Abstract
Microglia play critical roles in neural development, homeostasis and neuroinflammation and are increasingly implicated in age-related neurological dysfunction. Neurodegeneration often occurs in disease-specific spatially-restricted patterns, the origins of which are unknown. We performed the first genome-wide analysis of microglia from discrete brain regions across the adult lifespan of the mouse and reveal that microglia have distinct region-dependent transcriptional identities and age in a regionally variable manner. In the young adult brain, differences in bioenergetic and immunoregulatory pathways were the major sources of heterogeneity and suggested that cerebellar and hippocampal microglia exist in a more immune vigilant state. Immune function correlated with regional transcriptional patterns. Augmentation of the distinct cerebellar immunophenotype and a contrasting loss in distinction of the hippocampal phenotype among forebrain regions were key features during ageing. Microglial diversity may enable regionally localised homeostatic functions but could also underlie region-specific sensitivities to microglial dysregulation and involvement in age-related neurodegeneration.
Introduction
Microglia are a specialised population of tissue macrophages resident in the central nervous system (CNS) parenchyma and adapted to the unique properties of the CNS environment1. Recent studies have revealed an expanding array of functions for microglia during brain development and adult homeostasis and in neurodegeneration, infection and brain injury2. These studies have shown that the cellular activities of microglia extend beyond their well-established role as immune sentinels and effectors to include synaptic organisation3, 4, control of neuronal excitability5, phagocytic debris removal6 and trophic support for brain protection and repair7, 8.
The multifunctional roles of microglia may be considered part of a spectrum of environmental monitoring that is designed to sense perturbations and elicit appropriate microglial responses to maintain homeostasis. The local environment will therefore be a key influence shaping microglial phenotype. Notably, exposure to neuronal cell surface and soluble factors has been shown to maintain microglia in a comparatively quiescent immunophenotype (versus systemic macrophage populations)9, 10. Recent studies have extended these findings to describe key features of the genome-wide transcriptional profile of microglia that distinguishes their phenotype from non-CNS macrophages11, 12.
The microenvironment is not uniform throughout the various brain regions. Variations in neuronal subtypes, neurotransmitter profiles, haemodynamics and metabolism could all be an influence on and be influenced by local microglial phenotype. Moreover, the permeability of the blood-brain barrier and resultant exposure to systemic signals that can also modify microglial phenotype are regionally heterogeneous. It remains unclear if microglial phenotype is similarly diverse throughout the brain. Regional variations in microglial density13, surface expression of a small panel of immune molecules14, and dependency on maintenance by IL-3415, 16 suggest there could be differences.
Ageing is associated with alterations in the neuroinflammatory environment and recent studies have uncovered risk alleles in age-related neurodegenerative disease that implicate microglial dysfunction and neuroinflammatory processes as contributory factors17. The pathological targeting and progression of most neurodegenerative conditions occurs in region-specific patterns and regulatory mechanisms of gene expression in the human brain were recently shown to have regional differences18. This suggests that it is important to determine if ageing modifies any region-specific influences on microglial phenotype.
Here we have used genome-wide transcriptional profiling of adult microglia from discrete brain regions at three different ages in combination with network analyses to determine the nature of microglial diversity in the adult mouse brain and the impact of ageing. To our knowledge, these data provide the first account of the microglial regional transcriptome throughout the adult lifespan. Our data reveal microglia as richly diverse cells under steady-state conditions, show that microglial ageing occurs non-uniformly in a region-dependent manner, and define the transcriptional basis and major functional features responsible for this region- and age-related diversity. Our datasets provide an extensive and publically-accessible comparative resource for future studies exploring microglial function, dysfunction and contribution to age-related neurodegeneration.
Results
Isolation of adult microglia from discrete brain regions
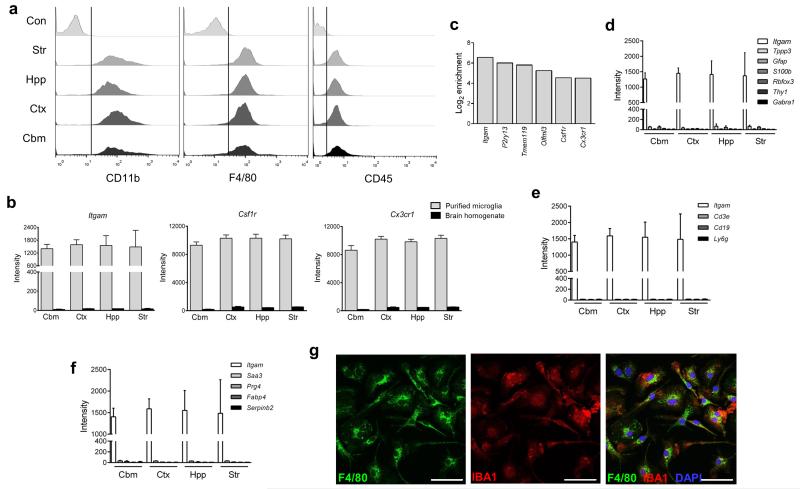
We refined established techniques to purify adult mouse microglia by density gradient and immuno-magnetic separation (Fig 1 and Supplementary Fig 1a). We first validated the consistency of microglial extraction from all regions of interest (cerebellum, cerebral cortex, hippocampus, striatum). The CD11b antigen is ubiquitously expressed on microglia throughout all brain regions as shown by colocalisation with GFP+ microglia in the Csf1r-EGFP “MacGreen” reporter mouse (Supplementary Fig 1b). The post-purified selected fraction consisted of a single population of CD11b+ and F4/80+ cells and there was no detectable CD11b or F4/80 staining in the post-purified non-selected fraction confirming CD11b as an efficient target for purification (Supplementary Fig 1c). Microglia obtained from all regions showed a uniform CD11b+F4/80+CD45lo profile (Fig 1a) characteristic of resident brain microglia and distinguishing them from CD45hi systemic macrophage populations. This indicated that we were extracting an equivalent microglial population from all brain regions. Expression of Itgam (encodes CD11b) and other established microglial/macrophage genes including Csf1r and Cx3cr1 was similarly enriched in purified microglia from each region in comparison to the respective mixed brain cell homogenates (Fig 1b). Additional genes recently reported as microglial “signature” genes (e.g. Tmem119, P2ry13)11, 12, 19 were also highly enriched in purified samples (Fig 1c) whereas markers of neurons, astrocytes and oligodendrocytes were expressed at negligible levels in purified microglia (Fig 1d). Genes highly expressed in blood leukocyte subsets including Cd3e (T lymphocytes), Cd19 (B lymphocytes) and Ly6g (granulocytes) were undetectable in purified microglia (Fig 1e) and there was no expression of systemic macrophage-specific genes identified from a recent study (e.g. Fabp4, Serpinb2)11 in microglial samples (Fig 1f). Immunostaining of isolated cells in culture showed that all cells stained positively for the microglial/macrophage antigens IBA1 and F4/80 (Fig 1g). Together these data verify the purity and consistency of microglial extraction across brain regions.
Figure 1.
Validation of multi-region microglial purification. (a) Microglia were purified from discrete brain regions and the profile of expression for indicated surface markers examined by flow cytometry. A consistent CD11b+F4/80+CD45lo profile was observed for all regions (Con denotes isotype control staining of whole brain microglia). Data are representative of four independent cell preparations, each from tissue pooled from eight mice. (b) Microarray expression profiles for selected genes in purified microglia and mixed brain cell homogenates from each brain region. Data show mean ± SD, n = 4 independent samples, each from tissue pooled from eight mice (c) The fold change (log2) in microarray expression level for purified microglia versus mixed cell brain homogenates for indicated genes. (d) Microarray expression profiles in purified microglia from discrete brain regions for established marker genes of neurons, astrocytes and oligodendrocytes, (e) T cells (Cd3e), B cells (Cd19), granulocytes (Ly6g), and (f) non-CNS macrophages with comparison to Itgam. Data show mean ± SD, n = 4 independent samples, each from tissue pooled from eight mice. (g) Immunofluorescence images of purified microglia cultured for 7d and immunostained for indicated markers. Images are representative of two independent cultures. Scale bar, 50μm. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum.
The microglial transcriptome is regionally heterogeneous
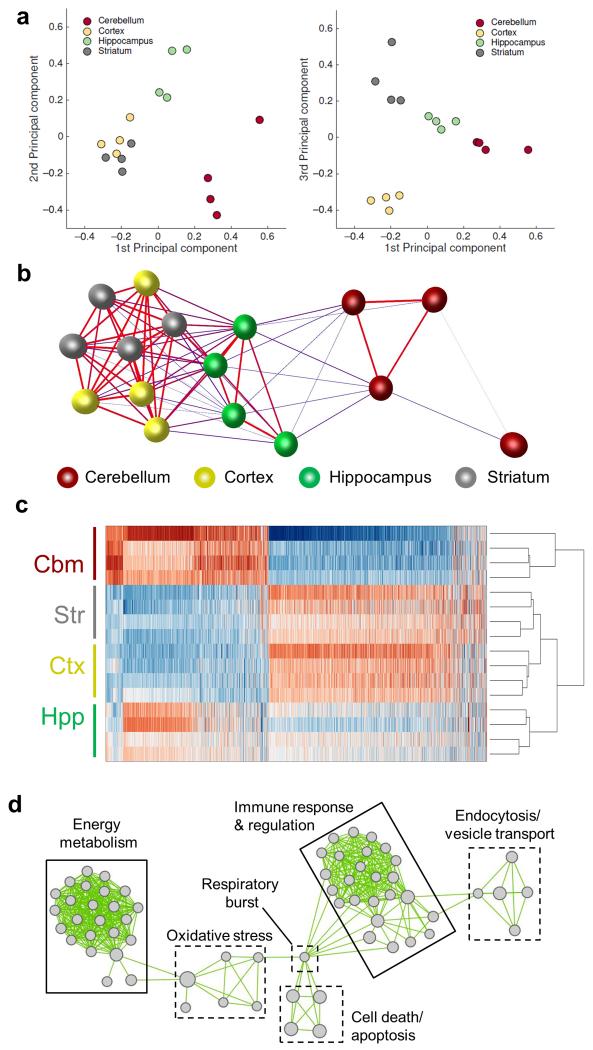
We initially determined whether the microglial transcriptome in the healthy young adult brain (4 months of age) is regionally heterogeneous. Principal components analysis (PCA) showed clustering of samples in a region-dependent manner and indicated a close relationship between microglial expression profiles of the cerebral cortex and striatum and relatively more distinct profiles in the cerebellum and hippocampus (Fig 2a). These relationships were validated non-subjectively using the network visualisation and analysis tool BioLayout Express3D (http://www.biolayout.org/). Sample-to-sample correlation analysis showed clustering according to brain region with similar inter-regional relationships as PCA (Fig 2b). Thus, the global gene expression profile of adult microglia in the healthy brain is regionally heterogeneous.
Figure 2.
The adult mouse microglial transcriptome is regionally heterogeneous. (a) Principal components analysis on microarray expression profiles for purified microglia from discrete brain regions. (b) Sample-to-sample correlation of microarray datasets was performed in BioLayout Express3D and a network graph generated (Pearson correlation threshold r ≥ 0.96). Nodes represent individual samples and edges the degree of correlation between them. (c) Heat map showing the expression pattern of probesets differentially expressed by brain region (p < 0.05 with FDR correction). The scaled expression value (row Z-score) is displayed in a blue-red colour scheme with red indicating high expression and blue low expression. (d) Differentially expressed probesets were analysed for enrichment of Gene Ontology (GO) Biological Processes in DAVID. Enriched GO terms were imported to Enrichment Map and a network graph generated. Nodes represent individual GO terms (gene sets) and edges the relatedness between them. Two major clusters defined by immunoregulatory and metabolic function were identified. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum.
Expression of 3,131 probesets (~7% of total) representing 2527 genes was differentially regulated by brain region (FDR q < 0.05) (Supplementary Table 1). Hierarchical clustering of samples based on differentially expressed probesets demonstrated the marked contrast in expression profile between cortical/striatal and cerebellar microglia and the intermediate profile in hippocampal microglia (Fig 2c). Genes involved in multiple aspects of immune function were among the most differentially expressed by region (Supplementary Table 1) including those with established function and others previously unexplored in microglia. Analysis of Gene Ontology (GO) biological processes using DAVID revealed “immune response” and “immune effector response” as significantly over-represented (Supplementary Table 2). There was also a striking over-representation of multiple processes associated with energy metabolism (Supplementary Table 2). We used the Enrichment Map (http://baderlab.org/Software/EnrichmentMap/) network visualisation tool to remove redundancy in GO enrichment annotation. The key feature of the network was the presence of two major clusters each comprising functionally-related and highly-connected enriched gene sets with roles in immune function and energy metabolism (Fig 2d). Full annotation of nodes within these clusters is presented in Supplementary Table 3. These data indicated immunoregulatory and bioenergetic/metabolic processes as the major contributors to regional diversity in microglial phenotype of the young adult brain.
Three major patterns of region-dependent microglial gene co-expression
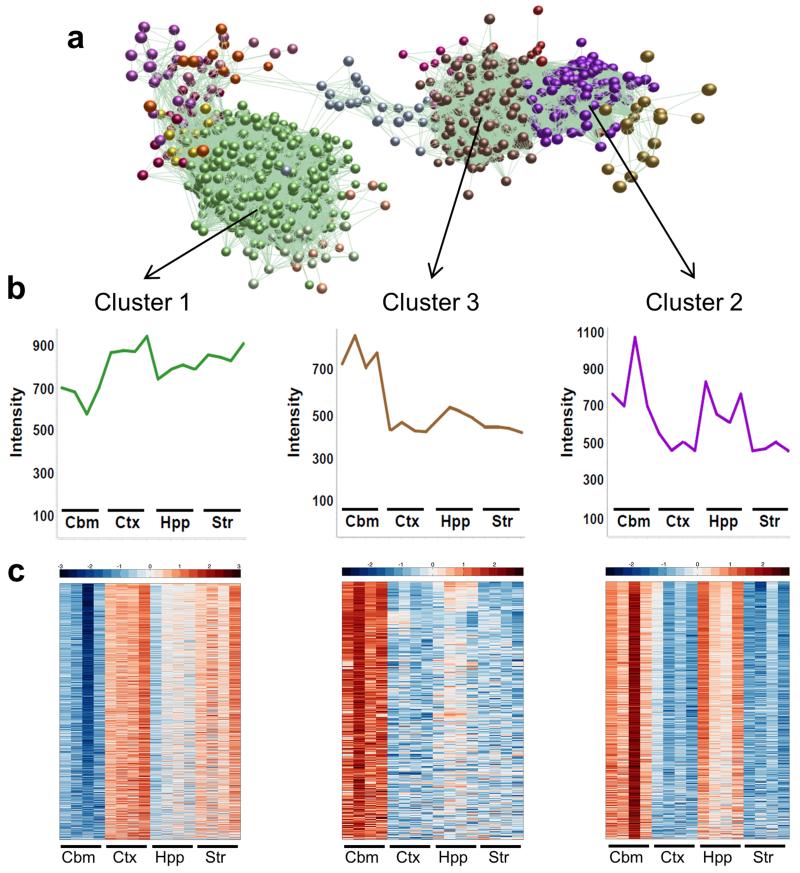
We next sought to define the region-specific microglial phenotypes by assessing patterns of gene co-expression using BioLayout Express3D (http://www.biolayout.org/)20. The utility of BioLayout Express3D for the identification of spatiotemporal patterns of gene expression and the discovery of transcriptional networks underpinning common functional pathways has been described previously20. A network graph constructed from the set of 3,131 regionally differentially expressed probesets was clustered using a Markov clustering algorithm to non-subjectively sub-divide the graph into discrete sets of co-expressed genes. Overall graph structure consisted of 14 clusters ranging in size from 10 to >1,000 nodes (Fig 3a and Supplementary Fig 2a). Three major clusters distributed across two distinct regions of the graph were evident (Fig 3a and Supplementary Table 4). The mean expression profiles of these three clusters showed that cluster 1 contained genes whose expression was relatively greater in cerebral cortex and lower in cerebellum (Fig 3b). In contrast, clusters 2 and 3, which were located together and distant from cluster 1, both contained genes with relatively greater expression in the cerebellum (with greater hippocampal expression in cluster 2). The expression profile for individual genes within each cluster generally followed the cluster mean (Fig 3c). Increasing the Pearson correlation threshold did not materially affect the overall graph or clustering structure (Supplementary Fig 2)
Figure 3.
Three major patterns of gene co-expression underpin regional microglial transcriptional heterogeneity. (a) A transcript-to-transcript correlation network graph of transcripts significantly differentially expressed by brain region was generated in BioLayout Express3D (Pearson correlation threshold r ≥ 0.80). Nodes represent transcripts (probesets) and edges the degree of correlation in expression pattern between them. The network graph was clustered using a Markov clustering algorithm and transcripts assigned a colour according to cluster membership. Three major clusters were identified. (b) Mean expression profile of all transcripts within clusters 1, 2 and 3. (c) Heat maps showing the expression profile of all transcripts contained within clusters 1, 2 and 3. Each probeset is represented in a blue-red row Z-score scale with red indicating high expression and blue low expression. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum.
Microglial immunophenotyppic and bioenergetic heterogeneity
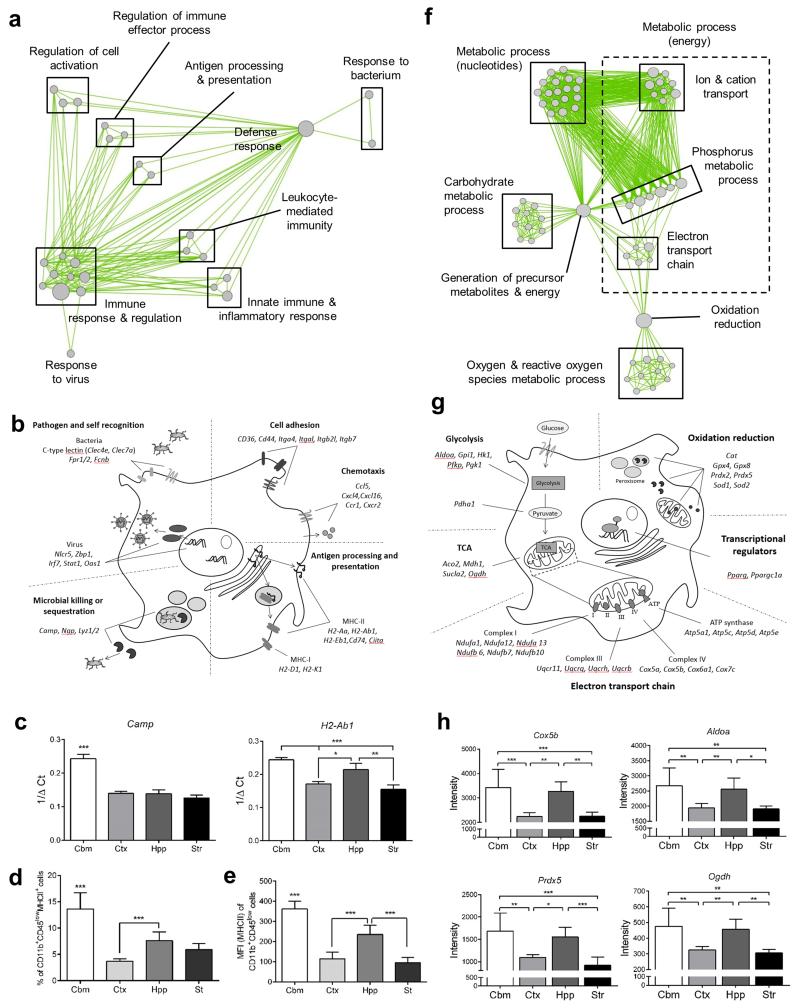
Genes which share highly correlated expression profiles across a range of experimental conditions (i.e. are co-expressed) are often distinct components of a common pathway or biological process21. A large number of cluster 3 genes (high expression in cerebellum) were immune-related. GO analysis revealed “immune response” and “defence response” as the most overrepresented biological processes (Supplementary Table 5) and clustering using Enrichment Map underlined the array of enriched immune-related processes (Fig 4a). To gain further insight to the molecular functions encoded in cluster 3, we manually annotated genes according to the following major categories: pathogen/self-recognition, cell adhesion and chemotaxis, signalling integration, antigen presentation, and microbial killing/sequestration (Fig 4b). Genes from multiple molecular classes involved in pathogen (or self) recognition were present in cluster 3. Bacterial recognition genes included the C type lectins (Clec4e (Mincle), Clec7a (Dectin 1), Cd209a (DC-SIGN)) and Fcnb (Ficolin B) and the formyl peptide receptors Fpr1 and Fpr2. Viral recognition was also evident in the high expression of the Zbp1 gene, which encodes cytoplasmic sensors of viral DNA22. Consistent with antiviral activity, there were a large number of interferon pathway genes (e.g. Stat1, Stat4, Ifit2, Ifitm3, Irf7, Oas1, Plscr1) and pathway analysis in Ingenuity identified an enriched interferon network and both interferon gamma and the type I interferon receptor (IFNAR) as top upstream regulators (Supplementary Fig 3 and Supplementary Table 6). A striking feature of cluster 3 was the presence of multiple genes involved in antigen processing and presentation, including both MHC-I (H2-D1, H2-K1) and MHC-II (H2-Aa, H2-Ab1, H2-Eb1, Cd74) pathways. Upstream regulation was evident through the presence of Ciita encoding the master regulator of MHC-II expression and Nlrc5, the master regulator of MHC-I gene expression. Pathway analysis in KEGG identified antigen processing and presentation as a significantly over-represented pathway (Supplementary Fig 4 and Supplementary Table 7). Genes encoding several classes of immune effector molecules, many involved in pathogen killing or sequestration, were present in this cluster. Camp and Ngp, genes encoding the anti-microbial peptides mCRAMP and neutrophilic granule protein respectively, were of particular note because a recent study identified these genes as unexpectedly highly expressed in microglia compared to non-CNS macrophages11. The regional expression profile of selected genes from cluster 3 was further assessed by quantitative PCR (Fig 4c) and at the protein level by flow cytometry with both demonstrating comparable profiles to the microarray data (Fig 4d, e). We also noted a panel of immunoregulatory molecules in cluster 1 (relatively high expression in cortex and low in cerebellum) indicating that some immunoregulatory pathways may be more active in regions other than the cerebellum (see below also). These were largely immune signalling genes (e.g. Cd47, Cd300a) encoding molecules that limit the strength of myeloid cell responses to external stimuli.
Figure 4.
Regional transcriptional heterogeneity in microglial immunophenotype and bioenergetics. (a) Cluster 3 transcripts were analysed for enrichment of Gene Ontology (GO) Biological Processes in DAVID (p < 0.05 with Benjamini correction) and a network graph of enriched GO terms generated in Enrichment Map. Nodes represent individual GO terms (gene sets) and edges the relatedness between them. (b) Examples of individual genes in cluster 3 manually annotated to functional categories of immunoregulatory function. (c) mRNA expression of selected genes in purified microglia measured by quantitative PCR. Data show mean ± SD, n = 4 independent samples, each from tissue pooled from eight mice. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA with Bonferroni correction. (d, e) Expression of MHC-II protein was measured by flow cytometry on freshly isolated adult microglia identified by CD11b+CD45lo profile in mixed brain cell suspensions from discrete brain regions. Data show (d) proportion of CD11b+CD45lo microglia positive for MHC-II and (e) mean fluorescence intensity of MHC-II expression on CD11b+CD45lo cells. Data show mean ± SD, n = 3 independent cell preparations. ***p < 0.001, one-way ANOVA with Bonferroni correction. (f) Cluster 2 transcripts were analysed for enrichment of GO Biological Processes (p < 0.05 with Benjamini correction) and a network graph of enriched GO terms generated in Enrichment Map. (g, h) Examples of individual genes in cluster 2 manually annotated to functional categories of bioenergetic function. Data show mean ± SD, n = 4 independent samples, each from tissue pooled from eight mice. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA with Bonferroni correction. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum. Specific p values for all statistical comparisons are presented in Supplementary Table 13.
The bioenergetic profile of myeloid cells is tightly linked to their immunophenotype and the environmental conditions they are exposed to (e.g. normoxia/hypoxia)23. We were therefore interested that cluster 2 (high expression in cerebellum and hippocampus) contained a large number of genes associated with key components of energy production systems and their regulation (Fig 4f-h). This was validated by GO analysis which revealed “generation of precursor metabolites and energy”, “electron transport chain” and “oxidative” phosphorylation” as among the most highly over-represented processes (Supplementary Table 8) and visualisation in Enrichment Map showed multiple clusters associated with glycolysis, the electron transport chain, ATP synthesis and redox metabolic activity (Fig 4f). Genes encoding most enzymes in the glycolytic pathway, the tricarboxylic acid (TCA) cycle, multiple subunit constituents of each of the proton pump complexes in the electron transport chain (I, III, IV) and the ATP synthase complex were present in cluster 2 (Fig 4g). Key regulators of energy metabolism were also present, notably peroxisome proliferator-activated receptor gamma (Pparg) and the associated co-activator peroxisome proliferator-activated receptor gamma coactivator 1-alpha (Ppargc1a), which co-operate to control transcription of an array of genes involved in mitochondrial function and energy metabolism. Antioxidant responses, important for counteracting oxygen radicals produced during oxidative phosphorylation, were also represented, including the superoxide dismutase (Sod1, Sod2), catalase (Cat), peroxiredoxin (Prdx2, Prdx5) and glutathione peroxidase (Gpx4, Gpx8) families (Fig 4g). Individual gene expression profiles of representative examples for each of the above classes are shown in Fig 4h.
Collectively, the above data support the suggestion that cerebellar and hippocampal microglia maintain a more immune-alert state than microglia in the striatum and cortex and this is accompanied by relatively greater expression of an extensive set of co-regulated genes involved in energy metabolism.
Microglial steady-state heterogeneity in immune alertness
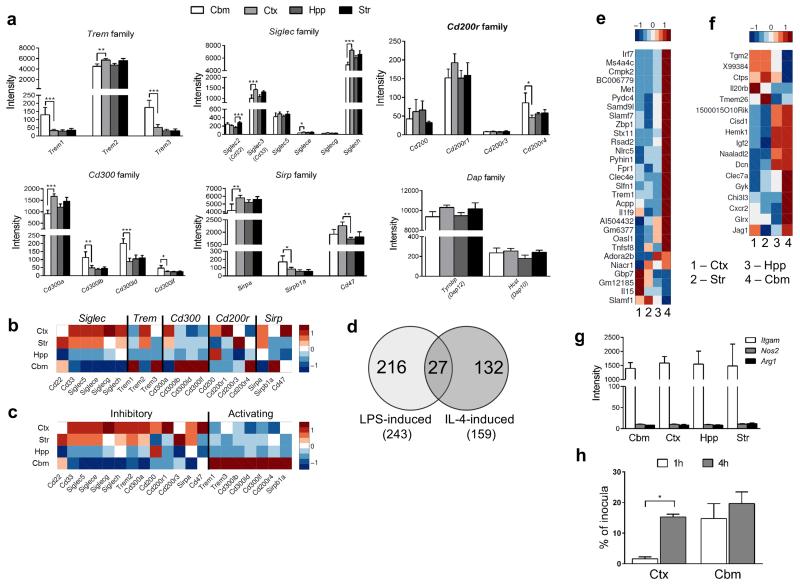
Immune cells maintain a balance between activating and inhibitory signals to fine-tune the strength of their responses in part through cell surface activating receptors that associate with immunoreceptor tyrosine-based activation motifs (ITAMs) and counteracting inhibitory receptors containing immunoreceptor tyrosine-based inhibition motifs (ITIMs)24. Given the more immune-alert state of cerebellar and hippocampal microglia suggested above, we explored whether there were regional differences in microglial expression of ITAM-associating and ITIM-containing immunoreceptors. We focussed on the triggering receptor expressed on myeloid cell (TREM), sialic acid-binding immunoglobulin-type lectins (Siglec), CD200R, CD300 and signal regulatory peptide (SIRP) families, each of which contains activating and inhibitory members (Fig 5a). In all families, we found inter-regional differences in expression of both ITAM and ITIM-signalling members and strikingly there was an opposing pattern of expression for activating and inhibitory receptors (Fig 5b, c). Activating ITAM-associating members were more highly expressed in cerebellar microglia whereas ITIM-containing inhibitory members showed the reverse pattern. For example, in the CD300 family, CD300a is the only member with a cytoplasmic tail containing an ITIM motif and Cd300a was expressed at lower levels in cerebellum and hippocampus. In contrast, other CD300 members have consensus sequences enabling association with ITAM-containing adapters such as DAP12 and all showed the opposite pattern of expression to Cd300a. One caveat relates to TREM2, which although associating with the ITAM-containing DAP12 adapter molecule, dampens microglial pro-inflammatory reactions, and thus the expression profile is consistent with other inhibitory immunoreceptors. Expression of the genes encoding the ITAM-containing adapter proteins DAP12 and DAP10 was consistent across brain regions, perhaps reflecting their common use for signalling by several receptors (Fig 5a).
Figure 5.
Regional microglial heterogeneity in immunophenotype suggests differences in immune vigilance. (a) Microarray expression levels in purified microglia of selected families of immunoreceptors containing activating and inhibitory members. Data show mean ± SD, n = 4 independent samples, each from tissue pooled from eight mice. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA with Bonferroni correction. (b,c) Heat maps showing microarray expression patterns of immunoreceptor genes arranged according to (b) family and (c) activating (A) or inhibitory (I) status. Column Z-score intensities represent the mean of four independent samples per region with red referring to a high probeset expression and blue low expression (d) Genes uniquely induced (>5-fold, p < 0.05 with FDR correction) by LPS or IL-4 in microglia were determined from publicly available microarray expression datasets25. (e,f) Heat maps showing microarray expression patterns for the subsets of unique (e) LPS- or (f) IL-4-inducible genes that were differentially expressed (p < 0.05 with FDR correction) according to brain region. Row Z-score intensities represent the mean of four independent samples per region with red referring to a high probeset expression and blue low expression. (g) Microarray expression levels of archetypal marker genes of M1 (Nos2) and M2 (Arg1) activation with Itgam as comparison. Data show mean ± SD, n = 4 independent samples, each from tissue pooled from eight mice. (h) Region-dependent variation of cortical and cerebellar microglia in response to the stimulation with E.coli, Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum. (h) Purified microglia were incubated with Escherichia coli strain K-12 and net replication of bacteria within microglia computed from counts of bacterial colonies derived from microglial cell lysates at indicated timepoints. Data are representative of two independent cell preparations and show mean ± SEM, n = 3 replicate samples from one cell preparation. p < 0.05, two-way ANOVA with Bonferroni correction. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum. Specific p values for all statistical comparisons are presented in Supplementary Table 13.
We next determined if the regional immunophenotypes of microglia extended to differences resembling overtly polarised states of microglia activation. We mined published microglial microarray datasets25 to establish a set of non-overlapping genes induced by classical (LPS) or alternative (IL-4) activation. This identified 216 LPS-induced genes and 132 IL-4-induced genes (Fig 5d). 17% of these LPS-induced genes and 18% of IL-4-induced genes were differentially expressed according to brain region in the present study. The LPS-inducible subset showed greater expression of the majority of genes in the cerebellum and hippocampus (Fig 5e). In contrast, greater expression of the differentially expressed subset of IL-4-inducible genes was not restricted to any particular brain region (Fig 5f). Furthermore, the majority of microglial genes associated with classical or alternative activation were expressed at almost undetectable levels in all brain regions, including the archetypal marker genes Nos2 and Arg1 (Fig 5g). Thus it appears the more “alert” phenotype of microglia in the cerebellum and hippocampus is distinct from conventional states of activation or polarisation.
We reasoned that steady-state differences in microglial immune alertness could predispose to region-dependent variations in function. To assess this directly, it was important to use an assay with freshly isolated microglia (prepared as for microarray) exposed to an equivalent challenge and over a short timeframe to avoid prolonged culturing which could result in de-differentiation from in vivo regional phenotypes. We achieved this using a bacterial phagocytosis and replication assay. Fewer bacteria were recovered from cortical microglia than cerebellar microglia 1 h after gentamicin treatment (Fig 5h), which may reflect distinct phagocytic or killing capacity, or both. Relative to the population of bacteria present inside microglia at 1 h after gentamicin treatment, there was a significant increase in intracellular net replication of bacteria in cortical but not cerebellar microglia by 4 h (Fig 5h). This suggested that cerebellar microglia were better able to control the net replication of internalized bacteria and support distinct functional responses of microglia to challenge that correlate with their region-specific immune alertness transcriptional profiles.
Transcriptional regulators of region-dependent co-expression networks
As described above, clusters 2 and 3 contained genes encoding known transcriptional regulators (e.g. Pparg, Nlrc5) of many of the respective cluster genes. To gain further insight to transcriptional control mechanisms that may drive microglial diversity we searched the annotated promoter regions of genes within these clusters for over-representation of transcription factor binding (TFB) motifs from the JASPAR collection26 using Clover27. Motifs recognised by the specificity protein (Sp), nuclear hormone receptor 4A (NR4A), estrogen related receptor (ERR) and RAR-related orphan receptor (ROR) were significantly over-represented in cluster 2 (Supplementary Table 9). Each of these families has established roles in regulating cellular energy metabolism28-31 which is consistent with the prominence of bioenergetic genes in cluster 2. The NR4A, ERR and ROR transcription factors are all members of the nuclear receptor family that act as both metabolic sensors and transcriptional regulators32, perhaps highlighting how the metabolic environment of microglia could direct region-dependent regulation of gene expression. TFB motifs over-represented in cluster 3 included those bound by early B-cell factor-1 (EBF1), forkhead box L1 (FOXL1), activator protein 1 (AP1) and c-Rel (REL) (Supplementary Table 9) all of which are known regulators of immune and inflammatory gene expression33-35 and therefore consistent with the immunoregulatory gene profile of cluster 3.
Brain region disproportionately affects cell surface gene expression
A subset of genes encoding microglial cell surface proteins was described recently11 and termed the microglial “sensome” with reference to their involvement in sensing the environment. We hypothesised that brain region would have a substantial impact on the expression of the sensome genes given that regional heterogeneity of microglial phenotype may in part arise from exposure to varying local environmental demands. GO terms associated with the cell surface were highly enriched in the set of regional differentially expressed genes (Supplementary Fig 5a) and 34 of the 100 sensome genes were differentially expressed according to brain region. The majority were expressed at greater levels in the striatum and cortex (Supplementary Fig 5b) and related to immune signalling (Supplementary Table 10). Moreover, of the differentially-regulated sensome genes involved in immune signalling, many encoded proteins involved in restricting over-activation of microglia, including Cx3cr1, Trem2, Cd33, Siglech and Fcgr2b (Supplementary Fig 5c). These data highlight that brain region has a disproportionately large effect on expression of genes encoding microglial cell surface proteins (compared to all genes) and that much of the heterogeneity affects receptors transducing microglial “off” signals10. Transmembrane pathways promoting microglial quiescence may therefore be active at differing levels depending on brain localisation and contribute to regional differences in immune alertness.
Inter-regional microglial heterogeneity mirrors macrophage tissue diversity
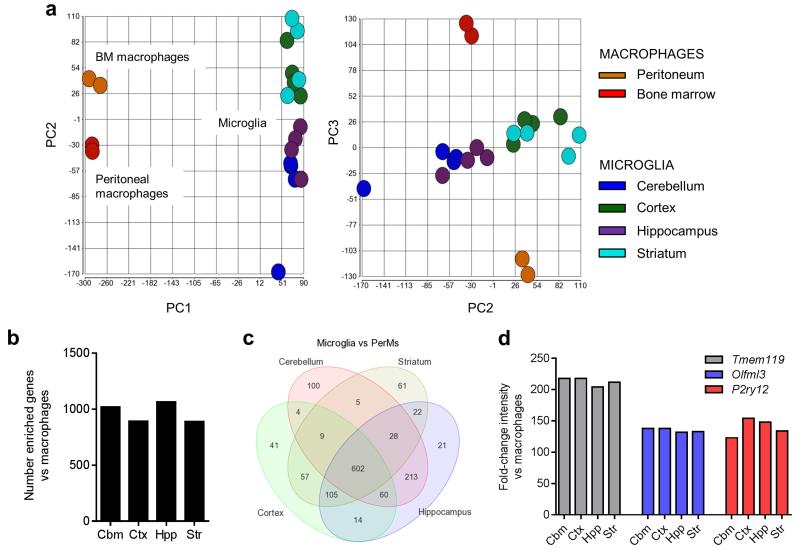
We assessed the extent of inter-regional microglial heterogeneity in the wider context of macrophage diversity by comparison of our regional microglial transcriptomes with selected purified macrophage datasets from the GNF Mouse GeneAtlas V3 (http://biogps.org/dataset/2394/gnf-mouse-geneatlas-v3/) acquired on the same microarray platform. As anticipated, using PCA the 1st principal component distinguished macrophage populations (peritoneal and bone marrow) from all microglial samples (Fig 6a). The 2nd principal component identified inter-regional heterogeneity in microglia that was comparable in magnitude to the differences between bone marrow and peritoneal macrophages. In view of the established diversity of tissue macrophages outside the brain, these data give an indication of the extent of inter-regional microglial heterogeneity discovered in the present study. We next determined the genes which were most highly expressed (>10-fold, FDR q < 0.05) in microglia compared to macrophages and if these were commonly found in microglia from all regions. Microglia from all regions each expressed a similar number of microglial-enriched genes (Fig 6b). Greater than 90% of these were common to at least two regions of the brain and approximately two-thirds to microglia from all regions (Fig 6c). Among these were genes recently identified as distinguishing microglia from macrophages including P2ry12, Tmem119 and Olfml311, 12, 19 (Fig 6d). Thus, in the healthy young adult brain, microglia express considerable regional heterogeneity yet retain a unifying core profile that, regardless of brain region, distinguishes them from tissue macrophages outside the brain.
Figure 6.
Regional microglial heterogeneity is comparable to inter-tissue macrophage diversity. (a) Principal components analysis of the present regional microglial expression datasets and systemic macrophage datasets shows the extent of regional microglial heterogeneity relative to macrophage tissue diversity. (b) The number of highly-enriched genes (>10-fold, p < 0.05 with FDR correction) in microglia compared to peritoneal macrophages was similar for microglia from each brain region. (c) Venn diagram showing regional overlap of the genes highly enriched in microglia versus peritoneal macrophages. (d) The fold-change (microglia versus peritoneal macrophages) in expression of selected genes recently identified as signature genes distinguishing microglia from systemic macrophages was comparable across brain regions. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum.
Ageing of microglia occurs in a region-dependent manner
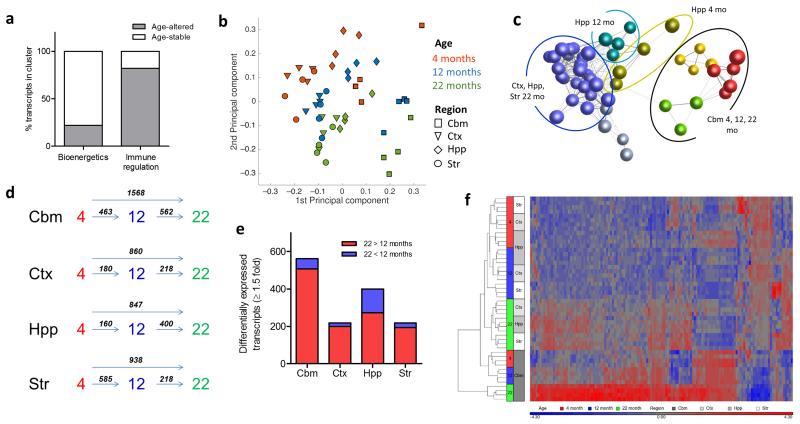
Ageing is associated with altered inflammatory status systemically and in the brain and involves marked changes in microglial morphology and phenotype. However, it is unclear if the impact of ageing on microglia is uniform throughout the brain. We first determined if the gene networks defining young adult regional heterogeneity were equally sensitive to ageing. Overall, approximately 50% of region-defining transcripts at 4 months were differentially-regulated during ageing, however there was an unequal distribution across the major 4 month-old region-defining clusters of gene co-expression (see Fig 3). Notably, the majority (>80%) of transcripts from the 4 month immune regulation cluster were age-regulated, but fewer than 25% of transcripts in the 4 month bioenergetics cluster were differentially expressed during ageing (Fig 7a). This shows that distinct modules of co-ordinated gene transcription that define microglial heterogeneity in the young adult brain are differentially sensitive to ageing.
Figure 7.
Region-specific microglial ageing. (a) Transcripts within the 4 months old immunoregulatory and bioenergetics clusters (see Fig 3) were assessed for age-regulated differential expression and the proportion of age-stable and age-altered transcripts determined. (b) Principal components analysis plot of microarray expression profiles for purified microglia from discrete brain regions at 4, 12 and 22 months of age. (c) Sample-to-sample correlation network graph of microarray datasets was performed in BioLayout Express3D and clustered using a Markov clustering algorithm. Nodes represent individual samples and edges the degree of correlation between their expression patterns. Colours denote discrete clusters. (d) Comparison of the number of differentially expressed transcripts (p < 0.05 with FDR correction, fold change ≥ 1.5) between different ages for each brain region. (e) Comparison of the number of up-regulated and down-regulated transcripts (p < 0.05 with FDR correction, fold change ≥ 1.5) at 22 vs 12 months in each brain region. (f) Hierarchical clustering and heat map of top transcripts with significant age-region interaction (p < 0.05, two-way ANOVA with FDR correction). The scaled expression value (row Z-score) is displayed in a blue-red colour scheme with red indicating higher expression and lower expression in blue. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum.
Principal components analysis showed that while the gross relative regional relationship identified at 4 months was generally preserved during ageing, there was an age-dependent progression suggesting an interaction between age and brain region (Fig 7b). First, cerebellar microglia were relatively more distant from the remaining brain regions at 12 and 22 months compared to 4 months. Second, the intermediate hippocampal microglia profile at 4 months was preserved at 12 months but largely converged with cortical and striatal samples at 22 months. We substantiated these observations non-subjectively using network analysis in Biolayout Express3D on the 13,741 transcripts regulated by age (FDR q < 0.05). Unbiased sample-to-sample correlation and clustering identified age-dependent unique cerebellar clusters and showed that hippocampal samples at 4 and 12 months clustered independently whereas at 22 months they formed a larger cluster together with striatal and cortical samples (Fig 7c). These data show a region-dependent influence on microglial ageing suggesting increased sensitivity of cerebellar microglia and a potential diminishing of the discrete hippocampal phenotype.
The kinetics of microglial ageing were also region-specific. Changes in gene expression profile occurred relatively consistently during “early” (4-12 months) and “late” (12-22 months) ageing in both cerebellar and cortical microglia (Fig 7d). In contrast, changes were most pronounced during early ageing in the striatum and during late ageing in the hippocampus. The hippocampal pattern was particularly interesting because, in contrast to other regions where only ~10% of gene alterations comprised decreased expression from 12 – 22 months, >30% declined in expression in hippocampal microglia (Fig 7e) supporting a diminishing distinction from other forebrain regions at the individual gene level. We also noted that twice the number of genes were differentially expressed (FDR q < 0.05, fold-change ≥ 1.5) at 22 versus 4 months in cerebellar microglia compared to other regions further reinforcing their greater sensitivity to age-related change (Fig 7d).
Unsupervised hierarchical clustering and visualisation of age-region interacting genes (FDR q < 0.05) demonstrated a number of striking patterns (Fig 7f) likely underpinning the gross age-region relationships above. First, a large group of genes increased in expression during ageing in all regions, however, in the cerebellum this occurred earlier (i.e. by 12 months) and/or to a greater magnitude by 22 months. Second, there was a cluster of genes that increased during ageing only in the cerebellum. Third, expression of a group of genes initially detected at relatively greater levels in hippocampal and cerebellar microglia at 4 months declined during ageing selectively in the hippocampus.
Collectively, these data show that the microglial transcriptome ages in a non-uniform manner across brain regions. Key observations are an accelerated and more amplified ageing trajectory in cerebellar microglia and a declining distinction of the hippocampal phenotype (relative to other forebrain regions).
Pathways underpinning region-specific microglial ageing profiles
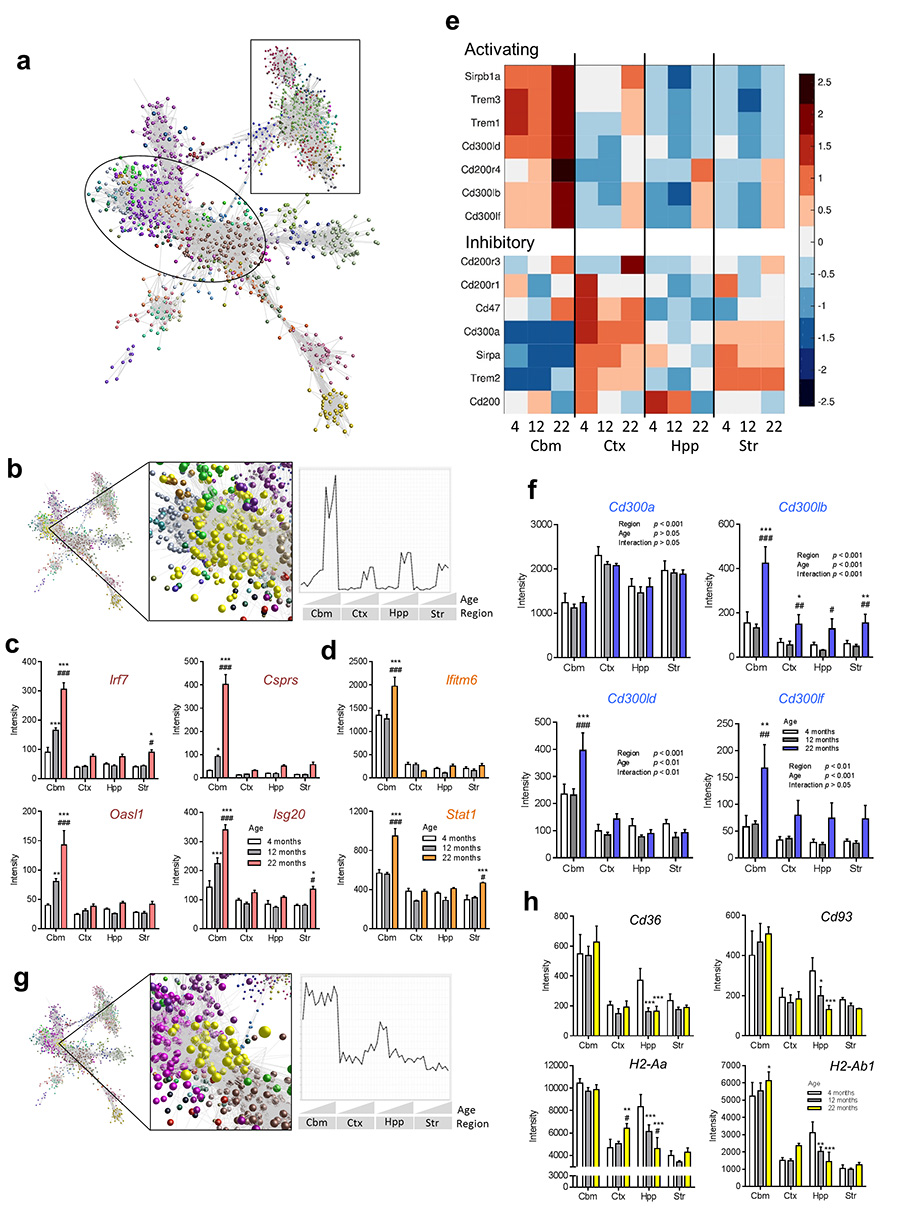
We next sought to establish the biological processes responsible for the age-region microglial interactions above. A correlation network graph of the age-regulated transcripts was clustered to non-subjectively sub-divide the graph into modules of highly co-expressed genes (Fig 8a). We focussed on clusters where interactions between age and region were most evident. In general, expression of cluster 2 transcripts increased with age but the most striking features were the greater and/or earlier age-regulated increased expression in cerebellar microglia (Fig 8b). The majority of genes were involved in immunoregulatory function and this was supported by GO analysis showing immune-related processes as the most over-represented in cluster 2 (Supplementary Table 11). Multiple families of molecules were represented including those involved in sensing of self and foreign ligands, immune cell adhesion and chemotaxis, cytokine signalling and anti-microbial effector responses. A large group of co-expressed genes involved in several aspects of the interferon pathway was particularly prominent. This included transcriptional regulatory factors (e.g. Irf7, Stat2, Oasl1) and interferon-regulated genes including those encoding effector proteins involved in anti-viral defence (e.g. Sp100, Csprs, Isg20, Ifit families, Bst2, Zbp1). Expression of the above genes was not only increased to a significantly greater extent (e.g. 5-fold) during ageing in the cerebellum but increased expression was also evident earlier (12 months) (Fig 8c). Further genes involved in the interferon pathway predominantly increased in cerebellar microglia although not until 22 months (e.g. Stat1, Ifitm family, Gbp family) (Fig 8d). Genes more sensitive to greater and/or earlier age-related changes in cerebellar microglial included both those already more highly expressed in cerebellar microglia and those expressed at negligible levels in all regions at 4 months.
Figure 8.
Biological pathways underlying region-specific microglial ageing. (a) Transcript-to-transcript correlation network graph of transcripts differentially expressed according to age (p < 0.05 with FDR correction) and clustered using a Markov clustering algorithm. Nodes represent individual transcripts and edges the degree of correlation in expression pattern between them. Colours denote discrete clusters. Circled region includes clusters with greater expression in cerebellum and/or increasing with age; square region includes clusters with greater expression in forebrain regions and/or declining expression with age. (b) Cluster position and mean expression profile of transcripts from cluster 2 indicating greater and/or earlier age-related changes. (c, d) Interferon pathway genes showing (c) earlier and/or (d) greater/selective increases in expression in cerebellar microglia compared to forebrain regions during ageing. Data show mean ± SD, n = 4 independent samples, each pooled from tissue from eight mice. *p < 0.05, **p < 0.01, ***p < 0.001 vs 4 month; #p < 0.05, ##p < 0.01, ###p < 0.001 vs 12 month, two-way ANOVA with Bonferroni correction. (e) Heat maps showing microarray expression patterns of selected immunoreceptor family genes during ageing arranged according to activating (A) or inhibitory (I) classification. Row Z-score intensities represent the mean of four independent samples per region and age with red indicating higher expression and lower expression in blue. (f) Expression patterns of Cd300 family genes show interaction between brain region and age for activating but not inhibitory members. Data show mean ± SD, n = 4 independent samples, each pooled from tissue from eight mice. *p < 0.05, **p < 0.01, ***p < 0.001 vs 4 month; #p < 0.05, ##p < 0.01, ###p < 0.001 vs 12 month, two-way ANOVA with Bonferroni correction. (g, h) Cluster position and mean expression profile of transcripts from cluster 14 indicating selective decline in expression during ageing in hippocampal microglia. (h) Expression profiles of selected genes from cluster 14. Data show mean ± SD, n = 4 independent samples, each pooled from tissue from eight mice. *p < 0.05, **p < 0.01, ***p < 0.001 vs 4 month; #p < 0.05, ##p < 0.01, ###p < 0.001 vs 12 month, two-way ANOVA with Bonferroni correction. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum. Specific p values for all statistical comparisons are presented in Supplementary Table 13.
Given the above data and regional differences in expression of ITAM and ITIM signalling immunoreceptors in microglia of young adult mice we assessed if there were region-dependent ageing responses of specific immunoreceptor families. There was a striking contrast in the ageing expression profile of amplifying and inhibitory immunoreceptors in all families examined (Fig 8e). Expression of inhibitory receptors from each family in general remained stable (e.g. Cd300a) or decreased in a largely region-independent manner (e.g. Cd200, Sirpa) during ageing. In contrast, expression of amplifying receptors increased with age and in a mostly region-dependent manner, affecting the cerebellar microglia selectively (e.g. Cd300ld, Trem1, Sirpb1a) or to a significantly greater extent (e.g. Cd200r4, Cd300lb) than other regions. The Cd300 family is presented as an example (Fig 8f) Thus, during ageing, alterations in immunoreceptor expression across multiple molecular families support a regionally-variable shift in balance towards immune amplification.
Genes significantly decreased in expression (FDR q < 0.05, fold-change ≥ 1.5) from 4-22 months in hippocampal microglia showed enrichment of GO processes related to cell adhesion/migration/motility, membrane organisation/endocytosis, immune/inflammatory function, and vascular development (Supplementary Table 12) suggestive of marked changes in the interaction of hippocampal microglia with their environment. Mining of these genes in the Biolayout Express3D network graph revealed their presence in several clusters each with a profile sharing reduced age-related hippocampal expression. Cluster 14 was particularly interesting because it contained genes expressed at relatively greater levels in both hippocampal and cerebellar microglia at 4 months that selectively declined in the hippocampus (and in the striatum for a subset) during ageing (Fig 8g, h). Among the genes in this cluster, many (e.g. Cd36, Cd93, Pf4, Lyve1) are involved in cell adhesion and motility pathways through interactions with matrix components and other extracellular ligands (Fig 8h). Consistent with the above functions, genes co-ordinating cross-regulation of endocytosis/phagocytosis and cytoskeletal reorganisation were present, notably Arhgef3, Dab2, Itsn1 and Vav3. Some of the above genes have overlapping roles in immune function through sensing and internalisation of microbial ligands and involvement in antigen processing and presentation (e.g. Cd36, Cd93, Pf4). Together with further genes in this cluster such as the mannose receptor gene Mrc1 and the MHC-II genes H2-Aa and H2-Ab1 (Fig 8h) it was evident that gene networks involved in certain aspects of ligand recognition, processing and presentation are relatively selectively suppressed during ageing in hippocampal microglia. More generally, the above data support a potential “disengagement” of aged hippocampal microglia with their environment compared to their young adult counterparts.
Regionally-variable depression of the “homeostatic” microglial signature during ageing
Microglial heterogeneity in the young adult brain was superimposed upon a core signature distinguishing microglia from systemic macrophages. Ageing resulted in a modest decline in expression across all forebrain regions of key signature genes and a significantly greater effect (e.g. 30% reduction in expression from 4 to 22 months old) in cerebellar microglia (e.g. Tmem119, P2ry12, P2ry13, Fcrls) (Supplementary Fig 6a). This supports data above that the greatest deviation from the “baseline” young adult homeostatic signature during ageing occurs in cerebellar microglia. Reduced expression of TGFβ receptor genes in aged microglia (Supplementary Fig 6b) may be important because expression of signature-defining genes is regulated by TGFβ12. In contrast, signature macrophage genes (e.g. Serpinb2, Alox15, Fabp4) were expressed at negligible levels at all ages (Supplementary Fig 6c) and genes commonly expressed on macrophages and microglia and upregulated on overtly activated microglia (e.g. Ptprc, Emr1) and Itgam were stably expressed during ageing in all regions (Supplementary Fig 6d). These data suggest that the age-related lessening of the young homeostatic microglial signature is regionally variable but is not accompanied by the gain of a macrophage-like signature.
Discussion
The data presented here provide compelling evidence of regional microglial phenotypic diversity in the healthy adult brain and the region-dependent impact of ageing on microglial phenotype. To our knowledge, this is the first demonstration that regional localisation of microglia influences their genome-wide expression profile across the adult lifespan, and notably we show that this extends beyond their immunophenotype. Key findings include that (1) transcriptional networks controlling microglial bioenergetic and immunoregulatory functions contribute prominently to heterogeneity in the young adult, (2) immunophenotypic variation suggests a more immune vigilant state of cerebellar microglia, (3) networks of gene co-expression underpinning heterogeneity in the young adult brain are differentially sensitive to ageing, (4) increasing distinction of cerebellar microglia and reduced distinction of hippocampal microglia (among forebrain regions) are key features of ageing, (5) microglial diversity is superimposed upon a core profile that distinguishes all microglia from macrophages, and (6) aged microglia display partial loss of the core young adult microglial identity in a regionally-variant manner but do not adopt a macrophage-like signature.
In the young adult brain (4 months old), the general relationship among the brain regions analysed showed that cerebellar microglia were most distinct, that cortical and striatal microglia were similar to each other, and that hippocampal microglia had an intermediate profile. This pattern suggests a microglial relatedness correlating with the relative positioning of brain regions along the rostro-caudal neuroaxis. Recent studies have reported that microglial responses to injury, disease or inflammatory challenge also vary according to neuroaxis location36, 37. Microglial morphology and density vary according to the relative composition of white and grey matter with a lower density of microglia reported in white matter of the adult mouse brain13. The extent to which white-to-grey matter ratios could influence regional differences in microglial transcriptomes is unclear, although the finding that the white matter-rich striatum and largely grey matter-dominant cerebral cortex have highly similar expression profiles suggests this may not be a major determinant. Forebrain microglia but not cerebellar microglia are dependent on IL-34, a ligand of the colony stimulating factor-1 receptor, for their maintenance15, 16. Furthermore, the involvement of enhancers in controlling tissue and cell identity is increasingly recognised, including in microglia38, and a recent study indicated that expression of brain region-specific enhancer RNAs may play a particularly important role in cerebellum-specific gene expression18.
Heterogeneity in microglial phenotypic markers other than immunophenotype has received negligible attention previously. The present data provide novel information on the transcriptional programmes controlling metabolism in microglia and show that regional differences in expression of these networks is a core feature of microglial diversity in the healthy young adult brain. Co-ordinated differences in regional expression of genes involved in all phases of the energy production pathway (mitochondrial production, glycolysis, TCA cycle, electron transport chain, ATP synthesis) was evident, emphasising the integrated nature of regional bioenergetic variation in microglia. The cluster profile revealed that hippocampal and cerebellar microglia have relatively greater expression of these genes suggesting greater energetic demands on microglia in these areas. Although there is little understanding of microglial bioenergetics, the routine cellular behaviour of microglia (e.g. process scanning, phagocytic activity) is likely to be metabolically demanding and regional differences in these activities could therefore influence energy demands39. In addition, the lower density of microglia in some areas, notably in the cerebellum, requires each microglial cell to survey a larger volume of tissue and would be expected to increase energy demands on an individual cell basis.
Our data suggest that microglia in some regions of the young adult brain exist in a more immune-vigilant state but one that does not equate to a conventional activated or primed microlgial/macrophage phenotype. Local differences in the physical and neurochemical environment, such as cellular and matrix composition, blood-brain barrier permeability, neurotransmitter profiles and heterogeneity in other cell types, may all be important. Consistent with this we found that a substantial proportion of genes encoding the microglial-enriched cell surface sensing apparatus were differentially expressed. Previous studies have suggested white matter microglia exist in a relatively less quiescent basal state than their grey matter counterparts37, which could contribute to the more vigilant profile of microglia in the white matter-enriched cerebellum. Another explanation is that the environment of some brain regions has evolved to support a more immune vigilant phenotype as a result of genomic integration of endogenous retroviruses (ERVs) and other retrotransposons. ERVs comprise ~10% of the murine genome40 and although normally inactive, deficiencies in innate immunity can predispose to reactivation41. Cerebellum-specific expression of the murine leukaemia virus (MuLV)-ERV has been shown previously42 and in mixed brain homogenates expression of the MuLV-ERV designated Mela (melanoma antigen) was restricted to the cerebellum (Supplementary Fig 7). The hippocampus is also more susceptible to retrotransposition in the human brain43. Thus, microglia, which we show have a more immune vigilant phenotype, including higher expression of anti-viral interferon networks, are found in areas where there may have been an evolutionary drive for development of greater immune vigilance.
Despite sharing similarities in their regional expression profile in the young adult brain, the major transcriptional networks (bioenergetics and immune) underlying regional heterogeneity at 4 months of age were affected differently by ageing. Genes within the immune networks were particularly sensitive whereas the majority within the bioenergetics cluster were unaffected by age. This indicates that regional differences are preserved during ageing for some functional pathways alongside marked divergence in others. Given the similar regional profiles of the immune and bioenergetics clusters at 4 months of age and the close relationship between immune function and metabolism, there may be some degree of immune-metabolic decoupling during ageing particularly in microglia from regions showing the greatest immunoregulatory deviation from the young adult (e.g. cerebellum). However, further work will be needed to determine if this divergence could allow for greater adaptation to the demands of ageing or could predispose to functional dysregulation.
The sensitivity of the immune network to ageing was largely responsible for the increasing distinction of the cerebellar phenotype in the aged brain. In contrast to ageing and direct immune stimulation ex vivo, however, we observed regionally comparable responses to acute systemic inflammatory challenge with bacterial lipopolysaccharide (data not shown) indicating that microglial heterogeneity encodes region-specific sensitivities in a stimulus/stressor-dependent manner. A selective or significantly greater induction in expression of many immune amplifying genes occurred in cerebellar microglia compared to other brain regions during ageing whereas genes involved in restraining excessive immune activity were generally stable across all brain regions. This implies that the more immune alert state of cerebellar microglia compared to other regions evident in the young adult is further augmented in the aged brain. The functional consequences of this are important to consider but may be complex. More caudal regions of the CNS such as the cerebellum may be more vulnerable to age- or disease-related inflammatory degeneration if this heightened alertness is poorly controlled. In support, age-related increases in inflammatory marker expression were predominant in the cerebellum and associated with functional cerebellar deficits37. However, the extra-alert phenotype may confer protective functions through increased vigilance and efficiency in removing potentially harmful agents. In this regard, it is pertinent to note the lower susceptibility of the cerebellum to amyloid deposition during ageing44.
Selective age-related alterations in gene networks were also evident in hippocampal microglia, however in contrast to the cerebellum, these resulted in a declining distinction from other forebrain regions. Decreasing expression of genes involved in matrix interactions and sampling the extracellular environment were the most prominent cause, suggesting that a declining engagement with their environment during ageing particularly affects hippocampal microglia. Although previous studies were not performed on a region-specific basis our data are consistent with reports of decreased process motility45 and reduced expression of cell surface sensing genes11 in aged microglia indicative of compromised environmental sampling capabilities. In contrast to the cerebellum, the hippocampus is vulnerable to age- and disease-related deposition of misfolded proteins which could in part relate to the age-driven divergence in environmental and immune alertness of microglia we describe in these brain regions. Dystrophic microglia in the aged human hippocampus have been described and postulated to represent a senescent state unable to carry out normal functions46; our data may provide a plausible transcriptomic basis for this hypothesis. More broadly, the loss of distinction in the overall hippocampal microglial phenotype at 22 compared to 4 months of age among forebrain regions is consistent with the concept of age-related loss of differentiation in neural function, cognitive performance and reorganisation of connectivity across brain regions47, 48. Notably, the weighting of regional connections of the hippocampus changes markedly during ageing48. Declining specialisation among neuronal populations has also been described in the aged brain49. It is therefore possible that age-related changes in the regional diversity of local signals derived from other neural components, altered inter-regional communication, and intrinsic microglial modifications in sensing pathways could all contribute to the diminished regional identity of hippocampal microglial in the aged brain.
Recent studies have revealed the distinctive transcriptional identity of microglia that distinguishes them from non-CNS tissue macrophages11, 12, 19. The present data now show that, although microglia have multiple transcriptional identities dependent on brain region, a core signature differentiating them from macrophages is retained across regions. Hence inter-regional microglial heterogeneity is superimposed upon a distinctive core profile. Microglial regional heterogeneity may be analogous in some respects to macrophage diversity observed within other tissues such as the spleen50. Despite reductions in expression of signature microglial genes with age, particularly in the cerebellum, highly macrophage-enriched genes (e.g. Fabp4, Alox15) were not expressed in any region at any age thus supporting that microglia also retain an overriding phenotypic individuality compared to macrophages in the aged brain.
In summary, regional microglial diversity described herein may be important for meeting the location-dependent demands of brain tissue under steady-state conditions. The impact of ageing on this diversity also suggests a basis for the regional variation in susceptibility to age-related neurodegenerative processes involving neuroinflammatory mechanisms. Further studies examining microglial diversity in the context of neurodegeneration are therefore warranted.
Methods
Accession codes
Microarray data are deposited in the NCBI GeoDatasets database with the accession number GSE62420.
Mice
Experiments were performed using male C57Bl/6J mice (Charles River Laboratories, UK) and Csf1r-EGFP reporter mice bred in-house. Mice were housed in individually-ventilated cages (five mice per cage) maintained under specific pathogen-free conditions and a standard 12 h light/dark cycle with unrestricted access to food and water. All experiments using live animals were conducted under the authority of UK Home Office project and personal licences and adhered to regulations specified in the Animals (Scientific Procedures) Act (1986) and Directive 2010/63/EU and were approved by both The Roslin Institute’s and the University of Edinburgh’s Animal Welfare and Ethics Committees.
Microglial purification and mixed brain cell/homogenate preparation
At 4, 12 and 22 months of age, mice were perfused transcardially with physiological saline and brains dissected into cerebellum, cortex, hippocampus and striatum. Tissue from eight mice was pooled for each regional replicate to obtain sufficient RNA for microarrays and the experiment was performed in quadruplicate for each region. Brain tissue was finely minced by scalpel blade in ice-cold Hanks Balanced Salt Solution HBSS (Sigma, UK), centrifuged (400 g, 5 min, 4°C) then resuspended and incubated for 1 h at 37°C using an enzyme cocktail containing 50U/ml collagenase, 8.5 U/ml dispase, 100 ug/ml Nα-Tosyl-L-lysine chloromethyl ketone hydrochloride and 5 U/ml DNaseI in 9.64 ml HBSS (Life Technologies, UK). Tissue was dissociated manually using a Dounce homogeniser and the enzymatic reaction terminated by addition of equal volume HBSS containing 10% fetal bovine serum. Homogenates were centrifuged (400 g, 5 min, 4°C) and pellets resuspended in 35% Percoll (GE Healthcare, Sweden), overlaid with HBSS then centrifuged (800 g, 45 min, 4°C). The supernatant and myelin layers were discarded and the cell pellet enriched with microglia resuspended in separation buffer (0.5% bovine serum albumin, 2 mM EDTA in PBS). The cell suspension was incubated with anti-CD11b microbeads (Miltenyi Biotec, UK) for 15 min at 4°C then applied to a magnetic LS column (Miltenyi Biotec) and cells retained on the column (microglia) were flushed and resuspended in appropriate buffer for downstream applications (see below). Unretained cells were also collected during initial validation for comparison. Mixed brain cell suspensions were prepared for flow cytometry according to the above protocol except the procedure was terminated before proceeding to centrifugation on Percoll gradient. For preparation of regional brain tissue homogenates for RNA extraction, mice were perfused and brain tissue dissected as above, and tissue snap-frozen and stored at −80°C. For validating that regional brain dissection did not result in cross-contamination of brain regions, the expression profile of established regionally enriched neuronal genes (Calb2, cerebellum-enriched; Rorb, cerebral cortex-enriched; Drd1a, striatum-enriched; Sstr4, hippocampus-enriched) was assessed in the present regional brain homogenates. This showed the expected enrichment of Calb2 in cerebellum, Rorb in cerebral cortex, Sstr4 in hippocampus, and Drd1 in striatum and was comparable to the regional pattern reported in the Allen Brain Atlas51 (http://mouse.brain-map.org/) (Supplementary Fig 1d). URLs for images shown in Supplementary Fig 1 are: Calb2, http://mouse.brain-map.org/experiment/show/79556662 (image 78); Drd1a, http://mouse.brain-map.org/experiment/show/352 (image 293); Rorb: http://mouse.brain-map.org/experiment/show/79360296 (image 61); Sstr4: http://mouse.brain-map.org/experiment/show/73636037 (image 234).
Flow cytometry
For routine verification of purified samples, cells resuspended in FACS buffer (0.1% BSA in PBS) were incubated with 1μg/ml anti-CD16/CD32 (Biolegend, UK, cat # 101301) to block Fc receptors and stained with anti-mouse CD11b-PE (Biolegend, cat # 101207, clone: M1/70), CD45-Pacific Blue (Biolegend, cat # 103125, clone: 30-F11) and F4/80-APC (Biolegend, cat # 123115, clone: BM8). To assess overlap between microglial CD11b and EGFP expression in mixed brain cell suspensions from Csf1r-EGFP mice, samples were stained as above and microglia identified according to their characteristic CD11b+F4/80+CD45lo profile. To measure MHC-II expression on microglia from mixed brain cell suspensions, cells were stained with RPE-Alexa Fluor 750 anti-mouse CD11b (AbD Serotec UK, cat # MCA74P750T), APC anti-mouse CD45 (Biolegend, cat # 103111) and eFluor 450 anti-mouse MHC Class II (eBioscience, UK, cat # 48-5321-80). Flow cytometry was performed using a FACS Aria IIIu or LSR Fortessa (Becton Dickenson, UK) and data analysed using FlowJo software (FlowJo, OR, USA).
Microglial culture and immunocytochemistry
1 × 105 purified microglial cells were cultured for 7 d in an 8-well chambered coverslip μ-slide (Ibidi, Germany) with DMEM/F12 (Life Technologies, UK) containing 10% FBS and 1% penicillin/streptomycin before fixation with 4% paraformaldehyde. The staining procedure was performed at room temperature. Fixed cells were permeabilised with 0.1% Triton X-100 for 5 min, washed and quenched by adding 0.25% NH4Cl for 5 min. Permeabilised cells were washed and blocked in PBS/0.1% BSA/5% donkey serum (Jackson ImmunoResearch, PA,USA) for 1 h before adding the primary antibodies prepared in 2.5% donkey serum in PBS. We used the following primary antibodies for staining: rabbit anti-IBA-1 (Wako Chemicals, Germany; 1:200) and biotinylated anti-mouse F4/80 (eBioscience, 1:200). Cells were washed and secondary antibodies anti-rabbit Alexa Fluor 594 (Life Technologies 1:1000) and Streptavidin Alexa Fluor 488 (Life Technologies; 1:1000) were incubated for 1 h before counter staining with DAPI (1μM) for 5 min. Imaging was performed using a Zeiss LSM 710 inverted confocal microscope. Maximum projection images of Z-stacks at 400x magnification objectives are presented.
RNA extraction
Microglia purified from individual brain regions were immediately processed for RNA extraction using the RNeasy Plus Micro Kit (Qiagen, UK). Preliminary experiments showed this method produced the highest yield and quality of RNA. RNA was extracted according to the manufacturer’s instructions with the exception of the final step where RNA elution was repeated twice with 10 μl RNase-free water. RNA quantities were determined by Nanodrop 1000 (Thermo Fisher Scientific, MA, USA) and RNA quality assessed using the Agilent Bioanalyzer (Agilent Technologies, CA, USA). RNA was also extracted from regional mixed brain cell homogenates using the RNeasy Midi Kit (Qiagen) following manufacturer’s instructions. All samples passed a quality control threshold (RIN ≥ 8) to proceed to microarray.
Transcriptional profiling using gene expression microarrays
Microarray assays were performed by Edinburgh Genomics, University of Edinburgh (https://genomics.ed.ac.uk/). Total RNA was labelled using the IVT Express Kit (Affymetrix). First-strand cDNA was synthesised and converted to double-stranded DNA template for transcription and synthesis of aRNA incorporating a biotin-conjugated nucleotide. aRNA was purified and fragmented prior to hybridisation on Affymetrix arrays. Biotin-labelled aRNA was hybridized to the whole mouse genome HT MG-430 PM array plate (Affymetrix, CA, USA) representing >39,000 transcripts, using the GeneTitan multi-channel instrument (Affymetrix).
qPCR
50ng of total RNA remaining from the microarray samples was reverse transcribed using Superscript III Reverse Transcriptase according to the manufacturer’s instructions (Life Technologies). The qPCR was performed in a Stratagene Mx3005P instrument (Agilent Technologies) using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) and primer pairs: B2m-f TGGCTCACACTGAATTCACCCCCA B2m-r TCTCGATCCCAGTAGACGGTCTTGG, Gapdh-f TGCATCCACTGGTGCTGCCAA, Gapdh-r ACTTGGCAGGTTTCTCCAGGCG, Camp-f AGGAACAGGGGGTGGTGA, Camp-r CACCTTTGCGGAGAAGTCCA, H2-d1-f TCCGAGATTGTAAAGCGTGAAGA, H2-d1-r GAACCCAAGCTCACAGGGAA. qPCR cycles were performed as followed: hot start denaturation cycle 95°C for 10 min, 40 cycles of amplification of 95°C for 15 sec, 60°C for 20sec and 72°C for 1 min.
Bacterial phagocytosis and replication assay
Purified cortical and cerebellar microglia (pooled from 8 mice, 4 months old) were infected with Escherichia coli (K-12 strain). Bacteria were grown in Luria-Bertani (LB) broth at 37°C, 190 rpm for 16 hours. Subsequently bacteria were subcultured at 1:1000 ratio into fresh LB. Bacteria were grown to mid-exponential phase at 37°C, 190 rpm for 3 hours and growth was monitored at OD600. Bacteria were resuspended in DMEM/F12 (no FBS/Penicillin/Streptomycin) at 1 × 107 bacteria/ml. 4 × 104 purified microglia were infected with 4 × 105 bacteria. 4 × 104 cells only and 4 × 105 bacteria only served as control. After incubation of microglia with bacteria for 2 hours to allow uptake, gentamicin was added to kill extracellular bacteria, enabling intracellular E. coli to be enumerated 1 and 4 hours later. Cells were then washed, lysed with 0.1% Triton X-100 and serial ten-fold dilutions plated on MacConkey agar. Bacterial colonies were counted after 17 h incubation at 37°C. Data are from triplicate microglial samples with each replicate pooled from eight mice. The microglia only control yielded no bacteria and extracellular E. coli treated with gentamicin under the same assay conditions was completed eliminated.
Computational analysis and bioinformatics
(i) Analysis of regional heterogeneity in the young adult (4 months old)
Microarray datasets were normalised by the Robust Multiarray Averaging (RMA) method in Affymetrix Expression Console (Affymetrix, CA, USA) prior to analysis in BioLayout Express3D (http://www.biolayout.org/)20 or directly during import for analysis in Partek Genomics Suite (Partek Inc., MO, USA. Analysis was perfomed using the ht_mg-430_pm.na33 annotation release (Affymetrix). 4 months datasets were first normalised and analysed independently from other ages. The gross inter-relationships among regional microglial transcriptomes were assessed by principal components analysis (PCA) on the log-transformed and Z-score transformed data matrix where every transcript had mean value zero and standard deviation one, using built-in functions of Matlab (MathWorks, MA, USA). Gross regional differences were also assessed in BioLayout Express3D by plotting a sample-to-sample correlation graph with the Pearson correlation threshold r = 0.96. Nodes represent individual samples (replicates) and edges between them show correlation of expression pattern with Pearson correlation coefficients above the selected threshold. The resulting network was clustered using the Markov clustering algorithm (MCL) (inflation 2.2) to non-subjectively sub-divide the graph into discrete clusters.
To assess if there were transcripts differentially expressed by region overall and between each individual region, normalised datasets were compared in Partek by ANOVA with false discovery rate (FDR) correction (q < 0.05). Data were visualised by heatmap with transcripts and samples organised by hierarchical clustering using average linkage with the Euclidean distance metric. Heatmap visualization and hierarchical clustering were performed on the log-transformed and Z-score transformed data matrix using built-in functions of Matlab. To assess gene co-expression relationships across brain regions, a pairwise transcript-to-transcript matrix was calculated in BioLayout Express3D from the set of regionally differentially expressed transcripts using a Pearson correlation threshold r = 0.80. A network graph was generated where nodes represent individual probesets (transcripts/genes) and edges between them correlation of expression pattern with Pearson correlation coefficients above the selected threshold. The graph was clustered into discrete groups of transcripts sharing similar expression profiles using the MCL algorithm (inflation 2.2, minimum cluster size 10 nodes). The composition and functional representation of the three major clusters were explored in more detail.
Enrichment analysis for Gene Ontology (GO) terms was performed in DAVID52 (http://david.abcc.ncifcrf.gov/) and visualised using the Enrichment Map plugin (http://www.baderlab.org/Software/EnrichmentMap)53 for Cytoscape (http://www.cytoscape.org/)54. In DAVID, gene lists were uploaded and the GOTERM_BP_FAT annotation category selected. Default settings were used for analysis with enrichment based on p < 0.05 with Benjamini correction. Enriched GO terms were uploaded to Enrichment Map and a network graph constructed. Nodes represent enriched GO terms and edges the degree of similarity between them using the overlap coefficient. Enrichment Map was also used to visualise the expression of all genes in the regionally differentially expressed dataset annotated with GO terms within the immune regulation cluster (Fig 2d). Results were visualised by heatmap with expression values normalised and transcripts organised by hierarchical clustering using default settings (Supplementary Fig 5). Visualisation of GO enrichment was also performed using the GOrilla tool55 (http://cbl-gorilla.cs.technion.ac.il/) with default settings applied. Pathway analysis on individual clusters from the BioLayout Express3D transcript-to-transcript network graph was performed in DAVID using the KEGG tool and Ingenuity Pathway Analysis (Qiagen).
To compare immunophenotypes in the present dataset with microglial activation profiles described previously, we mined published microarray datasets from microglia stimulated with either lipopolysaccharide (LPS) or interleukin-4 (IL-4)25. Raw expression data (.cel files) were downloaded from NCBI GEO DataSets (http://www.ncbi.nlm.nih.gov/gds/;GSE49329), imported to Partek and normalised with the microglia data. Non-overlapping genes significantly upregulated by LPS or IL-4 (>5-fold, FDR q < 0.05) were determined and these genes overlaid on the set of regional differentially expressed genes from the present study. The expression profile of overlapping transcripts for each stimulus was visualised by heat map using the log-transformed and Z-score transformed data matrix using built-in functions of Matlab.
To analyse a specific subset of microglial-enriched genes encoding cell surface proteins we assessed overlap between the 100 genes recently described as the microglial sensome and the set of regionally differentially expressed genes from the present study. The expression profile of overlapping transcripts was visualised by heat map using the log-transformed and Z-score transformed data matrix using built-in functions of Matlab.
To assess the extent of microglial regional heterogeneity in the context of general tissue macrophage diversity, we mined published peritoneal and bone marrow macrophage microarray datasets from the GNF MouseAtlas V3 (GEO DataSets GSE10246) which were generated on the same platform (MOE430 2.0) as the present arrays. All datasets were imported together and normalised in Partek as above. The gross relationships among microglia and macrophages were explored by PCA. Genes most highly expressed in microglia compared to macrophages (>10-fold, q < 0.05) were determined for each brain region and overlapping and unique genes among regions identified.
For the identification of transcriptional regulators which may act as contributing factors to the microglial regional diversity we used Clover27 to detect statistical over-representation of known TFB motifs in the promoter regions of co-expressed genes. Refseq IDs for each transcript on the Affymetrix ht_mg-430_pm.na33 array that was present in the immune regulatory and energy metabolism cluster were obtained from the NetAffx database (https://www.affymetrix.com/analysis/netaffx/index.affx). Promoter sequences 300bp upstream and 100bp downstream of the TSS were extracted from the mouse genome sequence (version mm9). Transcription factor binding site motifs were identified using the JASPAR CORE motif set26 (http://jaspar.cgb.ki.se) and Clover (p < 0.05, score threshold = 6) was used to detect over-represented motifs in promoters for each expression cluster compared with a background set27.
(ii) Effect of ageing and analysis of interactions between ageing and brain region
Analysis was performed on datasets from all ages normalised together and performed as above. Gross inter-relationships among regional microglial transcriptomes at different ages (4, 12, 22 months of age) were assessed by PCA as above and in BioLayout Express3D. For BioLayout Express3D, a sample-to-sample correlation network graph (Pearson correlation threshold r = 0.98) was generated from the transcripts differentially expressed according to age (FDR q < 0.05) and clustered using the MCL algorithm (inflation 2.2) to non-subjectively sub-divide the graph into discrete clusters. Nodes represent individual samples (replicates) and edges between them show correlation of expression pattern with Pearson correlation coefficients above the selected threshold. To determine effects of ageing and interactions between ageing and brain region on individual transcript expression, normalised datasets were analysed in Partek Genomics Suite using two-way ANOVA with FDR correction (q < 0.05) and appropriate post-hoc tests as indicated. Hierarchical clustering and visualisation of the top 150 transcripts with significant interaction between age and region on expression levels was performed in Partek.
Transcript-to-transcript co-expression relationships were assessed using BioLayout Express3D. A pairwise transcript-to-transcript matrix was calculated from the set of transcripts differentially expressed according to age using a Pearson correlation threshold r = 0.85. A network graph was generated where nodes represent individual probesets (transcripts/genes) and edges between them correlation of expression pattern with Pearson correlation coefficients above the selected threshold. The graph was clustered non-subjectively into discrete groups of transcripts sharing similar expression profiles using the MCL algorithm (inflation 2.2, minimum cluster size 10 nodes). The composition and functional representation of selected clusters were explored in more detail using approaches as above (e.g. GO and KEGG enrichment analysis).
Relative expression profiles of genes from selected immunoreceptor families were visualised by heat map using the log-transformed and Z-score transformed data matrix using built-in functions of Matlab. Genes were classified as activating or inhibitory according to known functional effects or prediction from presence of receptor ITAM-associating/ITIM domains.
Experimental design and statistical analysis
Experimental design, analysis and reporting followed the ARRIVE guidelines (https://www.nc3rs.org.uk/arrive-guidelines) where possible. Mice were randomised to treatment group (age) at cage level using using a computer-based random number generator (https://www.randomizer.org/). Microarray data are from n = 4 biological replicates with each replicate consisting of tissue pooled from 8 mice. No formal a priori statistical methods were used to pre-determine sample sizes due to insufficient previous data to enable this. However, sample sizes were chosen based on estimates of anticipated variability through previous general experience of microarray analysis and accounting for pooling of tissues reducing inter-replicate variance. To avoid potential confounding cage effects during pooling of tissue, each separate pool contained tissue derived from mice housed in all cages for each age group selected in a randomised manner using a computer-based random number generator (https://www.randomizer.org/). Data collection and analysis were performed with the assessor unaware of allocation to treatment group. Statistical tests for computational analysis are described above. Flow cytometry and qPCR data were analysed using one-way ANOVA with Bonferroni correction. Data from the bacterial uptake and replication assay were analysed by two-way ANOVA with Bonferroni correction. Data were checked for compliance with statistical assumptions for each test, including normal distribution and equal variances across groups. Tests were two-tailed throughout. Statistical significance was considered at p < 0.05 (or equivalent corrected for multiple comparisons). Data show mean ± SD unless otherwise stated.
Supplementary Material
Acknowledgments
We thank Edinburgh Genomics, The University of Edinburgh, for performing microarrays, Bob Fleming for technical assistance with microglial purification and Yu-Ting Lai for help optimising regional brain dissection. This work was funded by a PhD scholarship from the Darwin Trust of Edinburgh to K Grabert and grants from BBSRC (BB/J004332/1) and MRC (MR/L003384/1). The Roslin Institute and Edinburgh Genomics are partly supported through core grants from NERC (R8/H10/56), MRC (MR/K001744/1) and BBSRC (BB/J004243/1, BB/J004332/1).
Footnotes
Competing financial interests
The authors declare no competing interests.
A methods checklist is available with the supplementary materials.
References
- 1.Prinz M, Mildner A. Microglia in the CNS: immigrants from another world. Glia. 2011;59:177–187. doi: 10.1002/glia.21104. [DOI] [PubMed] [Google Scholar]
- 2.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 3.Paolicelli RC, et al. Synaptic Pruning by Microglia Is Necessary for Normal Brain Development. Science (New York, N.Y. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 4.Schafer Dorothy P., et al. Microglia Sculpt Postnatal Neural Circuits in an Activity and Complement-Dependent Manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coull JAM, et al. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature. 2005;438:1017–1021. doi: 10.1038/nature04223. [DOI] [PubMed] [Google Scholar]
- 6.Neumann H, Kotter MR, Franklin RJM. Debris clearance by microglia: an essential link between degeneration and regeneration. Brain. 2009;132:288–295. doi: 10.1093/brain/awn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalancette-Hébert M.l., Gowing G.v., Simard A, Weng YC, Kriz J. Selective Ablation of Proliferating Microglial Cells Exacerbates Ischemic Injury in the Brain. The Journal of Neuroscience. 2007;27:2596–2605. doi: 10.1523/JNEUROSCI.5360-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenn AM, Hall JCE, Gensel JC, Popovich PG, Godbout JP. IL-4 Signaling Drives a Unique Arginase+/IL-1β+ Microglia Phenotype and Recruits Macrophages to the Inflammatory CNS: Consequences of Age-Related Deficits in IL-4Rα after Traumatic Spinal Cord Injury. The Journal of Neuroscience. 2014;34:8904–8917. doi: 10.1523/JNEUROSCI.1146-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardona AE, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9:917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- 10.Biber K, Neumann H, Inoue K, Boddeke HW. Neuronal ‘On’ and ‘Off’ signals control microglia. Trends Neurosci. 2007;30:596–602. doi: 10.1016/j.tins.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butovsky O, et al. Identification of a unique TGF-[beta]-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–170. doi: 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- 14.de Haas AH, Boddeke HWGM, Biber K. Region-specific expression of immunoregulatory proteins on microglia in the healthy CNS. Glia. 2008;56:888–894. doi: 10.1002/glia.20663. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greter M, et al. Stroma-Derived Interleukin-34 Controls the Development and Maintenance of Langerhans Cells and the Maintenance of Microglia. Immunity. 2012;37:1050–1060. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karch CM, Goate AM. Alzheimer’s Disease Risk Genes and Mechanisms of Disease Pathogenesis. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao P, et al. Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat Neurosci. 2015;18:1168–1174. doi: 10.1038/nn.4063. [DOI] [PubMed] [Google Scholar]
- 19.Chiu Isaac M., et al. A Neurodegeneration-Specific Gene-Expression Signature of Acutely Isolated Microglia from an Amyotrophic Lateral Sclerosis Mouse Model. Cell Reports. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman TC, et al. Construction, Visualisation, and Clustering of Transcription Networks from Microarray Expression Data. PLoS Comput Biol. 2007;3:e206. doi: 10.1371/journal.pcbi.0030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hume DA, Summers KM, Raza S, Baillie JK, Freeman TC. Functional clustering and lineage markers: Insights into cellular differentiation and gene function from large-scale microarray studies of purified primary cell populations. Genomics. 2010;95:328–338. doi: 10.1016/j.ygeno.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 23.Ghesquiere B, Wong BW, Kuchnio A, Carmeliet P. Metabolism of stromal and immune cells in health and disease. Nature. 2014;511:167–176. doi: 10.1038/nature13312. [DOI] [PubMed] [Google Scholar]
- 24.Linnartz B, Wang Y, Neumann H. Microglial Immunoreceptor Tyrosine-Based Activation and Inhibition Motif Signaling in Neuroinflammation. International Journal of Alzheimer’s Disease. 2010;2010 doi: 10.4061/2010/587463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freilich RW, Woodbury ME, Ikezu T. Integrated Expression Profiles of mRNA and miRNA in Polarized Primary Murine Microglia. PLoS ONE. 2013;8:e79416. doi: 10.1371/journal.pone.0079416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathelier A, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Research. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frith MC, et al. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Research. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huss JM, Torra IP, Staels B, Giguère V, Kelly DP. Estrogen-Related Receptor α Directs Peroxisome Proliferator-Activated Receptor α Signaling in the Transcriptional Control of Energy Metabolism in Cardiac and Skeletal Muscle. Molecular and Cellular Biology. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mootha VK, et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearen MA, Muscat GEO. Minireview: Nuclear Hormone Receptor 4A Signaling: Implications for Metabolic Disease. Molecular Endocrinology. 2010;24:1891–1903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Archer MC. Role of Sp Transcription Factors in the Regulation of Cancer Cell Metabolism. Genes & Cancer. 2011;2:712–719. doi: 10.1177/1947601911423029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis GA, Fayard E, Picard F, Auwerx J. NUCLEAR RECEPTORS AND THE CONTROL OF METABOLISM. Annual Review of Physiology. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- 33.Griffin MJ, et al. Early B-cell Factor-1 (EBF1) Is a Key Regulator of Metabolic and Inflammatory Signaling Pathways in Mature Adipocytes. Journal of Biological Chemistry. 2013;288:35925–35939. doi: 10.1074/jbc.M113.491936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B-S, Yang S-K, Lan C-Y, Chuang Y-J. A systems biology approach to construct the gene regulatory network of systemic inflammation via microarray and databases mining. BMC Medical Genomics. 2008;1:46. doi: 10.1186/1755-8794-1-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Natoli G, Ghisletti S, Barozzi I. The genomic landscapes of inflammation. Genes & Development. 2011;25:101–106. doi: 10.1101/gad.2018811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnell L, Fearn S, Klassen H, Schwab ME, Perry VH. Acute inflammatory responses to mechanical lesions in the CNS: differences between brain and spinal cord. European Journal of Neuroscience. 1999;11:3648–3658. doi: 10.1046/j.1460-9568.1999.00792.x. [DOI] [PubMed] [Google Scholar]
- 37.Hart AD, Wyttenbach A, Hugh Perry V, Teeling JL. Age related changes in microglial phenotype vary between CNS regions: Grey versus white matter differences. Brain, Behavior, and Immunity. 2012;26:754–765. doi: 10.1016/j.bbi.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernhart E, et al. Lysophosphatidic acid receptor activation affects the C13NJ microglia cell line proteome leading to alterations in glycolysis, motility, and cytoskeletal architecture. PROTEOMICS. 2010;10:141–158. doi: 10.1002/pmic.200900195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stocking C, Kozak CA. Endogenous retroviruses. Cellular and Molecular Life Sciences. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu P, et al. Nucleic Acid-Sensing Toll-like Receptors Are Essential for the Control of Endogenous Retrovirus Viremia and ERV-Induced Tumors. Immunity. 37:867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Lee KH, Horiuchi M, Itoh T, Greenhalgh DG, Cho K. Cerebellum-specific and age-dependent expression of an endogenous retrovirus with intact coding potential. Retrovirology. 2011;8:82. doi: 10.1186/1742-4690-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson-Wood K, et al. Amyloid precursor protein processing and A beta42 deposition in a transgenic mouse model of Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:1550–1555. doi: 10.1073/pnas.94.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Damani MR, et al. Age-related alterations in the dynamic behavior of microglia. Aging Cell. 2011;10:263–276. doi: 10.1111/j.1474-9726.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Streit W, Braak H, Xue Q-S, Bechmann I. Dystrophic (senescent) rather than activated microglial cells are associated with tau pathology and likely precede neurodegeneration in Alzheimer’s disease. Acta Neuropathologica. 2009;118:475–485. doi: 10.1007/s00401-009-0556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sleimen-Malkoun R, Jean-Jacques T, Hong SL. Aging induced loss of complexity and dedifferentiation: Consequences for coordination dynamics within and between brain, muscular and behavioral levels. Frontiers in Aging Neuroscience. 2014;6 doi: 10.3389/fnagi.2014.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goh JOS. Functional Dedifferentiation and Altered Connectivity in Older Adults: Neural Accounts of Cognitive Aging. Aging Dis. 2011;2:30–48. [PMC free article] [PubMed] [Google Scholar]
- 49.Park DC, et al. Aging reduces neural specialization in ventral visual cortex. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13091–13095. doi: 10.1073/pnas.0405148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nature immunology. 2013;14:986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 52.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 53.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.