Abstract
Surface reactive nanostructures were fabricated using stimulated emission depletion (STED) lithography. The functionalization of the nanostructures was realized by copolymerization of a bifunctional metal oxo cluster in the presence of a triacrylate monomer. Ligands of the cluster surface cross-link to the monomer during the lithographic process, whereas unreacted mercapto functionalized ligands are transferred to the polymer and remain reactive after polymer formation of the surface of the nanostructure. The depletion efficiency in dependence of the cluster loading was investigated and full depletion of the STED effect was observed with a cluster loading exceeding 4 wt %. A feature size by λ/11 was achieved by using a donut-shaped depletion beam. The reactivity of the mercapto groups on the surface of the nanostructure was tested by incubation with mercapto-reactive fluorophores.
Keywords: STED lithography, functional polymers, zirconium oxo cluster polymerization, thiol polymers
In the midnineties, Hell and Wichmann proposed stimulated emission depletion (STED) microscopy1 as new concept of scanning fluorescence microscopy which allows to break the diffraction limit in optical imaging.2,3 Meanwhile, STED is a state of the art method with even sub-10 nm resolution.4 As proposed in the early work on STED microscopy,2,3 the STED concept was expected to be also apt to increase resolution in optical lithography. In three-dimensional optical lithography, cross-linked polymers are formed via multiphoton-induced polymerization by focusing an ultrashort pulsed laser into an acrylate monomer mixed with a photoinitiator.5 In STED-enhanced multiphoton lithography, a second laser locally inhibits the ability of the photoinitiator to start the polymerization in the outer rim of the point spread function (PSF).6 The polymerization volume is thus confined, which allows for structuring of nanometer-sized features. Recently realized STED- and STED-inspired lithography are the only methods allowing diffraction-unlimited structuring with visible light in three dimensions. STED lithography facilitates writing features with lateral structure sizes of 55 nm and a resolution of 120 nm.7 Some other STED-inspired techniques8−11 were proven to write features even with sub-50 nm resolution.12 Applications of STED lithography range from nanostructured photonic materials, 3D data storage, to life-science.13−18
STED lithography is currently limited to only one class of materials, which is radically polymerized (meth)acrylates. In fact, STED lithography has been realized with only a few different types of (meth)acrylate monomers, and true STED lithography has only been shown with a single photoinitiator, namely 7-diethylamino-3-thenoylcoumarin (DETC) (Figure 1c).6,19−21 Alternative STED-related techniques use other photoinitiators but still rely on (meth)acrylates as the photoresist. This severely restricts a covalent and targeted attachment of molecules (such as proteins) or metallization of the structures unless a further chemical modification of the polymer is performed.22−24 Lately, research on reactive photoresists, which enable facile subsequent covalent modification, was carried out. Functionalized polymers25 have been found to be particularly useful in fabrication of reactive hydrogels26,27 or thiol-ene-reactive structures,28 capable of binding proteins or metals.
Figure 1.
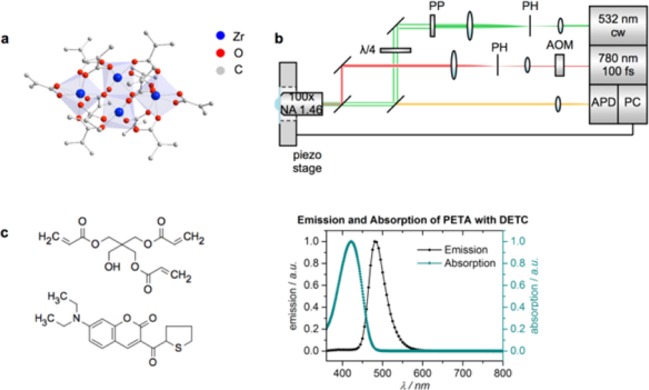
(a) Molecular structure of Zr4O2(OMc)12 (Zr4, OMc = methacrylate). Hydrogen atoms are omitted for clarity. (b) Scheme of the STED lithography setup. Green depicts the depletion beam (at 532 nm, continuous wave) and red the excitation beam (at 780 nm, 100 fs pulse duration). The depletion beam is shaped into a donut using a 2π spiral phase plate. Abbreviations: acousto-optic modulator (AOM), pinhole (PH), phase plate (PP), avalanche photo diode (APD), personal computer (PC). (c) Shows the chemical structure of pentaerythritol triacrylate (PETA, monomer), 7-diethylamino-3-thenoylcoumarin (DETC, photoinitiator) and the emission and absorption spectrum of DETC in PETA.
Thus, there is a need in the development of new functional monomers which can be applied in STED lithography. STED lithography, however, sets the following restrictions on the resin formulation: (I) The monomer must be strongly cross-linking to ensure the formation of polymer nanostructures with high mechanical strength. This is important to prevent breakdown of the structure during the development process. (II) To reduce aberrations of the point spread functions, the refractive indices of the monomer and the glass should match. (III) The resin must not show absorption in the wavelength regions of the excitation and depletion lasers.
In the last few years, copolymerization of organic monomers and transition metal oxo clusters with polymerizable ligands by a variety of polymerization techniques yielded cluster-cross-linked polymers with improved thermal and mechanical properties.29 Thereby, the clusters can be incorporated in the organic matrix without loss of their structural integrity. Exchanging part of the polymerizable methacrylate ligands of the clusters by ligands with a different organic functionality results in bifunctional clusters. By incorporation of transition metal oxo clusters in organic matrices, the intrinsic properties of the cluster (e.g., magnetical, optical, electronical properties) are transferred to the polymer.30,31 Furthermore, the great number of polymerizable ligands on the cluster surface leads to strong cross-linking and enhanced mechanical properties. Additionally, the facile preparation of bifunctional clusters makes this class of compounds interesting for the preparation of functional structures. Clusters are typically obtained in a one-step reaction as crystalline products with high yield and purity, and easy-to-perform protocols for ligand exchange reactions exist.32 On the contrary, purely organic based monomers which are bifunctional and strongly cross-linking are typically prepared by multi step synthesis protocols and show low yields over all synthesis steps.
Herein, we report structuring of multiphoton and STED lithographically fabricated structures employing a metal oxo cluster based resin with thiol functionality. We use the methacrylate-substituted zirconium oxo cluster Zr4O2(OMc)12 (Zr4, Figure 1a),33 whose methacrylate ligands (OMc) were partially exchanged against 3-mercaptopropionate ligands to introduce a mercapto-group as second functionality. The methacrylate ligands participate in the polymerization reaction and link the cluster covalently to the acrylate network. The mercapto-substituted ligands serve as anchor points for further functionalization after polymer formation. To the best of our knowledge, resins containing bifunctionalized clusters have not been applied in STED lithography before.
We are aware that parts of the mercapto functionalized ligands participate in thiol–ene polymerization and the underlying chemistry was described before.34−36 However, unreacted mercapto functionalized ligands remain reactive on the surface of the structure for further reactions, as will be outlined below.
Results
Specifically, 13% and 24% of the methacrylate ligands were exchanged against 3-mercaptopropionate ligands by treatment of the cluster Zr4 with 1.5 or 3 equiv of 3-mercaptopropionic acid. The NMR spectra of all clusters of this work are shown in the Supporting Information (Figure S1 and Figure S2). It was shown before that exchange of carboxylate ligands of Zr4 proceeds with retention of the cluster core structure.32 Determining the proportion of exchanged ligands, however, is not possible by NMR spectroscopy. The ligands on the cluster surface undergo fast dynamic exchange,37 which results in a broadening of the NMR signals and makes an integration of the signals ambiguous. However, exact determination of the number of ligands is possible when the cluster core is destroyed by treatment with a strong acid after the exchange reaction (see Methods section) and the NMR signals of the corresponding acids are integrated (see Figure S2). The composition of the bifunctional clusters was thus determined as Zr4O2(OMc)10.4(OPrSH)1.6 (ZrSH1, OPrSH = 3-mercapto propionate, 13% of the ligands exchanged) and Zr4O2(OMc)9.1(OPrSH)2.9 (ZrSH2, 24% of the ligands exchanged).
A scheme of the STED lithography setup is presented in Figure 1b and explained in detail in ref (7). Woodpile photonic crystals and lines were fabricated to demonstrate the applicability of the new resin for multiphoton and STED lithography. Cluster-based resins were obtained by mixing dichloromethane (DCM) solutions of metal oxo clusters with the monomer pentaerythritol triacrylate (PETA) and subsequent evaporation of the solvent, as described in the Methods section. 0.25 wt % 7-diethylamino-3-thenoylcoumarin (DETC) was used as photoinitiator in all experiments (chemical structures are shown in Figure 1c). The resin PETA with 0.25 wt % DETC showed good performance in STED lithography before.6,7 In order to test the functionality of the mercapto groups after polymerization, two- and three-dimensional structures with 1 wt % of ZrSH1 were covalently functionalized with Alexa647-maleimide as sketched in Figure 2a. Control structures with cluster-free resin and with 1 wt % Zr4 doped resin (without mercapto groups) were also fabricated. All structures were incubated for 20 min in an aqueous Alexa647-maleimide solution and subsequently washed as described in the Methods section. Confocal laser scanning microscopy (LSM) images of all structures were recorded before and after incubation. Figure 2b shows a three-dimensional reconstructed image of a woodpile structure. The DETC fluorescence is represented in blue (Figure 2b, central panel) and the specifically bound Alexa647-maleimide is shown in red (Figure 2b, right panel). The main panel in Figure 2c depicts an LSM fluorescence image of a mercapto functionalized slice of a woodpile structure after incubation as top view, evidencing the attachment of the Alexa647-maleimide. The control structures written with pure PETA/DETC, that is, without any clusters, or with resist containing Zr4 clusters, which carry only methacrylate but no thiol groups, (Figure 2c, small panels) both did not show a significant fluorescence signal in the red channel after incubation. Statistics of the intensity of the fluorescence signal for the mercapto-functionalized structure and both negative controls are shown in Figure 2d. The gray bars represent the emission of DETC (green channel, excitation at 488 nm), which is present in the same concentration in all photoresists. The fluorescence signals are therefore the same for all three analyzed structures.
Figure 2.
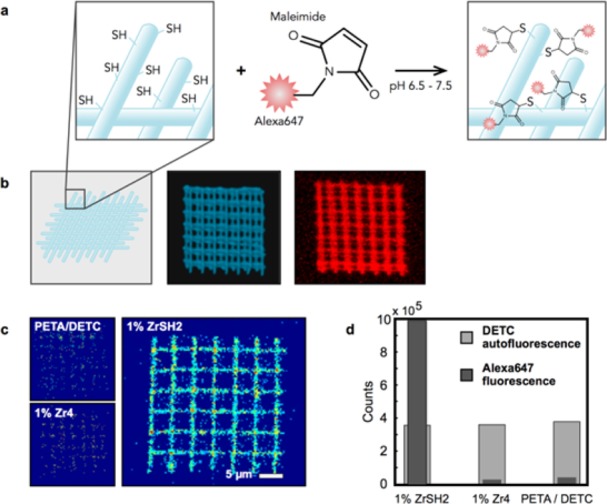
Functionality of SH bonds of Zr-clusters in copolymerized structures. (a) Sketch of the lithographically structured polymers and the incubation with 10 mM Alexa647-maleimide. (b) Confocal images of the woodpile structures. The central image represents the DETC fluorescence in the green channel of the microscope (excited at 488 nm). The right one shows the specific signal of Alexa647-maleimide taken with the red channel (excited at 647 nm). The photoresist used for structuring was composed of 1 wt % ZrSH2. (c) Confocal images of one layer of the three-dimensional woodpile structures. Three different photoresists were used: pure PETA with 0.25 wt % DETC as a negative probe and PETA/DETC doped with 1 wt % ZrSH2 and 1 wt % Zr4, respectively. The large confocal image shows the signal from the Alexa647-maleimide attached to the mercapto groups in the polymer. The upper small image shows unspecific binding of Alexa647-maleimid to a woodpile structure without any Zr cluster, the lower small image shows unspecific binding to acrylates containing 1 wt % Zr4 (LSM images were scaled identically). (d) Bar diagram showing the background corrected fluorescence signal in the green channel (DETC fluorescence) and the red channel (Alexa647).
Excitation at 647 nm (red channel of the LSM) shows that the fluorescence of the Alexa647 marker is increased 30-fold in the case of the mercapto functionalized structure containing 1 wt % ZrSH2 compared to the unreactive structures. The low residual signal of the unfunctionalized structures originates from unspecific binding of Alexa647-maleimide and is similar for all three samples.
Structures called “depletion patterns” were written to characterize the STED performance of the PETA/oxo cluster resists. Lines are written using varying excitation (at 780 nm) and depletion (at 532 nm) powers, whereas both the excitation and the STED PSFs were ordinarily shaped as shown in the top image of Figure 3a. The lines in one column of the depletion pattern are created with a constant excitation power while each line is written with different STED powers, which results in different line heights. In the case of the second column of the SEM image, the AFM profile is shown to the right of the SEM image. Within each column (constant excitation power), the depletion power was increased from 0–27 mW in 42 similar steps (bottom to top in Figure 3a). For left to right columns, the excitation power was increased, starting from 3.8 mW and increasing in 0.1 mW steps until an excitation power of 4.2 mW was reached (from left to right in Figure 3a). Depletion patterns were written for resins with concentrations of ZrSH1 or ZrSH2 ranging from 1–4 wt %. Figure 3b shows the results for the resists containing 1 wt % ZrSH1 as an example. Results for other compositions are shown in the Supporting Information. For all experiments, the first (bottom) and the last line (top) of the depletion curves were written without applying the depletion beam. Figure 3b shows line height profiles of depletion patterns determined from AFM scans. For instance, the orange stars in Figure 3b show the height of the lines as a function of ascending STED power. Up to 2 mW (taken in front of the objective lens), the line height is rapidly decreasing with increasing STED power. Lines are totally suppressed for STED intensities from 3 mW to 12 mW.
Figure 3.
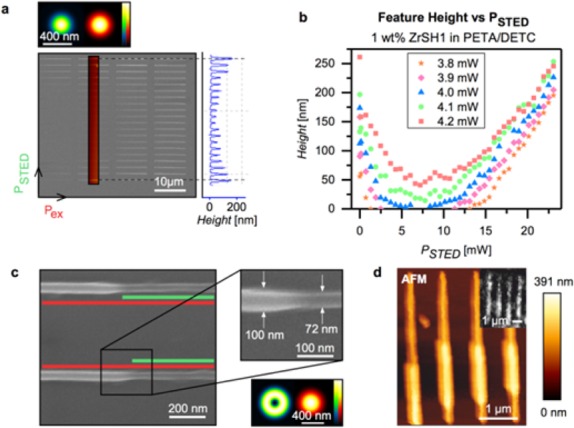
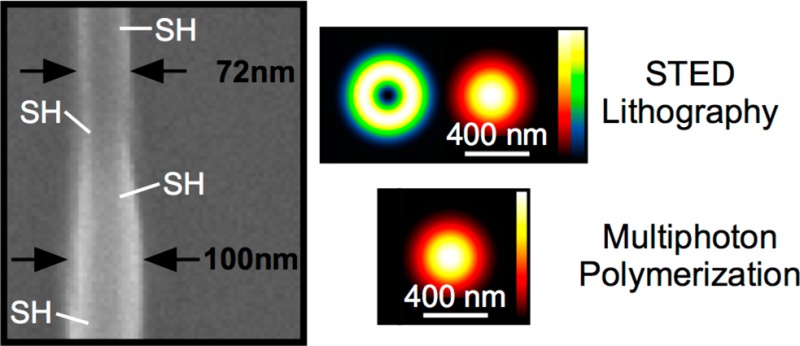
STED lithography experiments on cluster-doped photoresists. (a) SEM image and AFM line scan of a depletion test pattern written with ordinarily shaped STED PSFs (see inset). AFM profiles of these patterns allow for plotting the feature height versus the depletion power as a result of the STED induced inhibition of polymerization. (b) Height of lines for PETA with 0.25 wt % DETC doped with 1 wt % of ZrSH1 as a function of STED power for excitation powers from 3.8 to 4.2 mW as indicated. (c) SEM image of lines with ZrSH2 clusters. The minimum lateral feature size obtained by introducing a donut shaped STED beam (see inset) is 72 nm. The line was written with an excitation power of 3.2 mW (at 780 nm) and a depletion power of 7.8 mW (at 532 nm). (d) Multiphoton and STED lithographically fabricated lines using a photoresist doped with 1 wt % ZrSH1. The depletion beam was partially blocked while writing the features. Main image: AFM. Inset: fluorescence image of the lines after labeling with Alexa647-maleimide.
From 13 mW onward, lines reappear, however, they are now written due to either Anti-Stokes or two-photon absorption of 532 nm light from the STED beam.21,38 This region of STED intensity needs to be avoided. The depletion curves for the resin containing 3 wt % ZrSH2 are shown in Figure S3a and b shows a comparison of resins containing 1 wt % ZrSH1 and 1 wt % of ZrSH2.
From the depletion experiments, we determine a doping of the photoresist with 4 wt % of clusters (Zr4 and ZrSH1) as the maximum concentration which allows for complete structure suppression in the observed power range from 3.8 to 4.2 mW. The data indicate that higher cluster proportions increase residual absorption of the STED laser, which counteracts depletion. Similar trends were determined using clusters differing in the proportion of mercaptopropionate ligands (ZrSH1 and ZrSH2, Figure S3). The data show that depletion by STED depends on the amount of clusters in the polymers and also on the number of mercapto groups per cluster. The resin containing 1 wt % of ZrSH1 showed a lower parasitic polymerization due to large STED powers than the resin containing ZrSH2. Representative depletion curves for resins with 3 wt % ZrSH2 are shown in Figure S3 in the Supporting Information.
Next, experiments for characterizing the lateral restriction of the polymer structure were carried out. Lines were written in a resin containing 2.5 wt % ZrSH2 with an excitation power of 3.2 mW.
The green depletion laser with a power of 7.8 mW was switched on for a short sequence (see Figure 3c). The writing volume was thereby restricted by the donut-shaped PSF of the depletion laser. By switching on the depletion laser, the line diameter decreased from 100 to 72 nm, which corresponds to a restriction of 28%.
Figure 3d displays AFM images of lines written using a photoresist with 1 wt % of ZrSH1. While structuring, the donut-shaped depletion beam has been partially blocked. In the case where the STED beam was on, the lines became thinner. The inset shows the fluorescence of the STED lithographically written lines after covalent attachment of Alexa647-maleimide proving that the thiol functionalization is active also in the STED lithographically written structures.
Conclusions
In conclusion, a successful fabrication of nanostructures by combining STED lithography with functional polymers has been demonstrated. Using metal oxo clusters, mercapto-functionalized structures with feature sizes of 72 nm were realized. The reactivity of the thiol groups of the clusters in the polymerized structures has been demonstrated by incubation with Alexa647-maleimide. The combination of STED lithography with resists containing a second functional group offers a promising possibility for numerous applications, for example a covalent attachment of proteins, DNA or antigenes for cell behavior studies or the precise tuning of the physical and chemical surface properties of 2D and 3D structures.
Methods
Lithography Setup
The two-photon polymerization starters were excited with 780 nm ultrashort laser pulses (FemtoRay780, 50 MHz repetition rate, 100 fs pulse duration, Menlo Systems GmbH, Germany) and were locally depleted in the outer rim of the point spread function with a depletion beam (532 nm, continuous wave, Verdi-V5, Coherent, U.S.A.). The 532 nm depletion beam was shaped into a donut form using a 2π spiral phase mask (RPC Photonics, U.S.A.) and a λ/4 plate converting the depletion beam into a circularly polarized beam. Both beams were focused through an oil immersion objective lens (Zeiss α-plan Apochromat, 100×, numerical aperture NA = 1.46). Power adjustment of the excitation beam was provided by an acousto-optic modulator (Q1133, Isomet, U.S.A.). An avalanche photo diode (APD-SPCM-AQRH, PerkinElmer Optoelectronic Inc., U.S.A.) was used for aligning the foci. A three axes piezo stage (P-562.3CD, Physik Instrumente PI, Germany) with a bidirectional positioning accuracy of 2/2/4 nm and a travel range of 200 × 200 × 200 μm, is used for sample motion. The high-precision stage was mounted on top of a coarse x–y motor stage (M-686.D64, Physik Instrumente PI, Germany) with a travel range of 25 × 25 mm. The stages were driven in closed loop with two controllers (E710.3CD and C-867.260, both from Physik Instrumente PI, Germany). For sample positioning, recording of images and controlling the writing process, a LabView (LabView 2011, National Instruments Corporation, U.S.A.) program was used.
Confocal Laser Scanning Microscopy
Specific binding between Alexa647-maleimide and a woodpile structure was quantified by a confocal laser scanning microscope LSM 700 (Carl Zeiss AG, Germany). Images were taken using a 63×, NA = 1.4 oil DIC Plan-Apochromat. Generally, image sizes were 101.6 × 101.6 μm (line rate of 0.03 ms) by 3.6 μm (22 z-stacks). Samples were illuminated using 488 and 647 nm (Laser modules LSM 700) and after spectral filtering, the fluorescence was imaged on photomultiplier tubes (using the green or red channel of the LSM 700, respectively). The Zen data processing software (Carl Zeiss AG, Germany) was used for image visualization; additionally, images were analyzed using MATLAB (The MathWorks GmbH, Germany). For the analysis, the images were background corrected and fluorescence signals were averaged.
Combined Fluorescence Microscopy and Atomic Force Microscopy
The sample was sealed by a home-built camber and rinsed with phosphate-buffered saline (PBS). AFM measurements were performed using a NanoWizard 3 (JPK Instruments AG, Germany) system mounted on an Axiovert 200 inverted microscope (Carl Zeiss AG, Germany). The microscope is equipped with a 100× NA = 1.46 oil-immersion Plan-Apochromat TIRFM objective (Olympus, Japan) and a 20× NA = 0.8 Plan-Apochromat objective (Carl Zeiss AG, Germany). Samples were illuminated in epifluorescence configuration via the epiport using 488 nm (250 mW) and 647 nm (250 mW) light from diode lasers (Toptica Photonics, Germany), or 532 nm light from a solid state laser (Millennia X, Spectra Physics, U.S.A.), with intensities of 3–10 kW/cm2. After appropriate filtering, emitted signals were imaged on a back-illuminated, TE-cooled CCD-camera (Andor iXon Du-897, UK). Acousto-optical modulators (1205C, Isomet, U.S.A.) were used for the precise control of the illumination timing. Timing protocols were generated by an in-house program package implemented in LabView (LabView, National Instruments Corporation, U.S.A.). Illumination times were adjusted to values between 1 and 5 ms. The sample surface was first imaged with the fluorescence microscope to determine an appropriate site for the AFM measurements. Topographical images were recorded in Quantitative Imaging mode at room temperature in liquid (PBS) at a resolution of 128 × 128 pixels. The maximum force determined by the vertical deflection of the cantilever was set to 300 pN. Force distance cycles (scan rates) were controlled by the z length (250 nm), extension time (10 ms) and retraction time (50 ms). We used uncoated silicon cantilevers (MSNL-10, Bruker Corporation, U.S.A.) with a nominal spring constant in the range of 0.01–0.03 N/m. The spring constant for each cantilever was calibrated using the thermal noise method. JPK data processing (JPK Instrument, Germany) software was used for image analysis.39−42 The height, adhesion and slope of the force curve were collected simultaneously in both trace and retrace directions. Height images were line-fitted as required. Isolated scan lines were occasionally removed.
NMR Spectroscopy
Solution NMR spectra were recorded on a Bruker AVANCE 250 (250.13 MHz {1H}, 62.86 MHz {13C}). Gas-tight Young tubes were used for all experiments. CD2Cl2 was purchased from Euriso-Top and degassed by freeze–pump–thaw cycles.
IR Spectroscopy
Solid state ATR-IR spectra were recorded on a PerkinElmer Spectrum 400 FT-IR spectrometer equipped with a KBr window for MIR and a polyethylene window for FIR. A total of 128 scans were averaged for MIR and 256 scans were averaged for FIR measurements.
Photoresist
The used monomer was pentaerythritol triacrylate (PETA) (Sigma-Aldrich Co., U.S.A.) including 300–400 ppm monomethyl ether hydroquinone and 0.25 wt % 7-diethylamino-3-thenoylcoumarin (DETC) (Acros Organics, Belgium) as initiator. Zr-clusters were mixed into the resist as described below.
Preparation of the Clusters
All operations involving the preparation of the clusters and the ligand exchange reactions were performed in nitrogen atmosphere using standard Schlenk techniques. Hexane and toluene were dried over Na/benzophenone prior to use. DCM was distilled over CaH2. Methacrylic acid (99%, Sigma-Aldrich Co., U.S.A.) and 3-mercaptopropionic acid (99%, Sigma-Aldrich Co, U.S.A.) were freshly distilled over P2O5 prior to use. Zr(OPr)4 (70 wt % in propanol) was purchased from Sigma-Aldrich and used as received. Zr4O2(OMc)12 (Zr4) was prepared as previously reported.33
Ligand Exchange Reactions
Ligand exchange reactions were performed according to the procedure reported in ref (32). 3-Mercaptopropionic acid, 18.4 μL (0.22 mmol, 1.5 eq. in the case of ZrSH1) or 36.9 μL (0.44 mmol, 3 eq. in the case of ZrSH2), was added dropwise to a solution of 200 mg (0.14 mmol, 1 equiv) of Zr4 in 5 mL of CH2Cl2 and stirred for 30 min. After removing the solvent, the residue was washed multiple times with toluene, redissolved in 0.5 mL of CH2Cl2 (DCM) and precipitated with 50 mL of hexane. The white precipitate was separated, washed again three times with toluene, and dried under vacuum.
ZrSH1
1H NMR (250 MHz, 25 °C, CD2Cl2): δ = 1.28 (SH), 1.87 (CH3), 2.65 (—CH2CH2SH), 5.55 (=CH2), 6.17 (=CH2). 13C NMR (250 MHz, 25 °C, CD2Cl2): δ = 18.28 (—CH3), 20.39 (CH2—SH), 41.01 (CH2), 127.15 (=CH2), 138.87 (CH3—C=CH2) ppm. IR (ATR): ν = 2958 (w), 2924 (w), 2573 (w, SH), 1691 (m), 1638 (m), 1585 (w), 1521 (s), 1412 (s), 1371 (m), 1310 (m), 1284 (w), 1242 (s), 1102 (w), 1001 (s), 940 (s), 876 (s), 827 (s), 797 (w), 729 (w), 661 (w), 601 (s), 499 (s) cm–1.
ZrSH2
1H NMR (250 MHz, CD2Cl2): δ = 1.29 (SH), 1.87 (CH3), 2.65 (—CH2CH2SH), 5.56 (=CH2), 6.18 (=CH2). 13C NMR (250 MHz, 25 °C, CD2Cl2): δ = 18.28 (—CH3), 20.39 (CH2—SH), 41.01 (CH2), 127.15 (=CH2), 138.87 (CH3—C=CH2) ppm. IR (ATR): ν = 3932 (b), 2566 (w, SH), 1694 (s), 1589 (w), 1532 (s), 1419 (s), 1310 (s), 1283 (s), 1238 (s), 1204 (w), 1166 (m), 1099 (m), 1001 (s), 940 (s), 870 (s), 827 (s), 801 (w), 657 (w), 593 (w), 495 (w) cm–1.
Determination of the Number of Exchanged Ligands
A total of 50 mg of ZrSH1 or ZrSH2 was suspended in D2O and 10 μL of DCl was added. The solution was sonicated for 60 min before recording 1H NMR spectra. The ratio of methacrylic acid and 3-mercaptopropionic acid was determined according to the ratio of the integrals of the methyl group of methacrylic acid (1.77 ppm, 3H) and the —CH2CH2SH signal of mercaptopropionic acid (2.60 ppm, 4H) and was found to be 1:0.2 for ZrSH1 and 1:0.42 for ZrSH2. Thus, 13% of the methacrylate ligands were exchanged against mercapto propionate ligands in ZrSH1 and 24% in ZrSH2. The composition of ZrSH1 was therefore Zr4O2(OMc)10.4(OPrSH)1.6 and that of ZrSH2 Zr4O2(OMc)9.1(OPrSH)2.9.
Doping of the Photoresist with Clusters
Different proportions of Zr4, ZrSH1 or ZrSH2 dissolved in CH2Cl2 (100 μL of the solvent per 5 mg Zr-cluster) were used. The maximum amount of clusters, which can be dissolved in the photoresist was 5 wt %. However, complete depletion of the formation of structures by STED can be achieved only up to a 4 wt % proportion of clusters in the resin.
For structuring, 10 μL of the resist were pipetted onto untreated microscope coverslips (Menzel Gläser, Germany) with a thickness of ca. 0.17 mm. The solvent was removed under vacuum at room temperature for about 20 min. In the development process, DCM was used as solvent.
Maleimide Functionalization
The specific binding efficiency was assessed by using Alexa647-maleimide. The stock solution was dissolved with distilled water to obtain a final 10 mM concentration. The maleimide side of the linker binds covalently to accessible mercapto groups in the polymer structure (Figure 2a). The samples were incubated for 20 min and subsequently washed three times.
Acknowledgments
We would like to thank Heidi Piglmayer-Brezina for taking the SEM images and Habed Habibzadeh, Bernhard Fragner and Alfred Nimmervoll for technical support. The work was funded by the Austrian Science Fund (FWF), projects P 26461-N20 (3DS-PNA), P22915 and P 26337-B21. Funding from the European Research Council (ERC Starting Grant 257158 “Active NP”) is gratefully acknowledged.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.5b05863.
The 250 MHz 1H NMR spectra of Zr4, ZrSH1, and ZrSH2; depletion curves. (PDF)
Author Contributions
‡ The first two authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Hell S. W.; Wichmann J. Breaking the Diffraction Resolution Limit by Stimulated Emission: Stimulated-Emission-Depletion Fluorescence Microscopy. Opt. Lett. 1994, 19, 780–782. 10.1364/OL.19.000780. [DOI] [PubMed] [Google Scholar]
- Klar T. A.; Hell S. W. Subdiffraction Resolution In Far-Field Fluorescence Microscopy. Opt. Lett. 1999, 24, 954–956. 10.1364/OL.24.000954. [DOI] [PubMed] [Google Scholar]
- Klar T. A.; Jakobs S.; Dyba M.; Egner A.; Hell S. W. Fluorescence Microscopy with Diffraction Resolution Barrier Broken by Stimulated Emission. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 8206–8210. 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittweger E.; Han K. Y.; Irvine S. E.; Eggeling C.; Hell S. W. STED Microscopy Reveals Crystal Colour Centres with Nanometric Resolution. Nat. Photonics 2009, 3, 144–147. 10.1038/nphoton.2009.2. [DOI] [Google Scholar]
- Maruo S.; Nakamura O.; Kawata S. Three-Dimensional Microfabrication with Two-Photon-Absorbed Photopolymerization. Opt. Lett. 1997, 22, 132–134. 10.1364/OL.22.000132. [DOI] [PubMed] [Google Scholar]
- Wolf T. J.; Fischer J.; Wegener M.; Unterreiner A. N. Pump-Probe Spectroscopy on Photoinitiators for Stimulated-Emission-Depletion Optical Lithography. Opt. Lett. 2011, 36, 3188–3190. 10.1364/OL.36.003188. [DOI] [PubMed] [Google Scholar]
- Wollhofen R.; Katzmann J.; Hrelescu C.; Jacak J.; Klar T. A. 120 nm Resolution and 55 nm Structure Size in STED-Lithography. Opt. Express 2013, 21, 10831–10840. 10.1364/OE.21.010831. [DOI] [PubMed] [Google Scholar]
- Li L.; Gattass R. R.; Gershgoren E.; Hwang H.; Fourkas J. T. Achieving Lambda/20 Resolution by One-Color Initiation and Deactivation of Polymerization. Science 2009, 324, 910–913. 10.1126/science.1168996. [DOI] [PubMed] [Google Scholar]
- Scott T. F.; Kowalski B. A.; Sullivan A. C.; Bowman C. N.; McLeod R. R. Two-Color Single-Photon Photoinitiation and Photoinhibition for Subdiffraction Photolithography. Science 2009, 324, 913–917. 10.1126/science.1167610. [DOI] [PubMed] [Google Scholar]
- Harke B.; Bianchini P.; Brandi F.; Diaspro A. Photopolymerization Inhibition Dynamics for Sub-Diffraction Direct Laser Writing Lithography. ChemPhysChem 2012, 13, 1429–1434. 10.1002/cphc.201200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J.; von Freymann G.; Wegener M. The Materials Challenge in Diffraction-Unlimited Direct-Laser-Writing Optical Lithography. Adv. Mater. 2010, 22, 3578–3582. 10.1002/adma.201000892. [DOI] [PubMed] [Google Scholar]
- Gan Z.; Cao Y.; Evans R. A.; Gu M. Three-Dimensional Deep Sub-Diffraction Optical Beam Lithography with 9 nm Feature Size. Nat. Commun. 2013, 4, 2061. 10.1038/ncomms3061. [DOI] [PubMed] [Google Scholar]
- Klein F.; Richter B.; Striebel T.; Franz C. M.; von Freymann G.; Wegener M.; Bastmeyer M. Two-Component Polymer Scaffolds for Controlled Three-Dimensional Cell Culture. Adv. Mater. 2011, 23, 1341–1345. 10.1002/adma.201004060. [DOI] [PubMed] [Google Scholar]
- Scheiwe A. C.; Frank S. C.; Autenrieth T. J.; Bastmeyer M.; Wegener M. Subcellular Stretch-Induced Cytoskeletal Response of Single Fibroblasts Within 3D Designer Scaffolds. Biomaterials 2015, 44, 186–194. 10.1016/j.biomaterials.2014.12.018. [DOI] [PubMed] [Google Scholar]
- Wickberg A.; Mueller J. B.; Mange Y. J.; Fischer J.; Nann T.; Wegener M. Three-Dimensional Micro-Printing of Temperature Sensors Based on Up-Conversion Luminescence. Appl. Phys. Lett. 2015, 106, 133103. 10.1063/1.4916222. [DOI] [Google Scholar]
- Gu M.; Li X.; Cao Y. Optical Storage Arrays: A Perspective for Future Big Data Storage. Light. Light: Sci. Appl. 2014, 3, e177. 10.1038/lsa.2014.58. [DOI] [Google Scholar]
- Wiesbauer M.; Wollhofen R.; Vasic B.; Schilcher K.; Jacak J.; Klar T. A. Nano-Anchors with Single Protein Capacity Produced with STED Lithography. Nano Lett. 2013, 13, 5672–5678. 10.1021/nl4033523. [DOI] [PubMed] [Google Scholar]
- Wolfesberger C.; Wollhofen R.; Buchegger B.; Jacak J.; Klar T. A. Streptavidin Functionalized Polymer Nanodots Fabricated by Visible Light Lithography. J. Nanobiotechnol. 2015, 13, 27. 10.1186/s12951-015-0084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J.; Wegener M. Three-Dimensional Optical Laser Lithography Beyond the Diffraction Limit. Laser Phot. Rev. 2013, 7, 22–44. 10.1002/lpor.201100046. [DOI] [Google Scholar]
- Fischer J.; Mueller J. B.; Quick A. S.; Kaschke J.; Barner-Kowollik C.; Wegener M. Exploring the Mechanisms in STED-Enhanced Direct Laser Writing. Adv. Opt. Mater. 2015, 3, 221–232. 10.1002/adom.201400413. [DOI] [Google Scholar]
- Mueller J. B.; Fischer J.; Mayer F.; Kadic M.; Wegener M. Polymerization Kinetics in Three-Dimensional Direct Laser Writing. Adv. Mater. 2014, 26, 6566–6571. 10.1002/adma.201402366. [DOI] [PubMed] [Google Scholar]
- Terzaki K.; Vasilantonakis N.; Gaidukeviciute A.; Reinhardt C.; Fotakis C.; Vamvakaki M.; Farsari M. 3D Conducting Nanostructures Fabricated Using Direct Laser Writing. Opt. Mater. Express 2011, 1 (4), 586–597. 10.1364/OME.1.000586. [DOI] [Google Scholar]
- Vasilantonakis N.; Terzaki K.; Sakellari I.; Purlys V.; Gray D.; Soukoulis C. M.; Vamvakaki M.; Kafesaki M.; Farsari M. Three-Dimensional Metallic Photonic Crystals with Optical Bandgaps. Adv. Mater. 2012, 24, 1101–1105. 10.1002/adma.201104778. [DOI] [PubMed] [Google Scholar]
- Malinauskas M.; Baltriukiene D.; Kraniauskas A.; Danilevicius P.; Jarasiene R.; Sirmenis R.; Zukauskas A.; Balciunas E.; Purlys V.; Gadonas R.; Bukelskiene V.; Sirvydis V.; Piskarskas A. In Vitro and In Vivo Biocompatibility Study on Laser 3D Microstructurable Polymers. Appl. Phys. A: Mater. Sci. Process. 2012, 108, 751–759. 10.1007/s00339-012-6965-8. [DOI] [Google Scholar]
- Kumpfmueller J.; Stadlmann K.; Li Z.; Satzinger V.; Stampfl J.; Liska R. Two-Photon-Induced Thiol-Ene Polymerization as a Fabrication Tool for Flexible Optical Waveguides. Des. Monomers Polym. 2014, 17, 390–400. 10.1080/15685551.2013.840515. [DOI] [Google Scholar]
- Ovsianikov A.; Mironov V.; Stampfl J.; Liska R. Engineering 3D Cell-Culture Matrices: Multi-Photon Processing Technologies for Biological and Tissue Engineering Applications. Expert Rev. Med. Devices 2012, 9, 613–633. 10.1586/erd.12.48. [DOI] [PubMed] [Google Scholar]
- Selimovic S.; Oh J.; Bae H.; Dokmeci M.; Khademhosseini A. Microscale Strategies for Generating Cell-Encapsulating Hydrogels. Polymers (Basel, Switz.) 2012, 4, 1554–1579. 10.3390/polym4031554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick A. S.; Fischer J.; Richter B.; Pauloehrl T.; Trouillet V.; Wegener M.; Barner-Kowollik C. Preparation of Reactive Three-Dimensional Microstructures via Direct Laser Writing and Thiol-Ene Chemistry. Macromol. Rapid Commun. 2013, 34, 335–340. 10.1002/marc.201200796. [DOI] [PubMed] [Google Scholar]
- Schubert U. Polymers Reinforced by Covalently Bonded Inorganic Clusters. Chem. Mater. 2001, 13, 3487–3494. 10.1021/cm001258r. [DOI] [Google Scholar]
- Schubert U. Organofunctional Metal Oxide Clusters as Building Blocks for Inorganic-Organic Hybrid Materials. J. Sol-Gel Sci. Technol. 2004, 31, 19–24. 10.1023/B:JSST.0000047954.70820.dd. [DOI] [Google Scholar]
- Schubert U. Inorganic-Organic Hybrid Polymers Based on Surface-Modified Metal Oxide Clusters. Macromol. Symp. 2008, 267, 1–8. 10.1002/masy.200850701. [DOI] [Google Scholar]
- Kreutzer J.; Puchberger M.; Artner C.; Schubert U. Retention of the Cluster Core Structure During Ligand Exchange Reactions of Carboxylato-Substituted Metal Oxo Clusters. Eur. J. Inorg. Chem. 2015, 2015, 2145–2151. 10.1002/ejic.201403209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickelbick G.; Schubert U. Oxozirconium Methacrylate Clusters: Zr6(OH)4O4(OMc)12 and Zr4O2(OMc)12 (OMc = Methacrylate). Chem. Ber. 1997, 130, 473–478. 10.1002/cber.19971300406. [DOI] [Google Scholar]
- Bhagat S. D.; Filho E. B.; Da Silva; Stiegman A. E. High Refractive Index Polymer Composites Synthesized by Cross-Linking of Oxozirconium Clusters Through Thiol-Ene Polymerization. Macromol. Mater. Eng. 2015, 300, 580–585. 10.1002/mame.201500009. [DOI] [Google Scholar]
- Sangermano M.; Gross S.; Priola A.; Rizza G.; Sada C. Thiol-Ene Hybrid Organic/Inorganic Nanostructured Coatings Based on Thiol-Functionalized Zirconium Oxo Clusters. Macromol. Chem. Phys. 2007, 208, 2560–2568. 10.1002/macp.200700323. [DOI] [Google Scholar]
- Maggini S.; Cappelletto E.; Di Maggio R. High Temperature Resistant Silane/Zirconium-Oxocluster Hybrid Copolymers Containing “Free” Thiol/Ene Functionalities in the Polymer Matrix. J. Appl. Polym. Sci. 2013, 127, 2435–2441. 10.1002/app.37940. [DOI] [Google Scholar]
- Walther P.; Puchberger M.; Kogler F. R.; Schwarz K.; Schubert U. Ligand Dynamics on the Surface of Zirconium Oxo Clusters. Phys. Chem. Chem. Phys. 2009, 11, 3640–3647. 10.1039/b820731c. [DOI] [PubMed] [Google Scholar]
- Thiel M.; Fischer J.; von Freymann G.; Wegener M. Direct Laser Writing of Three Dimensional Submicron Structures Using a Continuous-Wave Laser at 532 nm. Appl. Phys. Lett. 2010, 97, 221102. 10.1063/1.3521464. [DOI] [Google Scholar]
- Hutter J. L.; Bechhoefer J. Calibration of Atomic-Force Microscope Tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. 10.1063/1.1143970. [DOI] [Google Scholar]
- Stark R. W.; Drobek T.; Heckl W. M. Thermomechanical Noise of a Free V-Shaped Cantilever for Atomic-Force Microscopy. Ultramicroscopy 2001, 86, 207–215. 10.1016/S0304-3991(00)00077-2. [DOI] [PubMed] [Google Scholar]
- Lévy R.; Maaloum M. Measuring the Spring Constant of Atomic Force Microscope Cantilevers: Thermal Fluctuations and Other Methods. Nanotechnology 2002, 13, 33–37. 10.1088/0957-4484/13/1/307. [DOI] [Google Scholar]
- Kim M.-S.; Choi J.-H.; Kim J.-H.; Park Y.-K. Accurate Determination of Spring Constant of Atomic Force Microscope Cantilevers and Comparison with other Methods. Measurement 2010, 43, 520–526. 10.1016/j.measurement.2009.12.020. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


