Abstract
Microbial pathogenesis studies are typically performed with reference strains, thereby overlooking microbial intra-species virulence heterogeneity. Here we integrated human epidemiological and clinical data with bacterial population genomics to harness the biodiversity of the model foodborne pathogen Listeria monocytogenes and decipher the basis of its neural and placental tropisms. Taking advantage of the clonal structure of this bacterial species, we identify clones epidemiologically associated with either food or human central nervous system (CNS) and maternal-neonatal (MN) listeriosis. The latter are also most prevalent in patients without immunosuppressive comorbidities. Strikingly, CNS and MN clones are hypervirulent in a humanized mouse model of listeriosis. By integrating epidemiological data and comparative genomics, we uncovered multiple novel putative virulence factors and demonstrated experimentally the contribution of the first gene cluster mediating Listeria monocytogenes neural and placental tropisms. This study illustrates the exceptional power of harnessing microbial biodiversity to identify clinically relevant microbial virulence attributes.
Introduction
Mechanistic studies in the field of microbial pathogenesis have greatly contributed to major discoveries in life sciences and led to key diagnostic and therapeutic advances in the field of infectious diseases. Most pathogenesis studies have been conducted with reference strains, which have been exchanged between investigators over the years and for which experimental tools have been developed. The systematic mutagenesis of reference strains filtered by phenotypic in vitro and in vivo screens has been an exceptionally fruitful approach that led to the discovery and characterization of key genes and gene products involved in microbial virulence1,2. As the use of reference strains purposely normalizes the inherent genetic heterogeneity of a given microbial species, this approach intrinsically misestimates the biodiversity and the ensuing virulence heterogeneity of microbial pathogenic species3-6.
Many clinical infectious diseases phenotypes remain poorly understood at the mechanistic level, and this results in part from the use of clinically irrelevant reference laboratory strains. We hypothesized that zooming out and considering (i) on the microbial side the species as a whole and (ii) on the host side detailed human epidemiological and clinical data, would offer the unique opportunity to uncover new virulence attributes associated with clinically relevant microbial phenotypes. We applied this approach to Listeria monocytogenes (Lm), a major foodborne pathogen and a widely recognized model microorganism7-9, which causes two deadly complications: central nervous system (CNS) and maternal-neonatal (MN) listeriosis.
Lm is a highly heterogeneous species: it can be divided into four evolutionary lineages10-12, 13 serotypes13 and four PCR serogroups14. Multilocus sequence typing (MLST) further subdivides the above categories into clones, which are geographically and temporally widespread15-17. Because of the very high morbidity and case fatality rates associated with human listeriosis, extensive surveillance programs that include food control and exhaustive investigation of human cases have been implemented. As of today, all Lm isolates are regarded as equally virulent by regulatory authorities, although there is evidence against this uniform view: lineage I or serotype 4b are more frequent among clinical isolates than lineage II or serotypes 1/2b, 1/2a and 1/2c, relative to the frequency of these categories in food12,18-21. Besides, strains with reduced pathogenicity displaying truncated and non-functional virulence factors such as internalin are commonly isolated from food21,22. However, a large-scale and systematic analysis integrating detailed molecular epidemiological data and comparative genomics has never been performed.
Results
Distribution of Lm clones in food and clinical sources
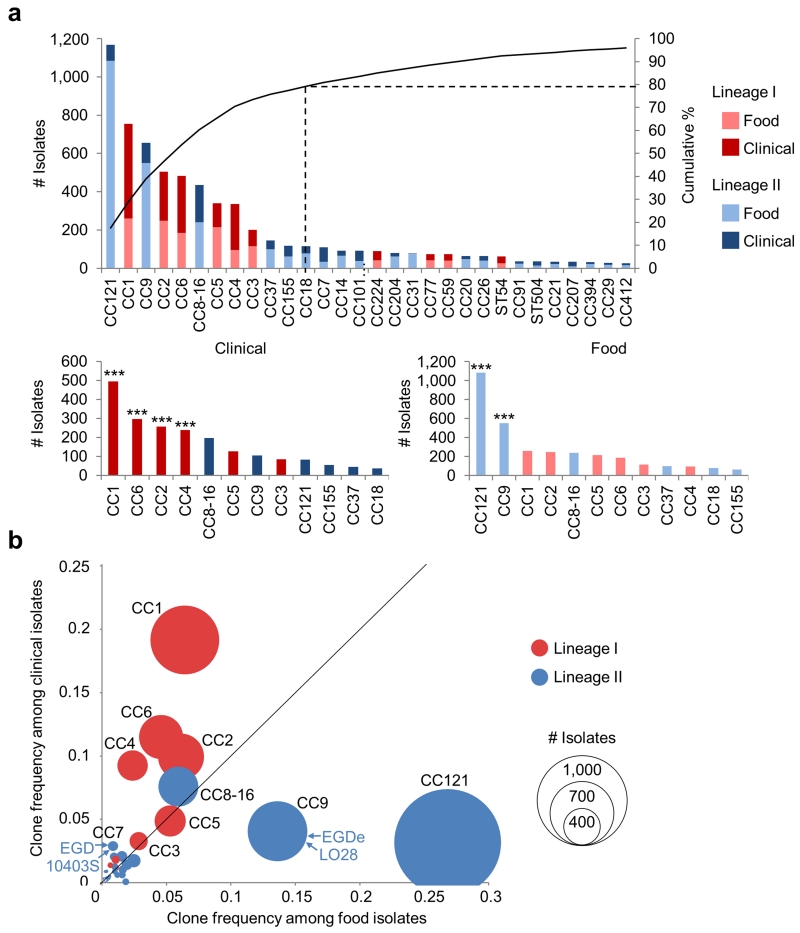
In France, listeriosis is a notifiable disease and a single national reference center receives prospectively all isolates of clinical and food origin to which the human population is exposed, ensuring the epidemiological representativeness of the resulting collection of Lm isolates. We conducted an exhaustive analysis of the epidemiological and microbiological data of 6,633 strains we collected prospectively over 9 consecutive years, including 2,584 clinical and 4,049 food isolates. We ascribed a clone (defined as an MLST clonal complex [CC]; see Online methods, Supplementary Note and Supplementary Figure 1) to each of these isolates: the 6,633 strains belong to 63 different clones, out of which the 12 most prevalent ones represent 79.2% of all strains of this study (Fig. 1a). Importantly, the frequency distribution of these clones is highly uneven (Fig. 1a): the most prevalent clones are CC121 (17.6% of total isolates), CC1 (11.4%), CC9 (9.9%), CC2 (7.6%), CC6 (7.3%), CC8 and CC16 (which were merged, 6.6%), CC5 (5.1%) and CC4 (5.1%). Remarkably, the clones that predominate in clinical or food samples are distinct (Fig. 1a), leading to striking differences in the relative prevalence of clones among clinical (Fig. 1b, y axis) and food isolates (Fig. 1b, x axis) (Fig. 1a and 1b; Supplementary Table 1). The proportion of clinical isolates per clone differs by one order of magnitude, ranging from 7.0% (CC121) to 71.3% (CC4) (Fig. 2a, x axis). Clones CC1, CC2, CC4 and CC6 are strongly associated with a clinical origin (p < 1.10−4, 62% of strains of these clones are of clinical origin), whereas clones CC121 and CC9 are strongly associated with a food origin (p < 1.10−4, only 10.3% of clinical origin) (Supplementary Table 1, Fig. 1a and 1b). These results therefore distinguish three categories of highly prevalent clones: infection-associated clones (CC1, CC2, CC4 and CC6), food-associated clones (CC9 and CC121), and intermediate clones (others) (Fig. 1a and 1b). Strikingly, the reference strains EGDe and LO28 belong to CC9, a food-associated clone very rarely causing human disease, whereas reference strains EGD and 10403S belong to CC7, which is very rarely isolated from food (0.9%) and infected patients (2.9%) (Fig. 1b).
Figure 1. Prevalence and distribution of MLST clones in food and clinical sources.
The analysis is based on 6,633 food and clinical isolates collected between 2005 and 2013. (a, top), Unequal prevalence of Lm MLST clones. The curve represents the cumulative percentage of isolates pertaining to clones, ordered by total number of isolates. Only clones with more than 10 isolates are shown. (a, bottom), Distribution of clones in food and clinical sources, ranked by number of isolates of each origin. The 12 major clones that represented 79.2% of all isolates are shown. Association to food or clinical origins (Chi2 tests): *, p < 0.01; **, p < 0.001; ***, p < 0.0001 (see Supplementary Table 1). (b) Frequencies of clones with > 10 isolates among food (x axis) and clinical (y axis) isolates. Circle size is proportional to the numbers of isolates. Positions of reference strains are shown.
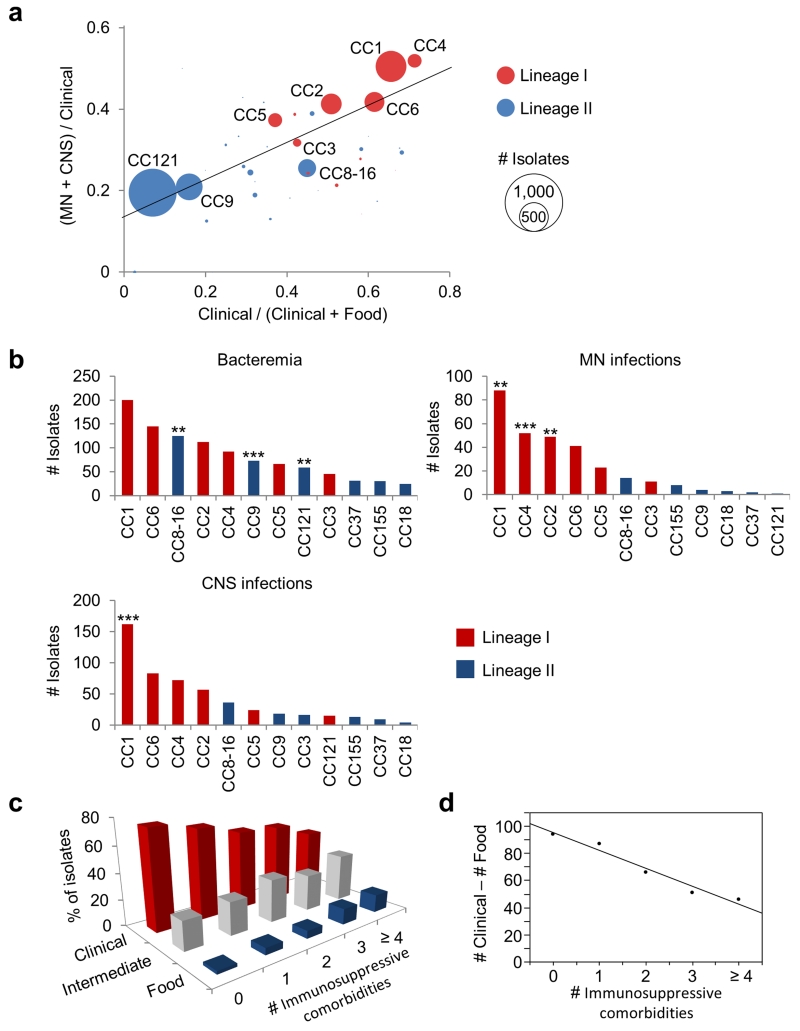
Figure 2. Infectious potential of MLST clones.
(a) Frequency of clones in CNS (Central Nervous System) and MN (Materno-Neonatal) infections as a function of clinical frequency. Linear regression was weighted based on number of isolates per clone: R2 = 0.62; p < 1.10−10. All clones with more than five isolates were included. (b) Distribution of the major clones in bacteremia (above, left), CNS (below) and MN (above, right). Absolute numbers of isolates per clone involved in each category of infection are shown. The 12 major clones were ranked by absolute number of isolates (Supplementary Table 2). Association to bacteremia, CNS and MN infections (Chi2 test): *, p < 0.01; **, p < 0.001; ***, p < 0.0001. (c) The histograms show the distribution of food-associated (food, CC9 and CC121, blue), infection-associated (clinical, CC1, CC2, CC4, and CC6, red) and intermediate clones (intermediate, CC8-16, CC5, CC3, CC37, CC155 and CC18, grey) in patient groups with 0, 1, 2, 3, and 4 or more immunosuppressive comorbidities. (d) Linear regression (R2 = 0.96; p = 0.0032) of the difference between the numbers of isolates belonging to infection-associated clones (#Clinical) and those belonging to food-associated clones (#Food) (y axis), against the number of immunosuppressive comorbidities of infected patients (# Immunosuppressive comorbidities, x axis).
Listeriosis can manifest as isolated bacteremia (where Lm has crossed the intestinal barrier), MN infection (where Lm has crossed the intestinal and placental barriers), and CNS infection (where Lm has crossed the intestinal and blood-brain barriers). Of note, the most clinically associated clones are also the most associated with MN and CNS infections as opposed to isolated bacteremia (Fig. 2a, linear regression: p < 10−10). Most notably, CC1 and CC4 are most strongly associated with MN and CNS infections (i.e., are negatively associated with bacteremia, p < 10−5, Supplementary Table 2), whereas clones CC121, CC9 and CC8-16 are associated with low frequencies of MN and CNS infections (p < 10−3). Considering next the three types of infection separately, CC1 is associated to CNS infections (p < 10−4), CC1, CC2 and CC4 are associated to MN infections (p < 10−3 for CC1 and CC2, p < 10−4 for CC4) and CC8-16, CC9 and CC121 are associated to bacteremia (p < 10−3 for CC121 and CC8-16, p < 10−4 for CC9) (Fig. 2b; Supplementary Table 2). Altogether, these results suggest that the strength of the association of a clone with clinical disease could be causally linked to its virulence.
Ecological distribution and virulence levels of clones
Lm is an opportunistic pathogen that infects mostly immunocompromised individuals. We collected detailed clinical and biological data for 812 infected patients enrolled in the MONALISA prospective study on Listeria and listeriosis (ClinicalTrials.gov Identifier NCT01520597) and analyzed the distribution of clones as a function of patients’ immunosuppressive comorbidities. Food-associated clones CC9 and CC121 are more often isolated in highly immunocompromised patients, whereas CC1, CC2, CC4 and CC6 are more prevalent among patients with little or no immunosuppressive comorbidities (Fig. 2c). Strikingly, there was an inverse linear relationship between the predominance of infection-associated clones and the number of immunosuppressive comorbidities (R2 = 0.96, p = 0.0032, Fig. 2d). As an infection results from the interplay between host and bacterial factors, these results indicate that specific virulence factors of these invasive clones may compensate for the absence of comorbidities to trigger disease, and are in support of the hypothesis that infection-associated clones are hypervirulent.
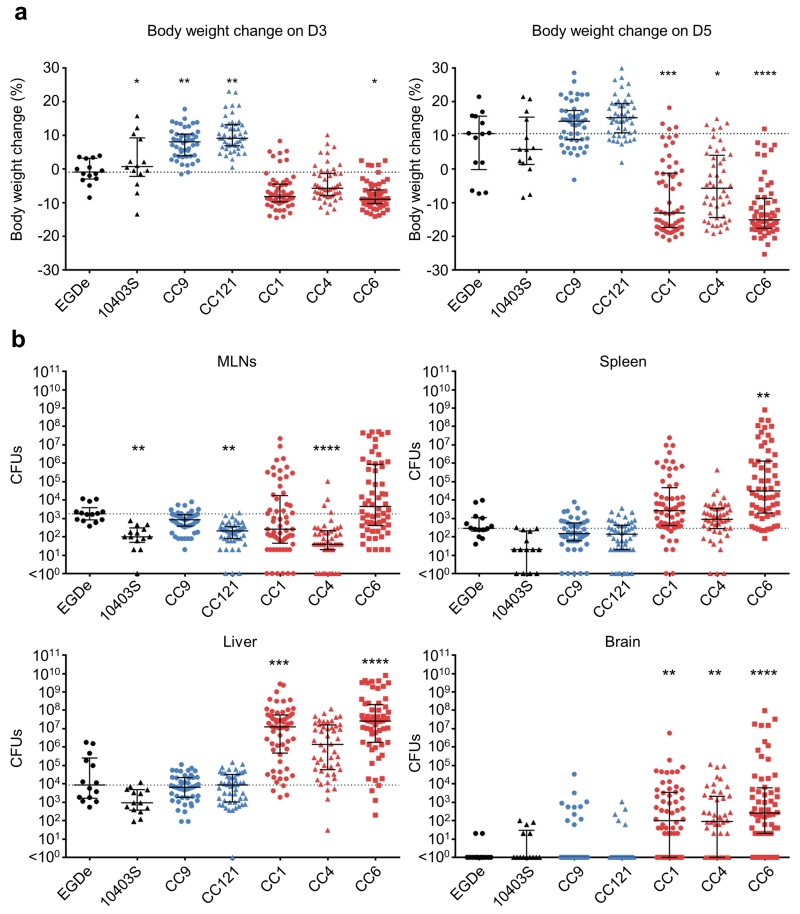
We therefore assessed the respective virulence of infection-associated and non-associated clones in a humanized mouse model of listeriosis23, relative to the reference strains EGDe (CC9) and 10403S (CC7). Isolates belonging to clones CC1, 4 and 6 (Supplementary Table 3) induced significantly more body weight loss (Fig. 3a) and infected more efficiently the liver (for CC1 and 6) and the brain (for CC1, 4 and 6), as compared to EGDe and 10403S, demonstrating that they are hypervirulent relative to the reference strains EGDe and 10403S, and, most importantly, are neurotropic in contrast to these reference strains (Fig. 3b). In contrast, CC9 and CC121 (Supplementary Table 3), which are epidemiologically strongly associated with food but not with clinical infection, did not induce body weight loss following infection, and were less invasive, demonstrating that they are hypovirulent (Fig. 3a and 3b). Remarkably, CFUs enumeration in infected organs and body weight change showed a very strong association with the clinical prevalence of the five clones (stepwise multiple regression: R2 = 0.9995, p < 0.03, Supplementary Table 4). Although lineage origin contributed in part to this association, these results suggest that the virulence level of Lm clones directly mirrors their epidemiological association with human listeriosis.
Figure 3. Compared virulence of the six major clonal complexes.
(a) Mouse body weight loss on day 3 and day 5 post infection. (b), Bacterial loads on day 5 post infection are shown as total colony forming units (CFUs) recovered from the entire organs. Humanized mice were orally inoculated with 2.108 CFUs. Results are shown as median ± interquartile range. The dotted line indicates the median value of EGDe infected mice. Two isolates from each origin: food, bacteremia, maternal-neonatal (MN) infection, and CNS infection were selected in each clone, except for CC1 and CC6, in which there are 4 human CNS isolates and 2 isolates in each other origin (food, bacteremia and MN infections). For CC121, there are 2 isolates from food, 2 from bacteremia, 1 from MN and 3 from CNS infection. Number of mice: n = 14 for EGDe and 10403S; n = 48 for CC9, CC121 and CC4; n = 60 for CC1 and CC6. Clinical-associated clones are represented in red, food-associated clones in blue and reference strains (EGDe and 10403S) in black. Dunn’s multiple comparison test relative to EGDe infected mice: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. The difference compared to EGDe infected mice was non-significant unless indicated. MLNs, mesenteric lymph nodes.
Genomic traits associated with Lm hypervirulence
To decipher the origin of virulence heterogeneity among clones, we analyzed the whole genome sequences of 104 strains representative of the major clonal complexes that compose Lm species15-17,24 (Supplementary Figure 2; Supplementary Table 5). In Lm, most virulence genes identified to date belong to its core genome25-28. We therefore computed Lm species core genome (Supplementary Figure 3; Supplementary Table 6) and analyzed the distribution and variation of known virulence gene products among clones. Distribution of InlA truncations was the main feature associated to the loss of virulence of hypovirulent clones (Supplementary Figure 4). Additional variation was observed in virulence genes (Supplementary Figures 4 and 5; Supplementary Table 7) and could account, at least in part, for differences of virulence among clones28.
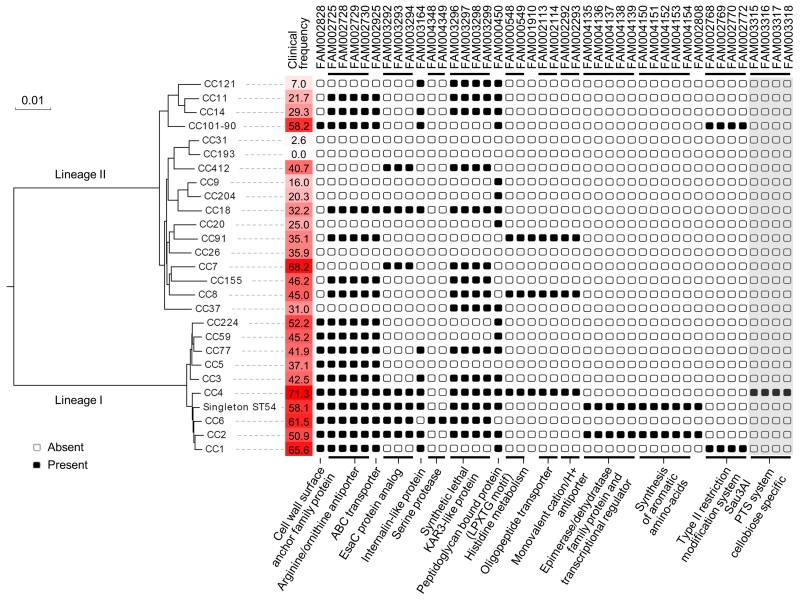
Putative virulence factors specific to hypervirulent clones (CC1, CC2, CC4 and CC6), including factors involved in Lm neuro-invasiveness, might have been overlooked by pathogenesis studies so far, as absent in reference strains which happen to belong to clones rarely responsible for human clinical cases (CC9, CC7). This hypothesis would fit with the observation that reference strains LO28, EGDe, EGD and 10403S, which all belong to these clones, are poorly neuro-invasive (Fig. 3b and our unpublished data). We therefore determined the distribution among clones of all gene families of Lm pan-genome (Supplementary Figure 3; Supplementary Table 8). To determine the evolutionary pattern of gene families and their correlation with the infection/food ratio of clones, the 1,791 genes of the core genome (Supplementary Table 6) were used to construct a recombination-purged phylogeny (see Supplementary Note). The clinical prevalence of clones displayed a strong phylogenetic inertia (λ = 0.9999, p < 0.001), indicating that the virulence potential of clones is determined, at least in part, by vertically transmitted features (Fig. 4). We therefore corrected for this evolutionary inertia29 to correlate the pattern of presence/absence of 6,867 gene families of the pan-genome with the infection/food ratio of clones (Supplementary Table 9). Strikingly, this analysis identified full-length InlA, Listeria pathogenicity island 3 (LIPI-3, or listeriolysin S cluster) and gene clusters responsible for teichoic acid biosynthesis in serotype 4b strains as being strongly associated with infectious potential at the population level (Supplementary Table 9). As these features were previously demonstrated to be involved in Lm virulence30-32, this validated our algorithm and indicated that other features strongly associated with infectious potential represent good candidates as novel virulence factors. They included protein families of defined putative function (Fig. 4) and many uncharacterized putative gene products. CC1- and CC4-specific features scored among the most strongly correlated with infection, as expected by the high infection/food ratio of these hypervirulent clones.
Figure 4. Phylogenetic distribution of novel putative virulence factors identified in this study.
Genes that were highly associated with high clinical frequency of clones (see GEE analysis in Online methods), and had an identified putative function are shown. The recombination-purged phylogeny within each lineage was based on its respective core genome. Percentages of clinical isolates in each clone are highlighted by a red gradient. Gene families are named as in Supplementary Table 9. Groups of syntenic genes are indicated by black horizontal lines. Putative functions or pathways are indicated below. Only the four LIPI-4 genes with no parologs in other genomes are shown (highlighted in grey).
LIPI-4, a locus involved in neural and placental infection
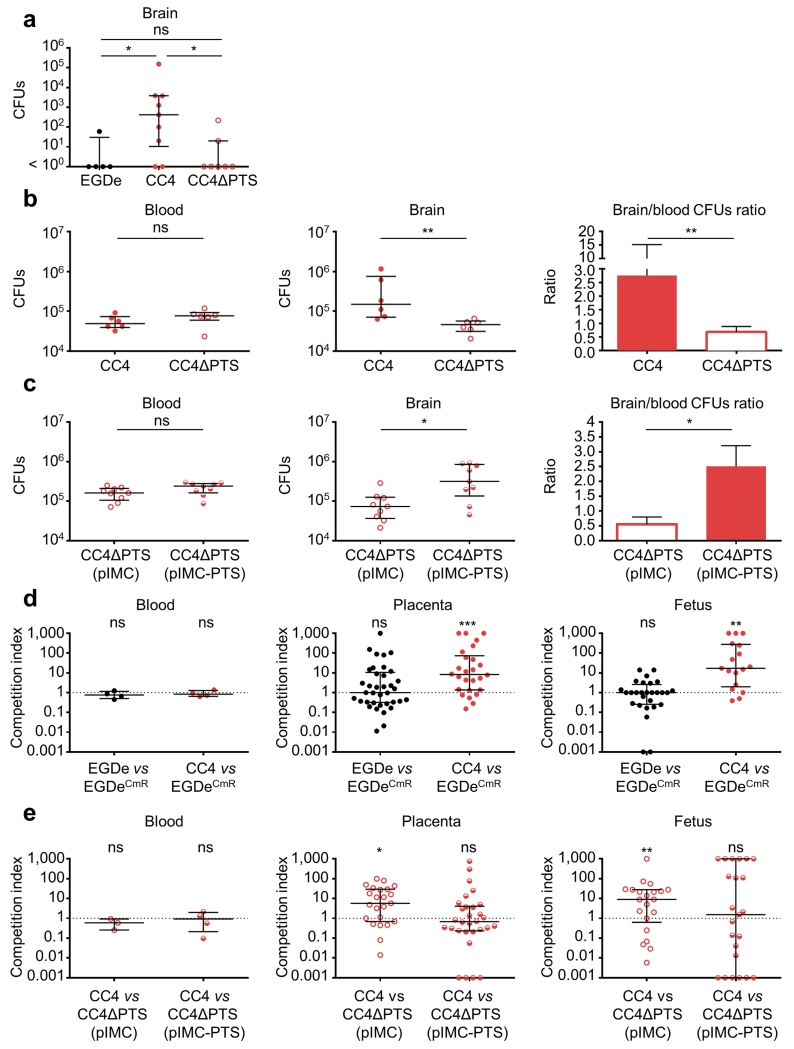
The above epidemiological and experimental data showed that CC4 comprises the highest proportion of clinical isolates of all Lm clones (71.3%, Fig. 2a and Fig. 4), is hypervirulent in vivo compared to EGDe and 10403S (Fig. 3), and is strongly associated with MN and CNS infections (p < 10−5, Supplementary Table 2). Prominent among 19 CC4-associated genes (Supplementary Tables 8 and 9) is a cluster of six genes annotated as a cellobiose-family PTS system (Supplementary Figure 6a). Carbon metabolism modulates virulence in Lm33. We therefore investigated the contribution of this putative sugar transport system to CC4 hypervirulence. A representative CC4 strain, LM09-00558, was chosen to construct a PTS deletion mutant (Supplementary Figure 6b). This mutant was not impaired for growth in culture medium (data not shown). In an oral humanized mouse model of infection, the CC4 strain was more able to infect the CNS than EGDe, whereas deletion of the entire PTS gene cluster considerably reduced CNS invasion in CC4 without affecting bacterial colonization in other tissues (Fig. 5a and Supplementary Figure 7a). In an intravenous infection model where the intestinal barrier is bypassed, we demonstrated that the PTS directly contributes to neuro-invasion (Fig. 5b and Supplementary Figure 7b). Reinserting a single copy of the PTS cluster onto the chromosome complemented this phenotype (Fig. 5c and Supplementary Figure 7c). These results demonstrate the role for this CC4-associated PTS in Lm neuro-invasiveness. We further tested CC4 infectivity in a mouse MN infection model, given its high association with human MN infection (Fig. 2b; Supplementary Table 2), using a competition index method23. Remarkably, the representative CC4 strain LM09-00558 infected better than EGDe the placentas and fetuses of pregnant humanized mice, but neither maternal organs nor maternal blood (Fig. 5d and Supplementary Figure 7d), mirroring the epidemiological association of CC4 with MN infection. To understand the involvement of CC4-associated PTS in MN infection, competitive index experiments were performed using either a ΔPTS or an isogenic PTS complemented strain compared to CC4 wild-type strain, respectively. While ΔPTS showed a significant defect in placental and fetal infection compared to its parental CC4 wild-type strain, the PTS complemented ΔPTS strain did not (Fig. 5e and Supplementary Figure 7e). These results therefore identify the CC4-associated PTS cluster as the first Lm virulence factor specifically implicated in CNS and MN infection. They indicate that the presence of this PTS locus accounts, at least in part, for the hypervirulence and enhanced CNS and MN tropism of CC4, which is strongly epidemiologically associated with both CNS and MN human infection. We propose to name this cluster Listeria pathogenicity island 4 (LIPI-4)31. The absence of LIPI-4 from CC1 and CC6 suggests that other factors contributing to their hypervirulence remain to be characterized (Supplementary Tables 8 and 9, Fig. 4).
Figure 5. Implication of hypervirulent clone CC4-associated PTS (LIPI-4) in CNS and placental infection.
Humanized mice were inoculated orally (dose 3.108) (a) or intravenously (dose 5.105) (b) with strain EGDe (n = 5 in a), CC4 strain LM09-00558 (CC4, n = 9 in a, n = 6 in b) or whole PTS cluster deletion mutant derived from LM09-00558 (CC4ΔPTS, n = 7 in a, n = 6 in b). (c) Humanized mice were intravenously infected (dose 5.105) by CC4ΔPTS containing either a single copy of pIMC (n = 9) or pIMC with PTS cluster under its native promoter (n = 9). (d) Competition index of WT EGDe (n = 4) or WT CC4 (n = 4) was tested against chloramphenicol-resistant EGDe (containing pIMC) in pregnant humanized mice. (e) Competition index of WT CC4 was tested against chloramphenicol-resistant CC4ΔPTS (pIMC) (n = 3) or CC4ΔPTS (pIMC-PTS) (n = 4) in pregnant humanized mice. Pregnant mice at day 14/21 of gestation were intravenously infected with a 1:1 mixture of the two strains as indicated (total dose 2.105). Mice were sacrificed on day 5 post infection for orally inoculated bacteria (a), or day 2 post infection when intravenously inoculated (b-e). Results are shown as median ± interquartile range. Each dot represents an organ (a-c) or blood (a-e) from one infected mouse, or one placenta or fetus (d and e). Statistical analyses were done by a Dunn’s multiple comparison test (a), Mann–Whitney U test (b and c), or Wilcoxon matched-pairs signed-rank test (d and e): *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
The epidemiological association of serotype 4b and lineage I of Lm with clinical disease has been documented18,19. Here, taking advantage of our fine-grained genetic analysis of Lm strain biodiversity filtered against exhaustive epidemiological and clinical data, we have been able to (i) establish the stratification of Lm virulence at the phylogenetic level of clones, (ii) identify a whole series of new putative virulence factors of Lm and (iii) demonstrate the involvement of one of them, encoding a putative PTS system (LIPI-4), in Lm hypervirulence. This CC4-associated gene cluster is the first Lm factor specifically implicated in its elective tropisms for the CNS and the fetal-placental unit and is therefore of high clinical relevance. These findings echo the pioneering comparative approach for Listeria at the inter-species level25 and demonstrate the power of integrating intra-species biodiversity, epidemiological, clinical and experimental approaches to discover novel virulence genes associated with specific and clinically relevant phenotypes34. This study establishes that Lm is a highly heterogeneous species with regards to pathogenicity, and is composed of hypervirulent and hypovirulent clones. Moreover, hypervirulent clones are those most likely to cause disease, and in particular CNS and MN listeriosis. This indicates that the clonal structure of Lm species should be taken into consideration for the surveillance of this major foodborne pathogen, which represents nearly half of deaths associated with foodborne infections in Western countries. This study opens a fresh perspective on Lm pathogenicity and also calls for the designation of novel reference strains, representative of those involved in actual human infections.
Online methods
Isolates selection for analysis of the source distribution of clones
Source distribution analysis was performed on a non-redundant collection of 7,342 isolates of food (n = 4,551) and clinical (n = 2,791) origins collected in France between January 2005 and October 2013. The 7,342 isolates were collected by the French National Reference Center (NRC) for Listeria (ntotal = 6,804; nfood = 4,013; nclinical = 2,791) and the French National Reference Laboratory (NRL) for Listeria (ntotal = nfood = 538).
The NRL for Listeria collects food isolates in the context of targeted controls in food industries. The NRC for Listeria collects nearly all isolates involved in human infection cases in France, amounting to an average of 360 strains per year. This high exhaustiveness is due to the fact that listeriosis is subjected to mandatory declaration in France. The NRC for Listeria also collects food isolates involved in nearly 80% of food alerts, which are triggered by a foodstuff being on the market and presenting a risk due to the presence of Listeria. Nearly 700 food or environmental strains are collected each year by the NRC for Listeria in the context of food alerts.
Among the food isolates of our collection, 3,143 (69.1% of all food isolates) were isolated from food alerts. Additional food strains were included: the NRC collected food strains in the context of investigations following neurological forms of listeriosis (n = 178, 3.9%), strains from own checks of food by industries (n = 692, 15.2%); and the NRL collected strains from food surveillance activities (n = 538, 11.8%). Absence of redundancy among strains involved in food alerts was obtained by keeping, from our initial collection, only one isolate out of those sharing the same date, food source of isolation, food alert number and MLST clone (after application of the PFGE-MLST dictionary, see below). Other food isolates were also de-duplicated based on date of isolation and food source. In total, 1,772 potential duplicate of food isolates were eliminated to obtain the final dataset, which was composed of 7,342 food and clinical isolates (no animal or environmental isolate was included).
Definition of cases
A MN infection is defined as a case of listeriosis in which Lm is isolated from blood or from a normally sterile site in a pregnant woman or a newborn of less than 28 days of age, or from a placenta, a fetus or a stillbirth. Only one isolate was considered when Lm was cultured from both the mother and the newborn. In all other cases, listeriosis was considered non-MN: bacteremia are cases with detection of Lm in blood with no evidence of CNS and MN infection, CNS infection are cases where Lm is isolated from the cerebrospinal fluid, or from blood in a patient with CNS clinical symptoms.
Pulsed-field gel electrophoresis (PFGE)
See Supplementary Note.
Multilocus sequence typing (MLST), clone definition and lineage assignation
MLST was performed as described by Ragon et al. (2008)15. Novel alleles and profiles were incorporated into the international MLST database (see URLs section). For clonal complexe assignations, we pooled MLST data from this study and from previous ones15-17,24,35. Clonal complexes were defined as groups of allelic profiles sharing 6 out of 7 genes with at least one other member of the group15. One exception was made for CC14 and CC91, as they corresponded to two separated clones that were merged due to a single intermediate allelic profile (ST206), and as the ApaI and AscI PFGE types of CC14 and CC91 differed largely from each other. ST206 was included in CC14. For public genomes, STs were deduced from the genomic sequences. The main phylogenetic lineage of each isolate was defined based on a phylogenetic tree inferred from the concatenated sequences of the seven MLST gene fragments.
PCR serogrouping
See Supplementary Note.
High confidence identification of MLST clones based on PFGE profiles
See Supplementary Note.
Statistical tests performed to identify associations of clones with food and clinical sources
In order to estimate the significance of associations of clones with food, infections or clinical sources, Chi2 tests were used. For each clone, a table of contingency was created (see Supplementary Note). The thresholds of significance were adapted to take into account multiple tests using the sequencial Bonferroni correction36. The principle is to perform the Chi2 test in order to obtain a p-value for each comparison. The tests are then ordered from the lowest to the highest p-value. The test with the lowest p-value is tested first with a Bonferroni correction taking into account all tests37. The second test is performed with a Bonferroni correction involving one fewer test, and so on for the remaining tests.
To quantify the relationships between the clinical frequency of Lm clones and their capacity to cause MN or CNS infections, linear regression analyses were used. These tests were conducted using the ‘lm’ function implemented in the basic R distribution by comparing, by clone, the frequency of clinical isolates among the total number of isolates, and the frequency of isolates involved in MN and CNS infections among the total number of clinical isolates. In order to avoid biases due to rare clones, we applied weights taking into account the total number of isolates in each clone.
Immunosuppressive comorbidities analysis
MONALISA is a French prospective national cohort that included microbiologically confirmed cases of invasive listeriosis from November 2009 to July 2013 (Clinical Trials NCT01520597). For each patient, clinical data including the medical background and samples including the clinical isolate were collected after written consent. The study received Institutional Review Board approval by a local ethical committee (Comité de Protection des Personnes Ile de France 3), according to the French legislation.
The immunosuppressive conditions taken into account were: reported daily alcohol uptake > 3 glasses on any day, cancer, congenital immune deficiency, diabetes, cirrhosis, hemodialysis for end-stage kidney disease, bone marrow transplantation, solid organ transplantation, hematological malignancies, pre-existing lymphopenia, pre-existing neutropenia, giant cell arteritis, systemic lupus erythematosus, rheumatoid arthritis, spondylarthritis, inflammatory bowel disease, other auto-immune disease, asplenia, age > 70 years, HIV infection, prescription of corticosteroids and prescription of other immunosuppressive treatments in the last 5 years.
In order to test for an association of numbers of immunosuppressive comorbidities with clones associated to clinical or food origins, we performed a linear regression between (i) the difference of number of isolates belonging to clinical associated clones (CC1, 2, 4 and 6) and to food associated clones (CC9 and 121), and (ii) the number of immunosuppressive conditions of patients infected by those isolates. We did the same analysis between the clinical associated clones (CC1, 2, 4 and 6) and the intermediate ones (CC8-16, 5, 3, 37, 155 and 18).
Strains used to assess the virulence levels of major clones in vivo
We assessed the virulence level of the three clones having the highest clinical frequencies (CC1, CC4 and CC6, see Fig. 2a) as well as the two clones having the lowest clinical frequencies (CC9 and CC121, see Fig. 2a) in a humanized mouse model. For each clone, we selected a minimum of 8 isolates, which included strains from each of these origins: food, CNS infections, MN infections and bacteremia. Isolates used for the in vivo experiments are indicated in the Supplementary Table 3.
Mouse infection
Animal experiments were performed as described38. We used 7- to 10-week old mEcad E16P KI female mice in a C57BL/6 genetic background. Six mice were used for each tested strain. As all mice were identical, the allocation of mice to experimental groups by randomization was not relevant, and no blinding was employed. Mice were food restricted overnight with free access to water. Lm culture was prepared as described and inoculated with a feeding needle intragastrically38. Mice were then immediately allowed free access to food and water. Pregnant mice were infected at day 14/21 of gestation as described23. Intravenous infections were performed by injecting 200 μL of bacterial suspension though tail vein. Competition index experiments were performed by mixing chloramphenicol-resistant pIMC containing strain and chloramphenicol-sensitive strain without pIMC in a 1:1 ratio. At indicated times, animals were euthanized and organs were homogenized in PBS. Serial dilutions of the homogenates were plated onto brain–heart infusion (BHI) agar plates with or without chloramphenicol. Competition indexes were calculated by dividing the number of chloramphenicol-sensitive CFUs by the number of chloramphenicol-resistant CFUs. All the procedures were in agreement with the guidelines of the European Commission for the handling of laboratory animals, directive 86/609/EEC (see URLs section) and were approved by the Animal Care and Use Committee of the Institut Pasteur, as well as by the ethical committee of “Paris Centre et Sud” under the number 2010-0020. Statistical analysis was performed by a Dunn’s multiple comparison test relative to EGDe infected mice. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Statistical tests of association between experimental data and clinical frequency
We performed a stepwise multiple regression of the average number of CFUs recovered in the mesenteric lymph nodes, the spleen, the liver, and the brain (transformed with LogX + 1) as well as the average body weight change on days three and five after infection of mice on the percentage of clinical isolates per clone. This regression was done by using the Bayesian Information Criterion (BIC) in order to identify which of these variables explains most the clinical frequency of clones.
Strain selection for whole genome sequencing
A total of 69 strains were selected for Illumina sequencing to represent the species diversity based on MLST and PFGE typing (Supplementary Figure 1; Supplementary Table 5). A minimum of two isolates with distinct PFGE profiles and isolated from distinct sources were selected per major clone, when it was possible. Thirty seven were isolated from human clinical infections, 16 from animal infections, 12 from food sources, 1 from environment and 3 were of unknown origin. Thirty five public genomes of good quality available at the time of the study were added in the analysis (Supplementary Table 5). Ten additional public draft genomes available at the time of the analysis were excluded because of unsatisfactory quality of the sequences, as assessed based on the number of contigs and of detected coding sequences (CDS), the average size of the CDS and the number of CDS common to EGDe. In total, 104 genomes were included, comprising 41 genomes of lineage I, 57 of lineage II, 5 of lineage III, and 1 of lineage IV. They represented 5 singletons and 34 clonal complexes.
Genome sequencing
Genomic DNA of the 69 strains was extracted using the Promega Wizard genomic DNA purification kit. Genomes were sequenced using the Illumina HiSeq 2000 system with the 2×100 nucleotides paired-end strategy. Quality trimming of reads and adapter clipping were performed using AlienTrimmer39. De novo assembly was performed on the final set of reads using CLCbio assembler (see URLs section) with a minimum contig size of 500 nt. Mauve Contig Mover program40 was used to re-order contigs using completely sequenced genomes as references: F2365 for lineage I and EGDe for lineage II. Genome sequences were submitted to the MicroScope/MaGe platform (Genoscope, Evry, France)41 for gene prediction and assignment of gene product functions. Missing genes from the 35 genomes from GenBank (see URLs section, last accessed in February 2013) were added using the MicroScope/MaGe platform in order to homogenize gene definition compared to the newly sequenced genomes.
Core genome definition
The core genome corresponds to the pool of genes ubiquitously found in all strains of the species. A core genome was defined first for the whole species using the 104 genomes, and then two additional core genomes were computed for lineage I (n = 41 genomes) and II (n = 57 genomes) separately. Orthologs were first identified as reciprocal best-hit, using end-gap free global alignments between the proteome of a reference genome and each of the proteomes of the genomes included in the analysis42. The reference genome used was EGDe for the core genome of the whole species and of lineage II, and F2365 for the core genome of lineage I. Hits with less than 60% similarity in amino acid sequence or more than 20% difference in protein length were discarded. In order to keep only orthologous genes in the final core genomes and delete all paralogs and xenologs, the genes outside blocks of synteny were removed. To do this, a gene was validated as part of the core genome only if among the five genes downstream and the five genes upstream, at least four were at the same location in all the genomes. The core genomes were defined as the intersection of pairwise lists of strict positional orthologs.
Distribution of virulence genes and detection of size variations of virulence gene products
The genome of EGDe25 strain was used as reference for the detection of size variability in virulence gene products encoded by all the other genomes. Homologs of all the genes of the reference genome were first searched using nucleotide blast against all the other genomes. Based on the distribution of all best e-values obtained for all blast analyses, a gene was considered absent if all the e-values were higher than 1.10−50 and if all matches were located in distinct syntenic blocks than in the EGDe. Only hits with e-values smaller than 1.10−50 or located in the same syntenic block (delimited by the core genes) than the reference gene were considered present and further analyzed for comparison with EGDe. To do that, detected homologous regions were extracted and translated into amino acid sequences. Size of the reference gene products in EGDe and of the translated matches from each genome were compared. A gene product was considered shorter than in the reference if it was shorter by at least 20 amino acids. Gene products that were smaller than in EGDe and encoded by genes located at the end of contigs were considered as present, as it was not possible to distinguish biological events from methodological artifacts. Virulence genes of the LIPI-1, LIPI-3 and SSI-1 (Stress Survival Islet 1) islands as well as inlA and inlB were analyzed in more details in order to detect the origins of size variations of gene products. To this aim, alignments of the nucleotide and amino-acid sequences of all detected matches were performed in order to detect nonsense mutations and internal deletions in the coding sequences.
Phylogenetic analyses based on the core genomes
See Supplementary Note.
Pan-genome definition
Homologous gene families (including paralogs, orthologs and xenologs) of the pan-genome were defined as previously in Touchon et al., 201443, except that Silix parameters were set such that a protein was considered homolog to another if the aligned part had at least 60% of similarity and represented more than 80% of the smallest protein. Chromosomal as well as plasmid genes were included in the pan-genome.
Gene presence and absence patterns among clones based on the pan-genome
The exhaustive collection of isolates that was used for the source distribution analysis and their assignment to clones based on PFGE and MLST correspondence allowed us defining a frequency of clinical isolates per clone. We thus needed the presence and absence patterns of genes of the pan-genome by clone. The presence or absence of gene families within a given clone was defined as the majority consensus (> 50%) of presence or absence among all genomes of each clone.
Phylogenetic inertia of clone clinical frequency and use of generalized Estimating Equations (GEE) to identify candidate virulence genes
Some of the genomes are very close whereas others are very distant. We therefore checked for phylogenetic inertia. The measurements of the phylogenetic inertia of the clinical frequency of clones were performed on the phylogeny of the fused lineages and of separated lineages by using the ‘phylosig’ tool, computed in R and implemented in the ‘phytools’ package using the lambda method44. Phylogenetic inertia was high for the complete dataset (Lineage I + II, Pagel’s lambda = 0.9999, p = 0.0005).
To identify gene families that are most associated with clones frequently involved in clinical infections, comparative analysis of the presence and absence patterns of gene families among clones according to the log10 of their clinical frequencies was performed taking into account their phylogenetic relationships. This was performed using generalized estimating equations (GEE) computed in R and implemented in the ‘ape’ package29. The estimates of the regression parameters from GEE were calculated for each gene family, reflecting their association with clones of high clinical frequency. The selection of candidate genes for further experimental analysis was made by identifying, among gene families with high GEE estimates, those that were specific of clones of interest, and by taking into account functional annotations of these gene families that we combined with data from the literature.
Listeria gene deletion mutant construction
Lm LM09-00558 ΔPTS mutant was obtained by deletion of entire putative PTS cluster by PCR-ligation and amplicon cloning in the suicide vector pMAD as previously described45. Briefly, primer pair 1 (Supplementary Table 10) was used to amplify the 5’ flanking region of PTS on Lm LM09-00558 genome, followed by PCR-ligation with the 3’ flanking region amplified with primer pair 2 (Supplementary Table 10). The amplified DNAs were subsequently cloned into SalI-BglII site in pMAD suicide vector. Obtained clones were isolated and sequenced to verify the sequence integrity. The plasmids with correct sequence were electroporated into electrocompetent cells prepared with LmLM09-00558 strain based on the method for Lm 4b strains46, followed by clone selection as described45. Obtained clones were isolated and the PTS flanking region was sequenced to verify the deletion.
PTS cloning
The entire PTS gene cluster of Lm LM09-00558 was amplified by primer pair 3 (Supplementary Table 10) followed by blunt-end cloning into pCR-Blunt (Invitrogen). Obtained clones were isolated and sequenced to verify the sequence integrity. The insert was digested, purified and cloned into SalI-NotI site of the plasmid pIMC, which integrates into Lm tRNAARG site in a single copy following conjugation thanks to the Listeriophage PSA integrase on the plasmid47.
Supplementary Material
Acknowledgments
We thank Carlos Soto Alvarez, Gwenaëlle Pontdeme, Thomas Cantinelli and Laure Diancourt for their contributions to MLST data production and analysis, and Sylvie Roche for providing low-virulence strains for genome sequencing. We also thank Damien Mornico from the Center of Bioinformatics, Biostatistics and Integrative Biology of the Institut Pasteur for his help with the submission of genome reads and assemblies. This study was funded by Institut Pasteur, Institut national de la santé et de la recherche médicale (INSERM), from the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (grant number ANR-10-LABX-62-IBEID), the European Research Council (ERC), the ERANET Proantilis, the Programme Hospitalier de Recherche Clinique MONALISA and the Programme de Recherche Translationnelle (PTR) ANSES – Institut Pasteur. Listeriosis surveillance in France is funded by Institut de Veille Sanitaire (InVS) and Institut Pasteur.
URLs
The international MLST database used in this study is available at http://www.pasteur.fr/mlst.
The guidelines of the European Commission for the handling of laboratory animals (directive 86/609/EEC) are available at http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm.
The CLCbio assembler software is available at www.clcbio.com/products/clc-genomics-workbench.
All genome sequences were submitted to EMBL-EBI, and are available at http://www.ebi.ac.uk/.
All public genomes used in this study are available at ftp://ftp.ncbi.nih.gov/genomes/ (last accessed in February 2013).
Footnotes
Accession codes
All genome sequences were submitted to EMBL-EBI (see URLs section). Accession numbers are listed in Supplementary Table 5. An umbrella project was created in order to group all reads and assemblies associated to the genomes sequenced in this study and is available by using this accession number: PRJEB10817.
Supplementary Note is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Falkow S, Isberg RR, Portnoy DA. The interaction of bacteria with mammalian cells. Annu Rev Cell Biol. 1992;8:333–363. doi: 10.1146/annurev.cb.08.110192.002001. [DOI] [PubMed] [Google Scholar]
- 2.Cossart P, Boquet P, Normark S, Rappuoli R. Cellular microbiology emerging. Science. 1996;271:315–316. doi: 10.1126/science.271.5247.315. [DOI] [PubMed] [Google Scholar]
- 3.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holden MT, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hensel M, et al. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 6.Parkhill J, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 7.Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lecuit M. Human listeriosis and animal models. Microbes Infect. 2007;9:1216–1225. doi: 10.1016/j.micinf.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci U S A. 2011;108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piffaretti JC, et al. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci U S A. 1989;86:3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiedmann M, et al. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun. 1997;65:2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orsi RH, den Bakker HC, Wiedmann M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol. 2011;301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Seeliger HPR, Jones D. Bergey’s Manual of Systematic Bacteriology. Vol. 2. Williams & Wilkins; 1986. pp. 1235–1245. [Google Scholar]
- 14.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol. 2004;42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ragon M, et al. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008;4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chenal-Francisque V, et al. Worldwide distribution of major clones of Listeria monocytogenes. Emerg Infect Dis. 2011;17:1110–1112. doi: 10.3201/eid1706.101778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haase JK, et al. The ubiquitous nature of Listeria monocytogenes clones: a large-scale Multilocus Sequence Typing study. Environ Microbiol. 2014;16:405–416. doi: 10.1111/1462-2920.12342. [DOI] [PubMed] [Google Scholar]
- 18.McLauchlin J. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur J Clin Microbiol Infect Dis. 1990;9:210–213. doi: 10.1007/BF01963840. [DOI] [PubMed] [Google Scholar]
- 19.Gray MJ, et al. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl Environ Microbiol. 2004;70:5833–5841. doi: 10.1128/AEM.70.10.5833-5841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward TJ, Ducey TF, Usgaard T, Dunn KA, Bielawski JP. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl Environ Microbiol. 2008;74:7629–7642. doi: 10.1128/AEM.01127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacquet C, et al. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J Infect Dis. 2004;189:2094–2100. doi: 10.1086/420853. [DOI] [PubMed] [Google Scholar]
- 22.Nightingale KK, Windham K, Martin KE, Yeung M, Wiedmann M. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl Environ Microbiol. 2005;71:8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Disson O, et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008;455:1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- 24.Chenal-Francisque V, et al. Optimized Multilocus variable-number tandem-repeat analysis assay and its complementarity with pulsed-field gel electrophoresis and multilocus sequence typing for Listeria monocytogenes clone identification and surveillance. J Clin Microbiol. 2013;51:1868–1880. doi: 10.1128/JCM.00606-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glaser P, et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 26.Hain T, et al. Pathogenomics of Listeria spp. Int J Med Microbiol. 2007;297:541–557. doi: 10.1016/j.ijmm.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 27.den Bakker HC, et al. Comparative genomics of the bacterial genus Listeria: Genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics. 2011;11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuenne C, et al. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics. 2013;14:47. doi: 10.1186/1471-2164-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paradis E, Claude J. Analysis of comparative data using generalized estimating equations. J Theor Biol. 2002;218:175–185. doi: 10.1006/jtbi.2002.3066. [DOI] [PubMed] [Google Scholar]
- 30.Lecuit M, et al. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science. 2001;292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 31.Cotter PD, et al. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008;4:e1000144. doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faith N, et al. The role of L. monocytogenes serotype 4b gtcA in gastrointestinal listeriosis in A/J mice. Foodborne Pathog Dis. 2009;6:39–48. doi: 10.1089/fpd.2008.0154. [DOI] [PubMed] [Google Scholar]
- 33.Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol. 2010;8:401–412. doi: 10.1038/nrmicro2351. [DOI] [PubMed] [Google Scholar]
- 34.Bille E, et al. A chromosomally integrated bacteriophage in invasive meningococci. J Exp Med. 2005;201:1905–1913. doi: 10.1084/jem.20050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for online methods
- 35.Cantinelli T, et al. “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J Clin Microbiol. 2013;51:3770–3779. doi: 10.1128/JCM.01874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand J Statist. 1979;6:65–70. [Google Scholar]
- 37.Bonferroni CE. Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 1936;8:3–62. [Google Scholar]
- 38.Disson O, et al. Modeling human listeriosis in natural and genetically engineered animals. Nat Protoc. 2009;4:799–810. doi: 10.1038/nprot.2009.66. [DOI] [PubMed] [Google Scholar]
- 39.Criscuolo A, Brisse S. AlienTrimmer: a tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics. 2013;102:500–506. doi: 10.1016/j.ygeno.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Rissman AI, et al. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vallenet D, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009;2009 doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Touchon M, et al. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol. 2014;6:2866–2882. doi: 10.1093/gbe/evu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- 45.Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monk IR, Gahan CG, Hill C. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl Environ Microbiol. 2008;74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monk IR, Casey PG, Cronin M, Gahan CG, Hill C. Development of multiple strain competitive index assays for Listeria monocytogenes using pIMC; a new site-specific integrative vector. BMC Microbiol. 2008;8:96. doi: 10.1186/1471-2180-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.