Mitochondria are the ‘power houses’ of eukaryotic cells, ensuring an adequate supply of energy for the myriad processes that must happen day in, day out, to keep the cell ticking over. In this guise, we often think about these organelles as machines that continuously supply energy to the cell, in the form of adenosine triphosphate (ATP). However, cellular physiology is not static, and in fact, over the day, the internal milieu of a cell waxes and wanes according to a ~24 hour (circadian) rhythm (1, 2). In this issue of Science, Peek et al. investigate a pathway by which mitochondria are regulated by the cellular clockwork that generates these circadian rhythms. They show that the clock-controlled bioavailability of nicotinamide adenine dinucleotide (NAD+), a coenzyme crucial for energy conversion within the cell, modulates oxidative metabolism in mammalian mitochondria (3).
Organisms as diverse as bacteria and humans use circadian clocks to temporally organise their physiology and behaviour in anticipation of the solar cycle. Such cellular timing mechanisms ensure that activity and feeding cycles resonate with the geophysical time and, in this way, guarantee efficient energy homeostasis by appropriately timed energy storage and utilisation through the day and night (4). Disruption of these rhythms, as experienced during shift work, is associated with long-term adverse health consequences including metabolic and mental disorders (5). In mammals, many metabolic functions, such as carbohydrate metabolism, are under circadian regulation, but the question of whether oxidative metabolism is regulated in a circadian fashion has remained unanswered until now, even though we know circadian redox oscillations are likely to be present in all oxygen-consuming organisms (6). Given the numerous links between oxidative stress and age-related diseases (7), measuring the temporal landscape of oxidative metabolism in health and disease is of emerging importance.
Our current view of the clockwork is modelled by interlocked transcriptional/translation feedback loops that generate daily rhythms at the genomic scale, extensively using post-transcriptional and post-translational modifications (8). In mammals, such cell-autonomous oscillators also encompass a cytosolic component consisting of accessory feedback loops that couple the transcriptional oscillator to cellular physiology (9). In particular, an NAD+ loop is composed of the circadian transcriptional factors BMAL1/CLOCK, which control the expression of the rate-limiting enzyme in NAD+ biosynthesis, nicotinamide phosphoribosyltransferase (NAMPT) (10, 11). NAD+ levels feed back onto the clockwork through the NAD+-dependent deacetylase SIRT1, which regulates several clock proteins including PER2 and CLOCK (12, 13). Moreover, in vitro studies suggest that NAD+:NADH ratio, a reflection of redox poise, might also directly regulate BMAL1/CLOCK DNA-binding affinity (14).
Using fasted mice, to remove the confounding effects of feeding rhythms upon metabolism, Peek et al. measured the rate of fatty acid oxidation and NAD+ levels in the liver and found that both of them displayed a circadian profile with a peak during the rest phase – ‘daytime’ for the ordinarily nocturnal mouse. Given that BMAL1/CLOCK regulates NAD+ biogenesis, the authors investigated the effect of Bmal1 deficiency on NAD+ levels, and on the oxidation of fatty acids – one of the mitochondrion’s fuel sources. They observed a significant decrease for both of them, thereby suggesting that BMAL1/CLOCK are necessary to maintain sufficient amounts of NAD+ for fatty acid oxidation to proceed.
In lines with this, detailed manipulation of mitochondrial respiratory activity in isolated mitochondria revealed that circadian mutant mice have decreased flux through the tricarboxylic acid (TCA) cycle, the churn within mitochondria that gathers the electrons required for the ongoing production of ATP (Figure 1). This was measured using fatty acids and pyruvate (the end-product of glucose breakdown by glycolysis), as energy input sources for their measurements. Their results were consistent with the fact that both fatty acid β-oxidation and pyruvate decarboxylation require sufficient levels of NAD+. Accordingly, when they bypassed the NAD+ ‘bottleneck’ using intermediary metabolites of the TCA cycle as alternative carbon sources for its reactions, normal oxygen consumption was restored. They went on to validate this further in live animals, employing nicotinamide mononucleotide (NMN), the precursor of NAD+, to replenish the NAD+ pool in Bmal1-deficient animals. This too restored oxidation rates close to normal levels.
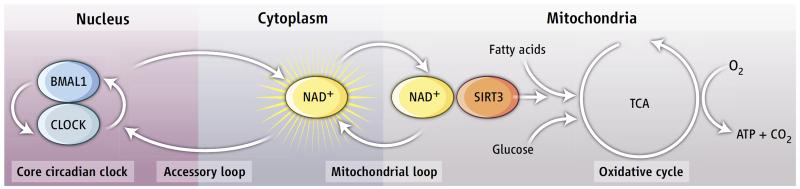
Figure 1. The circadian clock gives rhythm to mitochondrial respiration.
Mitochondria are the “power houses” of eukaryotic cells and produce energy in the form of ATP from glucose and fatty acids through a process called cellular respiration. The circadian transcriptional factors BMAL1/CLOCK control the rhythmic biosynthesis of the cofactor NAD+, which feedbacks onto the circadian oscillator through the NAD+-dependent deacetylase SIRT1. Peek et al. show that cytosolic NAD+ oscillations regulates mitochondrial oxidative metabolism in a circadian fashion (3). In particular, the mitochondrial NAD+-dependent deacetylase SIRT3 regulates fatty acid oxidation rate in response to daily variations in NAD+ and, by this way, generates rhythmic respiration in mammalian cells.
Cytosolic rhythms in NAD+ biosynthesis could conceivably drive redox rhythms by modulating NAD+:NADH ratio, and in this way, affect metabolic fluxes in mitochondria. However, Peek et al. demonstrated that the NAD+:NADH poise was not disturbed by Bmal1 deletion in cells, and therefore a mechanism sensing absolute NAD+ levels appears more likely to convey information from transcriptional cycles to mitochondrial oxidative metabolism. SIRT3, a mitochondrial NAD+-dependent deacetylase, seemed an reasonable candidate, given that it regulates mitochondrial fatty acid oxidation through reversible acetylation (15). Peek et al. accordingly observed a cluster of hyperacetylated proteins when they compared the mitochondrial proteome of wild type and Bmal1-deficient animals, supporting a recent study that reported circadian acetylation of mitochondrial enzymes (16). Moroever, many hyperacetylated proteins were bona fide SIRT3 targets, including manganese superoxide dismutase (MnSOD) that displayed circadian acetylation at a lysine residue specific to SIRT3.
The finding that mitochondrial oxidative metabolism is regulated by the circadian clock not simply extends further the catalogue of metabolic processes controlled by the circadian oscillator, but also underscores the importance of redox metabolism and accessory cytosolic loops in the mammalian clockwork. The crucial role of NAD+ in controlling mitochondrial metabolism indicates that so-called ‘accessory loops’ might be fundamental parts of the circadian oscillator and also suggests the existence of ‘mitochondrial loops’ – a possible new layer in the architecture of the mammalian circadian oscillator – given the growing evidence that reactive oxygen species and redox balance impinge the circadian clockwork (17). The striking attenuation of NAD+ levels in the absence of Bmal1 suggests that BMAL1/CLOCK not only modulate NAD+ biosynthesis in a circadian fashion, but would be essential to maintain NAD+ bioavailability in the cell. Such a critical role of circadian transcription factors in basal cytosolic processes indicates that BMAL1/CLOCK might also have important non-circadian functions in the cell.
A puzzling implication of circadian mitochondrial activity is the following: how can energy homeostasis be maintained throughout the circadian cycle, if respiration – and thus most ATP production – is segregated to a specific epoch of the circadian cycle? This could imply that there would be a burst in ATP consumption during a ‘respiration phase’, and most importantly, that cellular metabolism would be arranged around these respiratory rhythms, as occurs in the yeast metabolic oscillator (18). More generally, these results raise a vast array of questions about the circadian organization of metabolism and indicate that the influence of the circadian clock on mammalian metabolic cycles might have been previously underestimated, in the sense that the latter is likely to be regulated at the cellular level and not, as previously thought, to be solely a consequence of whole animal physiology. Understanding how these clock-driven metabolic cycles relate to non-transcriptional circadian redox oscillations (6, 19, 20) will surely help to delineate more clearly the origin and function of metabolic circadian rhythms. An exciting possibility is that mitochondrial respiratory rhythms, and possibly cycles observed in plastids (21, 22), could potentially be a ancient mechanism inherited from the endosymbiotic integration of bacterial oscillators into primitive eukaryotic cells.
References
- 1.Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 2.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Peek CB, et al. Circadian Clock NAD+ Cycle Drives Mitochondrial Oxidative Metabolism in Mice. Science. 2013 doi: 10.1126/science.1243417. (New York, NY) doi:10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. (New York, NY) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20:36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 8.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.07.002. doi:10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rey G, Reddy AB. Connecting cellular metabolism to circadian clocks. Trends Cell Biol. 2013;23:234–241. doi: 10.1016/j.tcb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. (New York, NY) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. (New York, NY) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 13.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. (New York, NY) [DOI] [PubMed] [Google Scholar]
- 15.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masri S, et al. The circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1217632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stangherlin A, Reddy AB. Regulation of circadian clocks by redox homeostasis. J Biol Chem. 2013;288:26505–26511. doi: 10.1074/jbc.R113.457564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. (New York, NY) [DOI] [PubMed] [Google Scholar]
- 19.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CH, et al. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science. 1995;269:1863–1865. doi: 10.1126/science.7569925. (New York, NY) [DOI] [PubMed] [Google Scholar]
- 22.Noordally ZB, et al. Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science. 2013;339:1316–1319. doi: 10.1126/science.1230397. (New York, NY) [DOI] [PubMed] [Google Scholar]