Abstract
Despite their fastidious nature, marine myxobacteria have considerable genetic potential to produce novel secondary metabolites. The marine myxobacterium Haliangium ochraceum SMP-2 produces the antifungal polyketide haliangicin (1), but its productivity is unsatisfactory. The biosynthetic gene cluster hli (47.8 kbp) associated with 1 was identified and heterologously expressed in Myxococcus xanthus to permit the production of 1 with high efficiency (tenfold greater amount and threefold faster in growth speed compared with the original producer), as well as the generation of bioactive unnatural analogues of 1 through gene manipulation. A unique acyl-CoA dehydrogenase was found to catalyse an unusual γ,δ-dehydrogenation of the diketide starter unit, leading to the formation of the terminal alkene moiety of 1. Biological evaluation of the analogues obtained through this study revealed that their bioactivities (anti-oomycete and cytotoxic activities) can be modified by manipulating the vinyl epoxide at the terminus opposite the β-methoxyacrylate pharmacophore.
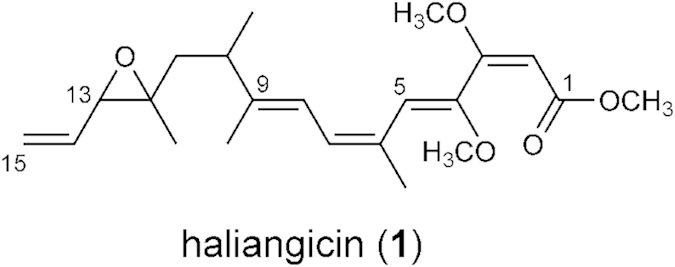
Natural products derived from microorganisms have a long history of use as antibiotics for clinical use. As an alternative to conventional antibiotic producers such as Streptomyces, myxobacteria have attracted considerable attention, especially since the remarkable discovery of epothilones as microtubule-stabilizing agent1,2. Myxobacteria are unique Gram-negative proteobacteria that are characterized by gliding and by the formation of multicellular fruiting bodies3. They have larger genomes than other taxa of bacteria, and they can produce structurally diverse bioactive secondary metabolites having novel modes of action, making them a promising alternative source of new drugs4,5. Marine myxobacteria are particularly rare and difficult to handle6,7,8,9; consequently, their products have only recently been examined. Several groups of antibiotics from the marine environment have been identified, including haliangicins10,11,12, miuraenamides13,14, salimabromides, salimyxins, and enhygrolides.15,16 Haliangicin (1), a polyene-type polyketide with a unique terminal epoxy alkene group and β-methoxyacrylate pharmacophore (Fig. 1), was isolated as the first marine myxobacterial secondary metabolite from the halophilic myxobacterium Haliangium ochraceum SMP-2. It has been reported to be a potent fungicide that interferes with the electron flow within the cytochrome b-c1 segment in fungal mitochondrial respiratory chains.11
Figure 1. Structure of haliangicin.
Most myxobacterial metabolites are polyketides, nonribosomal peptides, or their hybrids; these are known to be biosynthesized by megasynthase polyketide synthases (PKSs) and nonribosomal peptide synthases (NRPSs). Although it has been reported that the sequences of the PKS genes in marine myxobacterial isolates are highly novel,17 biochemical studies on their metabolites have been hampered by unfavourable factors, such as difficulties in isolation, slow growth rates, cell aggregation, and poor metabolite productivity. In the case of haliangicin (1), the producer requires culture periods in excess of two weeks to reach maximal production of 1, and typically produces only about 1 mg L−1. Furthermore, the lack of a well-developed genetic methodology applicable to the marine strains is another major obstacle. Therefore, biosynthetic studies on these fastidious marine myxobacteria are still at an early stage, and they remain quite challenging, although attractive.
Here, we describe the cloning and identification of the haliangicin biosynthetic machinery and its use as the first representative of a marine myxobacterial biosynthetic assembly line. The establishment of a heterologous expression system allowed us to optimize the production titre, to manufacture unnatural analogues, and to analyse the function of the biosynthetic enzymes, which would be difficult to achieve with the native producer of 1.
Results
Identification and heterologous expression of the haliangicin biosynthetic gene cluster (hli)
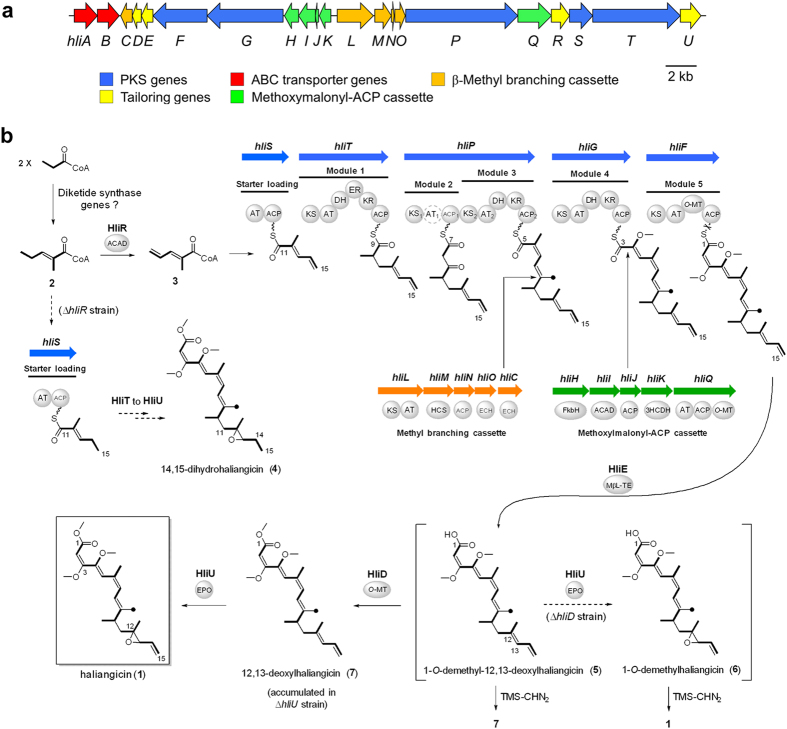
The polyene structure of haliangicin (1) suggests that its biosynthetic machinery is a pure PKS system that is accompanied by various enzymes involved in tailoring modifications. To clone the biosynthetic gene cluster responsible for formation of haliangicin, we first constructed a cosmid library of the H. ochraceum genome by using the E. coli/Streptomyces shuttle vector pDW10318. The resulting library, consisting of approximately 1700 cosmid clones, was then screened by colony hybridization using probes homologous to the ketosynthase (KS) genes of terrestrial myxobacteria (Supplementary Fig. S1). The end sequences of several KS-positive cosmids indicated that they might be responsible for the biosynthesis of 1 (Supplementary Fig. S2); these included HMG-CoA synthase for installing a β-branch methyl (9-Me) group and a 3-hydroxyacyl-CoA dehydrogenase for biosynthesizing methoxymalonyl-ACP, a precursor of the 1,2-dioxy unit at the C-3–C-4 position. Of the cosmids that were obtained, two partially overlapping cosmids, c7-6E and c10-11C, were sequenced, revealing a continuous 63-kbp segment that putatively harbours the entire haliangicin biosynthetic gene cluster (hli). Sequence annotation suggested that the hli gene cluster (GC content 67.2%) spans 47.8 kbp and contains 21 open reading frames (Fig. 2a, Table 1 and Supplementary Tables S1–S3). We assigned these genes to hliA-U.
Figure 2. Haliangicin biosynthetic machinery.
(a) Genetic organization of the haliangicin biosynthetic gene cluster (hli). (b) A proposed biosynthetic pathway for haliangicin (1). AT1 in module 2 is inactive, but no complementary trans-AT is found in hli. The dashed arrows indicate the unnatural pathways in the gene-disrupted strains of the heterologous host M. xanthus c10-11C/c7-6E. Abbreviations: ACAD, acyl-CoA dehydrogenase; ACP, acyl carrier protein; AT, acyl transferase; DH, dehydratase; ECH, enoyl-CoA hydratase; EPO, epoxidase; ER, enoyl reductase; FkbH, FkbH-like protein; HCS, HMG-CoA synthase; KR, ketoreductase; KS, ketosynthase; MβL-TE, metallo β-lactamase-type thioesterase; O-MT, O-methyltransferase; 3HCDH, 3-hydroxyacyl-CoA dehydrogenase.
Table 1. Proposed functions of the proteins involved in haliangicin biosynthesis.
| Protein | Size (aa) | Proposed function | Top-hit homologues (Origin) | Similarity/ identity (%) |
|---|---|---|---|---|
| HliA | 608 | ABC transporter | ABC transporter protein (Sorangium cellulosum), CCE88383 | 58/34 |
| HliB | 609 | ABC transporter | ABC transporter protein (Sorangium cellulosum), CCE88383 | 44/24 |
| HliC | 259 | enoyl-CoA hydratase | enoyl-CoA hydratase (Nostoc punctiforme PCC 73102), YP_001866568 | 74/53 |
| HliD | 217 | O-methyltransferase | JerF (Sorangium cellulosum), ABK32292 | 62/45 |
| HliE | 303 | metallo-β-lactamase type TE | β-lactamase (Rhodothermus marinus SG0.5JP17-172), YP_004824011 | 67/49 |
| HliF | 1372 | PKS (KS-AT-O-MT-ACP) | MtaF (Stigmatella aurantiaca DW4/3-1), YP_003953631 | 62/45 |
| HliG | 1966 | PKS (KS-AT-DH-KR-ACP) | MxaD (Stigmatella aurantiaca), AAK57188 | 57/42 |
| HliH | 367 | FkbH-like protein | HAD superfamily phosphatase (Sorangium cellulosum), CCE88385 | 78/62 |
| HliI | 387 | acyl-CoA dehydrogenase | acyl-CoA dehydrogenase (Sorangium cellulosum), CCE88384 | 73/57 |
| HliJ | 83 | ACP | hypothetical protein (Paenibacillus dendritiformis), WP_006675272 | 76/57 |
| HliK | 287 | 3-hydroxyacyl-CoA dehydrogenase | hydroxyacyl-CoA dehydrogenase (Sorangium cellulosum), CCE88386 | 79/66 |
| HliL | 950 | PKS (KS-AT) | polyketide synthase (Chondromyces crocatus), CBD77738 | 61/48 |
| HliM | 407 | HMG-CoA synthase | HMG-CoA synthase (Nostoc punctiforme PCC 73102), YP_001866566 | 65/48 |
| HliN | 83 | ACP | acyl carrier protein (Nostoc sp. 'Peltigera membranacea cyanobiont'), AGH69805 | 81/56 |
| HliO | 277 | enoyl-CoA hydratase | enoyl-CoA hydratase/isomerase (Nostoc punctiforme PCC 73102), YP_001866567 | 58/42 |
| HliP | 2890 | PKS (KS-ACP-KS-AT- DH-KR-ACP) | polyketide synthase (Myxococcus xanthus DK 1622), YP_632696 | 55/42 |
| HliQ | 861 | AT-ACP-O-MT | putative methoxymalonyl-CoA synthase (Sorangium cellulosum), AAK19884 | 59/44 |
| HliR | 392 | acyl-CoA dehydrogenase | acyl-CoA dehydrogenase (Herbidospora cretacea), WP_030450088 | 69/55 |
| HliS | 626 | AT-ACP | MxaF (Stigmatella aurantiaca), AAK57190 | 57/43 |
| HliT | 2225 | PKS (KS-AT-DH-ER- KR-ACP) | polyketide synthase (Sorangium cellulosum), CCE88378 | 61/44 |
| HliU | 463 | epoxidase | FAD-binding monooxygenase (Geodermatophilus obscurus DSM 43160), YP_003408356 | 56/42 |
Five Type-I PKS genes (hliF, G, P, S and T) were predicted to be responsible for the construction of the haliangicin backbone. Other genes could be assigned to a β-methyl branching cassette (hliL, M, N, O, and C), a methoxymalonyl-ACP cassette (hliQ, H, I, J, and K), and modification genes encoding an O-methyltransferase (HliD), a metallo-β-lactamase-type thioesterase (HliE), an acyl-CoA dehydrogenase (HliR), and an epoxidase (HliU). As discussed below, there were particular features in the gene cluster, for example, a rather disorganized gene arrangement (scattered PKS genes and segmented gene cassettes) and the presence of insufficient modules to construct the haliangicin backbone.
Demonstration that this gene cluster is responsible for the biosynthesis of haliangicin was hampered by the slow growth and a severe tendency for cell aggregation of the native producer. We therefore set out to express the biosynthetic machinery for 1 heterologously in a more readily cultivated and more tractable host. Our initial attempt to express the hli genes in a Streptomyces sp. resulted in transformants that showed no titre of haliangicin (1). Finally, we found that Myxococcus xanthus, a fast-growing terrestrial myxobacterium,19,20,21 is a suitable host. The aforementioned cosmids c7-6E and c10-11C, corresponding to the upstream and downstream parts of hli, respectively, were each modified through λ-Red recombineering22 (Supplementary Fig. S3 and S4) and integrated into the chromosome of M. xanthus ATCC25232 in a stepwise manner to reconstitute the haliangicin biosynthetic pathway (Supplementary Fig. S5–S7). The constructed strain, which we named M. xanthus c10-11C/c7-6E, was found to produce haliangicin (1) successfully, as revealed by HPLC (Fig. 3), verifying that the hli gene cluster is responsible for haliangicin biosynthesis. It is intriguing and worth mentioned that the heterologous host did not produce significant quantities of the 12,13-cis-epoxy isomer (cis-haliangicin), which usually accompanies 1 in the original producer (Fig. S8).
Figure 3. Production of haliangicin (1) by the heterologous host.
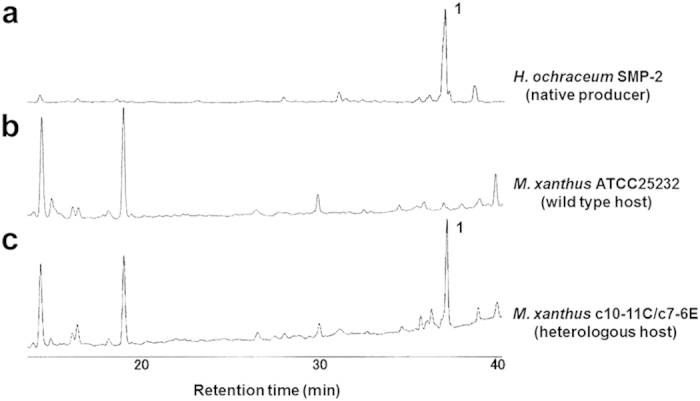
Reversed-phase HPLC of extracts of the native producer (a), the wild-type host M. xanthus (b), and the heterologous host harbouring the hli gene cluster (c).The chromatograms were recorded at 290 nm.
Next, we took advantage of the heterologous producer of 1, M. xanthus c10-11C/c7-6E, to settle several issues that were difficult to resolve with the native haliangicin producer.
Optimization of the heterologous production of haliangicin based on the identified biosynthetic precursors
The biosynthetic units for haliangicin (1) were first examined. The producer was fed various13C-labelled precursors, and the labelled haliangicins produced were analysed by NMR spectroscopy (Supplementary Fig. S9). This revealed that two acetate units, one acetate-derived methyl branch (9-Me), four propionate units, and three S-adenosylmethionine-derived O-methyl carbon atoms had been incorporated into 1 (Fig. 4a). The remaining two-carbon unit (C3–C4) bearing vicinal oxygen atoms was found to originate from glycerol, as supported by the incorporation of two carbon atoms from [U-13C3]glycerol (Supplementary Fig. S9e). Such a unit is known as a glycolate extender in some metabolites.23,24,25
Figure 4. Biosynthetic precursors and their effects on the production of haliangicin (1).
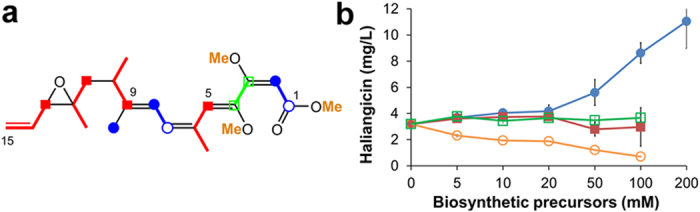
(a) Building blocks deduced from feeding experiments (red: [1-13C]propionate; blue: [1,2-13C2]acetate; green: [U-13C3]glycerol; orange: L-[methyl-13C]methionine). (b) Optimization of the production of 1 by feeding the appropriate biosynthetic precursors indicated by the same colours as in Fig. 4a. The fermentation was performed for 5 days.
Secondly, we evaluated the effects of these biosynthetic precursors on the productivity of haliangicin (1) (Fig. 4b). The haliangicin titre increased significantly when sodium acetate was added at a concentration higher than 50 mM and finally reached 11.0 ± 2.1 mg L–1 at 200 mM of sodium acetate after 5 days. The efficiency of haliangicin production was eleven times higher in quantity and three times shorter in culture period than that achieved with the native producer H. ochraceum.
Proposed mechanism for the biosynthesis of haliangicin
The use of the heterologous host for 1 also made gene manipulation much easier, allowing us not only to acquire a complete picture of the somewhat irregular biosynthetic machinery for 1, but also to generate several new analogues of haliangicin. On the basis of the deduced architecture of the hli gene cluster, we propose the haliangicin biosynthetic route shown in Fig. 2b. From the viewpoint of conventional PKS biosynthesis, the construction of the heptaketide backbone of 1 is likely to require six elongation steps following a C3 starter-loading step, but the hli machinery possesses only five elongation modules (modules 1–5 in Fig. 2b). An iterative function of any particular PKS is unlikely, because all the KS domains in hli are classified as modular not iterative (Supplementary Fig. S10). One possible solution to this problem of a missing module might be the direct use of a diketide (C6) starter, such as 2-methylpent-2-enoyl-CoA (2) (Fig. 2b and Supplementary Fig. S11), which is suggested by a putative diketide synthase found in the genomes of H. ochraceum and M. xanthus (Supplementary Fig. S12).
The diketide precursor 2 might undergo γ,δ-dehydrogenation by HliR, a particular acyl-CoA dehydrogenase, to form 2-methylpent-2,4-dienoyl CoA (3), which might serve as a starter for the formation of the unique 14,15-terminal olefin of 1. A detailed functional analysis of HliR will be described later. The diene starter 3, prepared by HliR, is next loaded into the typical starter-loading module encoded by hliS, and the five PKS-mediated elongation steps take place sequentially to generate the haliangicin backbone (Fig. 2b). Sequence analysis showed that the domain arrangements of the PKSs corresponded well with the chemical structure of haliangicin (1). The module 2-mediated elongation step is followed by the formation of β-methyl branch at the 9-position by the methyl-branching cassette26,27 (HliC and HliL–O) (Supplementary Fig. S13). Module 4 incorporates a glycolate extender, biosynthesized by the methoxymalonyl-ACP cassette24,28 (HliH–K and HliQ) (Supplementary Fig. S14), to construct the vicinal oxygen functionality at C3–C4. Finally, the nascent carboxylic acid released by HliE, which is homologous to a metallo-β-lactamase-type thioesterase,29,30 undergoes two post-assembly modification steps: O-methylation at the carboxyl terminus by HliD and epoxidation at the diene terminus by HliU (Fig. 2b).
Verification of some biosynthetic genes and generation of unnatural analogues of haliangicin
By using the genetically amenable host producer, we manipulated three genes involved in the formation of haliangicin to validate the proposed biosynthetic pathway discussed above and to produce several unnatural analogues of 1 (Fig. 5). First, we targeted the acyl-CoA dehydrogenase gene hliR, which plays an important role in the early stages of haliangicin biosynthesis. Disruption of hliR successfully generated the first unnatural analogue 14,15-dihydrohaliangicin (4), clearly demonstrating that hliR is responsible for the formation of the terminal olefin moiety (Supplementary Fig. S15). Subsequently, we inactivated two post-assembly tailoring enzymes: HliD (O-methyltransferase) and HliU (epoxidase). Inactivation of hliD led to the production of two free-acid intermediates, 1-O-demethyl-12,13-deoxyhaliangicin (5) and 1-O-demethylhaliangicin (6) as confirmed by LC-MS (Supplementary Fig. S16). However, because of their labile nature, these acids could not be purified for NMR analyses; consequently, their structures were chemically confirmed by methylation with trimethylsilyldiazomethane to give 12,13-deoxyhaliangicin (7) and haliangicin (1), respectively (Supplementary Fig. S17). The accumulation of 1-O-demethylhaliangicin (6) in the ΔhliD strain suggests that the free acid 1-O-demethyl-12,13-deoxyhaliangicin (5) also undergoes epoxidation by HliU. Finally, as expected, inactivation of hliU caused 12,13-deoxyhaliangicin (7) to accumulate without the formation of 1 (Supplementary Fig. S18). These results provide convincing confirmatory evidence for our proposed haliangicin biosynthetic machinery.
Figure 5. Generation of unnatural haliangicin analogues in the gene-disrupted heterologous hosts.
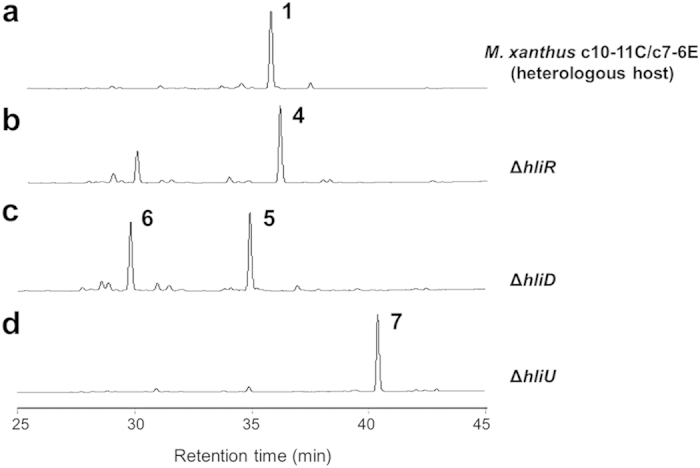
HPLC analyses of extracts of the heterologous producer M. xanthus c10-11C/c7-6E (a), ΔhliR (acyl-CoA dehydrogenase disrupted host, (b), ΔhliD (O-methyltransferase-disrupted host, (c), and ΔhliU (epoxidase disrupted host, (d). The chromatograms were recorded at 290 nm.
Functional analysis of dehydrogenase HliR responsible for terminal alkene formation
Terminal alkene structures are found in a few natural products, for example, curacin A,31,32 tautomycetin,33 and FK50634 The unique terminal vinyl epoxide alkene moiety in haliangicin (1) prompted us to elucidate the biosynthetic mechanism responsible for its formation. Because hliR is homologous to acyl-CoA dehydrogenases, the enzyme encoded by this gene might also be a good candidate for introducing the double bond between C-14 and C-15. Actually, the aforementioned gene disruption of hliR prevented the formation of 1 and resulted in the production of analogue 4 with a saturated terminus, thereby providing unequivocal evidence of the direct involvement of hliR. A detailed sequence and phylogenetic analysis indicated that HliR is homologous to short-chain acyl-CoA dehydrogenases and is closest to the propylmalonyl-ACP desaturase TcsD, which catalyses the desaturation of a propyl group to an allyl group in the side-chain biosynthesis of FK50635,36,37 (Fig. 6). Because functional analyses for such terminal dehydrogenases have been limited exclusively to FK506 and its related metabolites35,38,39, we next expressed HliR in Escherichia coli as a polyhistidine-tagged protein to analyse its biochemical features (Supplementary Fig. S19). With the full-length dihydro analogue 4 in hand, we first examined the possibility of HliR-mediated dehydrogenation being the ultimate step in the biosynthesis of haliangicin. The lack of conversion of 4 into 1 by HliR in vitro precluded this hypothesis (Supplementary Fig. S20). Next, we examined the possibility that formation of the terminal alkene might occur in the starter diketide 2. To prove this, a thio ester mimic of 2, S-(2-methylpent-2-enoyl)-N-acetylcysteamine (2a), was synthesized as the proposed substrate. The recombinant HliR converted 2a entirely into the corresponding dehydro product 3a (Fig. 7a). Preparation of 3a by feeding 2a to the HliR-expressing E. coli yielded a sufficient amount of 3a to permit comprehensive spectroscopic analysis (Supplementary Methods). The substrate specificity was then evaluated by using 14 additional short thio ester mimics, revealing that HliR is highly specific to α,β-unsaturated short acyl compounds, but does not consume saturated or γ,δ-unsaturated compounds (Fig. 7b and Supplementary Fig. S20). Among the substrates tested, S-(pent-2-enoyl)-N-acetylcysteamine (2b), like 2a, was also completely converted into 3b, whereas S-(hex-2-enoyl)-N-acetylcysteamine (2c) and S-(4-methylpent-2-enoyl)-N-acetylcysteamine (2d) underwent partial dehydrogenation. These results suggested that the presence of substituents at the γ- or δ-position might hinder the dehydrogenation reaction from the enzyme. In addition, unlike TcsD, which demands acyl-carrier protein-tethered substrates, HliR can utilize free small thio esters that mimic acyl-CoA substrates, offering a new strategy for preparing terminal alkenes.
Figure 6. Phylogenetic tree of bacterial acyl-CoA dehydrogenase (ACAD)-related oxidoreductases obtained by using the NJ method.
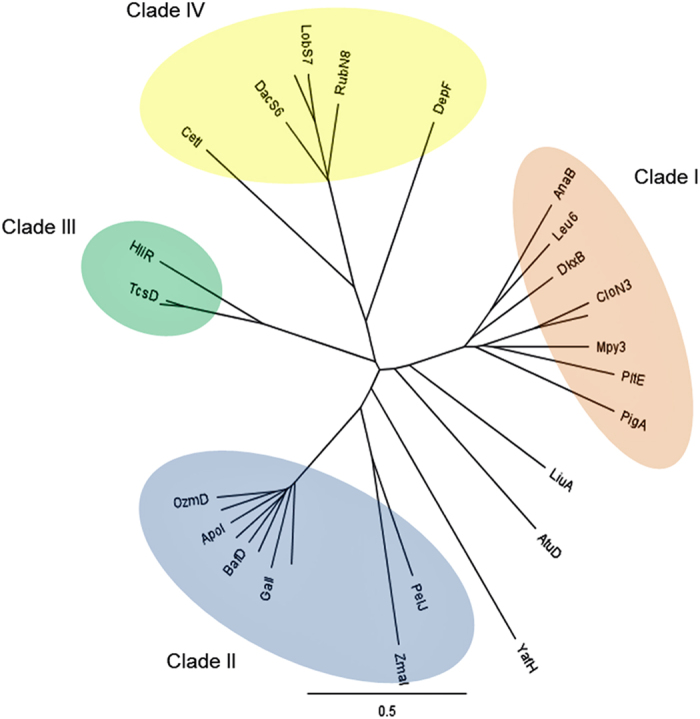
HliR is classified as Clade III with TcsD from the FK506 pathway. Clade I: l-prolyl-S-PCP dehydrogenase; Clade II: methoxymalonyl-ACP biosynthesis involved oxygenase; Clade III: propylmalonyl-ACP dehydrogenase; Clade IV: nitrosugar biosynthesis-related nitrososynthase.
Figure 7. Functional analyses of HliR, an acyl-CoA dehydrogenase.
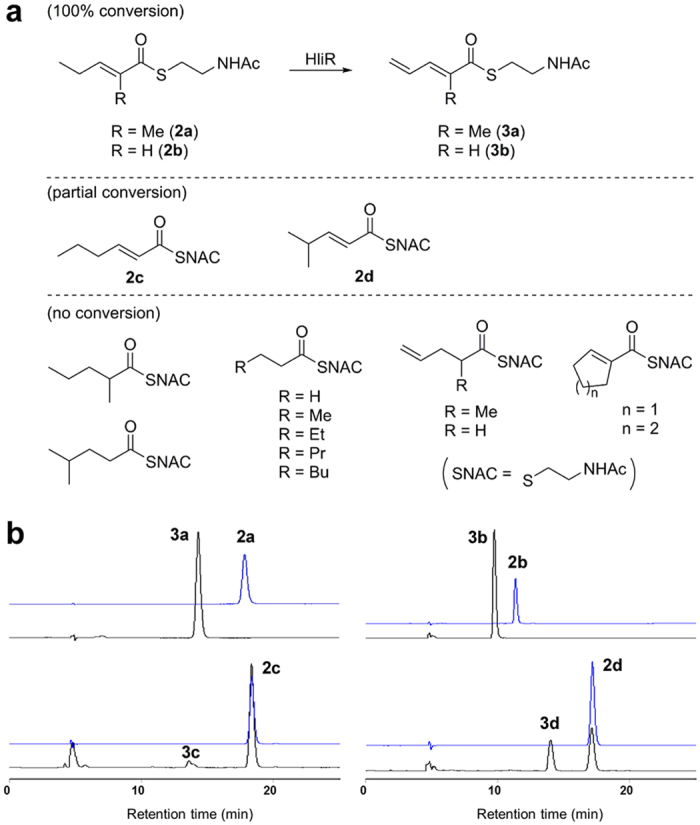
(a) Substrate specificity of recombinant acyl-CoA dehydrogenase HliR. (b) In vitro dehydrogenation of acyl-SNAC compounds by HliR. HPLC analyses of the enzymatic reaction mixtures (black lines) with the substrates 2a−d. The chromatograms were recorded at 254 nm.
Biological activities of haliangicin analogues
Haliangicin (1) is known to show potent inhibitory activity against the plant pathogenic oomycete Phytophthora capsici; in this activity, the terminal β-methoxyacrylate moiety serves as the pharmacophore12 Because the unnatural analogues 4 and 7 obtained in this study are modified at the terminus opposite the β-methoxyacrylate, additional biological information might be obtained. In a test using P. capsici, analogue 4, which is saturated at the 14,15-olefin, showed the same activity as 1 and the versatile fungicide metalaxyl, whereas analogue 7, lacking the 12,13-epoxide moiety, was 30-fold less active than the other analogues (Table 2), suggesting that the epoxide plays a key role in this activity. It is noteworthy that these compounds showed potent cytotoxicity against a tumour cell line and that analogue 7 was more active than the natural compound 1. These compounds showed no inhibitory effects on other microorganisms tested.
Table 2. Biological evaluation of haliangicin (1) and its analogues (4 and 7) obtained through gene disruption.
| Organisms | 1 | 4 | 7 | PC |
|---|---|---|---|---|
| Phytophthora capsici (μg disk–1) | 0.1 | 0.1 | 3.0 | 0.1 |
| Candida rugosa (MIC, μg mL–1) | >32 | >32 | >32 | 0.13 |
| Bacillus subtilis (MIC, μg mL–1) | >32 | >32 | >32 | 0.08 |
| Escherichia coli (MIC, μg mL–1) | >32 | >32 | >32 | 8.0 |
| HeLa S3 cells (IC50, nM) | 41 | 55 | 17 | 8.6 |
Growth inhibition of the oomycete P. capsici was evaluated by a disk diffusion test and denoted as minimum doses to form a definite inhibition zone. Metalaxyl was used as a positive control (PC). In MIC test, PCs were amphotericin B for C. rugosa and ampicillin for B. subtilis and E. coli. The cytotoxicity against HeLa cells was determined by MTT test and paclitaxel was used as PC.
Discussion
Since the isolation in 2002 of Haliangium ochraceum as the first marine myxobacterium, there have been few additional examples of this group. Although such a rare group of microorganisms might appear to be inconsequential, their secondary metabolites, once isolated, were found to be chemically and biologically novel. In addition, a report on Type I PKS genes of marine myxobacterial isolates suggested that the genes in marine myxobacteria were much more unusual than those in terrestrial strains. One of the greatest difficulties in studying marine myxobacteria is their fastidious nature (slow growth, low secondary metabolism, cell aggregation, etc.), which also makes it difficult to apply recently developed techniques for genetic manipulation. Therefore, the present work provides the first insights into biosynthesis in marine myxobacteria. The successful heterologous expression of the biosynthetic machinery for the PKS-type metabolite haliangicin (1) provided several benefits that would be difficult to achieve with the native producer, especially the improved productivity of the fungicide and the production of unnatural analogues, one of which was a more potent cytotoxin than the natural 1. The easier access to the biosynthetic machinery of 1 in a different organism also led to the functional characterization of a unique dehydrogenase that introduces the terminal alkene moiety of 1, thereby widening our understanding of the biosynthetic mechanism underlying the formation of terminal alkene groups. Such γ,δ-dehydrogenation reactions hold promise for generating novel structures in vitro in the future. Genomic information on the native antibiotic producer H. ochraceum, which suggests the presence of six other latent biosynthetic gene clusters, prompts us to analyse them by applying our present expertise to produce novel metabolites that are not obtained by cultivation of the native marine myxobacterium.
Methods
Bacterial strains, plasmids, primers and culture conditions
All bacterial strains, vectors, and oligonucleotides (STAR Oligo; Rikaken Co., Ltd., Nagoya) used in this study are listed in Supplementary Tables S4–S6. The marine myxobacterium Haliangium ochraceum SMP-2 was cultivated as previously described11. CTT medium40 was used for the preculture of Myxococcus xanthus ATCC25232 (wild type) and all its mutants at 30 °C. Production medium41 supplemented with 2–4% (w/v) Sepabeads SP207 resin (Mitsubishi Chemical Co., Tokyo) and a 0.1% (v/v) trace-elements solution (TES)42 was used for the heterologous production of haliangicin (1) and its related metabolites.
Construction and screening of a genomic cosmid library of Haliangium ochraceum SMP-2
The chromosomal DNA of H. ochraceum was prepared as previously described43. The chromosomal DNA was digested with Sau3AI (3 min at 37 °C) and then separated on 0.5% Certified™ Low Melt Agarose gel (Bio-Rad Laboratories, Berkeley, CA) by gel electrophoresis (20 V, overnight). The agarose gel containing DNA fragments of around 30–45 kb in size was melted at 68 °C for 10 min and subsequently digested with GELase™ (Epicentre, Madison, WI) at 45 °C for 1.5 h. The genomic DNA fragments were ligated into the BamHI-digested E. coli/Streptomyces shuttle vector pDW103, and the ligation mixture was packed using the Gigapack III Gold Packaging Extract kit (Stratagene California, San Diego, CA) to obtain about 530 μL of packaged phages, 25 μL of which were transferred into E. coli XL 1-Blue. Single colonies were transferred into eighteen 96-well microplates in LB medium.
A PCR amplification was carried out with the degenerate primers KS1UP/KSD144 by using the H. ochraceum genomic DNA as a template to obtain two KS fragments HKS1 and HKS2 (Supplementary Fig. S1). The genomic cosmid library containing about 1700 cosmids was then screened for PKS genes by colony hybridization with the above-mentioned KS probes HKS1 and HKS2. Five cosmids, c3-5F, c4-6B, c7-6E, c9-4F, and c10-11C, containing KS domain(s) were obtained (Supplementary Fig. S2). Each cosmid was end-sequenced (Supplementary Table S7) by forward-primer DWF1 and reverse-primer DWR. Of these five cosmids, c7-6E and c10-11C were subjected to shotgun sequencing (Takara Bio Inc., Tokyo). Two discrete contigs of 33,197 bp and 22,829 bp were obtained and the remaining 7,420 bp were further sequenced by the genome-walking approach. One continuous region of 63,446 bp was finally assembled and subjected to sequence analysis.
Analysis of the haliangicin biosynthetic gene cluster (hli)
The obtained continuous contig of 63.4 kbp was annotated by various sequence-analysis tools, including BLAST, CDD,45 and antiSMASH analysis46. The multiple alignments of amino acid sequences were generated by using the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/) provided by the European Molecular Biology Institute (Hinxton, UK) and neighbour-joining trees of proteins were generated by Geneious Tree Builder. KS domains in hli were extracted and analysed by using the NaPDoS Web tool (http://napdos.ucsd.edu/)47.
Heterologous expression of the hli gene cluster in M. xanthus
Cosmids c7-6E and c10-11C were modified through λ-Red recombineering. All techniques pertaining to the λ-Red recombineering are described in the published protocol22. For detailed modification procedures, refer to the Supplementary Methods and Figs S3 and S4. The resulting cosmids c7-6E TA-KanR and c10-11C TetR were introduced into the chromosome of M. xanthus ATCC25232 in a stepwise manner (Supplementary Fig. S5). Namely, the first cosmid c7-6E TA-KanR was integrated into the genome of M. xanthus through the TA fragment that is a 1.8 kbp internal fragment homologous to ta-1 responsible for the myxovirescin A biosynthesis48. The second cosmid c10-11C TetR was then introduced through the 14 kbp region shared with the first cosmid. Electrocompetent cells of M. xanthus were prepared according to the reported methods21,49. In the first stage, 1.5 μg of c7-6E TA-KanR was added to 100 μL of a chilled cell suspension of M. xanthus ATCC25232 and electroporated (Gene PulserTM, BIO-RAD) at 25 μF, 1.8 kV, and 200 Ω in 0.2 cm electroporation cuvettes. The kanamycin-resistant colonies (named as M. xanthus c7-6E TA-KanR) appeared after seven days and were verified by colony PCR using specific primers to detect the integration of c7-6E TA-KanR into the chromosome of M. xanthus ATCC25232 (Supplementary Fig. S6a). The expected homologous recombination of the TA fragment was also confirmed by PCR (Supplementary Fig. S6b). Subsequently, 1.7 μg of the second modified cosmid c10-11C TetR was electroporated into M. xanthus c7-6E TA-KanR under the same conditions as those for c7-6E TA-KanR through the 14 kbp region shared with cosmid c7-6E. Mutants were selected on CTT agar plate (oxytetracycline 12.5 μg/mL) for one week and checked by colony PCR using specific primers (Supplementary Fig. S7). The obtained construct carrying the entire length of hli was named M. xanthus c10-11C/c7-6E. The confirmation of the heterologous production of haliangicin (1) in M. xanthus c10-11C/c7-6E is described in the Supplementary Methods.
Feeding experiments with stable-isotope-labelled precursors
The heterologous host M. xanthus c10-11C/c7-6E was cultivated in three or four 100 mL aliquots of production medium supplemented with 2% (w/v) absorbent resin (Sepabeads SP207) and 0.1% TES. Filter-sterilized 0.2 M solutions of the isotope-labelled compounds [2-13C]acetate, [1, 2-13C2]acetate, [1-13C]propionate, L-[methyl-13C]methionine, [1-13C]glycolic acid, and [U-13C3]glycerol in water were separately added in two portions to the bacterial culture after 48 hours and 72 hours, respectively, to give a final concentration of 4 mM. After cultivation for six or seven days in total at 30 °C and 180 rpm, labelled haliangicin was extracted, purified, and analysed by 13C NMR spectroscopy (Supplementary Fig. S9).
Optimization of haliangicin production in the M. xanthus heterologous host
The heterologous host M. xanthus c10-11C/c7-6E was cultured in the production medium. 1 M Aqueous solutions of sodium acetate, sodium propionate, and glycerol were prepared, filter-sterilized, and added to the autoclaved culture medium. l-Methionine was added before autoclaving. For higher concentrations of sodium acetate (100 mM or 200 mM), an autoclaved 5 M stock solution was added in two portions to the medium on the second and third day, respectively, to avoid inhibition of the growth of M. xanthus in its early phase. After cultivation at 30 °C and 180 rpm for the scheduled time, the resins and cell pellets were separated from the medium by centrifugation (6000 rpm, 10 min) and extracted twice with acetone. The crude extracts were filtered, evaporated to dryness, and subjected to HPLC analysis [Develosil ODS-UG-5 (ϕ 4.6 × 250 mm), λ = 290 nm; 75% MeOH–H2O, flow rate: 1 mL/min]. The results are summarized in Supplementary Fig. 4b.
Inactivation of haliangicin biosynthetic genes in M. xanthus c10-11C/c7-6E
Disruption of the hli genes was accomplished by integration of a disruption vector into the genome of the heterologous host M. xanthus c10-11C/c7-6E through single crossover recombination. Plasmid pHSG398 (Takara Bio. Co., Ltd, Tokyo) was first linearized by inverse PCR using primers pHSG398 Inverse-F/pHSG398 Inverse-R; by infusion, the original chloramphenicol-resistance gene was replaced with the Tn5 promoter and a nourseothricin-resistance gene (NrsR) (amplified from pNR150 with primers Kan Nrs-f/Amp Nrs-r), resulting a plasmid named pHSG398 Infu NrsR. The internal fragments of the target genes hliD, hliR, and hliU were amplified from H. ochraceum genomic DNA with primers ORF4-MT-f1/ORF4-MT-r1, pHSG18ACAD Infu-f/pHSG18ACAD Infu-r, and pHSG 21Ox Infu F/pHSG 21Ox Infu R, respectively, and inserted into pHSG398 Infu NrsR. Each disruption vector (1.0–1.5 μg) was electroporated into the haliangicin-producing host M. xanthus c10-11/7-6E at 25 μF, 1.8 kV, and 200 Ω in 0.2 cm electroporation cuvettes. Transformants were selected on CTT agar plate (nourseothricin: 100 μg/mL) for about 5–7 days and then grown in 1 mL of CTT medium (kanamycin 200 μg/mL, oxytetracycline 12.5 μg/mL, nourseothricin 100 μg/mL) for an additional three days. The cell culture was directly subjected to PCR to detect integration of the corresponding plasmid into the genome (Supplementary Fig. S15, S16, S18). For the isolation of the unnatural analogues, please refer to the Supplementary Methods.
Cloning and expression of HliR (acyl-CoA dehydrogenase)
HliR was amplified from the H. ochraceum genome by PCR using high-fidelity PrimeSTAR Max DNA polymerase (Takara Bio Inc.) and the primers HliR Exp Infu f/HliR Exp Infu r, and subsequently cloned into BamHI/XhoI double-digested pET32a by using an In-Fusion® HD Cloning Kit (Clontech Laboratories, Inc., Mountain View, CA). The purified plasmid was transformed into Single Step (KRX) Competent Cells (Promega, Madison, WI). For the expression of HliR, when the OD600 reached 0.6, a final concentration of 0.1% rhamnose and 0.4 mM IPTG were added to induce protein expression. After induction for additional 20 h at 20 °C, the cells were harvested by centrifugation (6000 rpm, 5 min) and resuspended in 3 mL of extraction buffer (50 mM Tris-HCl, 0.4 M NaCl, pH 7.8). The resulting suspension was lysed by using a sonication homogenizer (50 W, five cycles) in the presence of benzonase nuclease (Novagen, Madison, WI) over ice for 30 min. The lysate was centrifuged (6000 rpm, 5 min) to remove insoluble cell debris. The crude protein extracts were stored at −30 °C for subsequent purification. A MagneHisTM Protein Purification System (Promega, Madison, WI) was used to purify the His6-tagged protein. The crude extract and purified HliR were analysed by SDS-PAGE (10% Tris-HCl gel) (Supplementary Fig. S19).
In vitro enzymatic characterization of HliR
A typical HliR assay contained 500 μM flavin adenine dinucleotide (FAD), 500 μM substrate (for chemical synthesis, see Supplementary Methods), and 1 μM recombinant HliR, in a pH 7.5 buffer (8 mM MgSO4, 10 mM Tris-HCl, and 1 mM potassium phosphate; total volume: 100 μL). The assay mixture was incubated at 30 °C overnight and then extracted with EtOAc. The extracts were analysed by HPLC under the conditions indicated in Supplementary Fig. S20.
Additional Information
How to cite this article: Sun, Y. et al. Heterologous Production of the Marine Myxobacterial Antibiotic Haliangicin and Its Unnatural Analogues Generated by Engineering of the Biochemical Pathway. Sci. Rep. 6, 22091; doi: 10.1038/srep22091 (2016).
Supplementary Material
Acknowledgments
This work was financially supported in part by Takeda Science Foundation. We thank the National Cancer Institute (USA) and the National BioResource Project (NIG, JAPAN): E. coli for providing bacterial strains. Y.S. is grateful for a scholarship provided by the China State-Funded Postgraduates Overseas Study Program from the Chinese Scholarship Council. J.S.’s work was carried out with the support of Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01128901), Rural Development Administration, Republic of Korea.
Footnotes
Author Contributions Y.S. deciphered the detailed mechanism of the haliangicin biosynthesis, performed its efficient production, and prepared the manuscript. Z.F. constructed the cosmid library. T.To. performed the biological tests. A.S. accomplished the heterologous expression. S.M. performed the incorporation experiments. T.Ts. and H.M. gave important advice. J.S. provided the cosmid vector and its associated information. T.I. and R.F. cultured the myxobacteria. M.O. designed the project and wrote the manuscript.
References
- Höfle G. H. et al. Epothilone A and B: Novel 16-membered macrolides with cytotoxic activity: Isolation, crystal structure, and conformation in solution. Angew. Chem. Int. Ed. 35, 1567–1569 (1996). [Google Scholar]
- Nettles J. H. et al. The binding mode of epothilone A on α,β-tubulin by electron crystallography. Science 305, 866–869 (2004). [DOI] [PubMed] [Google Scholar]
- Weissman K. J. & Müller R. A brief tour of myxobacterial secondary metabolism. Bioorg Med. Chem. 17, 2121–2136 (2009). [DOI] [PubMed] [Google Scholar]
- Wenzel S. C. & Müller R. Myxobacteria–‘microbial factories’ for the production of bioactive secondary metabolites. Mol. Biosyst . 5, 567–574 (2009). [DOI] [PubMed] [Google Scholar]
- Schäberle T. F., Lohr F., Schmitz A. & Konig G. M. Antibiotics from myxobacteria. Nat. Prod. Rep. 31, 953–972 (2014). [DOI] [PubMed] [Google Scholar]
- Iizuka T. et al. Enhygromyxa salina gen. nov., sp. nov., a slightly halophilic myxobacterium isolated from the coastal areas of Japan. Syst. Appl. Microbiol. 26, 189–196 (2003). [DOI] [PubMed] [Google Scholar]
- Iizuka T. et al. Pseudenhygromyxa salsuginis gen. nov., sp. nov., a myxobacterium isolated from an estuarine marsh. Int. J. Syst. Evol. Microbiol. 63, 1360–1369 (2013). [DOI] [PubMed] [Google Scholar]
- Fudou R., Jojima Y., Iizuka T. & Yamanaka S. Haliangium ochraceum gen. nov., sp. nov. and Haliangium tepidum sp. nov.: Novel moderately halophilic myxobacteria isolated from coastal saline environments. J. Gen. Appl. Microbiol. 48, 109–116 (2002). [DOI] [PubMed] [Google Scholar]
- Schaberle T. F. et al. Marine myxobacteria as a source of antibiotics: Comparison of physiology, polyketide-type genes and antibiotic production of three new isolates of Enhygromyxa salina. Mar. Drugs 8, 2466–2479 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudou R. et al. Haliangicin, a novel antifungal metabolite produced by a marine myxobacterium. 2. Isolation and structural elucidation. J. Antibiot. (Tokyo) 54, 153–156 (2001). [DOI] [PubMed] [Google Scholar]
- Fudou R. & Iizuka T. & Yamanaka S. Haliangicin, a novel antifungal metabolite produced by a marine myxobacterium. 1. Fermentation and biological characteristics. J. Antibiot. (Tokyo) 54, 149–152 (2001). [DOI] [PubMed] [Google Scholar]
- Kundim B. A. et al. New haliangicin isomers, potent antifungal metabolites produced by a marine myxobacterium. J. Antibiot. (Tokyo) 56, 630–638 (2003). [DOI] [PubMed] [Google Scholar]
- Iizuka T. et al. Miuraenamides A and B., novel antimicrobial cyclic depsipeptides from a new slightly halophilic myxobacterium: Taxonomy, production, and biological properties. J. Antibiot. (Tokyo) 59, 385–391 (2006). [DOI] [PubMed] [Google Scholar]
- Ojika M. et al. Miuraenamides: Antimicrobial cyclic depsipeptides isolated from a rare and slightly halophilic myxobacterium. Chem.–Asian J. 3, 126–133 (2008). [DOI] [PubMed] [Google Scholar]
- Felder S. et al. Salimabromide: Unexpected chemistry from the obligate marine myxobacterium Enhygromyxa salina. Chem.–Eur. J. 19, 9319–9324 (2013). [DOI] [PubMed] [Google Scholar]
- Felder S. et al. Salimyxins and enhygrolides: Antibiotic, sponge-related metabolites from the obligate marine myxobacterium Enhygromyxa salina. ChemBioChem 14, 1363–1371 (2013). [DOI] [PubMed] [Google Scholar]
- Komaki H. et al. PCR detection of type I polyketide synthase genes in myxobacteria. Appl. Environ. Microbiol. 74, 5571–5574 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun C. G. et al. An efficient approach for cloning the dNDP-glucose synthase gene from actinomycetes and its application in Streptomyces spectabilis, a spectinomycin producer. FEMS Microbiol. Lett. 183, 183–189 (2000). [DOI] [PubMed] [Google Scholar]
- Julien B. & Shah S. Heterologous expression of epothilone biosynthetic genes in Myxococcus xanthus. Antimicrob. Agents. Chemother. 46, 2772–2778 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D. C., Henry M. R., Murphy K. A. & Boddy C. N. Heterologous expression of the oxytetracycline biosynthetic pathway in Myxococcus xanthus. Appl. Environ. Microbiol. 76, 2681–2683 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlova O. et al. Reconstitution of the myxothiazol biosynthetic gene cluster by Red/ET recombination and heterologous expression in Myxococcus xanthus. Appl. Environ. Microbiol. 72, 7485–7494 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan S. K., Thomason L. C., Kuznetsov S. G. & Court D. L. Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T. W. et al. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. USA. 99, 7968–7973 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel S. C. et al. On the biosynthetic origin of methoxymalonyl-acyl carrier protein, the substrate for incorporation of “glycolate” units into ansamitocin and soraphen A. J. Am. Chem. Soc. 128, 14325–14336 (2006). [DOI] [PubMed] [Google Scholar]
- Wu K., Chung L., Revill W. P., Katz L. & Reeves C. D. The FK520 gene cluster of Streptomyces hygroscopicus var. ascomyceticus (ATCC 14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 251, 81–90 (2000). [DOI] [PubMed] [Google Scholar]
- Hertweck C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. 48, 4688–4716 (2009). [DOI] [PubMed] [Google Scholar]
- Calderone C. T. Isoprenoid-like alkylations in polyketide biosynthesis. Nat. Prod. Rep. 25, 845–853 (2008). [DOI] [PubMed] [Google Scholar]
- Carroll B. J. et al. Identification of a set of genes involved in the formation of the substrate for the incorporation of the unusual “glycolate” chain extension unit in ansamitocin biosynthesis. J. Am. Chem. Soc. 124, 4176–4177 (2002). [DOI] [PubMed] [Google Scholar]
- Li Y., Chooi Y.-H., Sheng Y., Valentine J. S. & Tang Y. Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an α-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganese thioesterase. J. Am. Chem. Soc. 133, 15773–15785 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi Y.-H. et al. Genome mining of a prenylated and immunosuppressive polyketide from pathogenic fungi. Org. Lett. 15, 780–783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z. X. et al. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 67, 1356–1367 (2004). [DOI] [PubMed] [Google Scholar]
- Gerwick W. H. et al. Structure of curacin-a, a novel antimitotic, antiproliferative, and brine shrimp toxic natural product from the marine cyanobacterium Lyngbya majuscula. J. Org. Chem. 59, 1243–1245 (1994). [Google Scholar]
- Ubukata M., Cheng X. C., Uzawa J. & Isono K. Biosynthesis of the dialkylmaleic anhydride-containing antibiotics, tautomycin and tautomycetin. J. Chem. Soc., Perkin Trans. 1, 2399–2404 (1995). [Google Scholar]
- Tanaka H. et al. Structure of FK506: A novel immunosuppressant isolated from Streptomyces. J. Am. Chem. Soc. 109, 5031–5033 (1987). [Google Scholar]
- Mo S. et al. Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J. Am. Chem. Soc. 133, 976–985 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. D. et al. Improvement of FK506 production in Streptomyces tsukubaensis by genetic enhancement of the supply of unusual polyketide extender units via utilization of two distinct site-specific recombination systems. Appl. Environ. Microbiol. 78, 5093–5103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andexer J. N. et al. Biosynthesis of the immunosuppressants FK506, FK520, and rapamycin involves a previously undescribed family of enzymes acting on chorismate. Proc. Natl. Acad. Sci. USA 108, 4776–4781 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner A. et al. Designed biosynthesis of 36-methyl-FK506 by polyketide precursor pathway engineering. ACS Synth. Biol. 2, 379–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranovič D. et al. Origin of the allyl group in FK506 biosynthesis. J. Biol. Chem. 285, 14292–14300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. & Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. USA. 74, 2938–2942 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojika M. et al. Cystothiazoles A and B., new bithiazole-type antibiotics from the myxobacterium Cystobacter fuscus. J. Antibiot. (Tokyo) 51, 275–281 (1998). [DOI] [PubMed] [Google Scholar]
- Reichenbach H. & Dworkin M. In: The prokaryotes 2nd edn, (eds Balows A., Truper H. G., Dworkin M., Harder W. & Schleifer K.) Ch. 118, 3416–3487 (Springer, 1992). [Google Scholar]
- Syn C. K. C. & Swarup S. A scalable protocol for the isolation of large-sized genomic DNA within an hour from several bacteria. Anal. Biochem. 278, 86–90 (2000). [DOI] [PubMed] [Google Scholar]
- Beyer S., Kunze B., Silakowski B. & Müller R. Metabolic diversity in myxobacteria: Identification of the myxalamid and the stigmatellin biosynthetic gene cluster of Stigmatella aurantiaca Sg a15 and a combined polyketide-(poly)peptide gene cluster from the epothilone producing strain Sorangium cellulosum So ce90. Biochim. Biophys. Acta 1445, 185–195 (1999). [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K. et al. antiSMASH 2.0: A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41, W204–W212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N. et al. The natural product domain seeker NaPDoS: A phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 7, e34064 (2012); doi: 10.1371/journal.pone.003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic V et al. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. ChemBioChem 7, 1206–1220 (2006). [DOI] [PubMed] [Google Scholar]
- Fu J. et al. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic Acids Res. 36, e113 (2008); doi: 10.1093/nar/gkn499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malonek S., Rojas M. C., Hedden P., Hopkins P. & Tudzynski B. Restoration of gibberellin production in Fusarium proliferatum by functional complementation of enzymatic blocks. Appl. Environ. Microbiol. 71, 6014–6025 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.