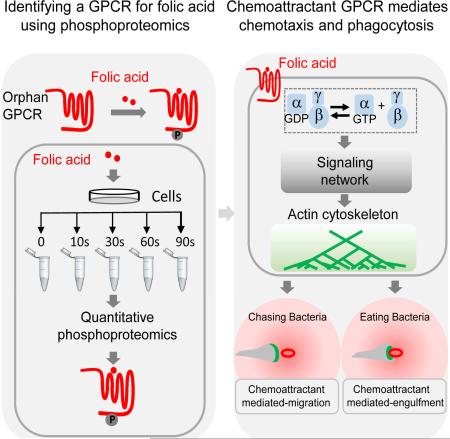
SUMMARY
Eukaryotic phagocytes search and destroy invading microorganisms via chemotaxis and phagocytosis. The social amoeba Dictyostelium discoideum is a professional phagocyte that chases bacteria through chemotaxis and engulfs them as food via phagocytosis. G-protein-coupled receptors (GPCRs) are known for detecting chemoattractants and directing cell migration, but their roles in phagocytosis are not clear. Here, we developed a quantitative phosphoproteomic technique to discover signaling components. Using this approach, we discovered the long-sought-after folic acid receptor, fAR1, in D. discoideum. We showed that the seven transmembrane receptor fAR1 is required for folic acid-mediated signaling events. Significantly, we discovered that fAR1 is essential for both chemotaxis and phagocytosis of bacteria, thereby representing a chemoattractant GPCR that mediates not only chasing but also ingesting bacteria. We revealed that a phagocyte is able to internalize particles via chemoattractant-mediated engulfment process. We propose that mammalian phagocytes may also use this mechanism to engulf and ingest bacterial pathogens.
Graphical Abstract
INTRODUCTION
Highly motile amoeboid cells, such as neutrophils, macrophages and Dictyostelium discoideum are professional phagocytes that track down bacteria by chemotaxis and capture and destroy them via phagocytosis (Freeman and Grinstein, 2014; Isik et al., 2008; Ravichandran and Lorenz, 2007). Both chemotaxis and phagocytosis require two processes: first, detection of external cues (chemoattractants or phagocytic targets), and second, activation of a signaling network that controls the actin cytoskeleton to generate pseudopods for cell migration or engulfment of the targets (Andrew and Insall, 2007; Flannagan et al., 2012; Insall and Machesky, 2009; Rougerie et al., 2013). G-protein-coupled receptors (GPCRs) for chemokines, detect chemoattractants generated by bacteria, and activate signaling pathways to regulate actin polymerization for cell migration towards the bacteria. After catching them, various receptors, such as Fcγ and C3a receptors, bind the targets and activate signaling pathways to generate actin polymerization around the bacteria to form phagocytic cups. While GPCRs are essential for chemotaxis, their roles in phagocytosis are not clear.
As an established model for the study of chemotaxis and phagocytosis, D. discodieum cells areknown to chase bacteria via chemotaxis to folic acid and ingest them as food through phagocytosis (Isik et al., 2008; Manahan et al., 2004). It is also acknowledged that chemotaxis towards secreted cAMP is essential for the development of D. discoideum into fruiting bodies under starvation conditions (Van Haastert and Devreotes, 2004; Veltman et al., 2014). The cAMP receptor, cAR1, was the first chemoattractant GPCR discovered in eukaryotic cells (Klein et al., 1988). However, although folic acid was shown to be a chemoattractant for D. discoideum to seek bacteria more than 40 years ago (Pan et al., 1972), a folic-acid receptor(s) has not been identified. Chemoattractant GPCRs are recognized to activate downstream signaling events, including phosphorylation or de-phosphorylation of key components in the signaling network (Cai et al., 2010; Charest et al., 2010; Chen et al., 2007; Kamimura et al., 2008; Klein et al., 1988; Swaney et al., 2010; Veltman et al., 2008; Xiao et al., 2010; Yi et al., 2014). A recentlydeveloped phosphoproteomic technique reveals the temporal dynamics of site-specificphosphorylation events in vivo upon receptor activation (Olsen et al., 2006). Significantly, massspectrometry (MS) based quantitative phosphoproteomics generates large-scale data sets, whichprovide a global view of signaling networks (Xiao et al., 2010; Yi et al., 2014).
Here we developed a phosphoproteomic approach for discovering missing components in a particular signaling network by combining the information from phosphoproteomic data sets with an annotated genome database. We used this approach to identify an orphan receptor as the long-sought-after folic acid receptor, fAR1. Our studies demonstrate that fAR1 controls both chemotaxis towards folic acid and phagocytosis of bacterial particles. Moreover, our data indicate that fAR1-mediated signaling events promote not only chemoattractant-mediated cell migration in a folic acid gradient but also chemoattractant-mediated engulfment of folic acid-coated particles.
RESULTS
Quantitative Phosphoproteomic Analysis Reveals Dynamic Phosphorylation upon Chemoattractant Stimulation
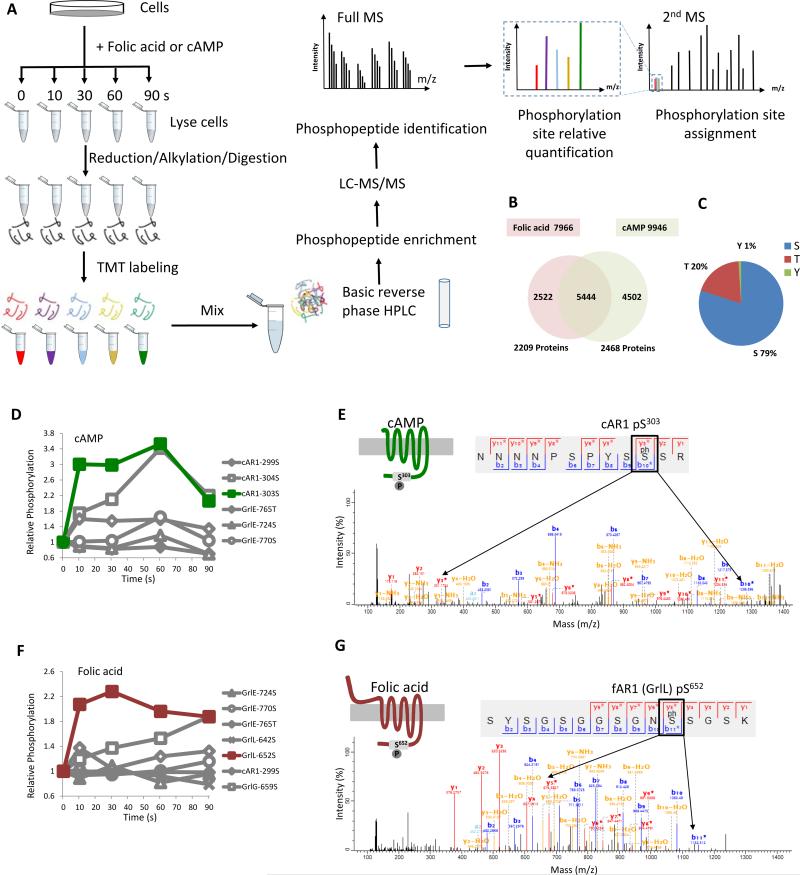
Our phosphoproteomic approach combines tandem mass tag (TMT) chemical labeling for time-point specific quantification, basic reverse-phase high-performance liquid chromatography (HPLC) and immobilized metal affinity chromatography (Fe-NTA) for phosphopeptide enrichment, and multistage MS for identification of phosphopeptides and quantification of phosphorylation sites (Eyrich et al., 2011) (Figure 1A). First, D. discoideum were stimulated with either cAMP or folic acid. Protein samples were collected at various time points, reduced, alkylated and enzymatically digested. The resulting peptides from each time point were then chemically linked to distinct TMT labeling reagents of identical mass. The TMT-labeled peptides from all time points were then mixed and separated into 12 fractions by basic reverse-phase HPLC, followed by enrichment of phosphopeptides on Fe-NTA columns. For phosphopeptide and phosphorylation site identification, each fraction was analyzed by online liquid chromatography (LC) MS/MS using an LTQ-Orbitrap Fusion mass spectrometer. Phosphopeptides of identical sequence but carrying different TMT tags were eluted as a single characteristic peak. Upon fragmentation, the position of the phosphorylated residue was determined by fragment ions, and each TMT tag releases a unique reporter ion, the intensity of which reflects the relative abundance of phosphorylation sites at each time point. Quantification of phosphorylation sites was processed and analyzed by Proteome Discoverer and MaxQuant software (Cox and Mann, 2008).
Figure 1. Ligand induced GPCR phosphorylation detected by quantitative mass spectrometry.
A. Experimental workflow for TMT-based quantitative phosphoproteomics of D. discoideum cells. Vegetative D.discoideum wild-type cells were treated with 100 μM folic acid or differentiated cells with 50 μM cAMP. At 0, 10, 30, 60 and 90 s, stimulated cells were harvested and lysed for reduction, alkylation and trypsin digestion. The peptides from each time point were specifically labeled by a single TMT labeling reagent. The labeled peptides were mixed and separated by basic reverse phase HPLC into 12 fractions. Phosphopeptides from each fraction were enriched by immobilized metal affinity chromatography and subjected to mass spectrometry for LC/MS/MS. Mass spectrometry data were analyzed and the dynamics of individual phosphorylation sites were determined. B. Number of phosphorylation sites and phosphorylated proteins detected by mass spectrometry under folic acid or cAMP stimulation. C. The phosphorylation sites detected were located on Ser, Thr and Tyr residues. D. cAMP induced cAR1 Ser303 and Ser304 phosphorylation. Six GPCR phosphorylation sites were detected, but only cAR1 Ser303 and Ser304 showed an increase in phosphorylation. E. MS/MS sequencing spectrum of the phospho-Ser303 peptide in cAR1. F. Folic acid induced an increase in GrlL Ser652 and GrlG Ser659 phosphorylation. Seven GPCR phosphorylation sites were detected, but only GrlL Ser652 and GrlG Ser659 showed an increase in phosphorylation. G. MS/MS sequencing spectrum of the phospho-Ser652 peptide in fAR1 (GrlL). Abbreviation for amino acids: S, Ser; T, Thr; Y, Tyr; N, Asn; P, Pro; R, Arg; G, Gly; K, Lys; ph S, phosphorylated Ser. See also Figure S1.
A total of 9946 phosphorylation sites corresponding to 2468 proteins were identified from cAMP-stimulated cells, and 7966 phosphorylation sites corresponding to 2209 proteins were identified from folic acid-stimulated cells (Figure 1B). Among those phosphopeptides, 79%, 20% and 1% were phosphorylated at serine (Ser), threonine (Thr) and tyrosine residues (Tyr) respectively (Figure 1C). This data set provided an opportunity for us to identify components in the cAMP and folic acid signaling networks based on phosphorylation sites that dynamically change in response to stimulation.
Folic-Acid-Induced Phosphorylation of GPCR Candidates
Previous studies indicated that folic-acid signaling functions through a GPCR pathway because downstream responses required both the heterotrimeric G-protein subunits Gα4 and Gβ (Hadwiger and Firtel, 1992; Hadwiger et al., 1994; Wu et al., 1995). Because chemoattractant GPCRs are typically phosphorylated at Ser or Thr residues on their C-terminal, cytoplasmic tails upon ligand stimulation (Caterina et al., 1995; Sibley et al., 1987), we used the D. discoideum genome data as a reference and searched the relevant global phosphoproteomic data for putative seven-transmembrane proteins that contain chemoattractant-induced phosphorylation at Ser or Thr within their C-terminal domains (Basu et al., 2013). To evaluate the sensitivity and specificity of this approach, we first searched for cAMP receptors using the data from cAMP-induced cells (Figure 1D). By searching the annotated genome of D. discoideum, we identified two seven-transmembrane proteins, including the cAMP receptor cAR1 and GrlE (GABA receptor like E), each of which yielded three C-terminal domain phosphorylation sites (Figure 1D), among 55 potential seven-transmembrane proteins encoded in the genome of D. discoideum (Prabhu and Eichinger, 2006). cAR1 displayed a clear increase in phosphorylation at Ser303 and Ser304 upon cAMP stimulation (Figure 1D), which corroborates earlier site-directed mutagenesis findings (Caterina et al., 1995). Figure 1E shows the topology of cAR1, the sequence of the identified phosphopeptide, and MS spectra of the phosphopeptide showing phosphorylation at Ser303. Thus, our phosphoproteomic approach has been validated because it clearly identified cAR1 as the cAMP receptor.
Using the same protocol, we searched the annotated genome of D. discoideum using the data from folic-acid stimulated cells, and found two seven-transmembrane proteins, GrlL and GrlG (GABA receptor like L and G) (Prabhu and Eichinger, 2006), whose phosphorylation at Ser652 and Ser659, respectively, increases upon stimulation (Figure 1F). The topology of GrlL, the identified phosphopeptide, and MS spectra are shown for GrlL in Figure 1G, and those for GrlG are shown in Figure S1. These results indicate that the two orphan receptors, GrlL and GrlG, are candidate folic-acid receptors (fARs). Thus, we designated GrlL as fAR1 and GrlG as fAR2, respectively.
fAR1 is Essential for Folic Acid Binding and Folic Acid-Mediated Signaling
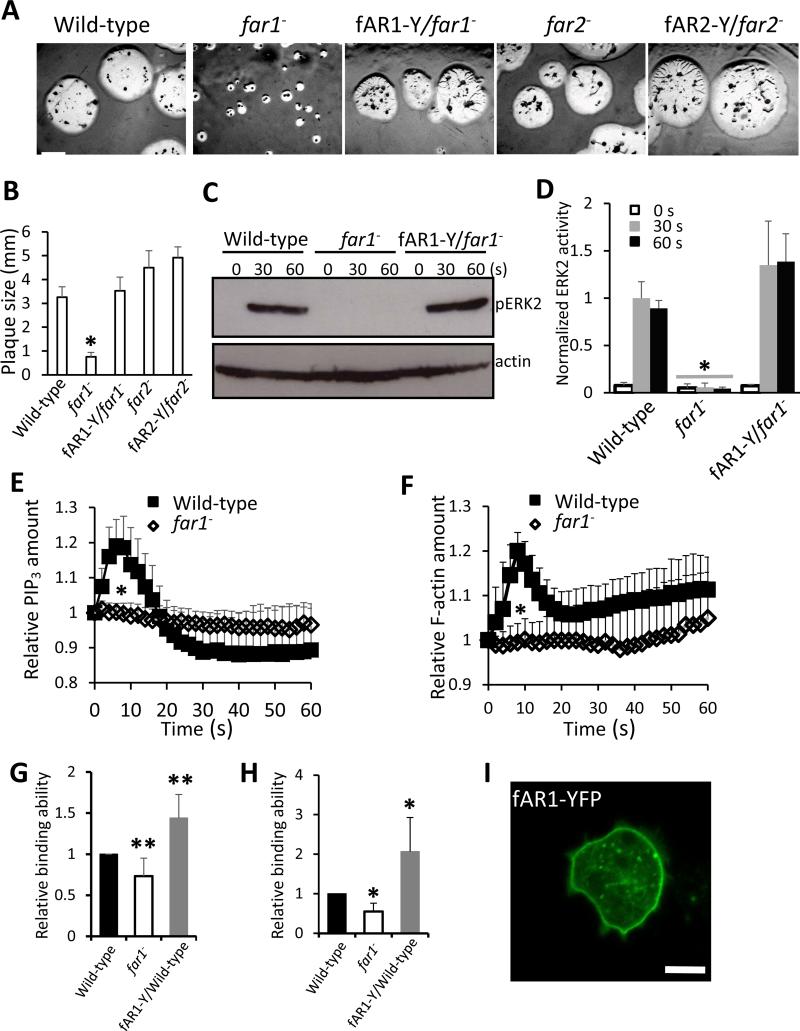
To examine the function of fAR1 and fAR2, we generated grlL-null (far1−) and grlG- null (far2−) strains by homologous recombination (Yan et al., 2012). The genotype of each null mutant was confirmed by PCR analyses (Figure S2A-2C). Both mutant strains exhibited a wild-type growth rate in axenic shaking medium (Figure S2D). While far2− mutants appeared normal when developed on lawns of Klebsiella aerogenes, the far1− mutant phenotype was characterized by the formation of small plaques. far1− mutants, however, did form aggregates, indicating that cAMP signaling remained intact (Figure 2A and 2B). Expression of C-terminal-tagged fAR1-YFP fusion protein (fAR1-Y) rescued the far1− mutant phenotype (Figure 2A and 2B). Interestingly, Gα4-null cells, which also remain responsive to cAMP and are able to aggregate, form small plaques on bacterial lawns likely due to their inability to respond to folic acid (Hadwiger et al., 1994). Gβ-null cells, which do not respond to either chemoattractant, also form small plaques and do not aggregate (Wu et al., 1995). These results are consistent with the notion that fAR1 is a folic-acid receptor linked to the Gα4βγ heterotrimer.
Figure 2. fAR1 is required for folic acid binding and folic acid-meditated signaling.
A. D.discoideum cells were mixed with K. aerogenes and plated on SM nutrient agar plates at 22 °C. Photographs were taken after 5 day of culture. Scale bar: 2 mm. B. The diameters of individual plaques in panel A were measured and graphed. Average plaque size and standard deviation (SD) of ten plaques from each cell line are plotted. A student t-test indicated a statistically significant difference compared with wild-type cells (*, P < 0.01). C. ERK2 signaling is impaired in far1− cells. ERK2 activation in vegetative wild-type, far1− and fAR1-Y/far1− cells in response to 100 μM folic acid stimulation was examined. ERK2 activation was determined by immunoblotting with anti–phospho-ERK2 antibody, using actin as a loading control. D. The mean and SD, resulting from quantification of three independent repetitions of the experiments exemplified in C are plotted. A student t-test indicated a statistically significant difference compared with wild-type cells (*, P < 0.01). E and F. Folic acid induced PHCRAC-GFP (E) and LimEΔcoil-GFP (F) translocation is abolished in far1− cells. Vegetative wild-type and far1− cells expressing PHCRAC-GFP were stimulated with 100 μM folic acid at 0 s. The transient increase in fluorescence intensity was measured at the plasma membrane and graphed. The intensity of the GFP signal was normalized to the first frame of each set of cells. Mean and SD from twenty cells are shown for the time course. A student t-test indicated a statistically significant difference compared with wild-type cells (*, P < 0.01). G and H. far1− has decreased folic acid binding ability. The folic acid binding was determined in the absence (G) or presence of 5 mM methotrexate (H) by measuring the fluorescent intensity of total amount bound ligand. Non-specific binding value was subtracted from all data. The fluorescent intensity ratio, which reflects the folic acid binding of wild-type, far1− and fAR1-Y/wild-type cells were graphed. The mean and SD resulting from quantification of five independent repetitions are graphed. A student t-test indicated a statistically significant difference compared with wild-type cells (*, P < 0.01; **, P<0.05). I. fAR1-YFP is localized on the cellular surface. fAR1-Y/far1− cells were visualized by confocal microscopy. Scale bar: 5 μm. See also Figures S2-4.
To examine the roles of fAR1 and fAR2 in folic-acid mediated signaling events, we first examined the role of fAR1 and fAR2 in folic acid-induced ERK2 signaling (Figure 2C, 2D and Figure S3). Stimulation induces ERK2 activation and can be detected by monitoring an increase in the phosphorylation state of ERK2 by Western blot (Maeda and Firtel, 1997; Maeda et al., 2004). We found that folic acid stimulation induced ERK2 phosphorylation in wild-type (Figure 2C and 2D) and far2− cells (Figure S3) but not in far1− cells (Figure 2C, 2D and Figure S3). Expressing fAR1-YFP in far1− cells restored folic acid-mediated ERK2 activation (Figure 2C, 2D and Figure S3). These results indicate that fAR1, but not fAR2, is required for folic acid-induced ERK2 signaling. Thus, our following study focused on fAR1. Previous studies showed that folic acid stimulation induced chemotactic signaling events, including PIP3 response and actin polymerization (Huang et al., 2013; Srinivasan et al., 2013; Veltman et al., 2014). We found that folic acid stimulation induced a clear PIP3 response and actin polymerization in wild-type cells but failed to trigger PIP3 production or actin polymerization in far1− cells (Figure 2E, 2F and Figure S4), indicating that fAR1 is required for folic acid-induced signaling events.
We then used fluorescence-labeled folic acid to measure ligand binding on the surface of wild-type, far1− and fAR1 overexpressing (fAR1-Y/wild-type, fAR1-Y/WT) cells (Figure 2G and 2H). Previous studies suggested that there may be two types of folic-acid receptors on the cell surface. One type that has a high affinity to methotrexate (a folic acid analogue) but does not mediate chemotactic responses; while the other type that has a low affinity to methotrexate mediates chemotaxis (De Wit and van Haastert, 1985; Segall et al., 1988). Using an established folic acid binding assay, we found that compared to wild-type cells, far1− cells exhibited a decreased folic acid binding ability and fAR1-Y/WT cells showed an increased folic acid binding ability in the absence of methotrexate (Figure 2G). In the presence of methotrexate, the far1− cells and fAR1-Y/WT cells displayed more significantly decreased and increased folic acid binding ability compared with wild-type cells, respectively (Figure 2H). Furthermore, fAR1-Y, like cAR1, localizes uniformly over the plasma membrane (Figure 2I). Taken together, our results indicate that the far1 gene encodes one type of folic acid receptor that mediates chemotactic signaling.
fAR1 Mediates Chemotaxis towards Folic Acid
Next, we examined the role of fAR1 in folic acid-mediated chemotaxis. We performed EZTAXIScan chemotaxis assays to test the ability of wild-type, far1− and fAR1-Y/far1− cells to crawl towards a linear gradient of folic acid (Liu et al., 2010) (Figure 3A). For all assays, wild-type or mutant cells were placed in the top well of the system and were allowed to migrate towards identical folic-acid gradients established from the bottom wells (Figure 3A). Chambers were imaged with bright-field illumination every 30 seconds to monitor cell movement (Figure 3A and Movie S1). As expected, wild-type cells migrated towards folic acid (Figure 3A); In contrast, far1− cells were unable to migrate efficiently towards folic acid (Figure 3A). Cell-tracing analysis was performed to quantify cell movement. The resulting cell tracks are shown in Figure 3B, and net path length, directionality and speed, of wild-type, far1− and fAR1-Y/far1− are reported in Figure 3C. far1− cells displayed significantly reduced movement (as judged by the values of net path length and speed) and decreased directionality. Importantly, all mutant phenotypes were rescued by expressing fAR1-Y in far1− cells (Figure 3A-3C). To further compare the chemotactic behavior of far1− and fAR1-Y/far1− cells under the same conditions, we mixed both cell types and recorded their migration to a micropipette releasing folic acid by fluorescence microscopy (Figure 3D, Movies S2 and S3). At time 0, there were eight far1− (transparent) cells and six fAR1-Y/far1− (green) cells in the view close to the micropipette. At 20 min, many more fAR1-Y/far1− (green) cells migrated from outside the original field of view to coalesce near the micropipette (Figure 3D, Movie S2 and S3). Quantitative analyses showed that movements of far1− cells displayed significantly reduced net path length, speed, and directionality compared to the movements of fAR1-Y/far1− cells (Figure 3E). We also compared the chemotaxis behaviors of wild-type and fAR1-overexpressing cells (fAR1-Y/WT) using a needle chemotaxis assay. No significant difference was found in chemotaxis behavior between these two cell types, suggesting that wild-type cells express a sufficient number of fAR1 receptors to carry out chemotaxis to folic acid (Figure S5, Movie S4). Taken together, our results indicate that fAR1 is required for folic-acid-mediated chemotaxis.
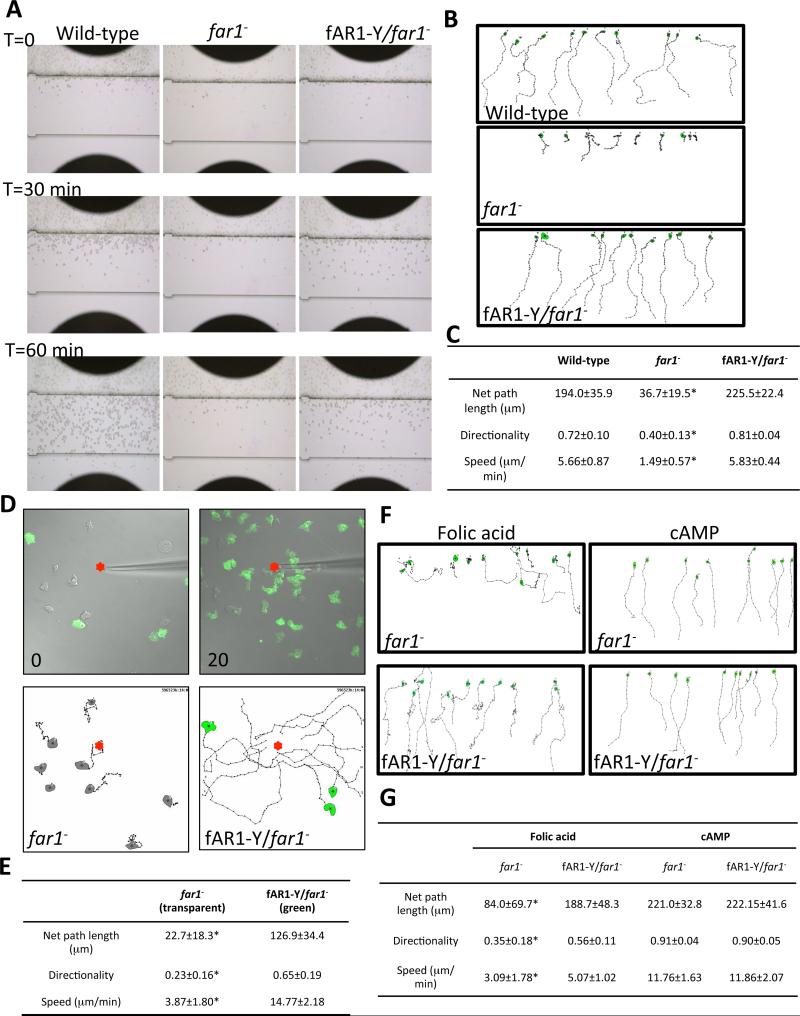
Figure 3. fAR1 is specifically required for folic acid-mediated chemotaxis in D.discoideum.
A. EZ-TAXIScan chemotaxis towards a linear folic acid gradient of vegetative wild-type, far1− and fAR1-Y/far1− cells. Images were recorded every 30 s. A linear concentration gradient of 0-100 μM folic acid in the channel formed from bottom to top in the figure. Images of each cell line at time 0, 30 and 60 min are shown. B. Migration paths towards folic acid of cells in panel A. Ten cells from each strain were used for single cell tracing. C. The mean and SD resulting from quantification of chemotaxis parameters from A and B are shown. A student t-test indicated a statistically significant difference compared with wild-type cells (*, P < 0.01). D. Chemotaxis analysis of far1− (transparent) and fAR1-Y/far1− (green) cells migrating to a micropipette filled with 100 μM folic acid (red dots). Images for each strain at time 0 and 20 min are shown. Migration paths towards folic acid are shown below. Eight or ten cells from each strain were used for single cell tracing. Scale bar: 20 μm. E. The mean and SD resulting from quantification of chemotaxis parameter from D are shown. F. Migration paths towards a linear gradient of 0-1 μM folic acid or cAMP of developed far1− and fAR1-Y/far1− cells determined by EZ-TAXIScan. Ten cells from each strain were used for single cell tracing. For folic acid gradient, cells were traced for 60 min; for cAMP gradient, cells were traced for 30 min. G. The mean and SD resulting from quantification of chemotaxis parameter from F. A student t-test indicated a statistically significant difference compared with fAR1-Y/far1− cells (*, P < 0.01). See also Figure S5, Movies S1-S6.
D. discoideum cells grown rich medium (axenic) and in bacterial culture are able to respond to folic acid but not cAMP due to the lack of cAMP receptors on the cell surface. Several hours after starvation, cells become chemotactically responsive to extracellular cAMP (Tang et al., 2014). First, we measured the level of far1 mRNA in axenic, bacterial culture and development stage using real-time PCR analysis. Compared to the cells grown in axenic medium, the level of far1 mRNA in bacteria cultured cells was about two-fold higher while that of developed cells was about 40% lower (Figure S2), which is consistent with previous reports (Parikh et al., 2010; Santhanam et al., 2015). Then, we examined chemotaxis of far1− and fAR1-Y/far1− cells to cAMP and folic acid after 6 hours of starvation using the EZ-TAXIScan system (Figure 3F). Our analyses showed, as expected, that starved far1− cells cannot efficiently migrate towards folic acid and the expression of fAR1-Y rescues the migratory defect. However, both far1− and fAR1-Y/far1− cells clearly chemotax to cAMP (Figure 3F, 3G, Movie S5 and S6), indicating that fAR1 is specifically required for folic-acid-mediated chemotaxis. Taken together, our results showed that fAR1 is a chemoattractant receptor that mediates cell migration to folic acid.
fAR1 Mediates Phagocytosis of Bacteria
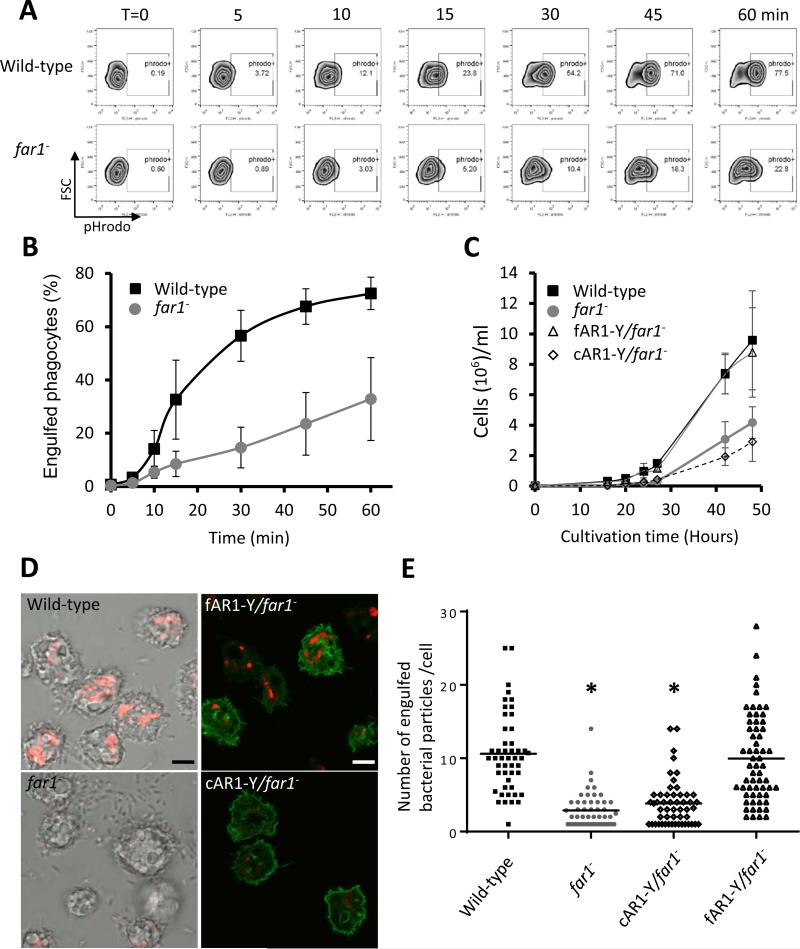
The far1− cells formed small plaques on a K. aerogenes bacterial lawn (Figure 2A), suggesting that cells lacking fAR1 receptors could not efficiently ingest bacteria as food. To explore fAR1's cellular function in phagocytosis, we quantitatively measured the internalization of bacteria by wild-type and far1− cells over the time course of one hour, using flow cytometry analysis (Figure 4A). Cells were mixed with pHrodo-labeled K. aerogenes, collected and suspended in a basic buffer to quench pHrodo fluorescence outside of the cells. This was followed by counting of pHrodo positive cells, which were the cells with internalized bacteria (Figure 4A). Compared to wild-type cells, far1− cells were still able to engulf, but significantly defective in bacteria uptake over time (Figure 4A and 4B). We then examined the growth of cells when nutrient liquid or bacteria was provided as food sources (Figure 4C). In liquid medium, both wild-type and far1− cells showed normal growth curve (Figure S2D), indicating that fAR1 is not essential for cell division or pinocytosis. However, when fed with bacteria, far1− cells did not grow as well as wild-type cells (Figure 4C). This growth defect can be rescued by expressing fAR1-Y (fAR1-Y/far1−) but not cAR1-YFP (cAR1-Y/far1−) (Figure 4C and Figure S6). We then imaged internalization of bacteria particles using confocal microscopy (Figure 4D). Relative to wild-type and fAR1-Y/far1− cells, far1− and cAR1-Y/far1− cells displayed a significant decrease in bacteria uptake, as measured by either bacterial number (red particles) (Figure 4E) or pHrodo intensity in a cell (Figure S6C). However, we did not detect any defects in phagosome maturation or acidification in far1− cells. Although the growth of far1− cells was significantly decreased compared to wild-type cells (Figure 4C), the far1− cells were still able to grow using only bacterial as their sole food source, suggesting that the cells are able to ingest bacteria. To monitor the pH value in phagosomes, we measured the intensity of pHrodo per bacterial particle inside cells, and found that the values of mean intensity per bacterial particle were similar in wild-type, far1−, cAR1-Y/far1− and fAR1-Y/far1− cells (Figure S6D), suggesting that phagosomal pH values are similar in both wild-type and far1− cells. In addition, we suggest that there is a correlation between the reduced engulfment of bacteria and the reduced growth in far1− cells (Figure 4B and 4C). Taken together, our results indicate that cells lacking fAR1 receptors are defective in the engulfment of bacteria, but not in phagosome maturation and acidification associated with bacteria ingestion.
Figure 4. fAR1 mediates phagocytosis of bacteria in D. discoideum.
A. Wild-type and far1− cells were mixed with pHrodo-labelled live K. aerogenes at a 1:50 ratio for the indicated time. Cells were suspended in basic pH buffer and analyzed for the percentage of pHrodo positive cells, which represents the cells that engulfed K. aerogenes. B. The mean and SD resulting from quantification of three independent repetitions of the experiments exemplified in A are plotted. C. far1− cells exhibited growth defect in live bacteria suspension culture. Wild-type and different mutant D. disocideum cells were cultured in suspension in association with live K. aerogenes in phosphate buffer. The growth curve with mean and SD is plotted, resulting from quantification of three independent repetitions of the experiments. D. Wild-type, far1− (transparent), fAR1-Y/far1− and cAR1-Y/far1− (green) cells, were mixed with pHrodo-labeled K. aerogenes at a 1:20 ratio. After 45 min, cells were mounted on a slide in basic pH buffer and analyzed by confocal microscopy. The engulfed pHrodo-labelled K. aerogenes are shown as red; Scale bars, 5 μm. E. Quantification of engulfed K. aerogenes is compared between different D. discoideum strains. The engulfed bacterial number in each cell was measured and the mean value of the pHrodo intensity over cell area was calculated and plotted for wild-type, far1−, cAR1-Y/far1− and fAR1-Y/far1− cells. A student t-test indicated a statistically significant difference compared with wild-type cells (*, P < 0.01). See also Figure S6.
fAR1 is not Essential for the Initial Binding between Bacteria and D. discoideum Cells
To determine the role of fAR1 in phagocytosis, we first examined the initial binding between bacteria and wild-type and fAR1-null cells (Figure 5). Wild-type and far1− cells were incubated with fluorescence-labeled K, aerogenes at 4 °C for 15 min, and then imaged using fluorescence microcopy in the presence of 2 mM NaN3, to inhibit bacterial internalization (Cohen et al., 1994). Under these conditions, bacteria bound to the surface of both wild-type and far1− cells and no defects in the bacteria-binding ability of far1− cells was observed relative to wild-type (Figure 5A and 5B). The binding ability of each cell lines was also analyzed by flow cytometry (Figure 5C and 5D). Wild-type and far1− cells were incubated with fluorescence-labeled bacteria at 4°C to block bacterial internalization (Cohen et al., 1994). Consistently, no defect in bacteria binding was observed in this assay (Figure 5D). Taken together, our results showed that fAR1 is not required for cells to make initial contact with bacteria.
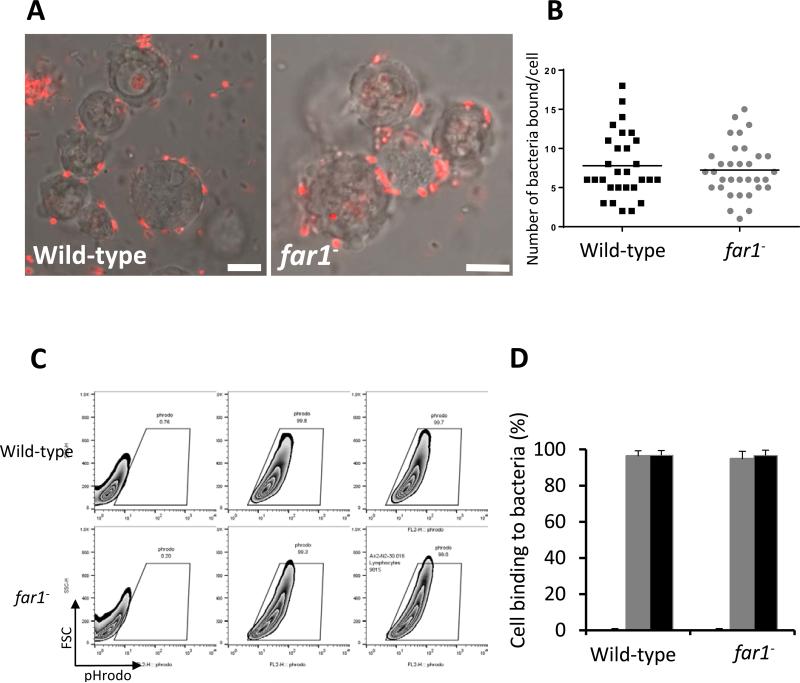
Figure 5. Bacterial binding is not impaired in far1− cells.
A. Wild-type and far1− cells were mixed with pHrodo-labelled K. aerogenes (red) at a 1:50 ratio on ice. After 15 min, the cells were mounted on a slide in the presence of 2 mM NaN3 and analyzed by confocal microscopy. Scale bars, 5 μm. B. Quantification of number of bound K. aerogenes is compared between wild-type and far1− cells. C. Vegetative wild-type and far1− cells were incubated with pHrodo-labelled K. aerogenes at a 1:50 ratio on ice for 15 min. Cells were analyzed by flow cytometry for the percentage of pHrodo positive cells, which represents the cells that bind K. aerogenes. D. The mean and SD resulting from quantification of three independent repetitions of the experiments exemplified in C are plotted.
fAR1 Mediates Bacteria-Induced Formation of Phagocytic Cups
We next examined the role of fAR1 in the bacteria-induced formation of phagocytic cups. Wild-type cells and far1− were transformed with an F-actin marker, ΔlimE-GFP, to visualize the actin-driven formation of the phagocytic cup (Gerisch et al., 2004; Schneider et al., 2003). Fluorescently labeled bacteria were added to a chamber containing wild-type or far1− cells expressing ΔlimE-GFP (ΔlimE-G/WT or ΔlimE-G/far1−), and the interaction process was imaged in real time using a confocal fluorescence microscope (Figure 6). Bacteria that bind to the cell surface induced actin polymerization and became surrounded by a phagocytic cup followed by engulfment in wild-type cells, as previously reported (Clarke and Maddera, 2006). We found that far1− cells were still able to bind bacteria, however the binding rarely induced either actin polymerization or formation of phagocytic cups (Figure 6A and 6B). Thus, the fAR1 receptor plays a role in bacteria-induced localized actin polymerization, following the initial interaction between bacteria and D. discoideum cells. Our results suggest that folic acid on the bacterial surface interact with fAR1 to induce localized chemotactic responses that promote engulfment of bacteria.
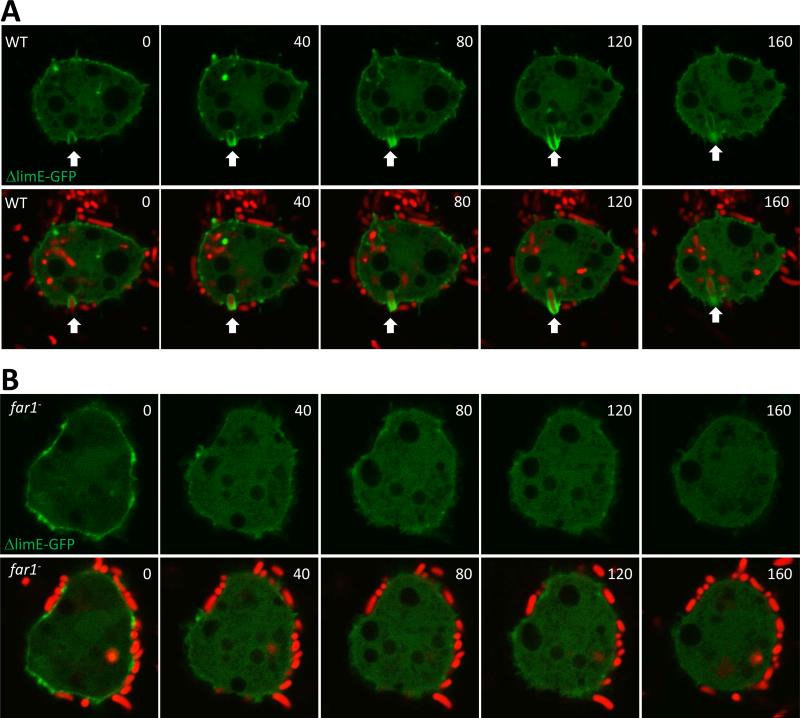
Figure 6.
Bacteria induced phagocytic cup formation in wild-type but not in far1− cells. Vegetative wild-type (A) and far1− (B) expressing LimEΔcoil-GFP (green) grown in D3-T medium were harvested, washed with PB buffer and settled in a 4 well chamber for 10 min. A 20 fold excess of pHrodo-labelled K. aerogenes (red) was added to the D. discoideum cells and the engulfment process was recorded by confocal microscopy.
Beads Coated with Folic Acid Triggered Localized Chemotactic Responses and Formation of Phagocytic Cups
To examine whether the binding of a chemoattractant to its GPCR promotes engulfment, we used latex beads with fluorescently-labeled folic acid crosslinked to the surface (Figure S7). Coated beads were added to chambers containing cells expressing fAR1 (fAR1-Y/far1−, PHGFP/WT or ΔlimE-G/WT) or cells lacking fAR1 (cAR1-Y/far1−, PH-G/far1− or ΔlimE-G/far1−), and imaged. In cells expressing fAR1 receptors (Figure 7A), the beads interacted the receptor (arrow in fAR1-Y at 0s), induced a localized PIP3 response (arrow in PH-G at 0s) or actin polymerization (arrow in ΔlimE-G at 0s) around the beads, and then followed by the engulfment of the beads (arrows in fAR1-Y at 80s, in PH-GFP at 24s, and in ΔlimE-GFP at 200s). In cells lacking fAR1, the beads made contact with cell surface (Figure 7B, arrows in cAR1-Y, PH-G and ΔlimE-G at 0s), but did not trigger accumulation of PH-G or ΔlimE-G around the beads, and were not internalized (Figure 7B, arrows in cAR1-Y at 80s, PH-G at 24s, and Δlime-G at 200s). In addition, beads without folic acid, as controls, did not trigger a PIP3 response, actin polymerization, or phagocytic cup formation in cells expressing fAR1 (Figure S7). Furthermore, when excess soluble folic acid (2 mM) was present, the folic acid coated-beads could no longer effectively trigger PIP3 response and actin polymerization in the cells around the beads for engulfment (Figure 7C). Our results indicate that the binding of immobile folic acid to fAR1 receptor induces localized PIP3 responses and actin polymerization to form phagocytic cups to promote the engulfment of the beads, a cell process that we have termed chemoattractant-mediated engulfment.
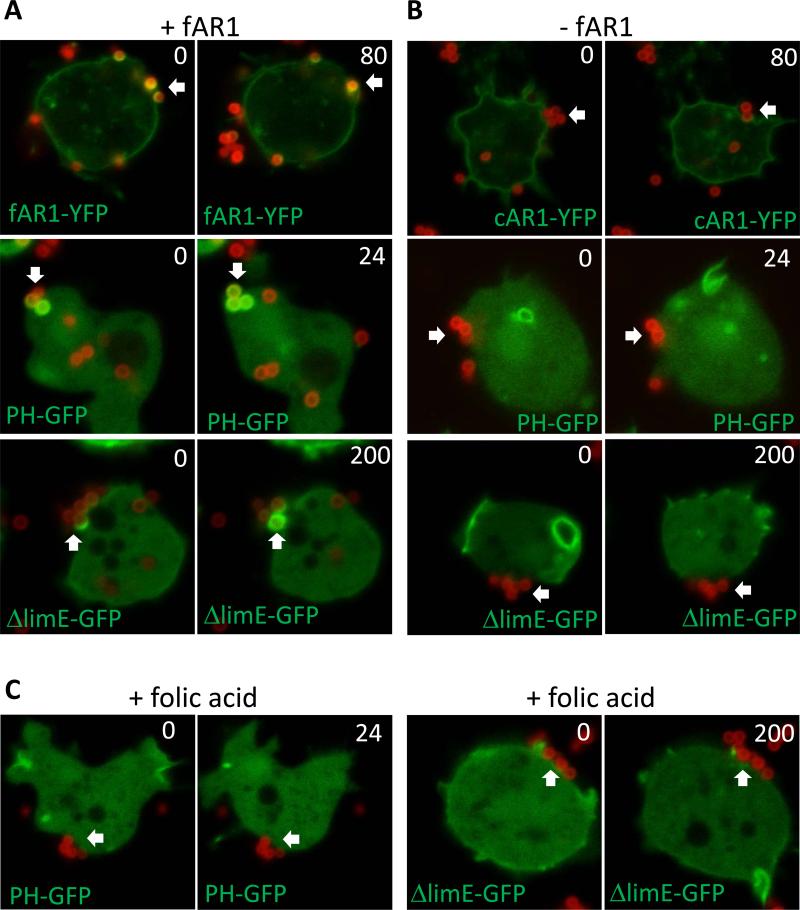
Figure 7.
Folic acid coated beads induce localized chemotaxis signaling and promoted phagocytosis. Vegetative wild-type and far1− expressing different protein markers grown in D3-T medium were harvested, washed with PB buffer, and settled in a 4 well chamber for 10 min. A 10 fold excess of beads was added to D. discoideum cells and the engulfment process was recorded by confocal microscopy. A. Phagocytosis of 1 μm folic acid coated latex beads (red) by wild-type expressing PHCRAC-GFP, LimEΔcoil-GFP, and far1− expressing fAR1-YFP (green). B. Phagocytosis of 1 μm folic acid coated latex beads by far1− expressing PHCRAC-GFP, LimEΔcoil-GFP or cAR1-YFP (green). C. Soluble folic acid (2 mM) inhibited PIP3 responses and actin polymerization induced by the folic-acid-coated beads (red), and wild-type cells expressing PHCRAC-GFP or LimEΔcoil-GFP failed to engulf the beads efficiently. See also Figure S7.
Discussion
In this work, we used a phosphoproteomic approach to uncover components involved in the chemoattractant GPCR-mediated signaling network. Specifically, we developed a mass spectrometry (MS)-based quantitative phosphoproteomic method to determine global and site-specific phosphorylation changes in D. discoideum cells upon stimulation with a specific chemoattractant (Figure 1). The large data set we generated provides an opportunity to identify signaling components based on dynamic changes in phosphorylation at specific sites in response to folic acid stimulation. It is well known that ligand binding to a GPCR induces phosphorylation on Ser or Thr residues on the C-terminus of the receptor (Hereld et al., 1994; Sibley et al., 1987). From our phosphoproteomic date set, we identified two orphan GPCRs as candidates of folic-acid receptors, fAR1 and fAR2. This phosphoproteomic approach for identifying orphan GPCRs can be applied to discovering orphan chemokine GPCRs in mammals and other missing components in any signaling network in which phosphorylation is regulated by a specific factor.
Combining this phosphoproteomic approach with other methods, we identified the long-sought-after folic acid receptor (fAR1) that controls D. discoideum chemotaxis towards bacteria as a food source. D. discoideum is a well-established model for studying eukaryotic chemotaxis. The cells respond and migrate to two major chemoattractants, cAMP and folic acid. Chemotaxis towards cAMP is essential for the development of D. discoideum, and chemotaxis towards folic acid is essential for amoebae to seek bacteria as food. The cAMP receptor, cAR1, is the first chemoattractant GPCR to be discovered in eukaryotic cells (Klein et al., 1988). However, despite the fact that folic acid was shown to be another chemoattractant for D. discoideum more than forty years ago (Pan et al., 1972), identification of a folic acid receptor(s) eluded discovery until now. It has been proposed that a GPCR coupled to the heterotrimeric G-protein consisting of Gα4 and Gβγ mediates chemotaxis towards folic acid (Hadwiger et al., 1994; Wu et al., 1995). We found that cells lacking the orphan GPCR, fAR1, displayed a developmental phenotype on bacterial lawn similar to of cells lacking Gα4 and Gβγ (Figure 2A). In addition, we showed that fAR1 receptors on the cell surface bind to extracellular folic acid, mediate folic acid-induced chemotactic signaling events (Figure 2), and control chemotaxis towards folic acid (Figure 3). Previous studies suggested that there may be two types of folic acid receptors, one type controlling chemotaxis and another type mediating unknown functions in D. discoideum. Our study indicates that fAR1, coupled with Gα4Gβγ, detects folic acid released from bacteria and mediates cell migration towards them, the chemoattractant sources.
We discovered a chemoattractant GPCR, fAR1 that also mediates phagocytosis. In macrophages and neutrophils, chemoattractant GPCRs are only recognized to mediate chemotaxis to chase microorganisms, while other receptors, such as Fcγ and C3a receptors, bind microorganisms for engulfment (Freeman and Grinstein, 2014)). Our work using the professional phagocyte D. discoideum demonstrates that a chemoattractant GPCR, can promote bacteria engulfment during phagocytosis. Our results show that the fAR1 is not required for the initial binding between bacteria and D. discoideum cells (Figure 5). Upon contact, bacteria induced the formation of phagocytic cups for engulfment in wild-type but not fAR1-null cells (Figure 6). Using folic acid-coated beads (Figure 7), we showed that immobilized folic acid interacts with fAR1 to trigger localized chemotactic responses such as PIP3 production and actin polymerization to promote the formation of phagocytic cups for engulfment. Cells lacking fAR1, Gα4 or Gβ subunits are still able to eat bacteria as food; however engulfment and ingestion occurs at a significantly lower rate (Figure 4). Therefore, it appears that cells eat bacteria by both fAR1-independent and fAR1-dependent modes. Future studies will aim to clarify whether the fAR1-independent mode of engulfment is mediated by random actin localization or through unknown factor interaction. We suggest that the fAR1/Gα4Gβγ machinery detects diffusible folic acid to mediate cell migration and immobile folic acid on the bacterial surface to promote bacterial engulfment. Our results support the notion that binding of folic acid to fAR1 activates heterotrimeric G-protein into Gα4-GTP and free Gβγ subunits, which in turn mediate signaling pathways to produce pseudopods for cell migration and engulfment. Thus, we have revealed a cellular process of “chemoattractant-mediated engulfment” in the model organism D. discoideum. Interestingly, in humans, formyl peptide receptors on the surface of neutrophils mediate chemotaxis towards bacteria by detecting N-formylmethionyl-leucyl-phenylalanine (fMLP), which is released by bacteria. Upon catching the bacteria, neutrophils engulf them. Future studies are needed to determine whether neutrophils also utilize a chemoattractant-mediated engulfment mechanism to promote phagocytosis to subdue bacterial infections.
EXPERIMENTAL PROCEDURES
ERK2 Activation Assay
Vegetative D. discoideum cells were harvested, washed with phosphate buffer (PB; 7.4 mM NaH2PO4·H2O, 4 mM Na2HPO4·7H2O, pH 6.5) and stimulated with 100 μM folic acid. At indicated time intervals after stimulation, 150 μl of the cell suspension was taken out, mixed with 50 μl 4 × sample buffer, and boiled for 3 min. Proteins were separated by SDS–PAGE, transferred to nitrocellulose membranes, and blotted with polyclonal anti–phospho-p44/p42 MAPK (pERK2) antibody and anti-actin antibodies.
PH-GFP and LimEΔcoil-GFP Translocation upon Folic Acid Stimulation
Vegetative D. discoideum cells expressing PHCRAC-GFP or LimEΔcoil-GFP were prepared using the same protocol for the ERK2 activation assay. Cells were plated in four well chambers and then imaged with a Zeiss Laser Scanning Microscope 780 with a 60×, 1.3 NA Plan-Neofluar objective lens. Fluorescent frames were acquired every 2 seconds and in 30 frames total. A final concentration of 100 μM folic acid was added to the cells to induce PHCRAC-GFP or LimEΔcoil-GFP translocation from cytosol to plasma membrane. The temporal-spatial intensity changes of PHCRAC-GFP LimEΔcoil-GFP in cells were directly imaged using a confocal microscope. For each cell, a region of interest (ROI) was drawn at the plasma membrane to measure the fluorescence intensity change over time. The intensity of the GFP signal was normalized to the first frame of each set of cells.
Folic Acid Binding Assays
Cells were harvested and resuspended in PB at 1×108 cells/ml. The suspension was incubated on ice for 15 min before use in the binding assay. 100 μl cell suspension was mixed with 50 μl binding mix containing 500 nM FolateRSense 680 and 0.33 mM 8-azaguanine with or without 5 μM methotrexate. The cells were separated from unbound folic acid by centrifugation at 10,000 g for 15 s through 100 μl silicon oil (AR20:AR200, 11:4) and the supernatant was aspirated without disturbing the cell pellet. The pellet was dissolved in 150 μl PB and the florescence intensity was measured by EnSpire Multimode Plate Reader. Blanks values were determined in the presence of 50 μM unlabeled folate and subtracted from all data.
EZ-TAXIScan Chemotaxis Assay
D. discoideum cells were harvested from SM plates, washed with PB, and resuspended. Cell migration was recorded at 30 s intervals at 22 °C for 40 min in the EZ-TAXIScan chamber. Chips used in the chamber were pre-coated with 1 % BSA at 22 °C for 30 min. A stable gradient of folic acid or cAMP as described in Figure 3 was established for the assay. Cell migration analysis was performed with DIAS software.
Micropipette Chemotaxis Assay
Washed cells were seeded into a one well chamber and allowed to adhere for 20 min. A micropipette filled with 100 μM folic acid was connected with a microinjector to generate a chemoattractant gradient. The micropipette was placed in the same focal plane as the cells and phase contrast and fluorescent frames were acquired with an interval of 30 s for 50 min using a Zeiss Laser Scanning Microscope 510, 40 × 1.3 NA Plan-Neofluar objective lens. Cell migration analysis was performed with DIAS software.
Folic Acid Conjugated Beads
To label the latex beads with folic acid, 500 μl 1 μm aliphatic amine latex beads (Thermo Fisher) were washed with PB three times and re-suspended into 1 ml PB. The beads were then incubated with 24 nmol FolateRSense 680 (Perkin Elmer) in the presence of 2 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide, hydrochloride (EDAC, Thermo Fisher) at 4 °C for 16 hours. After incubation, the coated beads were washed five times with pre-cold PB to remove excess free ligand and crosslinker. The schematic chemical reaction is shown in Supplemental Figure 7A.
Phagocytosis Assay and Flow Cytometry
K. aerogenes labeled with pHrodo Red were incubated with D. discoideum cells at a 50:1 ratio at 22°C. After incubation, the cells were washed and resuspended in basic buffer (50 mM Tris pH 8.8 and 150 mM NaCl). The phagocytes and K. aerogenes were distinguished by forward and side scatter (FSC). The appearance of pHrodo in the phagocyte population was monitored as an indicator of K. aerogenes engulfment. The phagocyte cell population characterized by high fluorescence of pHrodo was considered as the cells which engulfed K. aerogenes. Data acquisition and analysis were done using FACSort flow cytometer with Cell Quest software (v. 3.3) and analyzed using FlowJo (v. 10.0.8). The quantification of engulfed K. aerogenes was analyzed by confocal microscopy and ImageJ.
To visualize bead engulfment by D. discoideum cells, vegetative wild-type and far1− cells expressing different protein markers grown from D3-T medium were harvested, washed with PB buffer and settled in a 4 well chamber for 10 min. A 10 fold excess of beads was added to the D. discoideum cells and the engulfment process was recorded with a Zeiss Laser Scanning Microscope 780 with a 60×, 1.3 NA Plan-Neofluar objective lens. See Supplementary Experimental Procedures for more details.
Statistics
The statistical significance was assessed using analysis of variance with the two-tailed unpaired Student's t-test. Data are presented as mean ± sd., unless stated otherwise.
Supplementary Material
Highlights.
Using quantitative phosphoproteomics to de-orphan GPCRs.
Discovery of a folic-acid chemoattractant GPCR, fAR1.
The fAR1 GPCR mediates cell migration towards folic acid.
The fAR1 GPCR mediates engulfment of bacteria.
Acknowledgements
We thank D. Hereld, J. Brzostowski and X. Xiang for critical reading; M. Gucek, L. Olano, G. Nardone and C. Hammer for technical support on mass spectrometry analyses; P. van Haastert for suggestion on folic acid binding assay; C. Huang and P. Devreotes for providing plasmids; X. Wen, W. Quan, J. Lu and J. Manzella-Lapeira for help in the chemotaxis analyses; L. Tian, R. Wang and A. Grunfeld for phagocytosis assay. The work is supported by intramural funding from NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: M.P., X.X. and T.J. conceived the idea; M.P. and T.J. designed the study; M.P. and Y.C. carried out MS analyses; M.P. performed experiments; M.P. and T.J analyzed data and wrote the manuscript.
Supplementary Information includes seven figures, supplemental experiment procedures and six movies.
References
- Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- Basu S, Fey P, Pandit Y, Dodson R, Kibbe WA, Chisholm RL. DictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 2013;41:D676–683. doi: 10.1093/nar/gks1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Das S, Kamimura Y, Long Y, Parent CA, Devreotes PN. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol. 2010;190:233–245. doi: 10.1083/jcb.201001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina MJ, Devreotes PN, Borleis J, Hereld D. Agonist-induced loss of ligand binding is correlated with phosphorylation of cAR1, a G protein-coupled chemoattractant receptor from Dictyostelium. J Biol Chem. 1995;270:8667–8672. doi: 10.1074/jbc.270.15.8667. [DOI] [PubMed] [Google Scholar]
- Charest PG, Shen Z, Lakoduk A, Sasaki AT, Briggs SP, Firtel RA. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell. 2010;18:737–749. doi: 10.1016/j.devcel.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Iijima M, Tang M, Landree MA, Huang YE, Xiong Y, Iglesias PA, Devreotes PN. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev Cell. 2007;12:603–614. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Maddera L. Phagocyte meets prey: uptake, internalization, and killing of bacteria by Dictyostelium amoebae. Eur J Cell Biol. 2006;85:1001–1010. doi: 10.1016/j.ejcb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Cohen CJ, Bacon R, Clarke M, Joiner K, Mellman I. Dictyostelium discoideum mutants with conditional defects in phagocytosis. J Cell Biol. 1994;126:955–966. doi: 10.1083/jcb.126.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- De Wit RJ, van Haastert PJ. Binding of folates to Dictyostelium discoideum cells. Demonstration of five classes of binding sites and their interconversion Biochimica et Biophysica Acta. 1985;814:199–213. [Google Scholar]
- Eyrich B, Sickmann A, Zahedi RP. Catch me if you can: mass spectrometry-based phosphoproteomics and quantification strategies. Proteomics. 2011;11:554–570. doi: 10.1002/pmic.201000489. [DOI] [PubMed] [Google Scholar]
- Flannagan RS, Jaumouille V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol. 2012;7:61–98. doi: 10.1146/annurev-pathol-011811-132445. [DOI] [PubMed] [Google Scholar]
- Freeman SA, Grinstein S. Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunol Rev. 2014;262:193–215. doi: 10.1111/imr.12212. [DOI] [PubMed] [Google Scholar]
- Gerisch G, Bretschneider T, Muller-Taubenberger A, Simmeth E, Ecke M, Diez S, Anderson K. Mobile actin clusters and traveling waves in cells recovering from actin depolymerization. Biophys J. 2004;87:3493–3503. doi: 10.1529/biophysj.104.047589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger JA, Firtel RA. Analysis of G alpha 4, a G-protein subunit required for multicellular development in Dictyostelium. Genes Dev. 1992;6:38–49. doi: 10.1101/gad.6.1.38. [DOI] [PubMed] [Google Scholar]
- Hadwiger JA, Lee S, Firtel RA. The G alpha subunit G alpha 4 couples to pterin receptors and identifies a signaling pathway that is essential for multicellular development in Dictyostelium. Proc Natl Acad Sci U S A. 1994;91:10566–10570. doi: 10.1073/pnas.91.22.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereld D, Vaughan R, Kim JY, Borleis J, Devreotes P. Localization of ligand-induced phosphorylation sites to serine clusters in the C-terminal domain of the Dictyostelium cAMP receptor, cAR1. J Biol Chem. 1994;269:7036–7044. [PubMed] [Google Scholar]
- Huang CH, Tang M, Shi C, Iglesias PA, Devreotes PN. An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat Cell Biol. 2013;15:1307–1316. doi: 10.1038/ncb2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Isik N, Brzostowski JA, Jin T. An Elmo-like protein associated with myosin II restricts spurious F-actin events to coordinate phagocytosis and chemotaxis. Dev Cell. 2008;15:590–602. doi: 10.1016/j.devcel.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Xiong Y, Iglesias PA, Hoeller O, Bolourani P, Devreotes PN. PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr Biol. 2008;18:1034–1043. doi: 10.1016/j.cub.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein PS, Sun TJ, Saxe CL, 3rd, Kimmel AR, Johnson RL, Devreotes PN. A chemoattractant receptor controls development in Dictyostelium discoideum. Science. 1988;241:1467–1472. doi: 10.1126/science.3047871. [DOI] [PubMed] [Google Scholar]
- Liu L, Das S, Losert W, Parent CA. mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev Cell. 2010;19:845–857. doi: 10.1016/j.devcel.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Firtel RA. Activation of the mitogen-activated protein kinase ERK2 by the chemoattractant folic acid in Dictyostelium. J Biol Chem. 1997;272:23690–23695. doi: 10.1074/jbc.272.38.23690. [DOI] [PubMed] [Google Scholar]
- Maeda M, Lu S, Shaulsky G, Miyazaki Y, Kuwayama H, Tanaka Y, Kuspa A, Loomis WF. Periodic signaling controlled by an oscillatory circuit that includes protein kinases ERK2 and PKA. Science. 2004;304:875–878. doi: 10.1126/science.1094647. [DOI] [PubMed] [Google Scholar]
- Manahan CL, Iglesias PA, Long Y, Devreotes PN. Chemoattractant signaling in dictyostelium discoideum. Annu Rev Cell Dev Biol. 2004;20:223–253. doi: 10.1146/annurev.cellbio.20.011303.132633. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Pan P, Hall EM, Bonner JT. Folic acid as second chemotactic substance in the cellular slime moulds. Nat New Biol. 1972;237:181–182. doi: 10.1038/newbio237181a0. [DOI] [PubMed] [Google Scholar]
- Parikh A, Miranda ER, Katoh-Kurasawa M, Fuller D, Rot G, Zagar L, Curk T, Sucgang R, Chen R, Zupan B, et al. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010;11:R35. doi: 10.1186/gb-2010-11-3-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu Y, Eichinger L. The Dictyostelium repertoire of seven transmembrane domain receptors. Eur J Cell Biol. 2006;85:937–946. doi: 10.1016/j.ejcb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7:964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- Rougerie P, Miskolci V, Cox D. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: role and regulation of the actin cytoskeleton. Immunol Rev. 2013;256:222–239. doi: 10.1111/imr.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam B, Cai H, Devreotes PN, Shaulsky G, Katoh-Kurasawa M. The GATA transcription factor GtaC regulates early developmental gene expression dynamics in Dictyostelium. Nat Commun. 2015;6:7551. doi: 10.1038/ncomms8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider N, Weber I, Faix J, Prassler J, Muller-Taubenberger A, Kohler J, Burghardt E, Gerisch G, Marriott G. A Lim protein involved in the progression of cytokinesis and regulation of the mitotic spindle. Cell Motil Cytoskeleton. 2003;56:130–139. doi: 10.1002/cm.10139. [DOI] [PubMed] [Google Scholar]
- Segall JE, Bominaar AA, Wallraff E, De Wit RJ. Analysis of a Dictyostelium chemotaxis mutant with altered chemoattractant binding. J Cell Sci. 1988;91(Pt 4):479–489. doi: 10.1242/jcs.91.4.479. [DOI] [PubMed] [Google Scholar]
- Sibley DR, Benovic JL, Caron MG, Lefkowitz RJ. Regulation of transmembrane signaling by receptor phosphorylation. Cell. 1987;48:913–922. doi: 10.1016/0092-8674(87)90700-8. [DOI] [PubMed] [Google Scholar]
- Srinivasan K, Wright GA, Hames N, Housman M, Roberts A, Aufderheide KJ, Janetopoulos C. Delineating the core regulatory elements crucial for directed cell migration by examining folic-acid-mediated responses. J Cell Sci. 2013;126:221–233. doi: 10.1242/jcs.113415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Wang M, Shi C, Iglesias PA, Devreotes PN, Huang CH. Evolutionarily conserved coupling of adaptive and excitable networks mediates eukaryotic chemotaxis. Nat Commun. 2014;5:5175. doi: 10.1038/ncomms6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Keizer-Gunnik I, Van Haastert PJ. Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J Cell Biol. 2008;180:747–753. doi: 10.1083/jcb.200709180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, Lemieux MG, Knecht DA, Insall RH. PIP(3)-dependent macropinocytosis is incompatible with chemotaxis. J Cell Biol. 2014;204:497–505. doi: 10.1083/jcb.201309081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Valkema R, Van Haastert PJ, Devreotes PN. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J Cell Biol. 1995;129:1667–1675. doi: 10.1083/jcb.129.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Sun J, Kim J, Rajagopal S, Zhai B, Villen J, Haas W, Kovacs JJ, Shukla AK, Hara MR, et al. Global phosphorylation analysis of beta-arrestin-mediated signaling downstream of a seven transmembrane receptor (7TMR). Proc Natl Acad Sci U S A. 2010;107:15299–15304. doi: 10.1073/pnas.1008461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Mihaylov V, Xu X, Brzostowski JA, Li H, Liu L, Veenstra TD, Parent CA, Jin T. A Gbetagamma effector, ElmoE, transduces GPCR signaling to the actin network during chemotaxis. Dev Cell. 2012;22:92–103. doi: 10.1016/j.devcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T, Zhai B, Yu Y, Kiyotsugu Y, Raschle T, Etzkorn M, Seo HC, Nagiec M, Luna RE, Reinherz EL, et al. Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc Natl Acad Sci U S A. 2014;111:E2182–2190. doi: 10.1073/pnas.1404943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.