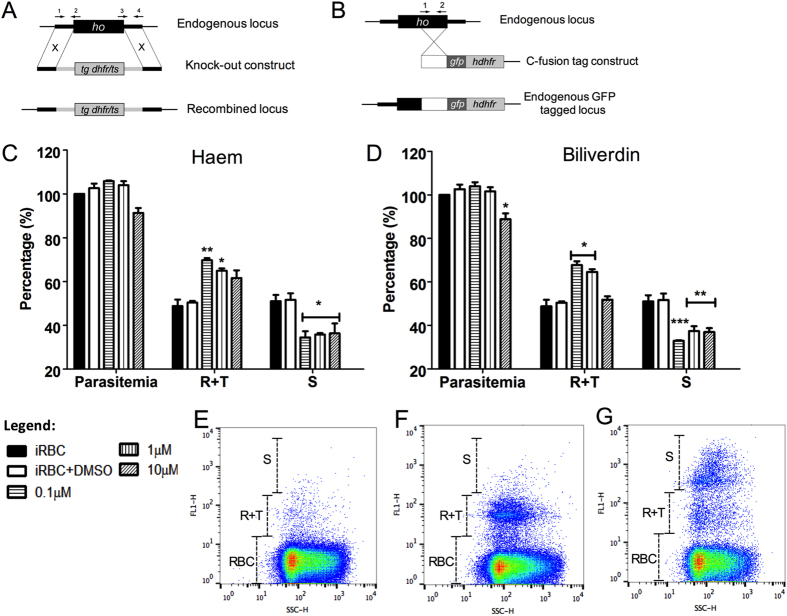
An alternative strategy to deplete the levels of haem within cells relies on the activity of haem oxygenase enzymes, which convert haem into BV, iron, and carbon monoxide. P. falciparum possess a haem oxygenase (PfHO)14,15; however, its cellular function within parasites remains unclear17. Therefore, to investigate the role of HO in Plasmodium, a strategy of knocking out the P. berghei haem oxygenase (PbHO) (Fig. 2A) and adding c-fusion GFP (green fluorescence protein) to PbHO (Fig. 2B) was performed. Both approaches were lethal to P. berghei, suggesting an unknown vital role for this enzyme in Plasmodium physiology.
Figure 2. Multiple strategies to investigate the function of haem oxygenase and effects of haem and biliverdin during erythrocytic development of Plasmodium.
(A) Schematic representation of the gene targeting strategy used for gene disruption via double homologous recombination in P. berghei. Primers 1 to 4 used for PCR of 5′ and 3′ fragments for knockout construct are indicated. (B) Schematic representation of the gene-tagging construct used for tagging of the endogenous pbho locus in P. berghei by single homologous recombination with gfp. Primers 1 and 2 used for amplifying the endogenous locus are indicated. (C) P. falciparum intraerythrocytic modulation during 48 hours of incubation with haem. Parasitaemia value of the control with no drugs or solvent (iRBC) was considered 100%. All treatments were compared with the control containing only solvent (iRBC + DMSO). iRBC + DMSO values: parasitaemia 103 ± 2%; R + T: 50 ± 1% and S: 52 ± 5%. Haem graphic, parasitaemia values: 0.1 μM (99 ± 3%); 1 μM (97 ± 5%); 10 μM (91 ± 2%). R + T values: 0.1 μM (70 ± 1%); 1 μM (64 ± 1%); 10 μM (62 ± 4%). S values: 0.1 μM (35 ± 3%); 1 μM (36 ± 1%); 10 μM (36 ± 4%). (D) BV graphic, parasitaemia values: 0.1 μM (104 ± 2%); 1 μM (98 ± 4%); 10 μM (89 ± 3%). R + T values: 0.1 μM (68 ± 1%); 1 μM (65 ± 1%); 10 μM (52 ± 2%). S values: 0.1 μM (33 ± 1%); 1 μM (37 ± 2%); 10 μM (37 ± 2%). The graphs in C and D represent means and standard error of three independent experiments. *P < 0.05%, **P < 0.01% and ***P < 0.001%. (E) Dot plot depicts a typical gating population of non-infected RBCs (RBC). (F) Dot plot depicts a typical gating population of RBCs and infected RBCs with mononucleated parasites (R + T). (G) Dot plot depicts a typical gating population of RBCs, R + T and infected RBCs with multinucleated parasites (S).
Moreover, the detection of BV at nanomolar concentrations in parasite culture led to the investigation of BV as a signal modulator during the P. falciparum life cycle forms R, T and S. For this purpose, synchronized infected RBCs in the ring stage were incubated for 48 hours in the absence and presence of 0.1, 1 and 10 μM of haem and BV (Fig. 2C,D, respectively). The distribution of P. falciparum in the R, T, and S forms was evaluated by counting 105 cells (Fig. E–G). Here, 10 μM BV exhibited a toxic effect after 48 hours (Fig. 2D). In addition, 0.1 and 1 μM BV and haem did not change the parasitaemia after 48 hours of incubation compared with the controls; however, at these concentrations, both compounds increase the R + T forms and decrease the S form.
Identification of BV targets in P. falciparum
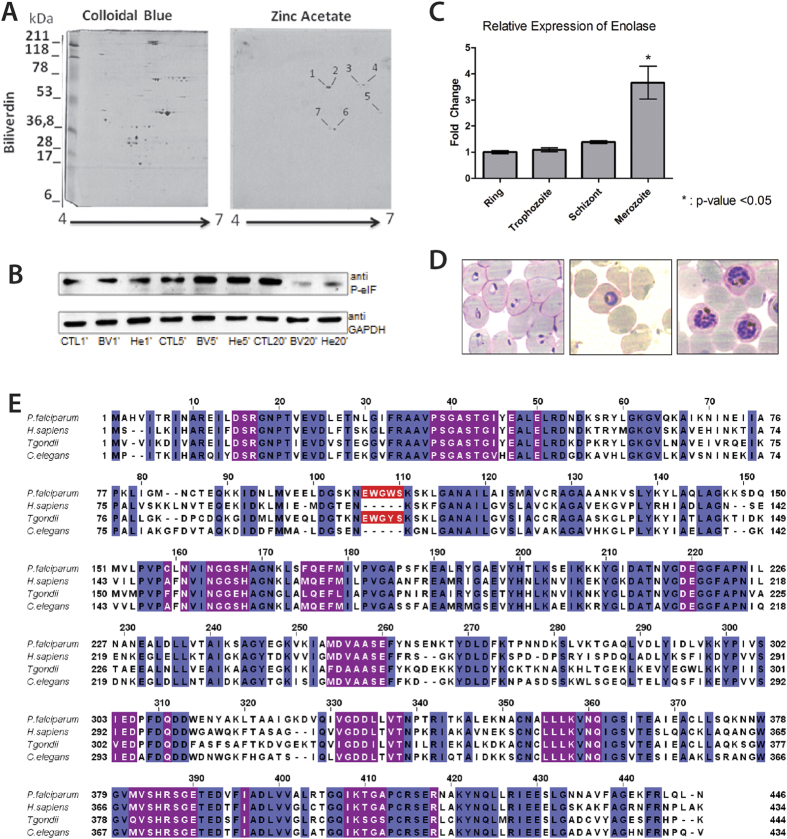
To investigate the mechanism of BV modulation in the P. falciparum cell cycle, a binding protein-BV assay was performed to identify candidate BV molecular target proteins (Fig. 3A). Zinc acetate was used to reveal protein-BV interactions, as described in Berkelman and Lagarias22. The results of mass spectrometry (MS) analysis of zinc acetate/Coomassie staining are shown in Table S1 in the supplemental material. The protein-BV binding assay using zinc acetate in a two-dimensional gel revealed seven positive spots (Fig. 3A) containing P. falciparum proteins (Table S1). As the number of peptides for enolase (access number: Pf10_0155) is considerably increased compared with the remaining peptides, we concluded that the BV-binding assay revealed P. falciparum enolase as the strongest candidate for a BV molecular target. Moreover, we have investigated the phosphorylation of eIF2α eukaryotic initiation factor as an indicator of stress factor for the cell after incubating P. falciparum-infected RBC at trophzoite stage with 1 μM haem and 1 μM BV by 1, 5 and 10 minutes followed by Western blot analysis using antibody anti-phospho eIF2α (anti P-eIF) (Fig. 3B). The results revealed that BV and haem promotes changes in the phosphorylation pattern of eIF2α and this could be part of their mechanism for reducing the growth of intraerythrocytic development of the malaria parasite Plasmodium falciparum.
Figure 3. BV binding protein assay in two-dimensional electrophoresis, phosphorylation of eIF2α, relative expression of enolase gene throughout 3D7 strain intraerythrocytic cycle, and enolase amino acid alignment.
(A) Total P. falciparum protein extract pre-treated with 20 μM BV for 30 minutes, revealing total protein staining (left colloidal Coomassie). On the right, the same gel reveals 7 positive spots (indicated by black lines) on zinc acetate assay treatment. (B) Western blot using antibody anti-phospho eIF2α (anti P-eIF) in P. falciparum-infected RBCs at trophozoite stage incubated with BV (BV) and haem (He) for 1, 5 or 20 minutes. GAPDH was used as a loading control. (C) Parasites were synchronized to isolate ring, trophozoite, schizont and merozoite stages using sorbitol. RNA samples were extracted, purified and subjected to qRT-PCR. Seryl-tRNA synthetase expression was quantified to normalize mRNA levels. The graphics reveal an increase in enolase expression in the merozoite stage. ★Statistically significant by t-test with P < 0.05. (D) Blood smears of P. falciparum culture indicating the stages (ring, trophozoite, schizont, respectively, from left to right) used to extract RNA for qRT-PCR analysis. (E) Amino acid alignment of enolase sequences of H. sapiens, P. falciparum, T. gondii and C. elegans. Identical residues are highlighted in blue, the pentapeptide that differentiates P. falciparum enolase from other organisms in red, and the residues that compose the catalytic site of the enzyme are indicated in purple.
Additionally, we have analysed changes in enolase expression throughout P. falciparum intraeritrocytic lifecycle at R, T and S stages. We detected an increase in enolase expression on merozoites, the RBC invasive form, (Fig. 3C). Fig. 3D illustrates R, T and S phases of the parasite lifecycle. Amino acid alignment with enolases from P. falciparum with other organisms (Fig. 3E) reveals that the enzyme has a peptide that is conserved only in apicomplexa phylum.
Recombinant purified Pf enolase-BV binding proprieties and in silico docking of BV-Pf enolase
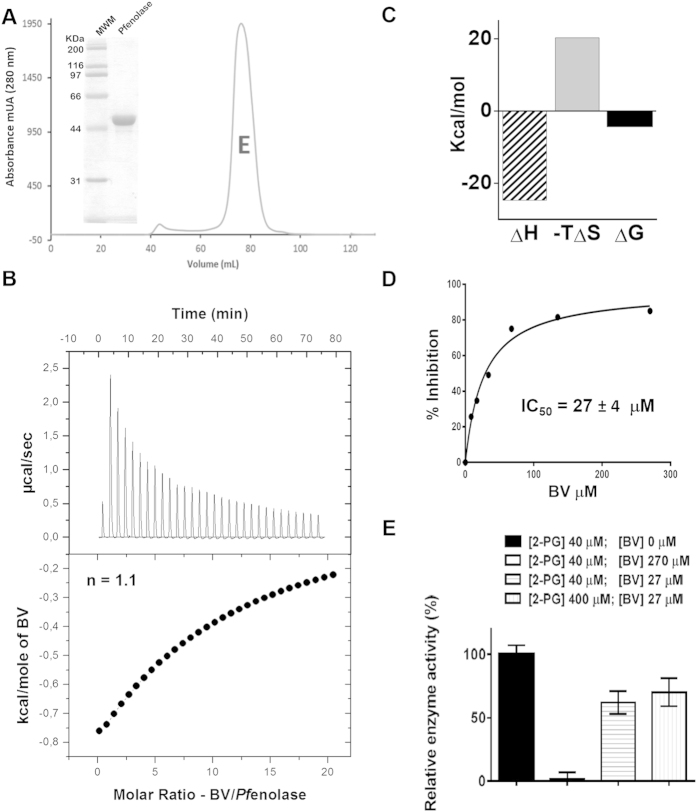
To assess the ability of Pf enolase to bind haem in vitro, we expressed recombinant Pf enolase in E. coli and purified it to homogeneity using a combination of chromatographic methods, including metal chelation and size-exclusion chromatography. We observed a single symmetric peak for a significant pure and folded dimeric 100-KDa protein in the gel filtration column (Fig. 4A). Next, we employed calorimetric analysis using isothermal titration calorimetry (ITC) to evaluate the stoichiometry and energetic components underlying the binding of BV to recombinant purified Pf enolase (Fig. 4B). The thermogram suggests an endothermic reaction (Fig. 4B), and the thermodynamic properties indicate that BV binding to Pf enolase is more enthalpically favoured (Fig. 4C, represented by larger ΔH bar in the histogram). The evaluated ΔG value of −4.5 Kcal/mol and the binding stoichiometry (n = 1.1) indicate that one BV binds to Pf enolase (Fig. 4B).
Figure 4. BV binding properties and inhibitory activity on Pf enolase.
(A) SDS–PAGE (10%) and gel filtration chromatogram of purified recombinant Pf enolase (MWM = molecular weight mass, E = enolase). (B) Isothermal titration calorimetry measurement of BV binding to Pf enolase. Top panel: changes in heat over time as BV is titrated into Pf enolase. Bottom panel: normalized change in heat after subtracting reference data of BV injections into buffer. The single-binding site model was used to fit the binding isotherms. (C) Thermodynamic signatures of BV binding to Pf enolase with Gibbs free energy of binding (ΔG; black), enthalpy (ΔH; black and white) and entropy (−TΔS; grey). (D) Inhibitory effect of BV on Pf enolase activity. (E) Non-competitive inhibitory profile of BV. Kinetic activity was assayed in the presence of a fixed dose of BV and increasing concentrations of substrate. First column: negative control [2-PG] = 40 μM (KM value); second column: [2-PG] = 40 μM (KM value) and positive control [BV] = 270 μM (10-fold IC50 value); third column: [2-PG] = 40 μM (KM value) and [BV] = 27 μM (IC50 value); fourth column: [2-PG] = 400 μM (10-fold KM value) and [BV] = 27 μM (IC50 value). Data were analysed from three different experiments.
Thus, we investigated whether BV might affect the catalytic activity of the enzyme. Hence, we evaluated the Pf enolase enzymatic activity in the presence of increasing doses of BV (Fig. 4D). BV inhibits the formation of phosphoenolpyruvate (PEP, enolase reaction product) with an IC50 value of 27 ± 4 μM. In addition, the effectiveness of BV as a Pf enolase inhibitor was evaluated by assessing the mechanism of inhibition. For this purpose, BV concentration was fixed at 27 μM (IC50 value), and the Pf enolase inhibition was compared in the presence of increasing doses of the substrate (2-phosphoglycerate, 2-PG). Fig. 4E demonstrates that BV inhibitory activity remains unchanged even at concentrations as high as 10-fold of the substrate KM value. This result suggests that BV does not act as an active site inhibitor but rather as a non-competitive inhibitor.
In silico docking of BV-Pf enolase
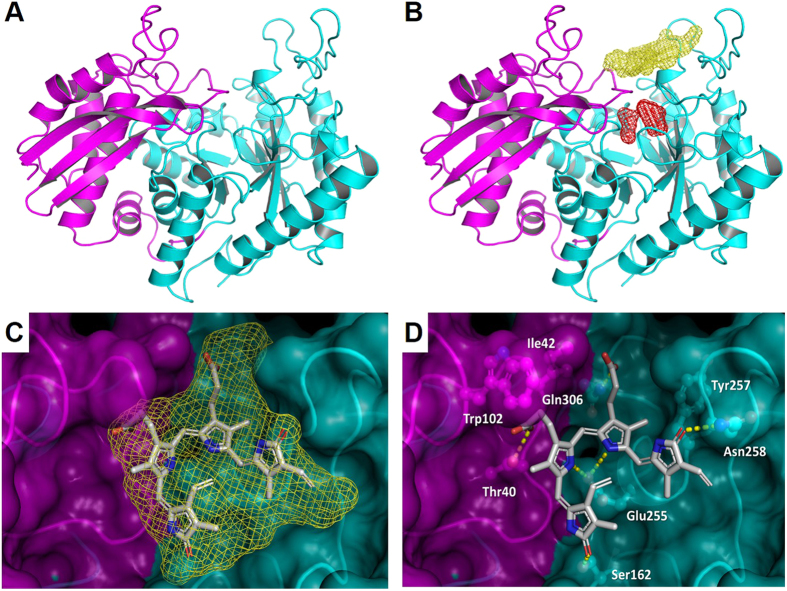
Based on the binding data collected and to shed light on the structural determinants underlying BV binding to enolase, molecular modelling approaches were employed to predict potential sites and investigate the BV binding mode. First, a homology-modelling study to predict the Pf enolase 3D structure was conducted. A combination of primary sequence and tertiary structure alignments against the known crystal structure of T. brucei enolase (PDB ID, 2PTZ) was used to construct a reliable three-dimensional (3D) model of the target protein. The best model comprised a monomeric structure of 439 residues, which exhibited a root-mean-square deviation (RMSD) of 1.6 Å over Cα for 429 equivalent residues of the T. brucei homologue. The Pf enolase monomer contains an eight fold β/α barrel domain preceded by an N-terminal α + β domain (Fig. 5A)23. According to the mechanism of inhibition, the BV binding site is somewhere other than the catalytic site. Therefore, SiteHound24 was employed to identify putative binding cavities in P. falciparum enolase. SiteHound is a web server that identifies ligand-binding sites by computing the interactions between a chemical probe and a protein structure. The top two scored binding cavities correspond to i. the catalytic site and ii. a cavity close to the catalytic site (Fig. 5B). Thus, the second best scored cavity was selected as a putative binding site for the molecular modelling of BV to Pf enolase. This putative binding site is located close to the active site and formed by polar and hydrophobic residues from the β/α barrel and α + β domains (Fig. 5B,C).
Figure 5. Pf enolase molecular modelling.
Pf enolase 3D modelling. (A) Pf enolase homology model (N-terminal domain = magenta; C-terminal domain = cyan). (B) Pf enolase envelopes for binding cavities identified by SiteHound (catalytic site = red mesh; dimer interface cavity = yellow mesh). (C) Modelled binding mode of BV (stick model) into Pf enolase putative cavity envelope (yellow mesh). (D) Detailed view of BV modelled binding mode (hydrogen bonds = yellow dashes; protein residues = ball and stick model).
The proposed binding mode of BV into the putative cavity indicates that the ligand occupies the central region of the putative binding site (Fig. 5D). In this conformation, the ligand establishes attractive polar and hydrophobic contacts with the Pf enolase binding site residues. The negatively charged carboxyl groups of BV are hydrogen bonded to the side-chains of Thr40 and Gln306. The electronegative carbonyl substituents of the pyrrolone groups are in a favourable orientation to accept hydrogen bonds from the side-chains of Asn258 and Ser162, whereas the nitrogen atoms of the central pyrrol groups are favourably oriented to donate hydrogen bonds to the side-chain of Glu255. Additionally, non-polar groups of BV make favourable van der Waals contacts with several structural elements of the protein, and some of these groups establish attractive hydrophobic interactions with their macromolecular counterparts that significantly contribute to the complex stability. In particular, the carbon atoms of the carboxyethyl substituent of the pyrrol groups are in van der Waals contact with the side-chains of Trp102 and Ile42; in addition, the 4-methyl substituents of the pyrrol groups are in close contact with the side-chains of Trp102 and Tyr257 (Fig. 5D).
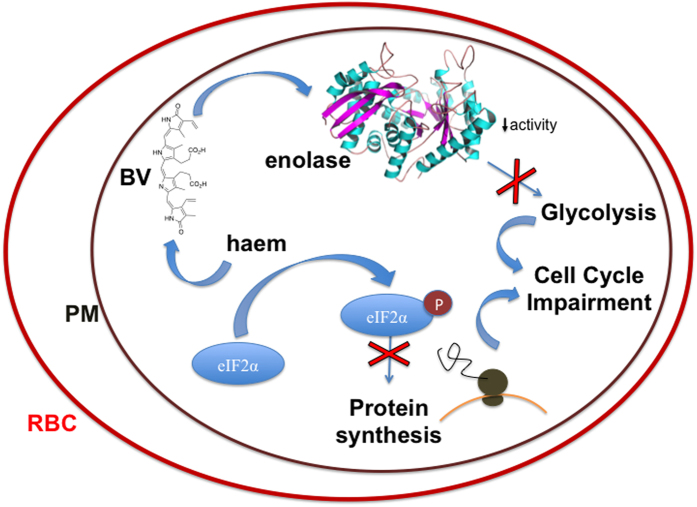
Finally, Fig. 6 depicts a proposed molecular mechanism for BV reducing the growth of Plasmodium falciparum cell cycle.
Figure 6. Proposed mechanism for BV reducing the growth of Plasmodium falciparum.
BV interacts with enolase decreasing its activity and glycolysis and, in turn, cell cycle progression. Moreover, BV and haem modulates phosphorylation of Eukaryotic Initiation Factor 2α (eIF2α). Phosphorylated eIF2α is a stress response factor which acts in the reduction of translation to a basal level and consequently the arrest of the cell cycle of P. falciparum. RBC: Red Blood Cell. PM: Plasmodium falciparum plasma membrane.
Discussion
During intraerythrocytic malaria infection, the millimolar amount of haem derivate from host haemoglobin digestion is sequestered as a haemozoin crystal within the digestive vacuole25,26. Okada 200914 reported the existence of PfHO, which exhibits in vitro HO and BVR activity. However, no canonical HO pathway was observed in vivo and in vitro17. The small difference observed in BV concentration in the supernatant of RBC cultures infected and not infected with P. falciparum (Fig. 1C) indicates that the parasite does not enzymatically produce or catabolize BV. BR induces oxidative damage and inhibits P. falciparum grown in micromolar concentrations16. In this study, the concentration of BV detected in P. falciparum-infected RBC culture was lower than 100 nM (Fig. 1H). Nevertheless, the levels of BV in human plasma can oscillate up to 40 μM in jaundice patients27. As haem catabolism products are toxic to Plasmodium, the perception of BV oscillation levels might be an important signal during malaria intraerythrocytic development. Accordingly, the addition of 0.1 and 1 μM BV during 48 hours increased the proportion of mononucleated forms (R and T) and decreased the multinucleated forms (S) without compromising parasitaemia (Fig. 2A). These data suggest that a delay in parasite development might serve as one strategy to address lower increases in BV concentration. Our data on the knockout of P. berghei HO indicated that the absence of the gene is lethal to the rodent parasite, suggesting greater vulnerability to haem and its catabolic products. Given that PfHO orthologues are noted in all Plasmodium species, HO activity must play a vital role within parasites.
The protein-BV binding assay using zinc acetate in a two-dimensional gel revealed P. falciparum enolase as a candidate BV molecular target. Enolase is the eighth enzyme in the glycolytic pathway. Interestingly, based on MS, Pf enolase was recently identified in several sub-cellular compartments, including the cytosol, nucleus, cell membrane and food vacuole, despite its lack of an organelle-targeting signal sequence28,29. This diversity in the sub-cellular localization of enolase suggests that the enzyme may function in cellular recognition/invasion, the formation/development of vacuoles, and transcription29 in addition to its role in glycolysis. This phenomenon is known as moonlighting function, and proteins of this class frequently exhibit distinct and physiologically important cellular functions. The multiple functions are not due to protein isoforms; that is, they do not include gene fusions, splicing variants or pleiotropic effects30. Enolase is a representative member of the moonlight proteins, acting as a plasminogen receptor31, neurotrophic factor32, heat-shock protein33, hypoxic stress protein34, DNA interaction factor35, ligand for mosquito gut epithelial receptor36, transcription factor in plants37 and cancer cells38. Another interesting feature of this protein is that enolase is a target for ubiquitination and three forms of the enzyme were identified (MW ~50, 65 and 75 KDa)39. In this work, the zinc acetate-binding assay revealed that Pf enolase has a MW of 50 KDa, which corresponds to the molecular weight of the non-modified monomer. In fact, the alignment between enolases of different organisms revealed that the proteins have almost the same MW, differing primarily by the presence of a pentapeptide in the apicomplexa phylum. This pentapeptide apparently stabilizes enolase in an active state through the formation of three hydrogen bonds between Ser108 and Leu49, allowing the enzyme to remain active even at high concentrations of ATP40. In addition, after RBC invasion by P. falciparum, the amount of enolase increases by 15-fold41, which is consistent with our quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) data. These data indicate a sudden increase in expression of this gene in the merozoite stage (Fig. 3C).
As haem and BV delayed the parasite life cycle, we aimed to investigate the molecular mechanism responsible for the observed impairment in the parasite cell cycle. Therefore, we investigated the relationship between the presence of haemin and BV and the phosphorylation state of eIF2α. This factor can be phosphorylated in various stress conditions; thus, protein production is reduced to a basal level in an attempt to circumvent stress. As previously reported18,19,20,21, eIF2α kinases possess haem binding sites, and their phosphorylating activity is modulated by haem. We analysed the phosphorylation state of eIF2α by Western blot and reported an increase in eIF2α phosphorylation levels after 5 minutes of treatment followed by a decrease in phospho-eIF2α up to 20 minutes after BV and haem treatment (Fig. 3B). In the same direction, it has been reported that PfPK4 that targets eIF2α as a substrate, has its phosphorylation activity reduced by 60% in the presence of haem at 10 μM42. Moreover, Surolia et al. 43 had reported a kinase activity in Plasmodium falciparum lisates, that resembles HRI activity.
The thermogram data suggest that BV-Pf enolase binding is endothermic (Fig. 4A); however, it must be noted that this result might be a consequence of BV dilution. That is, BV dilution is an extremely endothermic reaction and absorbs the heat released from the BV interaction with Pf enolase. The evaluated ΔG value of −4.5 Kcal/mol and the binding stoichiometry (n = 1.1) indicate that BV binds favourably to Pf enolase. The fact that BV can inhibit Pf enolase activity in vitro with an IC50 value of 27 ± 4 μM (Fig. 4C) is consistent with the observed toxicity in P. falciparum-infected RBC culture at 10 μM BV (Fig. 2A). The evaluated BV mechanism of inhibition indicated non-competitive behaviour; that is, Pf enolase has an allosteric binding site capable of modulating the enzyme activity. This finding might pave the way for the discovery of new inhibitors as antimalarial candidates. Additionally, Pf enolase may act as a sensor for extreme extracellular conditions (e.g., enhanced BV concentration in the cell). It is important to note that the inhibitory activity of Pf enolase does not occur at lower BV concentrations (e.g., nM); therefore, another mechanism in addition to the binding to Pf enolase must occur for BV to function as a signal molecule.
Conclusions
In this work, we detected BV in P. falciparum culture supernatant and investigated its role in the P. falciparum RBC cycle. BV affects the cell cycle of P. falciparum by increasing the abundance of the early stages (R and T) and reducing the proportion of S. Thus, we identified Pf enolase as a target for BV. The BV binding profile to Pf enolase was investigated, and a putative binding site was suggested. On that basis, we propose that BV binding to Pf enolase plays a role as a new moonlight function.
Finally, on Fig 6 we propose that the mechanism for the impairment of the cell cycle by BV is associated with a low activity of enolase and alterations to the cell cycle programme due to changes in eIF2α phosphorylation.
Materials and Methods
Cell culture, synchronization
The P. falciparum strain 3D7 was cultured at 37 °C in RPMI 1640 medium using 10% AB+ human serum44. Cultures were grown under a 5% CO2, 5% O2, and 90% N2 atmosphere. The culture was synchronized with 5% sorbitol45.
Cloning of P. falciparum enolase
E. coli Rosetta (DE3) carrying the P. falciparum enolase-pETTrx-1a plasmid were cultured at 37 °C with shaking at 150 rev/min in auto-induction medium supplemented with 15 μg/mL kanamycin and 34 μg/mL chloramphenicol for three hours until an OD600 of 0.8 was reached. The temperature was then reduced to 20 °C followed by expression for 20 hours. Cells were harvested by centrifugation (30 min; 3,500 g; 4 °C) prior to re-suspension in lysis buffer (buffer A: 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 20 mM imidazole and 10% glycerol) containing 4 mM dithiothreitol and 1 mM phenylmethanesulphonylfluoride. Cells were disrupted by sonication on ice. Insoluble debris was separated by centrifugation (50,000 g; 30 min; 4 °C), and the soluble fraction was filtered and loaded onto a HisTrap HP 5 mL column (GE Healthcare). The His-tagged protein was eluted using a 0 to 1 M imidazole gradient in the same buffer. The excess imidazole was removed by a desalting column (Hi-Trap column - GE Healthcare) followed by TEV protease digestion (1 mg per 20 mg Pf enolase). A second passage through the His-Trap column was employed to separate Pf enolase from TEV protease, cleaved His-tag-thioredoxin and uncleaved His-tag-thioredoxin Pf enolase. The sample containing Pf enolase was applied to a Superdex 200, 16/60 column (GE Healthcare), pre-equilibrated in 20 mM Tris-HCl buffer pH 8.0, 200 mM NaCl. The eluted protein was concentrated (10 KDa MWCO Amicon Ultra devices, Millipore) to 10 mg/mL. Protein concentrations were determined spectrophotometrically using a theoretical extinction coefficient of 43,235 mol L−1 cm−1 at 280 nm calculated using ProtParam. The high level of protein purity was confirmed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).
Sample preparation for HPLC analysis
A 30-mL supernatant sample collected from the desynchronized P. falciparum culture and a sample collected from the uninfected RBC culture were concentrated 2-fold by freeze-drying. The resulting suspension was acidified with 200 mL of concentrated hydrochloric acid (HCl(aq)36% w/w, Labsynth) and extracted 3 times with 20 mL dichloromethane (Labsynth). The organic solvent was removed under a N2(g) flux and the concentrate resuspended in 1 mL DMSO (Labsynth) and 2 mL methanol (Labsynth).
HPLC-UV-Vis analysis
HPLC analysis was performed on a Phenomenex Luna 5 m C18 column (250 × 4.60 mm) using a LC 20AT chromatograph equipped with a diode array UV-V is detector (SPD-M20A). The analyses were performed using 5% (v/v) acetonitrile (HPLC/spectro grade, Tedia) with 0.05% (v/v) formic acid (Merck) as solvent A and 100% acetonitrile as solvent B at a flow rate of 0.4 mL/min. Ten minutes after sample injection, a linear gradient of 40 minutes was applied to change solvent A to a mixed solvent A/B (2:8), which was maintained for 20 minutes. The initial condition was restored through a linear acetonitrile gradient (5 minutes).
HPLC-MS analysis
Mass spectrometry data were obtained in the positive ion mode using an Esquire 3000 Plus mass spectrometer (Bruker Daltonics), coupled with the HPLC instrument. The eluent was the same used in the HPLC analysis with the addition of 2% (v/v) formic acid to solvent A. Samples were introduced into the electrospray source at 70 μL/min. Capillary voltage was set at 4 kV, with a temperature of 250 °C. The nebulizer, dry gas, and dry temperature were set at 20 psi, 7.0 L/min, and 325 °C, respectively.
Generation of knockout and GFP tagging constructs
Knockout construct transfection was performed on P. berghei ANKA strain 2.34 parasites according to a previously published protocol46,47 with modifications. The pb HO knock-out vector was constructed for double crossover homologous recombination in plasmid pBS-DHFR, which contains a T. gondiidhfr/ts cassette conferring resistance to pyrimethamine, as described previously47,48. The knockout construct was generated by inserting the pbHO 5′ untranslated region (UTR) region upstream (as Apa1 + HindIII fragment) and the pbHO 3′UTR region (as EcoRI + Xba1 fragment) downstream of the dhfr cassette. Primers CCCCGGGCCCGGGCGATATGGAATGCACATTTTCTCCTC, N0501 and GGGGAAGCTTGCAATCATTTATTCTTTGTAATTG, N0502 were used to generate a 585 bp fragment of the 5′ upstream sequence of Pbho from genomic DNA, which was inserted into the ApaI and HindIII restriction sites upstream of the dhfr/ts cassette of pBS-DHFR. Primers CCCCGAATTCGAGCCGTAAATGGACACGATGGG (N0503)
and GGGGTCTAGAGTCATTTTAAGGTTGGCATTATATTAGC (N0504)
were used to generate a 418 bp fragment from the 3′ flanking region of Pbho, which was subsequently inserted downstream of the dhfr/ts cassette using EcoRI and XbaI restriction sites. The final knockout construct was digested with ApaI and XbaI to release the linearized fragment for transfection. To tag the endogenous locus with GFP, we generated a pbHO-GFP construct by single crossover homologous recombination. An 876-bp region coding for the C-terminus of PbHO without the stop codon was inserted in-frame and upstream of the GFP sequence in plasmid p277 containing the human DHFR cassette and conveying resistance to pyrimethamine, as previously described49. The primers used to amplify this fragment are provided below:
CCCCGGTACCGTAGAGATTATATTTACCATCTTGAAG, T1031;
CCCCGGGCCCTTTTTTTATATTTTCAAAATGTTTTGTCAAAATCATC, T1032.
Prior to transfection, the final construct was digested with Bsm1, which cuts the plasmid in the middle of the insert. This cut is optimal for the homologous recombination event. Briefly, electroporated parasites were mixed immediately with 200 μL of reticulocyte-rich blood from a phenylhydrazine (Sigma, UK)-treated naïve mouse and incubated at 37 °C for 30 minutes. Re-invaded parasites were then injected intraperitoneally. From day 1 post infection, pyrimethamine (Sigma, UK) was applied in the drinking water for four days. Mice were monitored for 15 days to allow any slow-growing mutants to reach patency. Drug selection was repeated after passage to a second mouse, and resistant parasites were used for cloning by limiting dilution and genotyping. Transfected P. berghei parasites were selected by pyrimethamine pressure in drinking water (7 mg/mL in DMSO dissolved in 500 mL water) or by intra-peritoneal injection (250 μg/100 μL/mouse in DMSO) according to a previously described protocol50.
Real-time qRT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA was quantified using the Nanodrop ND-1000 UV/V is spectrophotometer, and RNA integrity was assessed by electrophoresis in agarose gel. Random-primed RT was performed using 1.0 μg of total RNA according to the Super Script III kit protocol (Invitrogen). The relative transcriptional levels were determined through quantitative PCR (qPCR) with Sybr Green PCR Master Mix (Applied Biosystems) using the 7300 Real-Time PCR System (Applied Biosystems). Real-time data were normalized using the level of expression of seryl-tRNA synthetase and presented as relative expression compared with ring stage parasites. PCR primer sequences targeting enolase mRNA were synthesized as follows: left primer 5′-TTCAGAGCTGCCGTACCATC-3′ and right primer 5′-ACACCCTTTCCTAAGTACCTGC-3′. P-values were determined for the biological triplicates with Student’s t-test, using a one-tailed distribution and heteroscedastic variance.
Protein extraction and western blot
For detection of Phospho-eIF2α,in vitro P. falciparum-infected RBCs cultures were synchronized using 5% sorbitol to isolate trophozoites (24 h post invasion) and treated with either 1 μM of BV (Sigma-Aldrich, St Louis, MO, USA, E.U.A.), haem 1 μM (Sigma-Aldrich, St Louis, MO, USA, E.U.A.), and a control sample without any addition. Protein samples of each group were extracted after 1, 5 and 20 minutes of treatment. At each point, the cells were harvested, and the cell pellets were lysed for 60 s in ten volumes of 0.5% saponin prepared in PBS buffer. The parasite pellet was recovered by centrifugation at 8,000 g for 10 min at 4 °C, washed and resuspended in RIPA buffer (25 nM Tris-HCl pH7,4, 150 nM NaCl, 1% NP40, 1% SDS, 2 mM EDTA) with protease inhibitors (1 mM PMSF, 0.01 mM benzamidine, 10 μg /mL aprotinin, 10 μg/mL, leupeptin, 10 μg/mL pepstatin, 10 μg/mL chymostatin, 100 μM sodium orthovanadate, and 20 μM sodium fluoride) and fosfatase inhibitors (β-glicerofosfate, sodium fluoride and sodium pyrofosfate from Sigma-Aldrich, St Louis, MO, USA, E.U.A). Samples were kept on ice for 30 min and particulate material was removed by centrifugation at 14,000 g for 30 min. Protein fractions were separated on 12% SDS-PAGE gels under reducing conditions, transferred to PVDF membranes, blocked with 3% BSA, and probed with anti-Phospho-eIF2α 1:1000 dilution (Cell Signaling) overnight. After washing, the blots were incubated with HRP-conjugated 1:30000 anti IgG rabbit HRP (Ge Healthcare) at a dilution of 1:30000 for 1 h, washed, and visualized using ECL Plus (GE Healthcare). For protein loading control anti-GAPDH antibody was used in 1:5000 dilution (Sigma-Aldrich, St Louis, MO, USA E.U.A.) overnight, and visualized as described above.
Homology modelling of P. falciparum enolase
The 3D model of enolase from Plasmodium falciparum was built by homology modelling based on a high-resolution crystal structure of the closest homologous protein available on Protein Data Bank (PDB). The complete amino acid sequence database of the target protein was obtained in FASTA format from the UniProtKB database (Accession N. Q8IJN7). The atomic coordinates of the enolase crystallographic structure from Trypanosoma brucei (PDB ID 2PTZ) at 1.65 Å resolution and 62% sequence identity were used as template. The pairwise sequence alignment was performed using ClustalW251, and 10 models were constructed automatically using the MODELLER default parameters (version 9.10)52. The lowest global energy (DOPE) model was selected for the validation step, which included inspections of the Ramachandran plot and structural analysis with PROCHECK53 and MolProbity54. The compatibility of the model with its sequence was assessed by Verify-3D55, and the RMSD value between the main-chain atoms of the model and template was calculated by structural superposition of the template and the predicted structure with an optimized algorithm in Pymol 1.256 that performs independent sequence- and structure-based alignment.
Ligand site prediction of P. falciparum enolase
The putative BV binding site was identified by SiteHound, which identifies ligand-binding sites by computing interactions between a chemical probe (e.g., methyl) and a protein structure. The algorithm evaluates the interaction energy between the protein atoms and the molecular probe to identify energetically favourable regions within protein binding cavities. Subsequently, the grid points that exhibit favourable energy values are clustered according to their spatial proximity, and the final output lists the interaction energy clusters corresponding to putative binding sites. The most favourable (lowest energy) binding cavity for BV included residues 38–44, 102–103, 162–163, 253–259, 264–265, 268, 302–308, 312, and 330.
Molecular docking
Molecular docking and scoring protocols implemented in GOLD 4.1 (Cambridge Crystallographic Data Centre, Cambridge, UK)57 were used to investigate the possible binding conformations of the ligands within the P. falciparum enolase binding pocket. The P. falciparum enolase homology model generated in this work was used in the docking simulations. Hydrogen atoms were added in standard geometry using the Biopolymer module implemented in SYBYL 8.0. Histidine, glutamine, and asparagine residues within the binding site were manually assessed for possible flipped orientation, protonation, and tautomeric states with the Pymol 1.2 (DeLano Scientific, San Carlos, USA) side chain wizard script. The binding site was defined as all the amino acid residues encompassed within a 20-Å radius sphere centred on the Cβ atom of the Ser254 residue. The docking procedures were repeated 30 times for each inhibitor. The implemented GOLD scoring function and visual inspection were employed to select the representative conformation for BV.
Erythrocyte BV incubation
Erythrocytes infected with synchronized P. falciparum (3D7 strain) at the ring stage with 1% parasitaemia were incubated for 48 hours in flat bottom 48-well enzyme-linked immunosorbent assay (ELISA) plates containing different concentrations (0.1, 1 and 10 μM) of BV hydrochloride (Sigma). The controls were infected RBCs with and without DMSO. BV was solubilized in DMSO. After the incubation, the cells were centrifuged, and the pellets were fixed overnight with phosphate buffer solution (PBS) pH 7.4 containing 2% formaldehyde (v/v) (Labsynth). The fixed cells were resuspended in PBS pH 7.4 containing 0.1% Triton-100X (v/v) (Sigma) and 5 nM oxazole yellow homodimer (YOYO-1-labeled DNA, Molecular Probes) and incubated at 37 °C for 30 minutes. Parasitaemia was determined from dot plots (side scatter versus fluorescence) of 105 cells acquired on a FACSCalibur flow cytometer using the CELLQUEST software (Becton Dickinson). Fluorescence was excited with an argon laser at 488 nm, and the fluorescence emission was collected at 520 to 530 nm. Initial gating was performed with unstained, uninfected erythrocytes to account for the erythrocyte autofluorescence.
Statistical analyses
All results are expressed as the mean ± standard error of the mean (SEM) of at least three individual experiments. A repeated measure one-way analysis of variance (ANOVA) was used for comparisons among larger groups, followed by the Dunnet and Newman-Keuls post-tests. A P-value less than 0.05 was considered indicative of a statistically significant difference. GraphPad Prism software (San Diego, CA, USA) was used for all statistical tests.
Additional Information
How to cite this article: Alves, E. et al. Biliverdin targets enolase and eukaryotic initiation factor 2 (eIF2α) to reduce the growth of intraerythrocytic development of the malaria parasite Plasmodium falciparum. Sci. Rep. 6, 22093; doi: 10.1038/srep22093 (2016).
Supplementary Material
Acknowledgments
We thank FAPESP (processes 2011/51295-5, 2011/21442-6 and 2013/07600-3), CNPq INCT-INBEQMeDI 550514/2011-2 for funding the research and the MRC New Investigator grant and MRC project grant to Rita Tewari (G0900109 and MR/K011782/1).
Footnotes
Author Contributions E.A., F.V.M., V.B.B., M.S., M.N., D.B., P.H.S.P. and F.C.S.A. performed the experiments and contributed to the writing and ideas.R.V.C.G., G.O., R.S., L.H.C.; R.T. and C.R.S.G contributed to the writing and ideas.
References
- Garcia C. R. S. et al. Plasmodium in the postgenomic era: new insights into the molecular cell biology of malaria parasites. Int. Rev. Cell Mol. Biol. 266, 85–156 (2008). [DOI] [PubMed] [Google Scholar]
- Bannister L. & Mitchell G. The ins, outs and roundabouts of malaria. Trends Parasitol. 19, 209–213 (2003). [DOI] [PubMed] [Google Scholar]
- Elliott D. A. et al. Four distinct pathways of hemoglobin uptake in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 105, 2463–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater A. F. & Cerami A. Inhibition by chloroquine of a novel haem polymerase enzyme activity in malaria trophozoites. Nature 355, 167–9 (1992). [DOI] [PubMed] [Google Scholar]
- Docherty J. & Brown B. Haem degradation in human haemoglobin in vitro. Biochem. J. 222, 401–406 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutty R. K., Daniel R. F., Ryan D. E., Levin W. & Maines M. D. Rat liver cytochrome P-450b, P-420b, and P-420c are degraded to biliverdin by heme oxygenase. Arch Biochem Biophys 260, 638–644 (1988). [DOI] [PubMed] [Google Scholar]
- Maines M. D. Bile pigments: newcomers to the cell signaling arena. Toxicol. Sci. 71, 9–10 (2003). [DOI] [PubMed] [Google Scholar]
- Phelan D., Winter G. M., Rogers W. J., Lam J. C. & Denison M. S. Activation of the Ah receptor signal transduction pathway by bilirubin and biliverdin. Arch Biochem Biophys 357, 155–163 (1998). [DOI] [PubMed] [Google Scholar]
- Neuzil J. & Stocker R. Free and Albumin-bound Bilirubin are Efficient co-Antioxidants for a-Tocopherol, Inhibiting Plasma and Low Density Lipoprotein Lipid Peroxidation. J. Biol. Chem. 269, 16712–16719 (1994). [PubMed] [Google Scholar]
- Sinal C. J. & Bend J. R. Aryl hydrocarbon receptor-dependent induction of cyp1a1 by bilirubin in mouse hepatoma hepa 1c1c7 cells. Mol. Pharmacol. 52, 590–599 (1997). [DOI] [PubMed] [Google Scholar]
- Gardner M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Clough B., Coates L. & Wilson R. J. M. Enzymes for heme biosynthesis are found in both the mitochondrion and plastid of the malaria parasite Plasmodium falciparum. Protist 155, 117–125 (2004). [DOI] [PubMed] [Google Scholar]
- Surolia N. & Padmanaban G. De novo biosynthesis of heme offers a new chemotherapeutic target in the human malarial parasite. Biochem. Biophys. Res. Commun. 187, 744–50 (1992). [DOI] [PubMed] [Google Scholar]
- Okada K. The novel heme oxygenase-like protein from Plasmodiumfalciparum converts heme to bilirubin IXalpha in the apicoplast. FEBS Lett. 583, 313–9 (2009). [DOI] [PubMed] [Google Scholar]
- Sartorello R. et al. In vivo uptake of a haem analogue Zn protoporphyrin IX by the human malaria parasite P. falciparum-infected red blood cells. Cell Biol. Int. 34, 859–865 (2010). [DOI] [PubMed] [Google Scholar]
- Kumar S. et al. Bilirubin inhibits Plasmodium falciparum growth through the generation of reactive oxygen species. Free Radic. Biol. Med. 44, 602–13 (2008). [DOI] [PubMed] [Google Scholar]
- Sigala P. a., Crowley J. R., Hsieh S., Henderson J. P. & Goldberg D. E. Direct tests of enzymatic heme degradation by the malaria parasite Plasmodium falciparum. J. Biol. Chem. 287, 37793–37807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P., Equinet L., Packer J. & Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics 5, 79 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. et al. The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J. Exp. Med. 207, 1465–1474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A. P. et al. Heme-regulated eIF2?kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20, 6909–6918 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud C. G. eIF2 and the control of cell physiology. Semin. Cell Dev. Biol. 16, 3–12 (2005). [DOI] [PubMed] [Google Scholar]
- Berkelman T. R. & Lagarias J. C. Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal. Biochem. 156, 194–201 (1986). [DOI] [PubMed] [Google Scholar]
- Lebioda L., Stec B. & Brewer J. M. The structure of yeast enolase at 2.25-A resolution. An 8-fold beta + alpha-barrel with a novel beta beta alpha alpha (beta alpha)6 topology. J. Biol. Chem. 264, 3685–93 (1989). [DOI] [PubMed] [Google Scholar]
- Hernandez M., Ghersi D. & Sanchez R. SITEHOUND-web: A server for ligand binding site identification in protein structures. Nucleic Acids Res. 37, 413–416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis S. E., Sullivan D. J. & Goldberg D. E. Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu. Rev. Microbiol. 51, 97–123 (1997). [DOI] [PubMed] [Google Scholar]
- Egan T. J. et al. Fate of haem iron in the malaria parasite Plasmodium falciparum. Biochem. J. 365, 343–347 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickner T. & Gutteridge J. A simple colorimetric method for the estimation of plasma biliverdin. Clin. Chim. Acta 85, 125–129 (1978). [DOI] [PubMed] [Google Scholar]
- Das S., Shevade S., Lacount D. J. & Jarori G. K. Plasmodium falciparum enolase complements yeast enolase functions and associates with the parasite food vacuole. Mol. Biochem. Parasitol. 179, 8–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhowmick I., Kumar N., Sharma S., Coppens I. & Jarori G. K. Plasmodium falciparum enolase: stage-specific expression and sub-cellular localization. Malar. J. 8, 1–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amblee V. & Jeffery C. J. Physical Features of Intracellular Proteins that Moonlight on the Cell Surface. PLoS One 1–16 (2015). doi: 10.1371/journal.pone.0130575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles L. a et al. Role of cell-surface lysines in plasminogen binding to cells: identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry 30, 1682–1691 (1991). [DOI] [PubMed] [Google Scholar]
- Takei N. et al. Neuronal survival factor from bovine brain is identical to neuron-specific enolase. J Neurochem 57, 1178–1184 (1991). [DOI] [PubMed] [Google Scholar]
- Yahara H. I. and I. Yeast heat-shock protein of Mr 48,000 is an isoprotein of enolase. Nature 315, 688–690 (1984). [Google Scholar]
- Aaronson R. M., Graven K. K., Tucci M., McDonald R. J. & Farber H. W. Non-neuronal enolase is an endothelial hypoxic stress protein. J. Biol. Chem. 270, 27752–7 (1995). [DOI] [PubMed] [Google Scholar]
- Johnstone S. a., Waisman D. M. & Rattner J. B. Enolase is present at the centrosome of HeLa cells. Exp. Cell Res. 202, 458–463 (1992). [DOI] [PubMed] [Google Scholar]
- Hernández-Romano J. et al. Conserved peptide sequences bind to actin and enolase on the surface of Plasmodium berghei ookinetes. Parasitology 138, 1341–1353 (2011). [DOI] [PubMed] [Google Scholar]
- Lee H. et al. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 21, 2692–2702 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feo S., Arcuri D., Piddini E., Passantino R. & Giallongo A. ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: Relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett. 473, 47–52 (2000). [DOI] [PubMed] [Google Scholar]
- Shevade S., Jindal N., Dutta S. & Jarori G. K. Food Vacuole Associated Enolase in Plasmodium Undergoes Multiple Post-Translational Modifications: Evidence for Atypical Ubiquitination. PLoS One 8, 1–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S., Mukherjee D. & Jarori G. K. Replacement of Ser108 in P. falciparum enolase results in weak Mg(II) binding: Role of a parasite specific pentapeptide insert in stabilizing active conformation of enzyme. FEBS J. 282, 2296–2308 (2015). [DOI] [PubMed] [Google Scholar]
- Roth E. F. Jr., Calvin M. C., Max-Audit I., Rosa J. & Rosa R. The enzymes of the glycolytic pathway in erythrocytes infected with Plasmodium falciparum malaria parasites. Blood 72, 1922–1925 (1988). [PubMed] [Google Scholar]
- Mohrle J. J., Zhao Y., Wernli B., Franklin R. M. & Kappes B. Molecular cloning, characterization and localization of PfPK4, an eIF-2alpha kinase-related enzyme from the malarial parasite Plasmodium falciparum. Biochem J 328 (Pt 2, 677–687 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surolia N. & Padmanaban G. Chloroquine inhibits heme-dependent protein synthesis in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 88, 4786–4790 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W. & Jensen J. B. Human malaria parasites in continuous culture. Science 193, 673–675 (1976). [DOI] [PubMed] [Google Scholar]
- Lambros C. & Vanderberg J. P. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65, 418–420 (1979). [PubMed] [Google Scholar]
- Janse C. J., Ramesar J. & Waters A. P. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 1, 346–356 (2006). [DOI] [PubMed] [Google Scholar]
- Tewari R. et al. The systematic functional analysis of plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe 8, 377–387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reininger L. et al. An essential role for the Plasmodium Nek-2 Nima-related protein kinase in the sexual development of malaria parasites. J. Biol. Chem. 284, 20858–20868 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavic K. et al. Life cycle studies of the hexose transporter of Plasmodium species and genetic validation of their essentiality. Mol. Microbiol. 75, 1402–1413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse C. J. et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol. Biochem. Parasitol. 145, 60–70 (2006). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Eswar N. et al. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. Chapter 2, Unit 2.9 (2007). [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., Moss D. S. & Thornton J. M. Main-chain bond lengths and bond angles in protein structures. Journal of molecular biology 231, 1049–67 (1993). [DOI] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthy R., Bowie J. U. & Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature 356, 83–85 (1992). [DOI] [PubMed] [Google Scholar]
- DeLano W. L. PyMOL molecular viewer: Updates and refinements. Abstr. Pap. Am. Chem. Soc. 238, (2009). [Google Scholar]
- Jones G., Willett P., Glen R. C., Leach, A. R. & Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.