Abstract
Inconsistent conclusions have been drawn regarding the phylogenetic age of the Methuselah/Methuselah-like (Mth/Mthl) gene family of G protein-coupled receptors, the founding member of which regulates development and lifespan in Drosophila. Here we report the results from a targeted homolog search of 39 holozoan genomes and phylogenetic analysis of the conserved seven transmembrane domain. Our findings reveal that the Mth/Mthl gene family is ancient, has experienced numerous extinction and expansion events during metazoan evolution, and acquired the current definition of the Methuselah ectodomain during its exceptional expansion in arthropods. In addition, our findings identify Mthl1, Mthl5, Mthl14, and Mthl15 as the oldest Mth/Mthl gene family paralogs in Drosophila. Future studies of these genes have the potential to define ancestral functions of the Mth/Mthl gene family.
G protein-coupled receptors (GPCRs) represent one of the largest and most diverse receptor type protein families in animals1,2,3,4. Leveraging the access to complete genome sequences from all major branches of the tree of life, previous studies have elucidated many fundamental aspects of GPCR gene family diversification5,6. Important subfamilies, however, remain insufficiently defined. One such example is the Methuselah/Methuselah-like (Mth/Mthl) subfamily of GPCRs, named after the Drosophila GPCR gene methuselah (mth). Discovered due to its effects on lifespan and cellular stress resistance in adult flies7, the mth gene is also essential for embryonic development8,9. The molecular mechanisms underlying mth function have received major attention, leading to the discovery of candidate ligands10,11, the identification of Mth-specific small molecule inhibitors that reproduce the mth phenotype12,13, and the recent finding that Drosophila Mth signals through the TOR pathway13.
In contrast to this significant progress, evolutionary age and functional conservation of the Mth GPCR have remained subject to debate. Indeed, little is known yet about the functions of its 15 paralogs in Drosophila: Methuselah-like (Mthl) 1–15. Molecular phylogenetic studies revealed that most of these genes originated during two gene duplication surges in the past 60 million years of fruit fly species diversification9,14. Remarkably, mth itself represents one of the youngest descendants of this exceptional gene family expansion, raising the question of whether its roles in development and life history regulation are likewise of recent origin or represent conserved remnants of ancestral functions.
Further support of a more recent origin of Drosophila mth function comes from the challenge in detecting Mth/Mthl homologs outside insects. In line with the canonical organization of GPCRs, Mth/Mthl receptors are composed of an N-terminal ectodomain, a seven transmembrane domain (7TM), and a short intracellular C-terminal domain. Early studies identified Drosophila mth as a member of the Secretin/Adhesion Class B superclade of GPCRs based on the 7tm_2-specific configuration of its 7TM domain4,5,15. The Mth ectodomain, by contrast, was recognized to be novel and only conserved in higher Diptera5,7,9,16. Later studies identified GPCRs with detectable Mth ectodomains in other insect orders17, and possibly arthropods14, but not beyond.
Importantly, the Drosophila repertoire of Mth/Mthl gene family members also includes four paralogs that lack significant similarity to the Mth ectodomain, but were identified based on conserved sequence signatures in the 7TM domain: Mthl1, Mthl5, Mthl14, and Mthl159,16. The 7TM domain has thus been recognized as a more conserved sequence region of the Drosophila Mth/Mthl gene family, which includes both Mth ectodomain-positive and -negative members.
The significance of the 7TM domain is further underlined by the fact that some 7TM domain-based studies reported evidence of candidate Mth/Mthl homologs in metazoan species outside arthropods: the sea squirt Ciona intestinalis18, the lancelet Branchiostoma floridae, and the sea anemone Nematostella vectensis5. At the same time, no candidate Mth/Mthl homologs were noted in the first genome-wide survey of GPCRs in the lancelet19 or in similar studies of the acorn worm Saccoglossus kowalevskii20 and flatworms21, possibly due to incomplete genome sequence coverage or sampling. Finally, a recent study of GPCR diversity in insects expressed doubt about the monophyly of the Mth/Mthl subfamily22.
These confounding data leave three major questions: How deeply conserved is the Mth/Mthl gene family in the metazoan tree of life? Is the Mth ectodomain an ancestral or derived component of Mth/Mthl gene family members? And which are the most ancestral Mth/Mthl homologs in Drosophila that could give insights into the earliest functions of the Mth/Mthl gene family?
Results and Discussion
Early metazoan origin of the Mth/Mthl gene family
To improve our understanding of the Mth/Mthl gene family, we investigated the phylogenetic position of previously and newly identified candidate homologs. To this end, we searched a database of 39 genomes representing holozoan species diversity (Ichthyosporea + Corallochytrium + Filasterea + Choanoflagellatea + Animalia) (Supplementary Table S1) by BlastP with the 7TM domain of D. melanogaster Mthl1 as query and collected the 100 best hits as candidate homologs. We then used the best matching human candidate homolog (GPR98) from this pool as query for a second search, from which we collected an additional 100 best matching sequences after removing duplicates found in the first search. In parallel, we identified candidate homologs from species without complete genomes based on reciprocal BLAST evidence using the previously reported Mthl candidate homolog from N. vectensis as query5. Combined, these efforts resulted in a total of 278 GPCR sequences (Supplementary text file S1), which were used to build a multiple alignment of the 7TM domain (Supplementary text file S2) for molecular phylogenetic gene tree estimation using Bayesian and likelihood approaches.
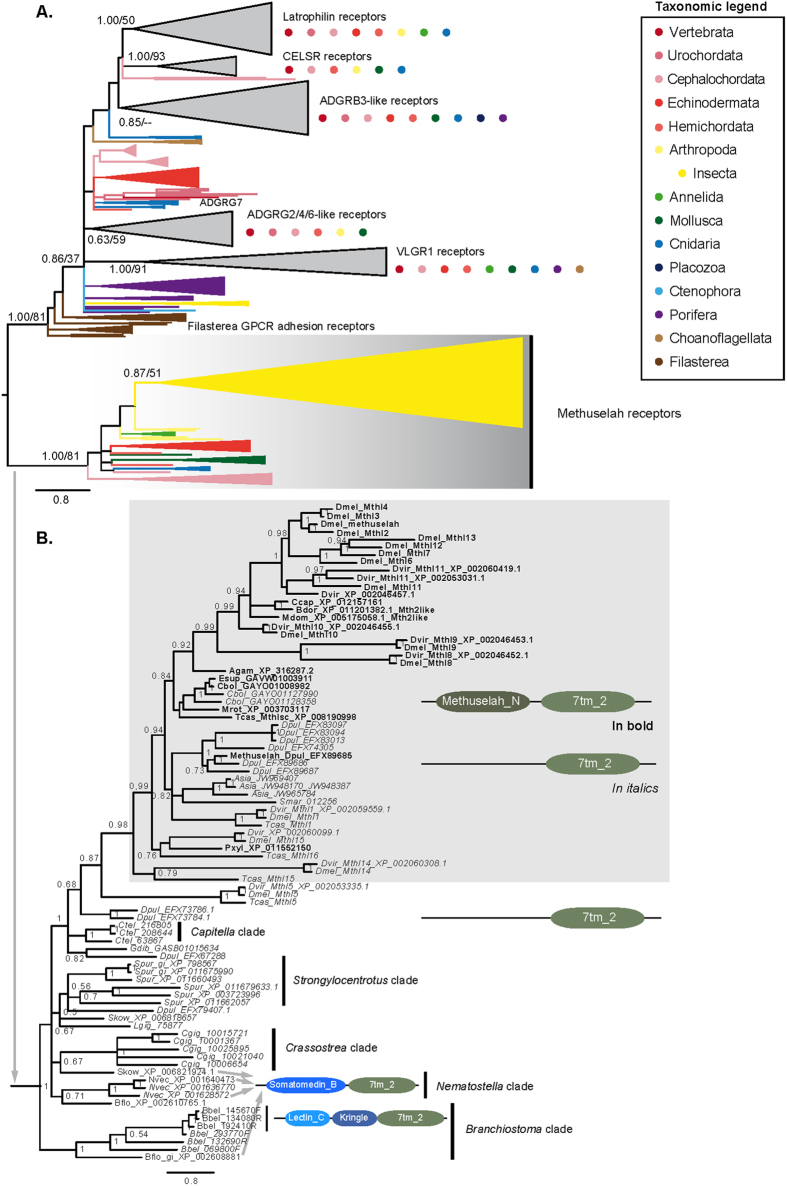
Consistent with previous studies, the resulting trees recovered a robustly supported clade of arthropod Mth/Mthl homologs (Fig. 1A and Supplementary text files S3 and S4). This clade was nested within a likewise robustly supported, more inclusive clade. Altogether, our trees identified 44 Mth/Mthl candidate homologs from diverse animal species outside insects. These included protostomes (Annelida, Mollusca), invertebrate deuterostomes (Echinodermata, Hemichordata, Cephalochordata), and the sea anemone N. vectensis.
Figure 1. Phylogenetic analysis of the relationships between Mth/Mthl and related holozoan GPCRs.
Nodal numbers represent Bayesian Posterior Probabilities and Maximum Likelihood bootstrap supports (100 replicates). (A) Collapsed Bayesian phylogenetic tree estimate. Clades were collapsed using congruent nodal supports by both tree estimation methodologies, defining stable clusters and conserved PFAM domain architectures30 as a secondary evidence for family classification. Orphan GPCR gene families were kept unclassified. The holozoan phyla represented in each GPCR group are indicated by the color described in the taxonomic legend box. (B) Uncollapsed view of the Mth/Mthl GPCR gene family cluster in panel (A). Grey background box denotes the expanded insect cluster of Mth/Mthl genes. Detectable Pfam domains in specific genes indicated to the right. Species abbreviations: Agam = Anopheles gambiae, Asia = Argulus siamensis, Bbel = Branchiostoma belcheri, Bdor = Bactrocera dorsalis, Bflo = Branchiostoma floridae, Cbol = Cordulegaster boltonii, Ccap = Ceratitis capitata, Cgig = Crassostrea gigas, Ctel = Capitella teleta, Dpul = Daphnia pulex, Dmel = Drosophila melanogaster, Dvir = Drosophila virilis, Esup = Epiophlebia superstes, Gdib = Glycera dibranchiata, Lgig = Lottia gigantea, Mdom = Musca domestica, Mrot = Megachile rotundata, Nvec = Nematostella vectensis, Pxyl = Plutella xylostella, Skow = Saccoglossus kowalevskii, Smar = Strigamia maritima, Spur = Strongylocentrotus purpuratus, Tcas = Tribolium castaneum.
To scrutinize the non-insect candidate homologs, we performed reciprocal BLAST searches against the NCBI nr protein sequences of D. melanogaster and B. floridae. This approach defined 24 high confidence Mth/Mthl homologs outside insects, which included 10 other arthropod, 4 mollusc, 7 hemichordate, and 3 cnidarian genes5 (Supplementary Table S2). Our targeted, double pronged approach thus corroborated the previously suggested presence of Mth/Mthl homologs in sea squirts18, lancelets, and cnidarians5 and, for the first time, detected Mth/Mthl homologs in the acorn worm S. kowalevskii20.
Mth/Mthl gene family extinctions and expansions
Within chordates, our analyses only detected Mth/Mthl homologs in the lancelet species B. floridae and B. belcheri (Fig. 1). To further explore the apparent absence of the Mth/Mthl gene family in other chordate clades, we searched the NCBI nr database by reciprocal BlastP for tunicate and vertebrate homologs using the B. floridae Mth/Mthl homologs XP_002610765 and XP_002608881 as queries (Fig. 1). Also this approach failed to detect Mth/Mthl homologs in either tunicates and vertebrates, leading to the conclusion that the Mth/Mthl gene family became extinct during early chordate evolution and was absent in the last common ancestor of urochordates and vertebrates.
Further losses of the Mth/Mthl gene family during metazoan evolution were indicated by absence of detectable Mth/Mthl homologs in genomes from Nematoda, Platyhelminthes, and Rotifera. A particularly prominent example in addition to vertebrates was the absence of Mth/Mthl homologs in the cnidarians Acropora digitifera and Hydra magnipapillata in contrast to the presence of three Mth/Mthl homologs in N. vectensis (Fig. 1 and Supplementary Tables S2 and S3).
Our trees also detected multiple independent expansions of the Mth/Mthl gene family in the metazoan tree of life. The most dramatic example of this continues to be the exceptionally enlarged Mth/Mthl gene family cluster of insects (Fig. 1)9,14. Other examples are found in Daphnia pulex (Crustacean), Capitella teleta (Annelid), Crassostrea gigas (Mollusc), Strongylocentrotus purpuratus (sea urchin), and N. vectensis.
While these repeated extinction and expansion events explain the previous difficulty of recognizing the ancientness of the Mth/Mthl gene family, they continue to pose a challenge to reconstructing the ancestral set of Mth/Mthl genes in the last common ancestor of Metazoans. At this point, the scattered conservation of Mth/Mthl homologs in distantly related lineages is best explained by the presence of a singleton homolog in the last common ancestor of Eumetazoans (Bilateria + Cnidaria).
Derived acquisition of the Mth ectodomain
The Mth ectodomain is defined by 10 cysteine residues, which form a total of five disulfide bonds7,16. Most of these residues were noted to be conserved in the Drosophila paralogs Mthl2–4, and Mthl 7–12 but not in Mthl1 and Mthl516. Consistent with this, we were able to confirm the presence of the Mth ectodomain for Mthl2–4, and Mthl 7–12 in Pfam database searches based on significant matches to the Methuselah_N domain PF06652 with e-values between 8.9E-18 to 3.1E-69 but not for Mthl1 and Mthl5 (Supplementary Table S3). Moreover, Pfam support for the presence of a Mth ectodomain was exceptionally low or non-significant for the more recently described Drosophila paralogs Mth15 (Pfam e-value: 0.00012) and Mthl14, respectively.
To address the question whether the presence of the Mth ectodomain is a derived or ancestral feature of Mth/Mthl gene family members, we searched all other members of the 7TM domain supported metazoan Mth/Mthl gene family clade for the presence of the Mth ectodomain. Consistent with the Pfam database listing of the Methuselah_N domain (PF06652) as arthropod-specific (Finn et al. 2014), this approach detected additional Mth ectodomains only in Mth/Mthl homologs from arthropod species (Supplementary Table S3). Almost all of these represented insects except for the previously reported custacean Mth/Mthl homolog EFX89685 from Daphnia (Pfam e-value: 0.00012)14.
Mapping the conservation of the Mth ectodomain onto the 7TM domain-based Mth/Mthl gene family tree further revealed that all Mth ectodomain-positive homologs are contained in the strongly expanded arthropod subcluster of Mth/Mthl homologs (Fig. 1B) and that this clade is sister to the Mth ectodomain-negative homolog Mthl5. Since Mth/Mthl gene family members outside arthropods are all Mth ectodomain-negative, outgroup rooting supports the model that the Mth ectodomain is a derived domain that has been acquired during arthropod evolution.
Interestingly, our Pfam searches detected evidence of additional ectodomain acquisition events in the Mth/Mthl gene family tree. This included the presence Somatomedin_B domains in the Mth/Mthl homologs XP_001628572 of N. vectensis, XP_002608881 of B. floridae, and XP_006821924 of S. kowalevskii (Fig. 1B). The evolution of the Mth/Mthl gene family may thus have been repeatedly shaped by ectodomain acquisition events.
Candidate ancestral Mth/Mthl receptors in Drosophila
51 Mth/Mthl gene family members in our trees possess ectodomains that lack significant similarities to any currently known protein domains. This includes Mthl1, Mthl5, and Mthl14, which, according to our trees, represent the oldest Mth/Mthl gene family members in Drosophila, together with Mthl 15, which is characterized by only marginal Mth ectodomain support (0.00012). The phylogenetic evidence thus suggests that defining the ligand binding properties and physiological functions for these Drosophila homologs has the potential to elucidate ancestral functions of the Mth/Mthl gene family.
Interestingly, expression studies in Drosophila point toward a role of Mthl5 in the development of the visceral and cardiac mesoderm9. Moreover, recent gene knockdown studies in the red flour beetle Tribolium castaneum produced evidence that Mthl5 is essential for embryonic and postembryonic survival23,24, interacting with several signaling pathways25. In combination, these first insights and the strong gene tree evidence of an ancestral status of Mthl5 prioritize this paralog for further study.
Conclusions
Taken together, our studies lead to four important conclusions: (I) Mth/Mthl homologs are present in cnidarians, protostomes, and deuterostomes, implying an early metazoan origin of this gene family. (II) The Mth/Mthl gene family is characterized by numerous extinction and expansion events, reflected most notably by its absence in vertebrates and abundance in insects. (III) The current definition of the Mth ectodomain most likely originated during the exceptional expansion of the Mth/Mthl gene family in arthropods. (IV) Mthl1, Mthl5, Mthl14, and Mthl15 represent the oldest Mth/Mthl paralogs in D. melanogaster, prioritizing these genes for the experimental characterization of potentially conserved ancestral functions of the Mth/Mthl gene family.
Methods
The multiple sequence alignment was built using MAFFT, applying the L-INS-i method26 and manually cleared of gaps and non-homologous regions outside the 7TM domain. The trimmed alignment of 365 amino acid positions was subjected to phylogenetic analysis with PhyloBayes 3.0 until 2 chains converged (maxdiff <0.3)27 and to estimate the 20 best RaxML 8.0 trees28 using the LG model and 8 categories for gamma distribution and 100 bootstrap replicates. Pfam searches were performed at default gathering threshold29. Species lists, sequences, alignments, and trees are available in Supplementary Information.
Additional Information
How to cite this article: de Mendoza, A. et al. Methuselah/Methuselah-like G protein-coupled receptors constitute an ancient metazoan gene family. Sci. Rep. 6, 21801; doi: 10.1038/srep21801 (2016).
Supplementary Material
Footnotes
Author Contributions J.W.J. and M.F. designed the study and performed the preliminary homolog collection. A.M. performed exhaustive homolog collection, all phylogenetic analyses, and Figure 1. M.F. generated supplementary Tables 2 and 3, and wrote the manuscript with contributions from the other authors.
References
- Stäubert C., Le Duc D. & Schöneberg T. In G Protein-Coupled Receptor Genetics (ed. Stevens C. W.) 23–43 (Humana Press, 2014). [Google Scholar]
- De Mendoza A., Sebé-Pedrós A. & Ruiz-Trillo I. The evolution of the GPCR signaling system in eukaryotes: modularity, conservation, and the transition to metazoan multicellularity. Genome Biol. Evol. 6, 606–619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson L. G. Evidence for kinship between diverse G-protein coupled receptors. Gene 239, 333–340 (1999). [DOI] [PubMed] [Google Scholar]
- Harmar A. J. Family-B G-protein-coupled receptors. Genome Biol. 2, reviews3031.1-3031.10 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K. J. V., Lagerström M. C., Wallér L. M. J., Fredriksson R. & Schiöth H. B. The Secretin GPCRs descended from the family of Adhesion GPCRs. Mol. Biol. Evol. 26, 71–84 (2009). [DOI] [PubMed] [Google Scholar]
- Fredriksson R. & Schiöth H. B. The Repertoire of G-Protein–Coupled Receptors in Fully Sequenced Genomes. Mol. Pharmacol. 67, 1414–1425 (2005). [DOI] [PubMed] [Google Scholar]
- Lin Y. J., Seroude L. & Benzer S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science 282, 943–946 (1998). [DOI] [PubMed] [Google Scholar]
- Song W. et al. Presynaptic regulation of neurotransmission in Drosophila by the g protein-coupled receptor methuselah. Neuron 36, 105–119 (2002). [DOI] [PubMed] [Google Scholar]
- Patel M. V. et al. Dramatic expansion and developmental expression diversification of the methuselah gene family during recent Drosophila evolution. J. Exp. Zool. B Mol. Dev. Evol. 318, 368–387 (2012). [DOI] [PubMed] [Google Scholar]
- Ja W. W., Carvalho G. B., Madrigal M., Roberts R. W. & Benzer S. The Drosophila G protein-coupled receptor, Methuselah, exhibits a promiscuous response to peptides. Protein Sci. 18, 2203–2208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvejic S., Zhu Z., Felice S. J., Berman Y. & Huang X.-Y. The endogenous ligand Stunted of the GPCR Methuselah extends lifespan in Drosophila. Nat. Cell Biol. 6, 540–546 (2004). [DOI] [PubMed] [Google Scholar]
- Ja W. W. et al. Extension of Drosophila melanogaster life span with a GPCR peptide inhibitor. Nat. Chem. Biol. 3, 415–419 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. Methuselah regulates longevity via dTOR: a pathway revealed by small-molecule ligands. J. Mol. Cell Biol. 7, 280–283 (2015). [DOI] [PubMed] [Google Scholar]
- Araújo A. R. et al. The Drosophila melanogaster methuselah gene: a novel gene with ancient functions. PLoS One 8, e63747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D. et al. Pfam: clans, web tools and services. Nucleic Acids Res. 34, D247–51 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A. P. Jr, Llamas L. L., Snow P. M., Benzer S. & Bjorkman P. J. Crystal structure of the ectodomain of Methuselah, a Drosophila G protein-coupled receptor associated with extended lifespan. Proc. Natl. Acad. Sci. USA 98, 3744–3749 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. et al. The G protein-coupled receptors in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 40, 581–591 (2010). [DOI] [PubMed] [Google Scholar]
- Kamesh N., Aradhyam G. K. & Manoj N. The repertoire of G protein-coupled receptors in the sea squirt Ciona intestinalis. BMC Evol. Biol. 8, 129 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K. J. V., Fredriksson R. & Schiöth H. B. The amphioxus (Branchiostoma floridae) genome contains a highly diversified set of G protein-coupled receptors. BMC Evol. Biol. 8, 9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A., Almén M. S., Fredriksson R. & Schiöth H. B. Remarkable similarities between the hemichordate (Saccoglossus kowalevskii) and vertebrate GPCR repertoire. Gene 526, 122–133 (2013). [DOI] [PubMed] [Google Scholar]
- Zamanian M. et al. The repertoire of G protein-coupled receptors in the human parasite Schistosoma mansoni and the model organism Schmidtea mediterranea. BMC Genomics 12, 596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K. J., Brown M. R. & Strand M. R. Phylogenetic investigation of peptide hormone and growth factor receptors in five dipteran genomes. Front. Endocrinol. 4, 193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. Methuselah-like genes affect development, stress resistance, lifespan and reproduction in Tribolium castaneum. Insect Mol. Biol. 23, 587–597 (2014). [DOI] [PubMed] [Google Scholar]
- Bai H., Zhu F., Shah K. & Palli S. R. Large-scale RNAi screen of G protein-coupled receptors involved in larval growth, molting and metamorphosis in the red flour beetle. BMC Genomics 12, 388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. Comparative RNA-sequencing analysis of mthl1 functions and signal transductions in Tribolium castaneum. Gene 547, 310–318 (2014). [DOI] [PubMed] [Google Scholar]
- Katoh K., Kuma K.-I., Miyata T. & Toh H. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform. 16, 22–33 (2005). [PubMed] [Google Scholar]
- Lartillot N., Lepage T. & Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009). [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.