Abstract
The study of biliary disease has been constrained by a lack of primary human cholangiocytes. Here we present an efficient, serum-free protocol for directed differentiation of human induced pluripotent stem cells into cholangiocyte-like cells (CLCs). CLCs show functional characteristics of cholangiocytes, including bile acids transfer, alkaline phosphatase activity, gamma-glutamyl-transpeptidase activity and physiological responses to secretin, somatostatin and VEGF. We use CLCs to model in vitro key features of Alagille syndrome, polycystic liver disease and cystic fibrosis (CF)-associated cholangiopathy. Furthermore, we use CLCs generated from healthy individuals and patients with polycystic liver disease to reproduce the effects of the drugs verapamil and octreotide, and we show that the experimental CF drug VX809 rescues the disease phenotype of CF cholangiopathy in vitro. Our differentiation protocol will facilitate the study of biological mechanisms controlling biliary development as well as disease modeling and drug screening.
Cholangiocytes are the main target of cholangiopathies, a diverse group of bile duct disorders that includes inherited diseases such as CF-associated cholangiopathy, developmental diseases such as Alagille Syndrome and autoimmune diseases such as primary biliary cirrhosis, as well as drug- and toxin-induced conditions (1). Cholangiopathies carry significant morbidity and mortality, accounting for up to a third of adult and >70% of pediatric liver transplantations (2). However, research on their pathophysiology has been limited by poor access to primary biliary tissue, difficulties in culturing primary cholangiocytes in vitro and inadequate animal disease models(3). The capacity of human induced pluripotent stem cells (hiPSCs) to proliferate indefinitely in culture and differentiate into a broad spectrum of cell types makes them well suited to in vitro disease modeling (5). Early methods for deriving cholangiocytes from hiPSCs (6) were based on spontaneous differentiation and had limited characterization of the resulting cells (7-8). Despite recent advances toward guided differentiation of hiPSCs to CLCs (9), current protocols show poor differentiation efficiency (<31%), and the derived cells differ considerably from primary biliary tissue in their transcriptional profiles. Furthermore, in vitro-generated cholangiocytes have not been shown to reproduce key functions of bona fide cholangiocytes, such as enzymatic activity (e.g., alkaline phosphatase (ALP) and gamma glutamyl transferase (GGT)), responses to hormonal stimuli (secretin and somatostatin) and chloride transfer through cystic fibrosis transmembrane regulator (CFTR) activity (7-9). Demonstration of these properties is essential for recapitulating cholangiopathies and studying the effects of therapeutic agents. Finally, current systems diverge from the physiological pathways controlling biliary development in vivo (7-9), limiting their value for developmental studies.
Here we report a stepwise method for cholangiocyte differentiation that recapitulates native biliary development (Fig. 1a). The quality, functionality and purity of the resulting CLCs is substantially higher compared to cells generated by previous methods (see Supplementary Note and Supplementary Figure 1 for detailed comparison).
Figure 1.
Generation of Cholangiocyte Progenitors (CP) from human Induced Pluripotent Stem Cells (hiPSCs). (a) Overview of the protocol used to differentiate hiPSCs to Cholangiocyte Like Cells (CLC). DE: Definitive endoderm, FP: Foregut progenitors, HB: Hepatoblasts, HC: Hepatocytes , BMP, bone morphogenetic protein; Ly294002 is a phosphatidylinositol-3-OH kinase inhibitor; CDM, chemically defined medium; RPMI, Roswell Park Memorial Institute medium; SB, SB-431542; HGF, hepatocyte growth factor; RA, retinoic acid; EGF, epidermal growth factor; FGF, fibroblast growth factor. (b) Immunofluorescence (IF) analyses demonstrating the expression of key hepatoblast markers as indicated, in day 12 hepatoblasts. Scale bars correspond to 100μm. (c) IF analyses demonstrating the expression of early biliary markers by immature cholangiocyte progenitors (day 16): Scale bars, 100μm. (d) Gene expression profile of hiPSC-derived cells at key stages of biliary differentiation and Primary Cholangiocytes (PCs). n=4 biological replicates for each stage of differentiation. n=3 independent samples for PCs. Center line, median; box, interquartile range (IQR); whiskers, range (minimum to maximum). IQR is not calculated for PCs, as n = 3. Asterisks represent statistical significance of differences between HBs, CPs and CLCs; # indicates lack of statistical significance (P > 0.05) (one-way ANOVA with Tukey correction for multiple comparisons). Values are relative to the housekeeping gene porphobilinogen deaminase (PBGD, also known as HMBS).
Results
Cholangiocyte progenitors generated from hiPSCs
We focused first on the generation of bipotent hepatoblasts, the common progenitor of hepatocytes and cholangiocytes (10). To achieve this goal, we adapted our established hepatic hiPSC differentiation protocol (11-12). Cells generated with the adapted protocol after 12 days of differentiation express hepatoblast markers, including AFP, HNF4A, HNF1B, TBX3, and CK19 (Figure 1b,1d), and have the potential to differentiate toward both the hepatic (Supplementary Figure 2a-2c) and biliary lineages (Figure 1c-1d, Supplementary table 1). To differentiate these hepatoblast-like cells into cholangiocyte progenitors (CPs), we interrogated pathways reported to control early biliary specification (10) (Supplementary Fig 3a-3c and data not shown) and found that activin in combination with retinoic acid suppressed the expression of the hepatoblast markers AFP, HNF4A and TBX3 (Supplementary Figure 3c). Addition of FGF10 along with activin and retinoic acid induced the expression of the early biliary specification markers SOX9, HNF1B and CK19 (Figure 1c-1d) (10), resulting in a population in which 75.1% of cells were CK19+/Sox9+ (Supplementary Figure 4a). Flow cytometry analyses identified the majority of the remaining cells as Sox9−/AFP+ hepatoblasts (Supplementary Figure 4a), explaining the presence of reduced but detectable AFP levels in our culture (Figure 1d). Mature biliary markers such as Secretin Receptor (SCR), Somatostatin Receptor 2 (SSTR2), Aquaporin1 and Anion Exchanger 2 (AE2) were not expressed (Figure 1d). Thus, activin, retinoic acid and FGF10 promote the differentiation of hepatoblast-like cells into early CLCs or cholangiocyte progenitors (CPs) in vitro.
To promote maturation of the CPs, we used 3D culture conditions known to induce cholangiocyte maturation through organoid formation (7-9). CPs grown in these conditions proliferated rapidly, organized into ring-like structures after 48-72 hours and within 5-7 days gave rise to cystic organoids and branching tubular structures (Figure 2a-2b) bearing primary cilia (Figure 2c) similar to those of primary cholangiocytes. The organoids expressed biliary markers, including CK7, CK18, CK19, HNF1B, Gamma Glutamyl-Transferase (GGT), Jagged1 (JAG1), NOTCH2, CFTR, SCR, SSTR2, Aquaporin1 and Anion Exchanger 2 (Figure 1d,2d) at levels similar to primary cholangiocytes (Figure 1d, Supplementary Figure 5). Transcriptomic analyses of common bile duct primary tissue and cells at key stages of our differentiation protocol (Supplementary Table 2, Figure 2e) using Euclidian hierarchical clustering revealed that CLCs are distinct from earlier developmental stages (Figure 2e, Supplementary Figure 6), cluster closely with primary common bile duct cholangiocytes (Pearson correlation coefficient for CLCs vs. CBD r = 0.747, CLCs vs. HBs r = 0.576, CLCs vs. hiPSCs: r = 0.474) and express both mature (SSTR2, ALP, KRT7) and fetal (SOX9) biliary markers, as shown by IF and QPCR analyses (Figure 2e, Supplementary Figure 6). These results confirm that CPs can differentiate into cells resembling biliary epithelial cells when grown in 3D culture.
Figure 2.
Generation of cholangiocyte-like cells (CLCs) from cholangiocyte progenitors (CPs). (a,b) Immunofluorescence (IF) (left) and light microscopy (right) images of CLC organoids (day 26) demonstrating the formation of cystic (a) and branching (arrows) tubular structures (b). Scale bars correspond to 100μm. (c) Transmitted electron microscopy images (right) and IF analyses (left) for acetylated alpha-tubulin demonstrating the presence of cilia (arrow). The IF image was acquired from the bottom of a large cystic organoid using confocal microscopy. Scale bars: 500 nm and 100μm respectively. L: Lumen. (d) IF analyses demonstrating the expression of early and mature biliary markers in day 25 CLCs as indicated. Scale bars: 100μm. (e) Euclidian hierarchical clustering analysis focusing on the genes that define the transcriptional signature of CLCs and HBs (4963 genes differentially expressed in CLCs vs. hiPSCs or in HBs vs. hiPSCs). For each probe, standard scores (z-scores) indicate the differential expression measured in number of standard deviations from the average level across all the samples. CLCs cluster closer and present higher correlation coefficient with Common Bile Duct cholangiocytes (CBD) used as a primary control, compared to HBs or hiPSCs (Pearson, CLCs vs. PCs r = 0.747, CLCs vs. HBs r = 0.576, CLCs vs. hiPSCs r = 0.474). Representative genes are indicated. The data corresponds to biological triplicates.
The efficiency of our differentiation protocol can be estimated from the observation that 1×106 hiPSCs produce by day 26 ~74.4×106 cells, of which ~57.28×106 express mature biliary markers (~74.4×106 cells generated X 77% (average differentiation efficiency across 3 lines, SD=6.5%) = ~57.28×106 CLCs) (Supplementary Figure 4a-b). More specifically, 74.5% of the resulting cells co-express the biliary marker Sox9 and the mature biliary marker CK7 (Supplementary Figure 4a). A further 7.5% of the cells express Sox9 but not CK7, consistent with immature cholangiocytes (Supplementary Figure 4a). 15% of the cells co-express AFP and Albumin (Supplementary Figure 4a), indicating the presence of a small fraction of hepatocytes in our culture conditions and explaining the detection of low AFP levels by QPCR analyses (Figure 1d). The remaining 3% of the cells were not characterized further. These results were confirmed on 3 independent hiPSC lines (Supplementary Figure 4a).
Next, we characterized the functionality of the generated organoids. In vivo, cholangiocytes re-absorb bile acids (13) and modify the composition of canalicular bile through a series of secretory and re-absorptive processes (14) regulated by intracellular calcium signaling (15). Native biliary epithelial cells have ALP and GGT activity and proliferate in response to stimuli such as Vascular Endothelial Growth Factor (VEGF). The secretory potential of CLCs generated in vitro was confirmed using Rhodamine123, a fluorescent substrate for the cholangiocyte surface glycoprotein multidrug resistance protein-1 (MDR1) (16-17). Rhodamine123 was actively secreted in the lumen of CLC organoids; however, luminal dye accumulation was prevented by the MDR1 inhibitor verapamil (Figure 3a-3c), confirming MDR1-dependent transfer of Rhodamine123. The capacity of CLCs for interacting with bile acids through the apical salt and bile transporter (ASBT) (13) was demonstrated by showing active export of the fluorescent bile acid cholyl-lysyl-fluorescein (CLF) from the lumen of CLF-loaded organoids compared to controls loaded with Fluorescein Isothiocyanate (FITC) (Figure 3d-3f, Supplementary Video 1,2). ASBT expression was confirmed by QPCR and IF analyses (Supplementary Figure 7a-7b). Furthermore, CLCs responded to acetylcholine and ATP stimuli by increasing intracellular calcium levels (Figure 3g, Supplementary video 3,4), demonstrated increased proliferation in response to VEGF stimulation (51% increase in fold expansion, P<0.0001, 2-tailed t-test) (Figure 3h-3i) and exhibited GGT and ALP activities similar to those of primary controls (GGT activity: 160% of human serum, P<0.0001, one-way ANOVA with Dunnett correction for multiple comparisons) (Figure 3j-3k). Together, these observations confirm that our hiPSC-derived CLCs display a range of functions of the native biliary epithelium.
Figure 3.
Functional characterization of CLC organoids (a) Representative images demonstrating the MDR1 fluorescent substrate Rhodamine123 detected in the lumen of CLC organoids, confirming MDR1 functionality. Scale bars, 50μm. (b) Fluorescence intensity measurements along the area indicated by the red line. (c) Mean intra-luminal fluorescence intensity normalized over background, in the presence (+VER) or absence (−VER) of verapamil, n=599 measurements, P=2.99×10−5 (2-tailed t-test). (d) Representative images demonstrating active export of the fluorescent bile acid cholyl-lysyl-fluorescein (CLF) from the lumen of CLC organoids compared to controls loaded with Fluorescein Isothiocyanate (FITC). (e) Fluorescence intensity along the area indicated by the red line. (f) Mean intraluminal fluorescence intensity normalized over background, n=1163 measurements, P<1×10−18 (2-tailed t-test). The data shown in panels a-f is representative of 3 different experiments. (g) Fluorescence intensity measurements (top) and representative fluorescence microscopy images (bottom) of CLC organoids loaded with the calcium indicator Fluo-4, demonstrating an increase in intracellular calcium levels following stimulation with ATP and acetylcholine. Plated primary cholangiocytes stimulated with ATP are used as a positive control. Grey area represents 1 SD, n=3. (h, i) Fold change over starting number of cells (h) and IF analyses for the proliferation marker Ki-67 (i) in the presence and absence of VEGF for 5 days, demonstrating that VEGF promotes CLC proliferation. Prim. Chol.: Plated primary cholangiocytes, n=10, p=4.77×10−17 (CLCs), p=4.63×10−17 (Prim. Chol.) (2-tailed t-test). (j) CLC organoids exhibit GGT activity. MEF: Mouse Embryonic Feeders, n=3, p<0.0001 for all comparisons (one-way ANOVA with Dunnett correction for multiple comparisons). (k) ALP staining revealing ALP activity in CLC organoids. Top: Photographs of stained wells (Scale bars:1cm). Bottom: Brightfield microscopy images, n=3. Scale bars:100μm. Data representative of multiple lines (Figure 6a-6b, Supplementary Figure 5). All error bars represent s.d. Asterisks (****) in panels c, f, h and j indicate statistical significance of the differences demonstrated (P < 0.0001).
CLCs model development of the human biliary system
To investigate potential applications of our system for developmental studies, we characterized signaling pathways that control organoid formation in vitro as compared with native duct development. First, we interrogated Activin/TGFβ signaling in view of its pivotal role in physiological biliary specification and tubulogenesis (10, 18-19). We blocked the activity of TGFβ, which is normally present in Matrigel, using the Activin receptor inhibitor SB-431542. SB-431542 completely abrogated organoid formation (Figure 4a, 4b) confirming the role of Activin/TGFβ signaling as a key regulator of organoid formation in our system.
Figure 4.
Activin and Notch signaling are essential for CLC organoid formation. (a) Number of CLC organoids following culture of CPs in matrigel in the presence and absence of SB-431542, demonstrating suppression of organoid formation secondary to inhibition of activin signaling. Error bars represent SD, n=4. (b) Live pictures demonstrating lack of organoid formation in response to SB-431542. Scale bars: 100μm. (c) QPCR analyses for the expression JAG1, NOTCH2, and the Notch downstream target HES1 in CLC organoids vs. CP cultured in matrigel in the presence of L-685,458, demonstrating reduced expression of this marker in response to L-685,458, n=4. Values are relative to the housekeeping gene PBGD). Center line, median; box, interquartile range (IQR); whiskers, range (minimum to maximum). ****P<0.0001 (two-tailed t-test). (d) IF analyses for NICD in CLC organoids vs. CP cultured in matrigel in the presence of L-685,458 demonstrating pathway activation and nuclear localization of NICD in CLC organoids vs. pathway inhibition in response to L-685,458. Scale bars:100μm, EL: extra-luminal space. (e) Number of CLC organoids following culture of CPs in matrigel in the presence and absence of L-685,458 demonstrating a significant reduction in organoid formation, following inhibition of Notch signaling. Error bars represent SD, n=4. (f) Live pictures demonstrating reduced organoid formation in response to L-685,458. The data shown in each panel is representative of 3 different experiments.
We performed similar analyses for Notch signaling during biliary specification of hepatoblasts to CPs and CLC organoid formation in vitro. Deregulation of Notch signaling is associated with Alagille Syndrome (AGS), characterized by a paucity of bile ducts (20). To explore the hypothesis that blocking Notch signaling would impair organoid formation in vitro, we first characterized the activity of the Notch pathway in our system. Notch activation results in cleavage and nuclear translocation of its intracellular domain (21-23). Immunofluorescence (IF) analysis with antibodies specific to the cleaved Notch intracellular domain (NICD) confirmed the presence of active NICD, with increased nuclear localization in CPs (Supplementary Figure 8) and CLC organoids (Figure 4d). The expression of NOTCH2, as well as its ligand JAG1 and its downstream target HES1, were also increased in both stages compared to hepatoblasts, consistent with pathway activation (CPs vs. HBs: NOTCH2: P<0.001, JAG1: P<0.001, HES1: P<0.05; CLCs vs. CPs or HBs: P<0.0001) (Figure 1d). Inhibition of Notch signaling in 3D culture conditions using the gamma-secretase inhibitor L-685,458 blocked cleavage of the NICD, suppressed HES1, NOTCH2 and JAG1 expression (Figure 4c) and blocked organoid formation (Figure 4e-4f), confirming the importance of Notch signaling for the generation of organoids incorporating a luminal space in our system.
These results reinforce previous findings obtained in mice by demonstrating the importance of Activin/TGFβ and Notch signaling pathways in human cholangiocyte specification (24) and underline the potential of our culture system for studying human biliary tree development in vitro.
CLCs validate drugs for polycystic liver diseases
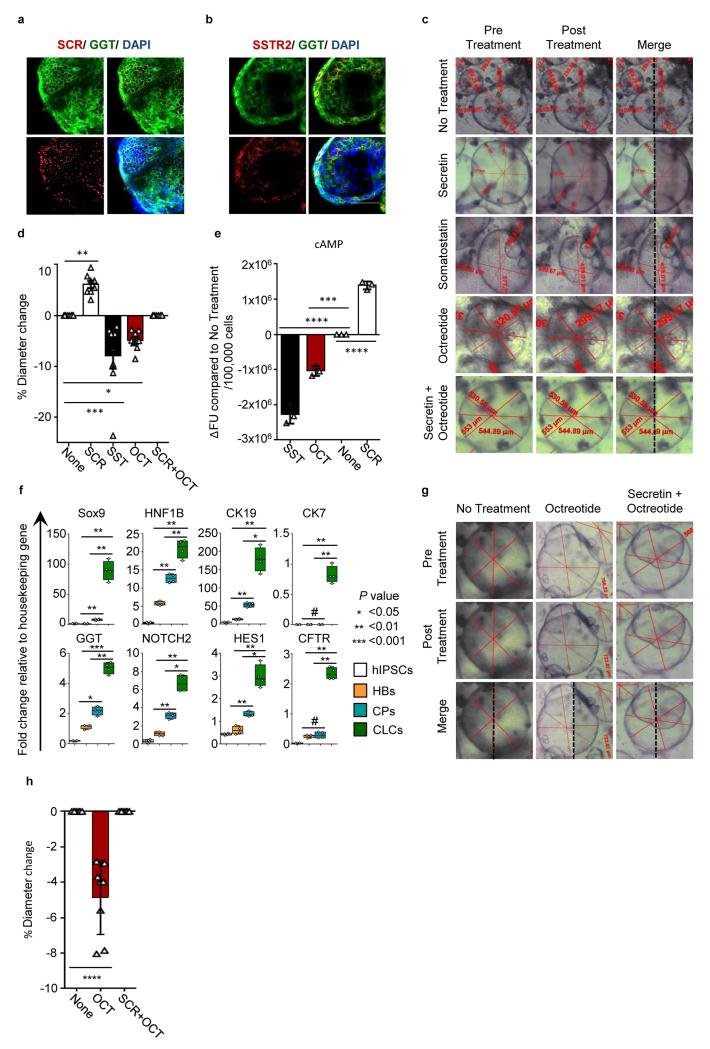
Polycystic liver diseases are characterized by multiple cystic lesions in the liver arising from fetal cholangiocytes (25-26). Intraluminal fluid secretion and cholangiocyte proliferation result in cyst expansion and liver impairment owing to space-occupying effects (27-28). We explored the use of CLC organoids to identify compounds that might reduce cyst size in PLD. The secretory activity of cholangiocytes is increased by the hormone secretin and reduced by the hormone somatostatin, resulting in changes in duct size. Octreotide, a synthetic analog of somatostatin, is used clinically to restrict cyst size in polycystic liver disease (28-32). CLC organoids express both SCR and SSTR2 (Figure 1d, 5a–5b) suggesting that these pathways may be functional in our cells. Accordingly, secretin increased organoid size (6.1% average diameter increase, P<0.01, one-way ANOVA with Dunnett correction for multiple comparisons), whereas somatostatin and octreotide decreased organoid size, compared to untreated controls (7.9% and 4.9% average diameter decrease respectively, p<0.001 and p<0.05 respectively, one-way ANOVA with Dunnett correction for multiple comparisons) (Figure 5c-5d, Supplementary video 5-7). Furthermore, octreotide negated the effects of secretin and decreased intracellular cAMP levels (45% of somatostatin response, P=0.001, one-way ANOVA with Dunnett correction for multiple comparisons), in keeping with previous studies (26,30) (Figure 5e). To further test the effects of octreotide on disease-specific CLCs, we differentiated hiPSCs derived from a patient with polycystic liver disease (Supplementary Figure 9) to CLCs (Figure 5f). Octreotide treatment reduced organoid size (4.86%, P<0.0001, one-way ANOVA with Dunnett correction for multiple comparisons) (Figure 5g-5h), reproducing the effects of the drug in vitro.
Figure 5.
CLC organoids respond to secretin and somatostatin stimuli and reproduce the effects of somatostatin analogues (octreotide) in Polycystic Liver Disease (PLD) in vitro. (a) IF analysis demonstrating the expression of secretin receptor (SCR) in CLCs. Scale bars: 100μm. (b) IF analysis demonstrating the expression of somatostatin receptor 2 (SSTR2) in CLCs. Scale bars: 100μm. (c) Live pictures demonstrating CLC organoids pre and post treatment with secretin, somatostatin, octreotide, secretin combined with octreotide. Diameter measurements are shown in each image. The images have been cropped to include a single cyst, but are representative of all the cysts measured. (d) Effect of secretin (SC), somatostatin (SST), octreotide (OCT) and the combination of secretin and octeotide on organoid diameter. Error bars represent SEM, n=8, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (one-way ANOVA with Dunnett correction for multiple comparisons. (e) Secretin treatment increases, while somatostatin and octreotide treatment decrease cAMP levels in CLC organoids. Error bars represent SEM, n=3, Asterisks represent statistically significant differences (one-way ANOVA with Dunnett correction for multiple comparisons). (f) QPCR demonstrating the expression of biliary markers in polycystic liver disease CLCs. Asterisks represent statistical significance in differences between HBs, CPs and CLCs (one-way ANOVA with Tuckey correction for multiple comparisons). (g,h) Live images (g) and diameter measurements (h) in PLD-CLC organoids pre and post treatment with octreotide or the combination of secretin and octreotide, ****:P<0.0001 (one-way ANOVA with Dunnett correction for multiple comparisons). The data shown is representative of 3 different experiments.
CLCs model CF Liver Disease
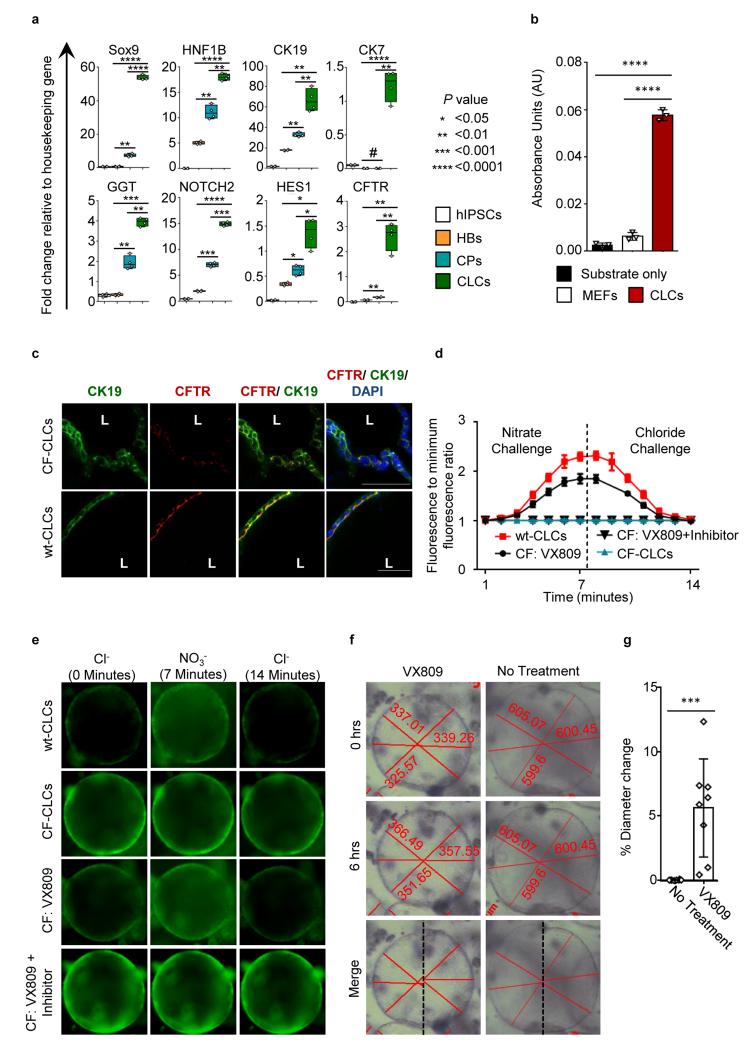
The autosomal recessive disorder CF is caused by mutations in the cystic fibrosis transmembrane conductance regulator gene (CFTR), a cell-surface chloride transporter (33-34). CFTR mutations in cholangiocytes result in reduced intra-luminal chloride secretion, increased bile viscosity, and focal biliary cirrhosis secondary to bile plugs occluding the intrahepatic bile ducts (35-36). To model CF biliary disease in vitro, we generated hiPSCs from skin fibroblasts of a patient homozygous for the most common CF mutation ΔF508 (CF- hiPSC) (37) and differentiated them into CLCs. CF-hiPSC-derived CLCs (CF-CLCs) expressed markers (Figure 6a) and displayed functional characteristic of biliary epithelial cells (Figure 6b). Transcription of the CFTR gene was confirmed using QPCR (Figure 6a), and IF analyses detected minimal CFTR protein expression (Figure 6c), in agreement with studies reporting very rapid ER degradation of the misfolded protein (38). We also used the fluorescent chloride indicator N-(6-methoxyquinolyl)acetoethyl ester (MQAE) (39) to monitor intracellular and intraluminal chloride concentration. Wild-type (WT) CLC organoids appropriately modified intracellular chloride in response to media with varying chloride concentration, whereas no change was observed in CF-CLCs (Figure 6d,6e), confirming the absence of functional CFTR in these cells.
Figure 6.
Modeling Cystic Fibrosis (CF) liver disease in vitro, using hiPSCs derived from patients with CF. (a) QPCR analyses of CLC organoids generated from CF-hiPSCs (CF-CLC), demonstrating the expression of biliary markers. Asterisks denote statistical significance in differences between HBs, CPs and CLCs (one-way ANOVA with Tuckey correction for multiple comparisons). (b) CFCLC organoids exhibit GGT activity. ****:P<0.0001 (one-way ANOVA with Dunnett correction for multiple comparisons). (c) IF analyses revealing very low CFTR protein expression in CF-CLC vs. wt-CLCs expressing CFTR. (d) MQAE fluorescence intensity normalized over the lowest intensity value. MQAE fluorescence is quenched in the presence of chloride, but not affected by nitrate. Changes in intracellular or intra-luminal chloride levels in response to extracellular chloride levels depend on the presence of CFTR functionality. MQAE fluorescence increases in response to a nitrate challenge depleting extracellular chloride and decreases in response to chloride in wt- and CFCLCs treated with VX809, however fails to respond to both challenges in CFCLCs and CF-CLCs treated with VX809 + CFTR inhibitor-172. (e) Live images demonstrating an increase in MQAE fluorescence in response to a nitrate challenge, followed by a decrease in response to a chloride challenge in wt-CLCs and CF-CLCs treated with VX809, however MQAE fluorescence remains unchanged in CF-CLCs and CF-CLCs treated with VX809 and CFTR inhibitor-172. (f) Live pictures demonstrating CLC organoids pre and post treatment with VX809. Diameter measurements are shown in each image. (g) Effect of VX809 treatment on mean organoid diameter. Error bars represent SD, n=8, P=0.001, (2-tailed t-test). Images cropped to include 1 cyst, but representative. All data shown is representative of 3 different experiments.
Next we investigated the effects of the experimental CF drug VX809 (41) in the context of biliary disease. VX809 stabilizes CFTR, corrects folding defects in patients with the ΔF508 mutation and increases CFTR functionality in lung cells (42). Incubation of CF-CLCs with VX809 for 48 hours increased CFTR function analyzed by MQAE to a level similar to that of WT-CLCs (Figure 6d, 6e). This effect was negated by the CFTR inhibitor-172, confirming that the phenotypic rescue of CF-CLCs by VX809 was dependent on improved CFTR function (Figure 6d, 6e).
Given the association between chloride and fluid secretion in cholangiocytes (33), we studied the impact of VX809 on organoid size. CF-CLC organoids treated with VX809 increased in size compared to their untreated counterparts (5.6% mean diameter increase, P=0.001, 2-tailed t-test) (Figure 6f, 6g). This observation confirmed that VX809 increases CFTR function and improves intraluminal fluid secretion, suggesting a previously unreported therapeutic effect for this drug in the context of CF liver disease.
Discussion
We present a protocol for the generation of CLCs from hiPSCs that addresses the limitations of previously published differentiation methods by more closely recapitulating natural bile duct development (Fig. 1a). Our results confirm the importance of FGF10 and activin/TGFβ for early biliary specification, as previously described in vivo (10,19,43), and reveal a role for retinoic acid in this process, at least in vitro. We show that the combination of these factors under chemically defined, serum-free conditions is sufficient to promote the specification of hiPSC-derived hepatoblasts into cholangiocyte progenitors with high efficiency. Production of cholangiocyte progenitors, mimicking physiological biliary development, has not been demonstrated previously and likely explains the efficiency of our culture system in generating CLCs that closely resemble bona fide cholangiocytes at the transcriptional (Supplementary Figure1, Supplementary table 3, 4) and functional levels (Supplementary table 3).
Our protocol will enable various applications, such as developmental studies, disease modeling, therapeutic target validation and drug screening. Accordingly, we used patient-derived hiPSCs to model polycystic and CF liver disease and applied these models to reproduce the effects of the therapeutic compounds verapamil, octreotide and VX809. VX809 has already completed phase IIa clinical trials (42) for CF, but its effects on CF-associated cholangiopathy have not been described, to our knowledge. Our results suggest that CLCs provide a suitable system for identifying new therapeutic agents, which is particularly important given the lack of high-throughput drug screening platforms for cholangiopathies. CLCs may also contribute to tissue engineering of livers or liver organoids that incorporate a biliary system for the treatment of end-stage disease.
Online Methods
Generation of hiPSC lines
All the hiPSC lines used here were derived and characterized previously by our lab (11, 37). Briefly, the lines used were generated from human skin fibroblasts and peripheral blood (ethics reference no. 08/H0311/201 and 09/H0304/77 respectively), using the Yamanaka approach (4, 9). The CF fibroblasts were obtained from the Coriell cell repository. The lines were authenticated using SNIP arrays and regularly tested negative for mycoplasma contamination.
Culture of hiPSCs
Human iPS cells were maintained in defined culture conditions as previously described (11-12, 44), using activin-A (10ng/ml) and b-FGF (12ng/ml).
Differentiation of hiPSCs into cholangiocyte progenitors
hiPSCs were differentiated into Foregut Progenitor cells (FP) as previously described (12, 44). Bipotent hepatoblasts were generated by culturing FPs in RPMI (Gibco, Invitrogen) + B27 supplemented with SB-431542 (10μM, Tocris Bioscience) and BMP4 (50ng/ml) for 4 days. To induce biliary specification, we cultured hepatoblasts for another 4 days in the presence of RPMI (Gibco, Invitrogen) + B27 supplemented with FGF10 (50ng/ml, Peprotech), activin-A (50ng/ml) and RA (3μM, Sigma-Aldrich).
Maturation of cholangiocyte progenitor cells to CLCs and organoid formation in 3D culture
Human CPs were passaged using Cell Dissociation Buffer (Gibco, Life Technologies) and suspended at a density of 8 × 104 cells/ml, in a mixture of 40% matrigel (BD Biosciences, catalogue number: 356237) and 60% William’s E medium (Gibco, Life Technologies) supplemented with 10mM nicotinamide (Sigma-Aldrich), 17mM sodium bicarbonate (Sigma Aldrich), 0.2mM 2-Phospho-L-ascorbic acid trisodium salt (Sigma-Aldrich), 6.3mM sodium pyruvate (Invitrogen), 14 mM glucose (Sigma-Aldrich), 20 mM HEPES (Invitrogen), ITS+ premix (BD Biosciences), 0.1uM dexamethasone (R&D Systems), 2mM Glutamax (Invitrogen),100U/ml penicillin per 100μg/ml streptomycin and 20ng/ml EGF (R&D Systems). A 50μL droplet of the cell suspension was added in the centre of each well of a 24-well plate; the gel was allowed 1 hour at 37°C to solidify and then overlaid with William’s E medium with supplements. The medium was changed every 48 hours and the cells were cultured for a total of 10 days.
Importantly, using the same methodology, we have been able to culture CLC organoids in multiple formats ranging from 6 to 96 well plates. To generate large numbers of CLC organoids, multiple 50μL droplets were added in a well of a 6 well plate or a 10cm dish. To provide a large number of wells compatible with high throughput screening and large scale experiments 30μL droplets were added in a well of a 96 well plate. In both cases, the gel was allowed 1 hour at 37°C to solidify and then overlaid with William’s E medium with supplements.
Inhibition of activin and Notch signaling in 3D culture and assessment of organoid formation
Human CPs were suspended at a density of 8 × 104 cells/ml, in a mixture of 40% matrigel and 60% William’s E medium (Gibco, Life Technologies) with supplements as described above. The cell suspension was distributed in 3 equal volume aliquots. One aliquot received no further supplementation and was used as a positive control. The second aliquot was further supplemented by 10μM SB-431542 for the inhibition of TGFβ/activin signaling. 50μM of L-685,458 (Tocris Biosciences) were added in the third aliquot for the inhibition of Notch signaling. Each aliquot was distributed in 24-well plate format. The same concentrations of inhibitors were added to the medium overlaying the matrigel on a daily basis. After a total of 10 days in 3D culture, the total number of cysts in 4 random wells of a 24 well plate was counted for each condition by a blinded researcher. Error bars represent SD.
Flow cytometry analyses
HiPSCs, FPs, HBs and CPs were dissociated to single cells using Cell Dissociation Buffer (Life Technologies). The cells were subsequently counted using a hemocytometer and fixed using 4% PFA for 20 minutes at 4°C. Cell staining and flow cytometry analyses were performed as previously described (44).
CLC organoids were washed once with PBS and 1ml of ice cold dispace was added per well of a 24 well plate. The matrigel was mechanically dissociated, transferred in a falcon tube and kept on ice to allow the combination of low temperature and dispase digestion to liquefy the matrigel. After 10 minutes the cells were centrifuged at 1600 rpm for 3 minutes and the supernatant was aspirated. The pellet was washed once with PBS and the centrifugation step repeated. The supernatant was aspirated and 1 ml of TrypLE (Life technologies) was added for 3-5 minutes until the organoids were dissociated to single cells. Finally, the single cell suspension was centrifuged at 1600 rpm for 3 minutes, and fixed using 4% PFA for 20 minutes at 4°C. Cell staining and flow cytometry analyses were performed as previously described (44).
Primary cholangiocytes
Frozen primary human cholangiocytes derived from common bile duct were obtained from Celprogen (Catalogue Number 36755-11). The cells were derived from donors negative for Hepatitis B Surface Antigen, HIV1 and 2, Syphilis, Hepatitis B Virus, Human T Lymphocyte Virus 1 and 2, Hepatitis C Virus, HIV 1, West Nile Virus and Trypanosoma cruzi. Each frozen ampule (1.2 × 106 cells) of 1 × 106 viable cells was thawed with gentle agitation in a 37°C water bath. The cells were transferred to a sterile centrifuge tube and 1 ml of pre-warmed Human Cholangiocyte Cell Culture Complete Growth Media (catalogue number M36755-11S) was added. The ampule was washed twice with Human Cholangiocyte Cell Culture Complete Growth Media and the cells were centrifuged at 100g for 7 minutes. The pellet was subsequently re-suspended in 500ul of Human Cholangiocyte Cell Culture Complete Growth Medium. The cells were distributed equally to 2 wells of a 6 well plate pre-coated with Human Cholangiocyte Cell Culture Extracellular Matrix (catalogue number M36755-11-6-wells) with 2ml of Human Cholangiocyte Cell Culture Complete Growth Medium per well. The cells were incubated at 37°C in a 5% CO2 humidified incubator for 48 hours and the media was changed once. After another 24 hours, when the wells were confluent the cells were lysed and RNA was extracted as previously described (11-12), or fixed with 4% PFA as previously described (11-12) for ALP staining. QPCR analyses for biliary markers (Figure 1d) and ALP staining (Figure 3k) confirmed the nature and cholangiocyte properties of the cells.
Primary biliary tissue
Primary biliary tissue (bile duct) was obtained from an organ donor. The liver and pancreas from the donor were being retrieved for transplantation. A section of the bile duct was excised during the multi-organ retrieval operation after obtaining informed consent from the donor's family (REC reference number: 09/H0306/73). The tissue was homogenized using a tissue homogenizer and RNA was extracted as previously described (11).
Immunofluorescence, RNA extraction and Quantitative Real Time PCR
IF, RNA extraction and QPCR were performed as previously described (9). A complete list of the primary and secondary antibodies used is provided in supplementary table 5. A complete list of the primers used is provided in supplementary table 6. All QPCR data are presented as mean values of four independent biological replicates, with the exception of the primary CDB cholangiocytes from Celprogen, where 3 independent samples were used. Error bars represent SD.
For IF in 3D matrigel cultures, the organoids were fixed in matrigel with 4% PFA for 20 minutes at room temperature, to avoid matrigel liquefaction. The samples were permeabilized and blocked with 0.1% Triton-X and 10% donkey serum respectively for 30 minutes and incubated with primary antibody in 1% donkey serum overnight at 4°C. The following day the samples were washed 3 times with PBS for 45 minutes per wash, incubated with secondary antibody in 1% donkey serum for 60 minutes at room temperature and washed again 3 times in PBS. Hoechst 33258 was added to the first wash.
All IF images were acquired using a Zeiss Axiovert 200M inverted microscope or a Zeiss LSM 700 confocal microscope. Imagej 1.48k software (Wayne Rasband, NIHR, USA, http://imagej.nih.gov/ij) was used for image processing. Changes in brightness or contrast during processing were applied equally across the entire image.
For RNA extraction in 3D matrigel cultures, the organoids were washed once with PBS and 1ml of ice cold dispace was added per well of a 24 well plate. The matrigel was mechanically dissociated, transferred in a falcon tube and kept on ice to allow the combination of low temperature and dispase digestion to liquefy the matrigel. After 10 minutes the cells were centrifuged at 1600 rpm for 3 minutes and the supernatant was aspirated. The pellet was washed once with PBS and the centrifugation step repeated. Finally, the supernatant was aspirated and 350μL of RNA lysis buffer were added to the pellet. RNA was extracted from the lysate using a kit (Sigma-Aldrich), according to the manufacturer’s instructions.
Microarrays
500ng of total cellular RNA was amplified and purified using the Illumina TotalPrep-96 RNA Amplification kit (Life Technologies) according to the manufacturer’s instructions. Three biological replicates for each condition were analysed. Biotin-Labelled cRNA was then normalized to a concentration of 150ng/μl and 750ng were hybridised to Illumina Human-12 v4 BeadChips for 16 hours (overnight) at 58 °C. Following hybridisation, BeadChips were washed and stained with streptavidin-Cy3 (GE Healthcare). BeadChips were then scanned using the BeadArray reader, and image data was then processed using Genome Studio software (Illumina). The raw and processed microarray data are available on ArrayExpress (Accession number: E-MTAB-2965, https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2965/).
Microarrays analysis
Probe summaries for all arrays were obtained from the raw data using the method “Making Probe Summary”. These values were transformed (variance stabilized) and quantile normalised using the R/Bioconductor package lumi (45). Standard lumi QC procedure was applied and no outliers were identified. Differential expression between pairs of conditions was evaluated using the R/Bioconductor package limma (46). A linear model fit was applied, and the top differentially expressed genes were tabulated for each contrast using the method of Benjamini and Hochberg to correct the p-values (47). Probes that failed to fluoresce above background in both conditions were removed. Differentially expressed probes were selected using a cutoff of adjusted p-value <0.01 and absolute fold-change > 2.
Probes differentially expressed between hiPSCs and CLCs or hiPSCs and HBs (representing the aggregate transcriptional “signature” of CLCs and HBs) were selected for Euclidean hierarchical clustering using Perseus software (MaxQuant). Standard scores (z-scores) of the log2 normalized probe expression values across the different conditions were calculated and used for this analysis.
Rhodamine123 transport assay
CLC organoids were incubated with 100μM of Rhodamine 123 (Sigma-Aldrich) for 5 minutes at 37°C and the washed with William’s E medium 3 times. Fresh William’s E medium with supplements was added following the third wash. The organoids were incubated at 37°C for another 40 minutes. To demonstrate that Rhodamine123 transfer indeed reflected the activity of the membrane channel MultiDrug Resistance Protein 1 (MDR1), CLCs were incubated with 10μM of Verapamil (Sigma-Aldrich) at 37°C for 30 minutes and the rhodamine assay was repeated. Following completion of each experiment, images were taken using a confocal microscope. Multiple fluorescence measurements were made (around 1000) between the organoid interior and exterior. Rhodamine123 fluorescence in the organoid lumen was normalized over background measured in the surrounding external area. Each experiment was repeated in triplicate. Error bars represent SD. Mean fluorescence intensity comparisons were performed using a two sided student’s t-test.
Cholyl-Lysyl-Fluorescein transport assay
CLC organoids were loaded with 5uM of Cholyl-Lysyl-Fluorescein (CLF, Corning Incorporated) for 30 minutes at 37°C and the washed with Leibovitz’s medium (Life technologies) 3 times. Following completion of the third wash, time lapse images were taken using a confocal microscope for 10 minutes. To demonstrate that the changes in CLF fluorescence intensity observed were secondary to active export of CLF from the organoid lumen, the experiment was repeated with 5μM of unconjucated Fluorescein Isothiocyanate (FITC) (Sigma-Aldrich) as a control. Multiple fluorescence measurements were made (around 1000) between the organoid interior and exterior. Fluorescence in the organoid lumen was normalized over background measured in the surrounding external area. Each experiment was repeated in triplicate. Error bars represent SD. Mean fluorescence intensity comparisons were performed using a two sided student’s t-test.
Measurement of intracellular calcium levels
Intracellular calcium signaling, regulated by stimuli such as acetylcholine and ATP constitutes a key second messenger for cholangiocytes (15). CLC organoids were incubated with 25μM of the calcium indicator Fluo-4 AM (Life technologies) for 60 minutes at 37°C and washed 3 times with William’s E medium. Fresh William’s E medium with supplements was added following the third wash. The organoids were stimulated with 1μM of Acetylcholine (Sigma-Aldrich) or 30μM of ATP (Sigma-Aldrich), while time lapse images were taken. Each measurement was repeated in triplicate. To calculate the number of cells responding to stimulation, the number of cells loaded with Fluo-4 AM was counted by 2 different researchers prior to the start of the experiment. Following stimulation with ATP or acetylcholine the number of responding cells (increase in fluorescence) was also counted and responsiveness was expressed as the ratio of responding cells over the total number of cells loaded with Fluo-4 AM. The statistical approach for smoothing the data and plotting bands for the confidence limits please see ‘Statistical analyses’.
Proliferation assays
20 50μL droplets of Matrigel, each containing 40,000 cells were distributed in 20 wells of a 24 well plate. VEGF at a concentration of 50ng/ml was added to half of the wells with every media change. Following 5 days of culture the matrigel was digested with dispase as described above (RNA extraction section) and the organoids were mechanically dissociated to single cells. The number of cells for each well was then counted using a haemocytometer. 20 different measurements were made by a blinded researcher. Primary cholangiocytes distributed in 6 wells of a 12 well plate were used as a positive control. 3 wells received VEGF at a concentration of 50ng/ml with every media change for 5 days after which, the number of cells in each well was counted as described above. Error bars represent SD. Mean cell number comparisons were performed using a two sided student’s t-test.
GGT activity
GGT activity was measured in triplicate using the MaxDiscovery™ gamma-Glutamyl Transferase (GGT) Enzymatic Assay Kit (Bioo scientific) based on the manufacturer’s instructions. Mouse embryonic feeders were used as a negative control. The equivalent serum GGT activity in IU/L was calculated following the manufacturer’s instructions by multiplying the average increase in absorbance over 10 minutes by 353. Error bars represent SD. Multiple mean absorbance comparisons (CLCs vs. substrate, CLCs vs. MEFs, CLCs vs. human serum) were performed using one-way ANOVA with Dunnett correction for multiple comparisons.
Alkaline Phosphatase staining
Alkaline phosphatase was carried out using the BCIP/NBT Color Development Substrate (5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium) (Promega) according to the manufacturer’s instructions.
Effect of Secretin, Somatostatin, Octreotide and VX809 on organoid size
Images of CLC organoids were taken using 5X magnification before and following the addition of secretin (100nM, Sigma Aldrich), somatostatin (100nM, Sigma Aldrich), octrotide (100nM, Sigma Aldrich) or embryo transfer water serving as a negative control, at 0.5 - 2 minute intervals until organoid size stabilized. To explore the impact of octreotide on the effect of secretin, cells were pre-incubated for 30 minutes with octreotide. 100nM of secretin (Sigma Aldrich) was subsequently added to the medium and the experiment was carried out as described above. To assess the effect of VX809 on organoid size images were taken before and 6 hours following the addition of VX809 (30mM, Selleck) or embryo transfer water, serving as a negative control. 3 random diameters were measured for 8 random organoids pre and post treatment. Graph measurements represent percentage differences in mean organoid diameter. Error bars represent SD. Statistical significance was calculated using one-way ANOVA with Dunnett correction for multiple comparisons. The videos available as online supplementary data were made by taking images pre and post treatment at 2 minute intervals, until organoid size stabilized.
cAMP levels
cAMP levels were measured in triplicate using the cAMP-Glo assay kit (Promega) based on the manufacturer’s instructions and a P450-Glomax 96 microplate luminometer (Promega). Error bars represent SD. Statistical significance was calculated using one-way ANOVA with Dunnett correction for multiple comparisons.
CFTR activity
CFTR activity was measured as previously described (37). Briefly, MQAE is a fluorescent dye quenched by the presence of chloride but not affected by other anions or pH changes (39). Chloride transfer across the cell membrane is mainly regulated by CFTR in cholangiocytes. Therefore, cells with a functional CFTR will respond to a chloride challenge by rapidly increasing intracellular (and intraluminal in case of organoids) chloride concentration thereby quenching MQAE fluorescence. Chloride depletion using a nitrate solution will have the opposite effect.
Cells were incubated with 8mM MQAE fluorescent dye (Life Technologies) and 5μM forskolin for 4 hours at 37°C. MQAE fluorescence is quenched in the presence chloride. Standard Ringers solution containing NaCl, KCl, CaCl, MgCl, glucose and hepes was used to provide a chloride challenge expected to increase intracellular chloride levels in the presence of functional CFTR. Modified Ringers solution consisting of NaNO3, KNO3, CaNO3, MgNO3, glucose and hepes was used to promote chloride efflux and deplete intracellular chloride. Live pictures were captured every minute as each solution was added. To demonstrate the effect of VX809 on CFTR functionality, CLC organoids were incubated with 30mM of VX809 (Selleck) for 48 hours. The assay was repeated as described above in the presence and absence of 7uM CFTR inhibitor-172 (Sigma-Aldrich) to confirm the specificity of the compound for CFTR. Intracellular fluorescence intensity was measured in 3 random areas from the wall of each organoid using ImageJ software and normalized over the minimum fluorescence value for each area. Error bars represent SD.
Cytochrome p450 activity
Cyp3A4 activity was measured using the p450-Glo assay kit (Promega) according to the manufacturer’s instructions and a P450-Glomax 96 microplate luminometer (Promega).
Timing of experiments on CLC organoids
All the experiments and characterization with regards to CLCs were performed on CLC organoids, following 10 days of 3D culture unless stated otherwise
Statistical analyses
All statistical analyses were performed using GraphPad Prism 6 or the R statistical environment. For comparison between 2 mean values a 2-sided student’s t-test was used to calculate statistical significance. For comparison between multiple values one-way ANOVA was used with Tuckey correction for multiple comparisons when comparing multiple values to each other (e.g. QPCR plots) or Dunnett correction for multiple comparisons when comparing multiple values to a single value (e.g. functional assays where the values are compared to a negative control). The normal distribution of our values was confirmed using the Kolmogorov-Smirnov test where appropriate. Further information on the statistical analysis of our data is provided in Supplementary table 7 (test used for each experiment/ analysis, test statistic, degrees of freedom, P value).
To smooth our data for generating the curves in figure 3g we used functional data analysis theory (48) implemented in the R package 'fda' (http://cran.r-project.org/web/packages/fda/index.html). First, we represented our data values (3 replications at each fluorescence intensity measurement) using 60 equidistant B-spline basis functions, and roughness penalties in the second derivative (lambda=1). We used the functions create.bspline.basis and smooth.fd in the interval 1-100 seconds. Then, we evaluated the mean and the standard deviation of the functional data objects using the R functions mean.fd and sd.fd.
Supplementary Material
Acknowledgments
This work was funded by ERC starting grant Relieve IMDs (L.V., N.H.), the Cambridge Hospitals National Institute for Health Research Biomedical Research Center (L.V., N.H., F.S.), the Evelyn trust (N.H.) and the EU Fp7 grant TissuGEN (M.CDB.). FS has been supported by an Addenbrooke’s Charitable Trust Clinical Research Training Fellowship and a joint MRC-Sparks Clinical Research Training Fellowship.
The authors would like to thank the Cambridge BRC hiPSCs core facility for the derivation of the Cystic Fibrosis hiPSC line, Dr Petroula-Anastasia Tsagkaraki for her help with the generation of the manuscript figures and statistical analyses, Dr Jeremy Skepper, Ms Lyn Carter and the University of Cambridge Advanced Imaging Centre for their help with electron microscopy, Dr Chris McGee and the Wellcome Trust Sanger Institute for their help with microarray data processing and analysis, Mr Barlow McLeod for IT support and Dr Stephanie Brown for technical support and advice.
Footnotes
Competing interests: LV is a founder and shareholder of DefiniGEN. The remaining authors have nothing to disclose.
References
- 1.Lazaridis KN, Strazzabosco M, Larusso NF. The cholangiopathies: disorders of biliary epithelia. Gastroenterology. 2004;127:1565–77. doi: 10.1053/j.gastro.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Murray KF, Carithers RL, Jr, AASLD AASLD practice guidelines: Evaluation of the patient for liver transplantation. Hepatology. 2005;41:1407–32. doi: 10.1002/hep.20704. [DOI] [PubMed] [Google Scholar]
- 3.Pollheimer MJ, Trauner M, Fickert P. Will we ever model PSC? - “it's hard to be a PSC model!”. Clin Res Hepatol Gastroenterol. 2011;35:792–804. doi: 10.1016/j.clinre.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampaziotis F, Segeritz CP, Vallier L. Potential of human induced pluripotent stem cells in studies of liver disease. Hepatology. 2014 doi: 10.1002/hep.27651. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 7.Zhao D, Chen S, Cai J, Guo Y, Song Z, et al. Derivation and characterization of hepatic progenitor cells from human embryonic stem cells. PLoS One. 2009;4(7):e6468. doi: 10.1371/journal.pone.0006468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanimizu N, Miyajima A, Mostov KE. Liver progenitor cells develop cholangiocyte-type epithelial polarity in three-dimensional culture. Mol Biol Cell. 2007;18:1472–9. doi: 10.1091/mbc.E06-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dianat N, Dubois-Pot-Schneider H, Steichen C, Desterke C, Leclerc P, et al. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology. 2014 doi: 10.1002/hep.27165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–89. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–36. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannan NR, Segeritz CP, Touboul T, Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat Protoc. 2013;8:430–7. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia X, Francis H, Glaser S, Alpini G, LeSage G. Bile acid interactions with cholangiocytes. World J Gastroenterol. 2006;12:3553–63. doi: 10.3748/wjg.v12.i22.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanno N, LeSage G, Glaser S, Alvaro D, Alpini G. Functional heterogeneity of the intrahepatic biliary epithelium. Hepatology. 2000;31:555–61. doi: 10.1002/hep.510310302. [DOI] [PubMed] [Google Scholar]
- 15.Minagawa N, Ehrlich BE, Nathanson MH. Calcium signaling in cholangiocytes. World J Gastroenterol. 2006;12:3466–70. doi: 10.3748/wjg.v12.i22.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gigliozzi A, Fraioli F, Sundaram P, Lee J, Mennone A, Alvaro D, Boyer JL. Molecular identification and functional characterization of Mdr1a in rat cholangiocytes. Gastroenterology. 2000;119:1113–22. doi: 10.1053/gast.2000.18156. [DOI] [PubMed] [Google Scholar]
- 17.Cízková D, Morký J, Micuda S, Osterreicher J, Martínková J. Expression of MRP2 and MDR1 transporters and other hepatic markers in rat and human liver and in WRL 68 cell line. Physiol Res. 2005;54:419–28. [PubMed] [Google Scholar]
- 18.Antoniou A, Raynaud P, Cordi S, Zong Y, Tronche F, et al. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136:2325–33. doi: 10.1053/j.gastro.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, et al. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849–54. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turnpenny PD, Ellard S. Alagille syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet. 2012;20:251–7. doi: 10.1038/ejhg.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 22.Saravanamuthu SS, Gao CY, Zelenka PS. Notch signaling is required for lateral induction of Jagged1 during FGF-induced lens fiber differentiation. Dev Biol. 2009;332:166–76. doi: 10.1016/j.ydbio.2009.05.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisler F, Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382–92. doi: 10.1002/hep.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, et al. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–39. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raynaud P, Tate J, Callens C, Cordi S, Vandersmissen P, et al. A classification of ductal plate malformations based on distinct pathogenic mechanisms of biliary dysmorphogenesis. Hepatology. 2011;53:1959–66. doi: 10.1002/hep.24292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandok N. Polycystic liver disease: a clinical review. Ann Hepatol. 2012;11:819–26. [PubMed] [Google Scholar]
- 27.Temmerman F, Missiaen L, Bammens B, Laleman W, Cassiman D, et al. Systematic review: the pathophysiology and management of polycystic liver disease. Aliment Pharmacol Ther. 2011;34:702–13. doi: 10.1111/j.1365-2036.2011.04783.x. [DOI] [PubMed] [Google Scholar]
- 28.Caroli A, Antiga L, Cafaro M, Fasolini G, Remuzzi A, et al. Reducing polycystic liver volume in ADPKD: effects of somatostatin analogue octreotide. Clin J Am Soc Nephrol. 2010;5:783–9. doi: 10.2215/CJN.05380709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marinelli RA, Tietz PS, Pham LD, Rueckert L, Agre P, et al. Secretin induces the apical insertion of aquaporin-1 water channels in rat cholangiocytes. Am J Physiol. 1999;276:G280–6. doi: 10.1152/ajpgi.1999.276.1.G280. [DOI] [PubMed] [Google Scholar]
- 30.Gong AY, Tietz PS, Muff MA, Splinter PL, Huebert RC, Strowski MZ, Chen XM, LaRusso NF. Somatostatin stimulates ductal bile absorption and inhibits ductal bile secretion in mice via SSTR2 on cholangiocytes. Am J Physiol Cell Physiol. 2003;284:C1205–14. doi: 10.1152/ajpcell.00313.2002. [DOI] [PubMed] [Google Scholar]
- 31.Caperna TJ, Blomberg le A, Garrett WM, Talbot NC. Culture of porcine hepatocytes or bile duct epithelial cells by inductive serum-free media. In Vitro Cell Dev Biol Anim. 2011;47:218–33. doi: 10.1007/s11626-010-9382-3. [DOI] [PubMed] [Google Scholar]
- 32.Masyuk TV, Masyuk AI, Torres VE, Harris PC, Larusso NF. Octreotide inhibits hepatic cystogenesis in a rodent model of polycystic liver disease by reducing cholangiocyte adenosine 3′,5′-cyclic monophosphate. Gastroenterology. 2007;132:1104–16. doi: 10.1053/j.gastro.2006.12.039. [DOI] [PubMed] [Google Scholar]
- 33.Rowe SM, Miller SB, Sorscher EJ. Cystic Fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 34.Davies JC, Alton EW, Bush A. Cystic fibrosis. BMJ. 2007;335:1255–9. doi: 10.1136/bmj.39391.713229.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colombo C. Liver disease in cystic fibrosis. Curr Opin Pulm Med. 2007;13:529–36. doi: 10.1097/MCP.0b013e3282f10a16. [DOI] [PubMed] [Google Scholar]
- 36.Staufer K, Halilbasic E, Trauner M, Kazemi-Shirazi L. Cystic fibrosis related liver disease--another black box in hepatology. Int J Mol Sci. 2007;15:13529–49. doi: 10.3390/ijms150813529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hannan NR, Sampaziotis F, Segeritz C, Hanley N, Vallier L. Generation of Distal Airway Epithelium from Multipotent Human Foregut Stem Cells. Stem Cells Dev. 2015 doi: 10.1089/scd.2014.0512. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem. 1994;269:25710–8. [PubMed] [Google Scholar]
- 39.Shenoy A, Kopic S, Murek M, Caputo C, Geibel JP, et al. Calcium-modulated chloride pathways contribute to chloride flux in murine cystic fibrosis-affected macrophages. Pediatr Res. 2011;70:447–52. doi: 10.1203/PDR.0b013e31822f2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haack A, Aragão GG, Novaes MR. Pathophysiology of cystic fibrosis and drugs used in associated digestive tract diseases. World J Gastroenterol. 2013;19:8552–8561. doi: 10.3748/wjg.v19.i46.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci U S A. 2011;108:18843–8. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clancy JP, Rowe SM, Accurso FJ, Aitken ML, Amin RS, et al. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax. 2012;67:12–8. doi: 10.1136/thoraxjnl-2011-200393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanai M, Tatsumi N, Hasunuma N, Katsu K, Endo F, et al. FGF signaling segregates biliary cell-lineage from chick hepatoblasts cooperatively with BMP4 and ECM components in vitro. Dev Dyn. 2008;237:1268–83. doi: 10.1002/dvdy.21520. [DOI] [PubMed] [Google Scholar]
- 44.Hannan NR, Fordham RP, Syed YA, Moignard V, Berry A, et al. Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Reports. 2013;1:293–306. doi: 10.1016/j.stemcr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du P, Kibbe W, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 46.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 47.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate : A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 48.Ramsay JO, Silverman BW. Functional Data Analysis. 2nd ed Springer; New York, USA: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.