Abstract
Circadian clocks are cellular timekeeping mechanisms that coordinate behavior and physiology around the 24-h day in most living organisms. Misalignment of an organism’s clock with its environment is associated with long-term adverse fitness consequences, as exemplified by the link between circadian disruption and various age-related diseases in humans. Current eukaryotic models of the circadian oscillator rely on transcription/translation feedback loop mechanisms, supplemented with accessory cytosolic loops that connect them to cellular physiology. However, there is mounting evidence questioning the absolute necessity of transcription-based oscillators for circadian rhythmicity, supported by the recent discovery of oxidation-reduction cycles of peroxiredoxin proteins, which persist even in the absence of transcription. A more fundamental mechanism based on metabolic cycles could thus underlie circadian transcriptional and cytosolic rhythms, thereby promoting circadian oscillations to integral properties of cellular metabolism.
Keywords: peroxiredoxin, oxidation-reduction, redox, oscillator, posttranslational, posttranscriptional
INTRODUCTION
Biological clocks are ubiquitous timekeeping mechanisms that provide living organisms with temporal organization over timescales ranging from seconds to years. Organisms have evolved such rhythms to adapt to repetitive environmental and geophysical events, such as solar cycles, tides, the rotation of the Moon, and the recurring seasons. Among biological oscillators, the circadian clock holds a preeminent role because it allows organisms from bacteria to humans to anticipate the alternation of our days and nights.
The Latin etymology of the word circadian---circa, about; diem, a day---reflects that the intrinsic period of these rhythms is not exactly 24 h but, for example in humans, closer to 25 h. These clocks do not, however, need to be extremely accurate, because in their natural setting they are reset daily by external cues, such as light-dark cycles, that ensure their synchrony with environmental cycles (a process sometimes termed entrainment). When kept in constant conditions (that is, when the cycle is removed, such as in constant darkness), an organism relies only on its intrinsic clock to keep time and thus exhibits a so-called free-run rhythm because it is no longer directly constrained by the external driving cycle.
These observations are exemplified by an experiment performed in 1962 by the French cave-explorer Michel Siffre (1), when he revealed the existence of such an intrinsic timekeeping mechanism in humans by isolating himself for two months in a 130-m-deep cave in the French Alps. He recorded his sleep-wake activity during the study and found that, even though he had completely lost conscious perception of the passing of time, his activity pattern clearly demonstrated a persistent 24.5-h rhythm for the duration of the experiment. A few years later, Jürgen Aschoff (2) confirmed Siffre’s discovery by conducting experiments in more controlled laboratory conditions and observed the influence of the circadian clock on the daily pattern of numerous physiological functions, including body temperature, blood pressure, cognitive performance, and hormone plasma concentration. Since then, intensive research in humans and in a wide variety of prokaryotic and eukaryotic model organisms have established the pervasive role of the circadian clock in regulating physiological and behavioral processes (3--5). Moreover, studies in cyanobacteria and plants have revealed the clear adaptive significance of circadian clocks, given that strains with a period matched to the external cycle---that is, in resonance with their environment---outcompeted the others (6--8). Similarly, in humans, there is mounting evidence that circadian misalignment, a mismatch between external and internal cycles, as it occurs in shift work and jet lag, has adverse health consequences and can increase the risk of cancer, metabolic, and mental diseases (9).
In the quest for the origin of circadian rhythms in higher species, researchers have attempted to locate anatomical loci that would be responsible for the generation of behavioral rhythms. In mammals, such approaches led to the identification of a paired structure in the brain’s hypothalamus called the suprachiasmatic nuclei (SCN) and subsequently revealed a hierarchical organization of the mammalian circadian system (10). Working as a central pacemaker, the SCN are entrained by the environment through photic input from the retina and, in turn, synchronize peripheral organs using a variety of systemic signals (4). However, peripheral organs are not merely forced to oscillate by the master SCN clock because most cells (and, indeed, tissues) in our body harbor self-sustained circadian oscillators in their own right. Thus, in the body, there is a pervasive network of cellular oscillators, all kept in synchrony by the central clock in the SCN (11).
Suprachiasmatic nuclei (SCN): in mammals, a paired structure in the brain’s hypothalamus working as a master pacemaker
In the past 20 years, our understanding of circadian timekeeping has experienced tremendous advances stemming from the successful identification of such cell-autonomous mechanisms. The current model for circadian timekeeping in all species is thought to incorporate similar transcription/translation feedback loop (TTFL) mechanisms at their core. However, each species’ clock employs a different set of “gears” in its timing mechanism, given that so-called clock genes are not phylogenetically conserved among vertebrates, fungi, plants, and bacteria (12).
Transcription/translation feedback loop (TTFL): designates current models of transcription-based circadian oscillators
Transcription-based models, however, were only one of the several models proposed before the successful identification of circadian genes. Notably, molecular (in vitro), biochemical network, and membrane models were developed before the advances of molecular genetics (13). These models were thereafter discarded, probably not because of their inaccuracy, but rather because of the difficulty in identifying bona fide clock components and therefore in constructing detailed molecular models.
Recently, posttranscriptional and posttranslational mechanisms have gained considerable attention owing not only to their importance in transcription-based feedback models (14--16), but also to the discovery of nontranscriptional clocks in various species. The most notable evidence of nontranscriptional circadian oscillations was published in 2005, when Kondo and colleagues reconstituted a molecular clock in a test tube, composed of only the three Kai proteins from the cyanobacteria Synechococcus elongatus, supplemented with ATP as an energy source (17). A few years later, rhythmic oxidation-reduction of peroxiredoxins (PRDXs), antioxidant proteins involved in scavenging intracellular hydrogen peroxide, was demonstrated in human red blood cells and also in conditions when transcription is arrested in the unicellular alga Ostreococcus tauri (18, 19). Furthermore, a subsequent study revealed the deep phylogenetic conservation of PRDX redox oscillations from Archaea to humans (20), suggesting that nontranscriptional oscillators may be common to all circadian systems.
Peroxiredoxins (PRDXs): a family of thiol-specific antioxidant enzymes involved in scavenging hydrogen peroxide inside the cell
In this review, we discuss the importance of metabolic and nontranscriptional rhythms for circadian timekeeping in eukaryotes. To this end, we first recapitulate the main results and concepts underlying current transcription-based models and highlight the numerous challenges encountered in attempts to reconcile these models with experimental observations. We then summarize current knowledge about the two most prominent examples of nontranscriptional rhythms: the KaiC oscillator in cyanobacteria and PRDX oxidation-reduction rhythms. Finally, we discuss how metabolic and circadian cycles may be coupled, the potential role of PRDXs and other metabolic oscillations in circadian timekeeping, and perspectives on the evolution of 24-h rhythms in living organisms.
THE PRINCIPLE OF TRANSCRIPTION/TRANSLATION FEEDBACK MODELS
The Core Transcriptional Oscillator
The first step in constructing current transcriptional models was the identification of a genomic locus involved in behavioral rhythms in Drosophila in the early 1970s. Konopka & Benzer (21) performed a mutagenesis screen for abnormal eclosion rhythms in these fruit flies and found three mutations that all mapped to the same genomic locus, which they termed “period.” The three mutant alleles had dramatic effects on behavioral rhythms, because they caused arrhythmic, short- (19 h) or long-period (28 h) phenotypes. The cloning of period (per) independently by the Young and Rosbash laboratories in the 1980s allowed its subsequent molecular characterization (22, 23), which revealed that both per mRNA and its protein are expressed in a circadian fashion (24, 25), with a delay between the production of RNA and protein. These results had already suggested that per would be part of a negative-feedback-loop mechanism in which PER protein represses its own expression (24).
A few years later, studies in the fungus Neurospora crassa offered proof of the principle that a transcriptional feedback loop could indeed be the molecular basis of behavioral rhythms, because the protein product of frequency negatively regulates its own transcription (26). The loop was later closed similarly in flies in the late 1990s, when the heterodimeric basic-helix--loop--helix transcription factors CLOCK and CYCLE were shown to activate transcription of per and timeless (tim), and their expression was found to be repressed by their protein products (27--29). The potential involvement of Clock in circadian timekeeping had been previously suggested in mice, because a mutant allele in this gene caused a lengthening of the circadian period (30). Interestingly, however, deletion of this gene does not have appreciable consequences for behavioral and molecular rhythmicity (31), thus causing researchers to question its central importance in the core transcriptional clockwork.
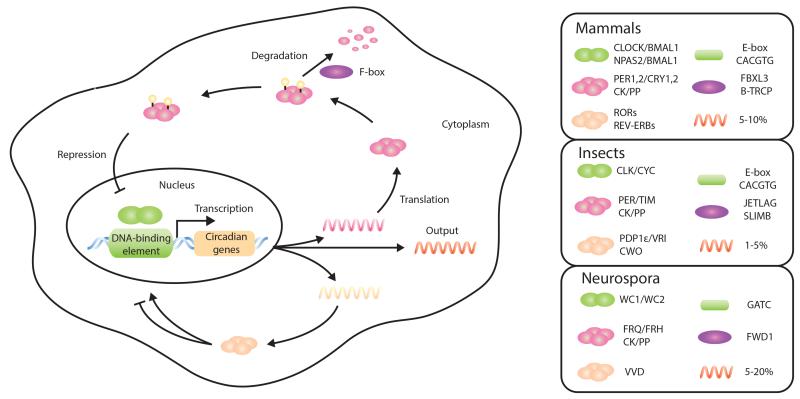
Extensive research has further dissected the genetic basis of circadian rhythmicity and has permitted the construction of detailed TTFL models in all species that exhibit overt circadian rhythms (32). The fundamental principle relies on a transcriptional activator inducing the transcription of a repressor, which results in the accumulation of the latter over time, until it reaches a sufficient level to repress its own activation (Figure 1). In mammals, the “core” transcriptional machinery is composed of two interlocked feedback loops, in which the transcription factors CLOCK/BMAL1 (or NPAS2/BMAL1) occupy a central role, acting as heterodimers. These activate Period (Per) and Cryptochrome (Cry) gene families through interaction with E-box regulatory sequences (33). As a result, the PER and CRY protein products form heterotypic complexes that accumulate over time in the cytoplasm. The level of PER/CRY complexes increases until it reaches a threshold, marking the beginning of the repressive phase, in which PER/CRY complexes translocate back to the nucleus and repress CLOCK/BMAL1 transactivation.
Figure 1.
The transcriptional feedback loop model in eukaryotes, showing the principle of the TTFL model common to mammals, insects, and fungi. Only key components are represented to highlight the similarity of the feedback mechanism, despite the divergence of molecular components in the different phyla (12). Legend shows the core clock components for the three different model organisms together with the estimated fraction of the transcriptome under circadian control (bottom right of each panel) (39, 184). Abbreviations: CK, casein kinases; CWO, clockwork orange; FRQ, frequency; FRH, FRQ-interacting RNA helicase; FWD1, F-box and WD40 repeat-containing protein 1; PDP1ε, PAR-domain protein 1 epsilon; PP, protein phosphatases; RORs, RAR-related orphan receptors; TTFL, transcription/translation feedback loop; VVD, vivid; VRI, vrille; WC1/2, white collar 1,2.
In a second loop, CLOCK/BMAL1 positively regulate the transcription of the heme-sensing nuclear hormone receptors REV-ERBα (NR1D1) and REV-ERBβ (NR1D2), which in turn repress the transcription of Bmal1 (34). The REV-ERB transcriptional repressors are also involved in transcriptional control of Per and Cry, likely modulating the phase of expression of the latter (35, 36). This simplistic description exemplifies the principle of interlocked negative-feedback loops, but CLOCK/BMAL1 and REV-ERBs are also directly involved in global regulation of circadian transcription (35--38), which constitutes a significant proportion of the total transcriptome. Approximately 5--10% of the transcriptome in mouse and 1--5% in flies is estimated to be under clock control (39). Moreover, virtually every step from transcription factor binding to protein degradation may have a circadian component in its orchestration, given that epigenetic, posttranscriptional, and posttranslational regulatory mechanisms have important roles in current transcriptional models (14--16, 40, 41).
Posttranscriptional and Posttranslational Mechanisms in the Clockwork
Posttranslational regulation is an essential mechanism within the circadian clockwork, because it modulates a myriad of processes, including protein turnover, intracellular localization, and DNA-binding affinity. As such, it contributes to setting the pace of the oscillator (14, 15, 42). Phosphorylation is the most well-documented posttranslational modification in the clockwork. In particular, regulation of protein stability by phosphorylation has emerged as a strong determinant of circadian period in the different taxa (15), but many other modifications such as acetylation (43, 44), methylation (45, 46), sumoylation (47), and glycosylation (48--51) have been implicated in regulating circadian processes. The phosphorylation status of circadian repressors, such as the mammalian PER and CRY proteins, increases daily owing to the concerted action of kinases and phosphatases, which subsequently leads to degradation of repressors via F-box proteins. Moreover, enzymes such as casein kinase 1 and 2 (CK1 and CK2, respectively) and protein phosphatase 2A (PP2A) serve phylogenetically conserved roles in species-specific TTFL models (14), despite clear differences in their clock gene components.
In mice, the nuclear F-box protein FBXL3 controls the half-lives of CRY proteins by targeting phosphorylated substrates for proteasomal degradation (52--54), and its action is counteracted by the cytoplasmic F-box protein FBXL21 (55, 56). In accordance with a model in which an increase in the stability of the repressors would lead to longer periods, Fbxl3-mutant mice have a behavioral period of ~27 h (52, 54), whereas Fbxl21-mutant mice exhibit a period shortening of more than 1 h (56). Similarly, the F-box protein beta-transducin repeat containing protein 1 (β-TrCP1) regulates the ubiquitylation-mediated degradation of PERIOD2 (57). The importance of F-box protein--mediated degradation is also observed in other organisms, as Drosophila PER and TIM proteins are regulated by SLIMB (58) and JETLAG (59), respectively. Similar mechanisms have also been proposed in Neurospora (60) and in plants (61). The importance of the proteasomal degradation machinery alone is supported by the fact that Ostreococcus tauri requires a functional proteasome for the generation of circadian rhythms (62). That F-box proteins, which are so intimately involved in protein turnover in general within cells, may impact mechanisms that require protein degradation to oscillate is perhaps not surprising.
The purely posttranslational oscillator described in cyanobacteria also strongly suggests that posttranslational mechanisms may be fundamental to the clockwork in higher species (see next section for details). This is implied by the results of global approaches that reveal posttranslational control as a key mechanism not only in the core transcriptional oscillator, but also in the sculpting of the circadian proteome. An examination of cycling proteins in mice showed that only half of the circadian proteome had a corresponding oscillation at the transcriptional level in the liver (40) and that this proportion went down even further down (11--38%) in the SCN (63). In addition, reversible lysine acetylation shapes the hepatic proteome in a circadian fashion, targeting cytosolic and mitochondrial metabolic enzymes preferentially (64).
Additionally, a growing momentum implicates posttranscriptional mechanisms in the circadian clockwork, including microRNA-mediated degradation and RNA-binding proteins involved in transcript stability and translational regulation (16). Moreover, recent studies highlight the role of nuclear export (65), splicing (66), and polyadenylation (67) in the clockwork. The general importance of circadian posttranscriptional regulation is further supported by recent genome-wide studies showing that correlation between rhythmic transcription and mRNA accumulation is much lower than expected (38, 68--70).
Secondary Loops Connect the Core Oscillator to Cellular Physiology
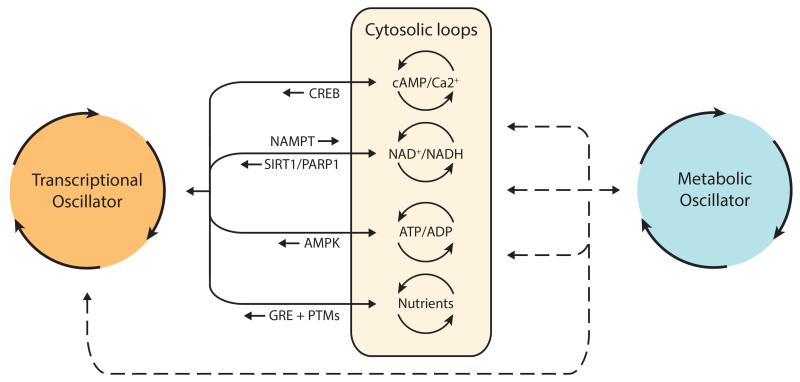
In addition to the posttranscriptional and posttranslational mechanisms described above, there is strong evidence that cytosolic processes have important roles in broadcasting transcriptional oscillations to downstream cellular machinery to control cellular physiology (42, 71). This picture has recently emerged in response to observations that a number of clock-controlled cytosolic processes can feed back on the transcriptional oscillator. Accordingly, such processes form accessory loops that tightly connect the clockwork to cellular metabolism (Figure 2).
Figure 2.
Transcriptional, cytosolic, and metabolic cycles. The discovery of cytosolic processes as integral to the TTFL models extended the concept of genetic timekeeping to include cytosolic components (42, 71). The latter are mainly involved in redox and energy metabolism and form accessory loops that are controlled by the TTFL oscillator and in turn feedback to it. The reported resilience of current TTFL models to transcriptional perturbations (80, 81, 83, 91, 92) together with the discovery of nontranscriptional rhythms both in prokaryotic (17) and eukaryotic species (20, 107, 111) suggest that a nontranscriptional oscillator underlies transcriptional and cytosolic rhythms. The molecular identity of the latter has not yet been resolved, but the deep phylogenetic conservation of peroxiredoxins’ oxidation cycles suggests that it is of metabolic origin (20). Abbreviations: CREB, cAMP response element--binding protein; GRE, glucose response element; PTMs, posttranslational modifications; TTFL, transcription/translation feedback loop.
An exemplar of this metabolic feedback is nicotinamide adenine dinucleotide (NAD+) biosynthesis and its salvage pathway. The rate-limiting enzyme in NAD+ biosynthesis, nicotinamide phosphoribosyltransferase (NAMPT), is transcriptionally controlled by CLOCK/BMAL1 in mice, and NAD+ levels exhibit circadian oscillation in the cytoplasm (72, 73). NAD+/NADH redox poise may also directly regulate NPAS2/BMAL1 DNA-binding affinity (74), and thus close this accessory loop. NAD+ levels can also be indirectly transduced back to the clockwork through the NAD+-dependent deacetylase SIRT1, which deacetylates PER2 (44) and balances CLOCK-mediated acetylation (43). Although its activity is driven by feeding cycles, poly(ADP-ribose) polymerase 1 (PARP-1), an NAD+-dependent ADP-ribosyltransferase, is also involved in transmitting NAD+ rhythms because it mediates ADP-ribosylation of CLOCK in a daily manner, thus modulating CLOCK/BMAL1 DNA-binding affinity (75). Similarly, other cytosolic pathways that may loop back into the transcriptional oscillator include cyclic adenosine monophosphate (cAMP) and Ca+2 signaling (76), AMP kinase (77), and glucose levels (78).
Cyclic adenosine monophosphate (cAMP): a second messenger important for regulation of cellular metabolism in eukaryotes
Nicotinamide adenine dinucleotide (NAD+): a coenzyme working as an electron carrier that is involved in redox reactions
The fact that all these accessory loops are involved in maintaining energy homeostasis inside the cell further strengthens the existing links between circadian clocks and metabolic pathways and shows that the boundaries between cellular timekeeping and metabolism are becoming blurred as new knowledge is acquired. Importantly, all these observations have been made in the presence of a transcriptional oscillator. Thus, it is difficult to tease apart the relative dependence of so-called accessory loops and the timekeeping mechanism, if this distinction is actually meaningful in view of the current evidence. Indeed, cellular metabolic cycles may be coupled to 24-h rhythms within the cells, and they may still continue to oscillate, perhaps with a different period, even in absence of clock genes (79). Accordingly, NAD(P)H levels can exhibit circadian rhythms in the absence of input from the transcriptional oscillator, as exemplified in human red blood cells (18).
Challenging Transcriptional Models
Current transcription/translation models are sufficient to explain most of the fundamental features of the circadian clock, such as its persistence in constant conditions, the ability to entrain to external cues, and coordination of output functions such as behavior and physiology. However, an increasing number of results question the necessity of a functional transcriptional oscillator for cellular rhythmicity. For example, circadian cycles are relatively insensitive to cell division in mammalian cells, and mother cells transmit circadian-phase information to their daughters (80). A similar phase coherence between mother and daughter cells has been reported in cyanobacteria (81). Given the stochastic nature of circadian transcription in mammalian cells (82), the perturbation of transcription during cell division is expected to result in greater phase variability. In line with this hypothesis, global inhibition of transcription with actinomycin D and α-amanitin has revealed the robustness of circadian oscillators to such severe perturbations, as single cells continue to exhibit bioluminescence rhythms even when the transcription rate is reduced by up to 70% (83).
Furthermore, studies showing that constitutive expression, or deletion, of clock genes does not abolish circadian rhythms call into question the importance of transcription in current clock models. In flies, expression of both per and tim under the control of a constitutive promoter can affect circadian rhythms, but approximately half of the flies still exhibit robust behavioral rhythms (84). Moreover, fungi can exhibit conidiation (spore formation) rhythms even in the absence of central components of the feedback loop, leading to questions about the very definition of clock genes (85). Furthermore, in some organisms, the dominant mechanism regulating circadian rhythms seems to be posttranscriptional, as evidenced in circadian control of translation of luciferin binding protein in the unicellular alga Gonyaulax polyedra (86, 87).
The situation is somewhat trickier to dissect in mammals because circadian genes often have multiple homologs. Therefore, double-mutant animals are generally needed to observe a behavioral phenotype. Bmal1 was thought to be the only exception to this, with its suppression leading to clear behavioral arrhythmicity (88). Constitutive brain-specific expression of Bmal1 in knock-out animals, however, can restore behavioral rhythmicity, casting doubt on the necessity of rhythmic Bmal1 transcription (89). In addition, brain-specific knockout of Bmal1 expression produces gross pathology, with a striking abundance of activated microglia in the brains of mice, which gets progressively worse over the first 6 months of life. This makes it extremely difficult to dissociate the effects of these pathological findings with those specifically related to the malfunction of a biological clock (90).
Even more importantly, imaging of SCN slices from arrhythmic Bmal1−/− and Cry1−/−Cry2−/− animals with bioluminescence reporters has revealed the persistence of low-amplitude rhythms in individual neurons (91, 92). Critically, developmental effects may well belie the apparent arrhythmicity that is observed when adult animals are assayed, as is the case in most experimental paradigms (93). Interestingly, the period of such rhythms tends to be shorter, ~20 h instead of 24 h. Such phenomena may stem from a cytoplasmic oscillator, because cAMP signaling is thought to be linked to such “clockless” oscillations (91). The methamphetamine-sensitive circadian oscillator is another 24-h oscillator that appears to function independently of the known circadian clockwork. When methamphetamine is supplied to rodents in drinking water, it has the peculiar property of restoring behavioral rhythms in arrhythmic animals caused by either lesion of the SCN (94, 95) or genetic defects in clock genes (96). Together, such overwhelming evidence indicates that current TTFLs cannot possibly account for the multiple lines of experimental evidence that have revealed circadian oscillations in the presence of inactivated feedback loops, or, indeed, in their absence, as discussed further below.
NONTRANSCRIPTIONAL CLOCK MECHANISMS
Early Evidence and Models
The numerous experimental anomalies surrounding the current transcriptional/translational model of the circadian clock suggest that other mechanisms are required to explain fully the molecular basis of circadian timekeeping. In this respect, it is worth highlighting that transcriptional mechanisms were regarded as only one of several possibilities investigated before the discovery of clock genes (13). Of note, models based on feedback control in biochemical networks were examined in great detail as a mechanistic basis for the generation of self-sustained oscillations. These models successfully explained ultradian (that is, with a period less than 24 h) oscillators such as the glycolytic oscillator in yeast (97, 98) and cAMP oscillations in slime molds (99,100). In addition, mitochondrial calcium cycles were proposed to explain the circadian oscillation of NAD+ and NADP+ in Euglena through the opposing effects of the Ca2+-activated calmodulin complex on NAD+ kinase and NADP+ phosphatase (101). Indeed, rhythms of Ca2+ in another plastid, the chloroplast, and in the cytoplasm of plant cells have been observed, suggesting that rhythmic ion fluctuations may be a common feature in organisms in the green lineage (102).
Another class of models postulated that membranes and cellular compartments could be involved in the generation of circadian rhythmicity, given that disruption of ion transport and other membrane properties perturbs overt rhythms (103, 104). Such models proposed that ion gradients influence active ion transport systems in an autoregulatory fashion that provides the nonlinearity required for self-sustained oscillations. The supposedly slow passive diffusion across membrane was thought to be responsible for the relatively long circadian period. Later models incorporated protein synthesis as a component on which ion concentration may impinge to stimulate active ion transport (105, 106).
The enthusiasm for membrane and biochemical models in the 1970s was stimulated by experimental evidence supporting a limited contribution by the cell’s nucleus to normal circadian rhythms. The macroscopic unicellular alga Acetabularia can, for example, maintain self-sustained circadian rhythmicity in photosynthetic activity, even when its nucleus is removed by cutting off its nucleus-containing rhizoid process (107). Intriguingly, its nucleus is able to dictate the phase of oscillation but is dispensable for entrainment and phase shifting (108). Moreover, inhibition of transcription with actinomycin D did not suppress rhythms in both nucleated and enucleated Acetabularia cells, although the former lost rhythmicity after two weeks under these conditions (109). In addition, studies in erythrocytes, which lack a nucleus and are therefore unable to perform transcription, revealed daily rhythms in the membrane-bound Mg2+-ATPase (110). Similarly, platelets were used to show that glutathione exhibited circadian oscillations relying on de novo synthesis of this tripeptide (111), again in the absence a nucleus, but in the presence of mitochondria that are normal inhabitants of platelets. The examples above point to the fact that current circadian models are not able to explain many early observations that, in some cases, were made more than 40 years ago and that had already suggested the existence of nontranscriptional rhythms.
To reconcile these seemingly paradoxical points of view---current transcriptional networks and nontranscriptional oscillators---one can envision the following two major possibilities. On the one hand, transcription and translation in the current models may have only a limited role in setting the pace of the oscillator and may be needed mainly for maintaining the levels of clock proteins and for controlling circadian output functions. In this view, posttranslational modifications of known clock proteins could be the fundamental oscillators, but the transcriptional oscillator would be important for robustness and may amplify posttranslational oscillations. A prototype for such a model exists in cyanobacteria, in which the master transcriptional regulator KaiC is also part of its posttranslational oscillator. On the other hand, circadian timekeeping may have evolved more than one clock in the cell to meet the requirements of precision, robustness, and stability. Accordingly, the known transcriptional oscillator may be coupled to an uncharacterized posttranslational oscillator. The recent discovery of circadian oxidation-reduction cycles in PRDX proteins is supportive of such a hypothesis (18, 19) and, moreover, immediately suggests a common phylogenetic origin for circadian timekeeping mechanisms in virtually all species relying on oxygen as the terminal electron acceptor for energy metabolism (20).
The KaiC Oscillator
Although this review focuses mainly on nontranscriptional mechanisms in eukaryotic species, it is worth briefly describing the purely posttranslational oscillator discovered in the cyanobacterium Synechococcus elongatus. The finding that phosphorylation of a substrate could exhibit 24-h rhythms on its own offered the first mechanistic basis for nontranscriptional rhythms. It also triggered an intense debate about the roles of transcriptional and nontranscriptional oscillators in both prokaryotic and eukaryotic species.
The core cyanobacterial oscillator is composed of three proteins (KaiA, KaiB, and KaiC), and negative-feedback regulation of the expression of the kaiBC promoter was initially proposed to be part of a TTFL model similar to those described in eukaryotes (5). However, the fact that the phosphorylation state of KaiC robustly oscillates in the cell with a 24-h period, even under conditions, where neither transcription nor translation was permitted, raised strong doubts about the transcription/translation hypothesis in cyanobacteria (112). Kondo and colleagues subsequently showed that KaiC circadian oscillations could be reconstituted in vitro using only the three purified Kai proteins supplemented with ATP as a source of energy, providing definitive evidence of a purely posttranslational oscillator (17).
KaiC is a phosphoprotein with both autokinase and autophosphatase activities at the serine 431 and threonine 432 residues. It forms an hexamer complex that is regulated by KaiA. The latter dimerizes to enhance the autophosphorylation of KaiC and drives it toward a hyperphosphorylated state, whereas KaiB associates with KaiC to inhibit KaiA’s activity. Detailed dissection of the phosphorylation cycle of KaiC has shown that four forms of KaiC phosphorylation states appear and disappear in an ordered pattern and that the relative abundance of each phosphorylated form determines the phase of the oscillator (113--115). Recent studies have also shed light on the entrainment properties of the cyanobacterial posttranslational oscillator by showing that it can directly sense intracellular metabolic states through the ADP:ATP ratio (116). Moreover, the Kai oscillator can also be entrained by light-driven redox changes, because the redox-sensitive protein LdpA modulates the levels of KaiA and CikA, a key input-pathway component (117). In addition, KaiA binding to a quinone reduces its own stability and diminishes KaiC autophosphorylation activity (118), providing another input pathway.
The relative contribution of transcriptional and posttranslational cycles is currently under intense investigation in the cyanobacterial circadian system. Emerging consensus supports a model in which both the posttranslational and transcriptional oscillators are needed for physiological timing, given that disruption of one or the other leads to defects in timekeeping in the whole organism (119--122). The question of whether the transcriptional components constitute a self-sustained or damped oscillator is still under debate (119, 120), but the extraordinary robustness of the cyanobacterial posttranslational oscillator, even in the absence of a cell, indicates that a biochemical oscillator is at the core of cellular timekeeping. In any case, these results imply that eukaryotic models are also likely to have similar posttranslational components that provide them with their reported resilience to transcriptional perturbations (80, 83, 84, 91, 92). An autonomous posttranslational mechanism may indeed exist in higher species, as an in silico model suggests that an oscillator consisting of a substrate reacting with two counteracting enzymes---for example, a phosphoprotein controlled by the activity of a kinase and a phosphatase---would be sufficient to recapitulate the fundamental properties of circadian oscillators (123).
Peroxiredoxin Rhythms
Posttranslational modifications are an integral feature of current TTFL models, but a definitive posttranslational oscillator has not yet been identified in eukaryotic species, likely owing to the difficulty of teasing apart the contribution of transcriptional and nontranscriptional mechanisms in their complex, multiorganellar cells. In this context, the recent discovery of oxidation cycles in PRDX proteins offers a new perspective on nontranscriptional rhythms in higher organisms (18--20, 124).
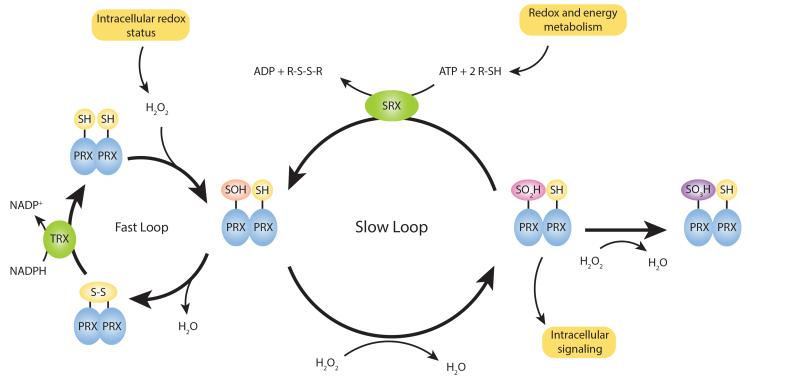
PRDXs are an antioxidant protein family involved in hydrogen peroxide metabolism and signaling (125). Their catalytic mechanism involves the oxidation of a catalytic cysteine residue in the enzymes’ active site to sulfenic acid (Cys-SOH), which then forms a disulfide bond with another noncatalytic (so-called resolving) cysteine residue. In most organisms, the thioredoxin system completes the cycle by reducing this disulfide bond at the expense of oxidizing a molecule of NADPH (Figure 3). This catalytic loop has rapid turnover and allows the maintenance of low levels of intracellular hydrogen peroxide. The so-called typical 2-Cys PRDXs, a subclass of PRDX whose basic functional unit is a homodimer in which catalytic and resolving cysteines belong to different PRDX molecules, can undergo further oxidation of their catalytic cysteine to sulfinic and sulfonic acid forms (Cys-SO2/3H). Over-oxidized Cys-SO2H residues can be slowly recycled through ATP-dependent reduction by sulfiredoxin (126), whereas further oxidation to Cys-SO3H (termed hyperoxidation) is thought to be irreversible. Hyperoxidation of catalytic cysteines is thought to be involved in peroxide signaling through disulfide bond exchange and the chaperone activity of hyperoxidized oligomers (125). Importantly, typical 2-Cys PRDXs have the property of shifting from a homodimer to a doughnut-shaped decameric structure depending on the redox status (127).
Figure 3.
The typical 2-Cys PRDX system. The catalytic mechanism of typical 2-Cys PRDXs is composed of two interlocked cycles. In the first cycle, PRDXs undergo peroxidation of their catalytic cysteine to sulfenic acid form (Cys-SOH), followed by the disulfide bond formation with the resolving cysteine (resolution) and the recycling step catalyzed by TRX. The sulfenic moiety of PRDXs (Cys-SOH) can be further oxidized to sulfinic and sulfonic acid forms (Cys-SO2/3H), thereby entering the second cycle in which overoxidized Cys-SO2H residues can be slowly recycled through ATP-dependent reduction by sulfiredoxin (126). Further oxidation to Cys-SO3H (termed hyperoxidation) is thought to be irreversible. In vitro measurement of the turnover rate of each cycle shows that the peroxidatic cycle is fast (185), whereas the overperoxidatic cycle is much slower (158). Abbreviations: PRDX, peroxiredoxin; SH, sulfhydryl group; SOH, sulfenic group; SO2H, sulfinic group; SO3H, sulfonic group; SRX, sulfiredoxin system; S-S, disulfide bond; TRX, thioredoxin system.
In human red blood cells, where transcription is naturally absent, PRDXs exhibit circadian accumulation of their dimeric overoxidized form (Cys-SO2/3H) over several days (18). Such rhythms fulfill the criteria of bona fide circadian rhythms: persistence in constant conditions, the ability to entrain (via temperature cycles in this case), and temperature compensation (the clock does not run faster in higher temperatures and vice versa). Moreover, these redox rhythms are accompanied by oscillations in hemoglobin oxidation and metabolic variables including NAD(P)H and ATP levels. Similar rhythms are present in the unicellular alga Ostreococcus tauri, even when transcription is inhibited by prolonged darkness (19), lending strong support to nontranscriptional circadian processes in these diverse organisms separated by 1 billion years of evolution. Strikingly, the deep phylogenetic conservation of PRDX redox rhythms extends to include fungal, plant, bacterial, and even archaeal species (20). Critically, such rhythms are not dependent on previously identified clock genes, because mutants lacking circadian components maintain redox oscillations, albeit slightly phase shifted.
The phylogenetic conservation of PRDX rhythms suggests that primordial redox oscillators probably evolved following the Great Oxidation Event 2.5 billion years ago, when photosynthetic bacteria acquired the ability to evolve oxygen from water and caused a dramatic increase in atmospheric oxygen. Rhythmic production of oxygen and reactive oxygen species (ROS) by sunlight may have been a key driving force in the coevolution of clock mechanisms and ROS removal systems that could anticipate and resonate with externally driven redox cycles (20).
Reactive oxygen species (ROS): oxygen-containing molecules with oxidant activity, whose accumulation can lead to harmful oxidative stress in cells
REDOX AND METABOLIC CLOCKS IN EUKARYOTES
Linking Metabolic and Redox Rhythms to Transcriptional Clocks
The interplay between circadian and metabolic cycles is a recurring theme in chronobiology, and the reciprocal effects that disruption of one cycle has on the other is clear both at physiological and molecular levels (71, 128). For example, a high-fat diet lengthens the behavioral period of rhythms in mice and changes the expression pattern of clock genes (129). Conversely, a recent long-term laboratory study conducted with healthy patients showed that three weeks of circadian dyssynchrony is sufficient to cause prediabetic symptoms (130). Growing evidence suggesting that circadian rhythms are fundamentally metabolic requires that current transcriptional oscillations be tightly coupled to metabolic cycles, in particular redox oscillations. This hypothesis is strongly supported by numerous examples of accessory loops embedding the circadian transcriptional clock within cellular metabolism (Figure 2).
The NAD+/NADH redox pair is likely to play an important role in connecting cytosolic and compartment-specific redox states to transcriptional clock components such as PER2 (44) and CLOCK/BMAL1 (43, 74, 75). In addition, other redox-sensitive mechanisms have been identified in the clockwork, in particular, the heme-sensing transcriptional regulators. For example, heme controls the DNA-binding activity of BMAL1/NPAS2 in vitro by inhibiting DNA binding in response to carbon monoxide (131). The nuclear receptors REV-ERBα and -β are heme-binding transcriptional repressors (132, 133) that, at least in the case of REV-ERBβ, employ reactive cysteine residues in their ligand-binding domain (133). Similarly, PER2 interaction with CRY proteins is inhibited by ferric heme (134).
Regulation of protein function by small metabolites covalently modifying their amino acid residues may represent another class of coupling mechanisms, as the availability of reactive metabolites could feedback on circadian and metabolic proteins via associated posttranslational modifications. In a similar fashion, the metabolic status of the cell is thought to regulate gene expression through the epigenetic modification of histones (135). In particular, nonenzymatic acetylation of lysine residues has been described for the latter (136), suggesting that a local increase in the concentration of acetyl-CoA could favor the acetylation of surrounding proteins (137). Importantly, acetylation has been associated with the regulation of many metabolic enzymes, especially in mitochondria, (138, 139). In this way, metabolic flux producing acetyl-CoA may be controlled by a feedback mechanism using acetylation of metabolic proteins, thereby leading to metabolic oscillations. Such an acetylation system would be readily coupled to the clockwork because acetylation regulates PER2 stability (44) and CLOCK/BMAL1 transactivation, counteracted by the deacetylase activity of SIRT1 (43).
In a similar fashion, intracellular nutrient levels can be sensed through O-linked N-acetylglucosamine (O-GlcNAc) glycosylation, which uses uridine diphosphate (UDP)-GlcNAc as a donor, the end product of the hexosamine biosynthesis pathway. The latter integrates input from glucose, amino acid, and fatty acid metabolism pathways; therefore, UDP-GlcNAc concentration is indicative of general nutrient availability. Such nutrient-responsive modification has been implicated in regulating a large range of cellular processes including signaling, transcription, and the stress response (140). A number of core oscillator proteins are glycosylated, both in flies and in mammals, underlining the importance of O-GlcNAc glycosylation to the clockwork (48--51). For instance, both BMAL1 and CLOCK are rhythmically modified by O-GlcNAc, and this moiety stabilizes CLOCK/BMAL1 by inhibiting their ubiquitylation (51). Moreover, disruption of O-linked GlcNAc transferase perturbs glucose homeostasis in mice, probably via improper control of gluconeogenic genes by the liver clock. Thus, these results emphasize that metabolite and nutrient levels can be sensed and retroactively act on the clockwork and metabolic enzymes, exploiting metabolites as donors for posttranslational modifications.
Circadian Redox Rhythms
Even in early molecular studies of the circadian clock, before clock genes had been discovered in any model organism, rhythms in redox had been reported. In plants, the NADP+-to-NADPH ratio exhibits circadian cycles in seedlings kept in constant darkness (141). In rodents, several studies showed that key redox parameters including pyridine and glutathione redox ratios were diurnally regulated in the liver, with the proviso that these oscillations may have been partially driven by food intake (142--146). Nevertheless, human platelets kept in vitro showed circadian rhythms in glutathione content (111), suggesting that the aforementioned feeding cycles may resonate with these cell-autonomous biochemical rhythms. This may also have effects on platelet function, which has been reported recently in humans (147).
The hypothesis that metabolic cycles may be a fundamental mechanism underlying biological clocks has been proposed on the basis of both theoretical and experimental observations (148). Potential evidence for this in mammals came from McKnight and colleagues when they discovered that BMAL1/CLOCK DNA-binding activity can be modulated in vitro by the redox poise of NAD(P)+/NAD(P)H coenzymes (74, 149). Moreover, the concerted action of BMAL1/CLOCK on the expression of genes encoding NAD+-producing enzymes including lactate dehydrogenase and NAMPT could potentially feed back onto intracellular redox balance (72--74). These results, however, still lack in vivo confirmation, given the relatively high concentration (millimolar range) of the coenzymes used in previous in vitro assays. Nevertheless, the recent discovery of PRDX oscillations in nontranscriptional systems offers supportive evidence that redox cycles can function as circadian oscillators in their own right.
This line of thinking is substantiated by several recent studies illustrating the connection of redox balance, as well as the regulation of intracellular peroxide levels, with cellular timekeeping. For instance, hydrogen peroxide is able to induce circadian gene expression in zebrafish cells through the mitogen-activated protein kinase pathway (150). In Drosophila, both glutathione biogenesis and the response of flies subjected to hydrogen peroxide are regulated over the circadian cycle (151, 152). In mice, redox cycles in the SCN probably play a role in the daily variation of neuronal firing (153). Such rhythms may be driven by BMAL1, given that slices from Bmal1-deficient animals, which exhibit low-amplitude circadian oscillations (91), fail to exhibit redox oscillations.
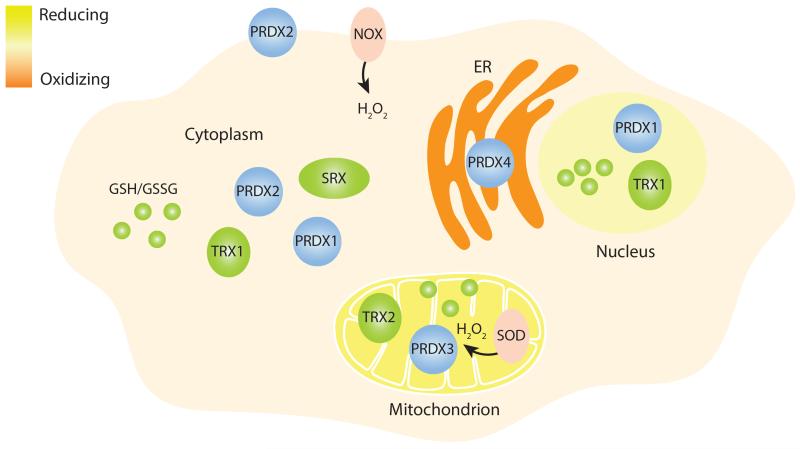
Redox systems clearly affect downstream cyclical physiology as well as influence circadian molecular oscillations. In mammals, for example, the mitochondrial isoform of the PRDX family, PRDX3, has a physiological role in the generation of rhythms of corticosterone production in the mouse adrenal cortex, as its circadian inactivation is required for normal hormone secretion (154). Surprisingly, under normal laboratory conditions, the rhythmic transition of PRDX3 to its over/hyperoxidized state was apparent only in the adrenal cortex and not in other tissues, suggesting that other compartment-specific PRDX isoforms likely participate in PRDX oxidation cycles observed in mouse liver and the SCN (20) (Figure 4). In the fungus N. crassa, ROS homeostasis also seems to be required for normal output rhythms, as the use of mutants for superoxide dismutase or NADPH oxidase to manipulate genetically ROS levels results in enhanced oscillations or arrhythmicity, respectively (155). In addition, superoxide dismutase suppression leads to slightly shorter conidiation (spore formation) rhythms (156), whereas its overexpression has the opposite effect (157). Even if the in vitro DNA-binding activity of the fungal transcriptional activator complex is sensitive to H2O2 levels, and may be involved in transducing peroxide levels (155), it is equally likely that intracellular H2O2 levels are a key variable of a putative redox oscillator, whose dynamic properties would also be affected by the manipulation of peroxide levels.
Figure 4.
PRDXs and redox compartmentalization. Given the compartmentalization of redox balances in eukaryotic cells (186), a putative redox oscillator may be localized in specific cellular organelles. Accordingly, the various isoforms of mammalian typical 2-Cys PRDXs have different subcellular localizations, as shown in this schematic representation of a cell with its nucleus, ER, mitochondrion, cytoplasm, and membrane. The SRX is mainly localized in the cytoplasm (158), whereas the TRX has two isoforms found mainly in the cytoplasm and nucleus (TRX1) and in the mitochondrion (TRX2). Compartment-specific pools of reduced and oxidized glutathione are also represented (GSH/GSSG). Two notable examples of ROS-producing enzymes, NOX and SOD, are shown. Each organelle is represented with a color corresponding to its relative reductive or oxidative environment as shown by the scale (top left). Abbreviations: ER, endoplasmic reticulum; NOX, NADPH oxidase; PRDXs, peroxiredoxins; ROS, reactive oxygen species; SOD, superoxide dismutase; SRX, sulfiredoxin system; TRX, thioredoxin system.
Peroxiredoxins: A Biochemical Clock or a Conserved Biomarker?
Although evidence supporting a metabolic oscillator is increasingly convincing, an underlying mechanism has yet to be described. The PRDX system may well be part of this oscillator, given the striking phylogenetic conservation of its circadian oxidation rhythm, extending to all domains of life. In such a model, sulfiredoxin, which recycles the sulfinic form (Cys-SO2H) of PRDX to its sulfenic form (Cys-SOH), is likely to play an important role (Figures 3 and 4). This ATP-dependent enzyme has the peculiar property of being poorly efficient, with a turnover rate in the range of only 6 to 11 molecules per hour (158). Interestingly, the converse and spontaneous reaction, PRDX oxidation to its sulfinic form, also proceeds relatively slowly: Only approximately 1 in every 1,000 PRDX1 molecules per turnover undergoes inactivation when H2O2 is maintained at low steady-state levels (159). By comparison, reaction rates of phosphorylation in the KaiC oscillator are even slower, in the range of 0.1 to 0.5 per hour (113). Therefore, PRDX hyperoxidation kinetics may be compatible with the generation of redox cycles but may require interactions with other redox or metabolic systems to achieve 24-h periodicity. However, it is not clear which multimeric form is measured when performing in vitro activity assays using purified sulfinic acid PRDX (158). Therefore, the transition between dimeric and decameric conformations of 2-Cys PRDXs (and, indeed, any intermediate forms) may have an important effect on kinetic parameters. In this respect, there may be parallels with the KaiC molecule, because it also forms multimers (hexamers) during its catalytic cycling, which has important biophysical effects that contribute to its oscillatory properties (160).
An alternative hypothesis is that PRDX are a conserved biomarker of underlying biochemical oscillations. Accordingly, deletion of PRDX in cyanobacteria and plants does not abolish rhythmicity (20). However, a clear argument for the maintenance of rhythms is that a putative PRDX oscillator and existing (known) clockwork could be two complementary timekeeping mechanisms that can exist independently of each other. The hypothesis of independent metabolic and transcriptional oscillators is supported by recent experiments showing that circadian glucose uptake in undifferentiated stem cells precedes the rhythmic expression of clock genes appearing in differentiated cells (161). In this context, note that the unicellular alga Gonyaulax polyedra can exhibit independent rhythms of aggregation and bioluminescence and that the phasing of two putative oscillators can be teased apart under special conditions (79). This also seems to be the case in other single-celled species, including Neurospora (162), but the molecular basis of these oscillators remains obscure.
In organisms in which metabolic oscillations have been found, but conventional TTFLs have not been discovered (such as in the worm Caenorhabditis elegans), insights into such oscillatory mechanisms may be more forthcoming. However, a search for a role for a period homolog in this species was not fruitful, and, ultimately, the lin-42 gene was found to be a developmentally regulated gene, but not a clock gene (163). Thus, it is eminently plausible that more fundamental metabolic oscillations could drive PRDX oscillations in the absence of known transcriptional feedback oscillators (124). So-called accessory loops including NAD+/NADH and cAMP cycles are potential candidates for self-sustained metabolic oscillators, but further studies of their oscillatory properties in clock mutant backgrounds will be of great interest to researchers attempting to identify bona fide components of metabolic oscillators.
Metabolic and Redox Oscillators as Prototypes of a Circadian Metabolic Clock
Since the 1960s, biochemical clocks have been investigated as potential underlying mechanisms of behavioral rhythms. The well-known example of the glycolytic oscillator, first described in yeast (164, 165), has been extensively studied, and detailed mathematical models have been successfully built to recapitulate its dynamic properties (166). Notably, such ultradian glycolytic oscillations are not restricted to yeast, because many mammalian cells including muscle (167), pancreatic β (168), and heart cells (169) can generate such short-period oscillations. These models were considered prototypes for the circadian clock, and several theoretical studies proposed mechanisms to generate 24-h rhythms from these short-period oscillations, including frequency demultiplication (170), interaction with a large storage pool (171), and coupling between a collection of oscillators (172, 173; for reviews, see References 13,166).
Glycolytic oscillations, however, lack the feature of temperature compensation, which is normally expected of circadian clocks. On the contrary, the yeast metabolic cycle, which generates a metabolic oscillation with periods ranging from dozens of minutes to a few hours, depending on the growth conditions, exhibits this peculiar property of circadian systems (174, 175). Under nutrient-limiting conditions, the budding yeast Saccharomyces cerevisiae exhibits ultradian respiratory rhythms that drive gene expression of approximately half of the yeast transcriptome (174). These metabolic rhythms are characterized by a striking temporal organization that separates reductive and oxidative processes in successive phases. Such a metabolic program is visible at both the transcriptome and metabolome levels (174, 176, 177), with a long reducing phase, where nonrespiratory modes predominate, followed by a short period of intense respiration in the oxidative phase.
Similar metabolic rhythms have recently been described in the nitrogen-fixing cyanobacteria Cyanothece (178). However, in this system, the period is circadian under nutrient-limiting conditions (low light and low CO2), whereas ultradian oscillations prevail under saturating conditions. Importantly, the short-period rhythms were not temperature compensated, possibly suggesting different underlying mechanisms.
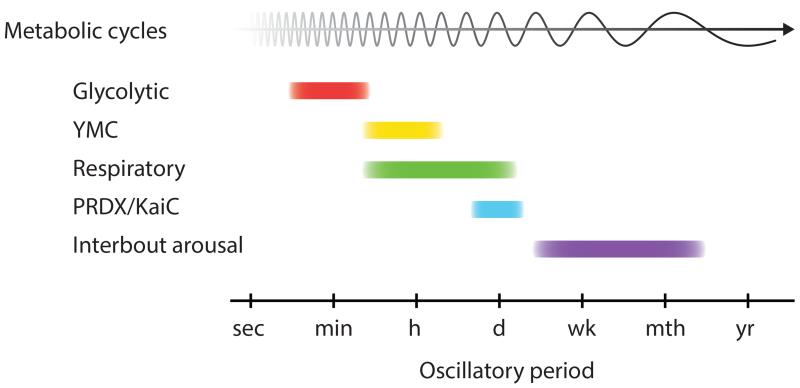
A final interesting example is the phenomenon of periodic increases in body temperature during hibernation. Animals oscillate between periods of torpor, when body temperature is low and metabolism is depressed, and interbout arousal, when internal temperature is maintained and metabolic processes are more active. Such a phenomenon may result from long-period (from days to weeks) metabolic cycles similar to those described in yeast and cyanobacteria (179, 180). Accordingly, it has been proposed that interbout arousals could consist of a respiration burst following a long reductive phase (torpor). The different examples of metabolic oscillators described in this section span period lengths from minutes to weeks and suggest that circadian metabolic oscillators may be only one example among many others of oscillators that are specifically adapted to the 24-h day (Figure 5).
Figure 5.
Metabolic oscillators over timescales ranging from minutes to months. Metabolic oscillators spanning the oscillatory period range from minutes to months have been described in various species. Glycolytic oscillations have been reported in yeast (164) as well as mammalian cells (166); they have a short period in the order of minutes. The YMC is a metabolic oscillator with a period ranging from dozens of minutes to a few hours that is composed of temporally separated reductive and oxidative phases characterized, respectively, by low and high oxygen consumption (174, 175). Similar respiratory cycles have recently been reported in cyanobacteria with a period from 10 h to 24 h, depending on the conditions (178). KaiC phosphorylation and peroxiredoxin oxidation cycles are two of the most preeminent examples of nontranscriptional rhythms in the circadian range. Periodic increase in body temperature during hibernation (interbout arousal) is an example of metabolic oscillation with a period longer than 24 h (from few days to several weeks) (179, 180). Abbreviations: PRDX, peroxiredoxin; YMC, yeast metabolic cycle.
Evolution of Metabolic Clocks
The results and ideas presented here propose that the requirements of robust timekeeping selected a mechanism based on several clocks. It is likely that the identified TTFLs are mainly dedicated to controlling the output of the clock, that is, coordinating behavior and physiology around the circadian day. By contrast, actual timekeeping would come from an underlying biochemical oscillator.
The fact that PRDX oxidation cycles are conserved among all domains of life, whereas TTFL models are generally restricted to a given phylum, supports a model wherein rhythmic transcriptional processes have evolved in addition to an underlying metabolic oscillator to fulfill the particular needs of different species in their particular environmental niche. A long-standing hypothesis about the origin of circadian clocks is the “escape from light” hypothesis, which proposes that primordial organisms have evolved timekeeping mechanisms to segregate UV-sensitive processes, such as DNA replication, into the dark period of the day (181). However, circadian gating of cell division during the day in modern cyanobacteria contradicts such a hypothesis (182) and suggests that other selective agents may have predominated. One such agent could be another source of cellular stress, such as ROS-induced damage, as intimated by the role PRDXs play in maintaining levels of ROS homeostatically. The existence of metabolic rhythms ranging from minutes to weeks would indicate that temporal organization may be the fundamental feature of such biochemical oscillations rather than a by-product of diverse transcriptional oscillators. In this view, circadian metabolic oscillations may have principally evolved to anticipate and separate processes needed during the energy-harvesting (day) and utilization (night) phases (183), thereby alleviating energy-consuming futile cycles in metabolic pathways (9, 180). The soundness of such a hypothesis seems clear for unicellular photosynthetic organisms growing in conditions of limited resources, which would have crucially needed to manage their energetic reserves optimally. However, for multicellular mammals, the complex organization of physiology, which guarantees an almost-constant supply of energy to nonmetabolic tissues, makes the interpretation of such a hypothesis more challenging.
CONCLUSION
Perhaps one of the most exciting features of circadian clocks is the possibility of linking overt behavioral and physiological rhythms in health and disease states to specific molecular processes and their perturbations. Current TTFL networks, together with their cytosolic accessory loops, offer detailed models that explain how pervasive orchestration of cellular physiology is achieved. All cytosolic loops identified so far are involved in energy and redox homeostasis, suggesting an important role for metabolic cycles. The recent discovery of metabolic nontranscriptional cycles further extends this line of thinking and supports the hypothesis of a circadian metabolic oscillator underlying transcriptional and cytosolic rhythms (Figure 2). The PRDX system may be part of this uncharacterized metabolic oscillator, given its broad phylogenetic conservation and its slow kinetics, which could be compatible with 24-h rhythmicity. It may, as well, be a reporter of more fundamental oscillations. Nevertheless, redox cycles seem to be an important component of circadian timekeeping.
The various examples of metabolic clocks spanning the oscillatory period range from minutes to months offer supportive evidence that temporal separation of reductive and oxidative phases may be a key principle of a circadian metabolic oscillator (Figure 5). Given the ubiquitous influence of redox poise on metabolic reactions, dissection of precise molecular components may be a challenging problem, as already experienced by those investigating the yeast metabolic oscillator.
Taken together, a wealth of experimental evidence points toward the view that living organisms probably evolved multiple coupled oscillators to fulfill the many requirements of a clock, including mechanisms to “keep” time and to “tell” time. The deep phylogenetic conservation of the circadian redox oscillations suggests that metabolic oscillators may be more dedicated to keep track of time and organize underlying biochemical networks, whereas the transcription-based oscillators may have evolved in addition to this biochemical clock, with a predominant role in dictating temporal programs specific to the physiology of each species. Therefore, given that time-telling mechanisms may have been primarily uncovered in eukaryotes, a fundamental understanding of a circadian timekeeping mechanism is within our grasp.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Siffre M. Hors du temps. Julliard; Paris: 1963. [Google Scholar]
- 2.Aschoff J. Circadian rhythms in man. Science. 1965;148:1427–32. doi: 10.1126/science.148.3676.1427. [DOI] [PubMed] [Google Scholar]
- 3.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 2006;40:409–48. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 4.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010;72:517–49. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 5.Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annu. Rev. Biophys. 2011;40:143–67. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. USA. 1998;95:8660–64. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol. 2004;14:1481–86. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Dodd AN, Salathia N, Hall A, Kevei E, Tóth R, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–33. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 9.Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20:36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 2003;4:649–61. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 11.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 2002;12:540–50. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 12.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–15. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 13.Edmunds LNJ. Cellular and Molecular Bases of Biological Clocks. Springer; New York: 1988. [Google Scholar]
- 14.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 2007;8:139–48. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 15.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem. Sci. 2009;34:483–90. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J. Cell Sci. 2011;124:311–20. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–15. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 18.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–58. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–64. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1971;68:2112–16. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bargiello TA, Young MW. Molecular genetics of a biological clock in Drosophila. Proc. Natl. Acad. Sci. USA. 1984;81:2142–46. doi: 10.1073/pnas.81.7.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, et al. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38:701–10. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- 24.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–40. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 25.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci. 1990;10:2749–62. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–84. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 27.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 28.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 29.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93(5):805–14. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 30.Vitaterna M, King D, Chang A, Kornhauser J, Lowrey P, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–25. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeBruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–77. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 32.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–15. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 33.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA. 1998;95:5474–79. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 35.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–27. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, et al. REV-ERBα and REV-ERBβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–67. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koike N, Yoo S-H, Huang H-C, Kumar V, Lee C, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–54. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doherty CJ, Kay SA. Circadian control of global gene expression patterns. Annu. Rev. Genet. 2010;44:419–44. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, et al. Circadian orchestration of the hepatic proteome. Curr. Biol. 2006;16:1107–15. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 41.Ripperger JA, Merrow M. Perfect timing: epigenetic regulation of the circadian clock. FEBS Lett. 2011;585:1406–11. doi: 10.1016/j.febslet.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 42.Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr. Biol. 2008;18:R805–15. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 43.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 45.Etchegaray J-P, Yang X, DeBruyne JP, Peters AHFM, Weaver DR, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 2006;281:21209–15. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 46.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38:369–74. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 47.Cardone L, Hirayama J, Giordano F, Tamaru T, Palvimo JJ, Sassone-Corsi P. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309:1390–94. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 48.Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai J-Y, et al. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J. Biol. Chem. 2011;286:44606–19. doi: 10.1074/jbc.M111.278903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim EY, Jeong EH, Park S, Jeong H-J, Edery I, Cho JW. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012;26:490–502. doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaasik K, Kivimäe S, Allen JJ, Chalkley RJ, Huang Y, et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013;17:291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M-D, Ruan H-B, Hughes ME, Lee J-S, Singh JP, et al. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 2013;17:303–10. doi: 10.1016/j.cmet.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Godinho SIH, Maywood ES, Shaw L, Tucci V, Barnard AR, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 53.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–4. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 54.Siepka SM, Yoo S-H, Park J, Song W, Kumar V, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–23. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell. 2013;152:1106–18. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 56.Yoo S-H, Mohawk JA, Siepka SM, Shan Y, Huh SK, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, et al. β-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J. Biol. Rhythms. 2007;22:375–86. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 58.Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–78. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- 59.Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–12. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He Q, Cheng P, Yang Y, He Q, Yu H, Liu Y. FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J. 2003;22:4421–30. doi: 10.1093/emboj/cdg425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim WY, Fujiwara S, Suh S-S, Kim J, Kim Y, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–60. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 62.van Ooijen G, Dixon LE, Troein C, Millar AJ. Proteasome function is required for biological timing throughout the twenty-four hour cycle. Curr. Biol. 2011;21:869–75. doi: 10.1016/j.cub.2011.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deery MJ, Maywood ES, Chesham JE, Sládek M, Karp NA, et al. Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr. Biol. 2009;19:2031–36. doi: 10.1016/j.cub.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 64.Masri S, Patel VR, Eckel-Mahan KL, Peleg S, Forne I, et al. The circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc. Natl. Acad. Sci. USA. 2013;110:3339–44. doi: 10.1073/pnas.1217632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morf J, Rey G, Schneider K, Stratmann M, Fujita J, et al. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338:379–83. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- 66.McGlincy NJ, Valomon A, Chesham JE, Maywood ES, Hastings MH, Ule J. Regulation of alternative splicing by the circadian clock and food-related cues. Genome Biol. 2012;13:R54. doi: 10.1186/gb-2012-13-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26:2724–36. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le Martelot G, Canella D, Symul L, Migliavacca E, Gilardi F, et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10:e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valekunja UK, Edgar RS, Oklejewicz M, van der Horst GTJ, O’Neill JS, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc. Natl. Acad. Sci. USA. 2013;110:1554–59. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rey G, Reddy AB. Connecting cellular metabolism to circadian clocks. Trends Cell Biol. 2013;23:234–41. doi: 10.1016/j.tcb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 72.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–54. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–57. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–14. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 75.Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–53. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 76.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–53. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–40. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hirota T, Kon N, Itagaki T, Hoshina N, Okano T, Fukada Y. Transcriptional repressor TIEG1 regulates Bmal1 gene through GC box and controls circadian clockwork. Genes Cells. 2010;15:111–21. doi: 10.1111/j.1365-2443.2009.01371.x. [DOI] [PubMed] [Google Scholar]
- 79.Roenneberg T, Morse D. Two circadian oscillators in one cell. Nature. 1993;362:362–64. doi: 10.1038/362362a0. [DOI] [PubMed] [Google Scholar]
- 80.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 81.Kondo T, Mori T, Lebedeva NV, Aoki S, Ishiura M, Golden SS. Circadian rhythms in rapidly dividing cyanobacteria. Science. 1997;275:224–27. doi: 10.1126/science.275.5297.224. [DOI] [PubMed] [Google Scholar]
- 82.Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–74. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 83.Dibner C, Sage D, Unser M, Bauer C, d’Eysmond T, et al. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 2009;28:123–34. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–67. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 85.Lakin-Thomas PL. Transcriptional feedback oscillators: maybe, maybe not. J. Biol. Rhythms. 2006;21:83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- 86.Morse D, Milos PM, Roux E, Hastings JW. Circadian regulation of bioluminescence in Gonyaulax involves translational control. Proc. Natl. Acad. Sci. USA. 1989;86:172–76. doi: 10.1073/pnas.86.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mittag M, Lee DH, Hastings JW. Circadian expression of the luciferin-binding protein correlates with the binding of a protein to the 3′ untranslated region of its mRNA. Proc. Natl. Acad. Sci. USA. 1994;91:5257–61. doi: 10.1073/pnas.91.12.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006;15:R271–77. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 89.McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–8. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J. Clin. Investig. 2013;123(12):5389–5400. doi: 10.1172/JCI70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, et al. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 2010;8:e1000513. doi: 10.1371/journal.pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maywood ES, Chesham JE, O’Brien JA, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc. Natl. Acad. Sci. USA. 2011;108:14306–11. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ono D, Honma S, Honma K-I. Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat. Commun. 2013;4:1666. doi: 10.1038/ncomms2670. [DOI] [PubMed] [Google Scholar]
- 94.Honma K, Honma S, Hiroshige T. Activity rhythms in the circadian domain appear in suprachiasmatic nuclei lesioned rats given methamphetamine. Physiol. Behav. 1987;40:767–74. doi: 10.1016/0031-9384(87)90281-2. [DOI] [PubMed] [Google Scholar]
- 95.Tataroğlu Ö , Davidson AJ, Benvenuto LJ, Menaker M. The methamphetamine-sensitive circadian oscillator (MASCO) in mice. J. Biol. Rhythms. 2006;21:185–94. doi: 10.1177/0748730406287529. [DOI] [PubMed] [Google Scholar]
- 96.Mohawk JA, Baer ML, Menaker M. The methamphetamine-sensitive circadian oscillator does not employ canonical clock genes. Proc. Natl. Acad. Sci. USA. 2009;106:3519–24. doi: 10.1073/pnas.0813366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sel’kov EE. Self-oscillations in glycolysis. 1. A simple kinetic model. Eur. J. Biochem. 1968;4:79–86. doi: 10.1111/j.1432-1033.1968.tb00175.x. [DOI] [PubMed] [Google Scholar]
- 98.Goldbeter A, Lefever R. Dissipative structures for an allosteric model. Application to glycolytic oscillations. Biophys. J. 1972;12:1302–15. doi: 10.1016/S0006-3495(72)86164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martiel JL, Goldbeter A. A model based on receptor desensitization for cyclic AMP signaling in dictyostelium cells. Biophys. J. 1987;52:807–28. doi: 10.1016/S0006-3495(87)83275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tyson JJ, Alexander KA, Manoranjan VS. Spiral waves of cyclic AMP in a model of slime mold aggregation. Physica D. 1989;34:193–207. [Google Scholar]
- 101.Goto K, Laval-Martin DL, Edmunds LN. Biochemical modeling of an autonomously oscillatory circadian clock in Euglena. Science. 1985;228:1284–88. doi: 10.1126/science.2988128. [DOI] [PubMed] [Google Scholar]
- 102.Johnson CH, Knight MR, Kondo T, Masson P, Sedbrook J, et al. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science. 1995;269:1863–65. doi: 10.1126/science.7569925. [DOI] [PubMed] [Google Scholar]
- 103.Sweeney BM. A physiological model for circadian rhythms derived from the acetabularia rhythm paradoxes. Int. J. Chronobiol. 1974;2:25–33. [PubMed] [Google Scholar]
- 104.Njus D, Sulzman FM, Hastings JW. Membrane model for the circadian clock. Nature. 1974;248:116–20. doi: 10.1038/248116a0. [DOI] [PubMed] [Google Scholar]
- 105.Schweiger HG, Schweiger M. Circadian rhythms in unicellular organisms: an endeavor to explain the molecular mechanism. Int. Rev. Cytol. 1977;51:315–42. doi: 10.1016/s0074-7696(08)60230-2. [DOI] [PubMed] [Google Scholar]
- 106.Burgoyne RD. A model for the molecular basis of circadian rhythm involving monovalent ion-mediated translational control. FEBS Lett. 1978;94:17–19. doi: 10.1016/0014-5793(78)80896-5. [DOI] [PubMed] [Google Scholar]
- 107.Sweeney BM, Haxo FT. Persistence of a photosynthetic rhythm in enucleated Acetabularia. Science. 1961;134:1361–63. doi: 10.1126/science.134.3487.1361. [DOI] [PubMed] [Google Scholar]
- 108.Schweiger E, Wallraff HG, Schweiger HG. Endogenous circadian rhythm in cytoplasm of Acetabularia: influence of the nucleus. Science. 1964;146:658–59. doi: 10.1126/science.146.3644.658. [DOI] [PubMed] [Google Scholar]
- 109.Mergenhagen D, Schweiger HG. The effect of different inhibitors of transcription and translation on the expression and control of circadian rhythm in individual cells of Acetabularia. Exp. Cell Res. 1975;94:321–26. doi: 10.1016/0014-4827(75)90499-1. [DOI] [PubMed] [Google Scholar]
- 110.Cornelius G, Rensing L. Daily rhythmic changes in Mg2+-dependent ATPase activity in human red blood cell membranes in vitro. Biochem. Biophys. Res. Commun. 1976;71:1269–72. doi: 10.1016/0006-291x(76)90791-9. [DOI] [PubMed] [Google Scholar]
- 111.Radha E, Hill TD, Rao GH, White JG. Glutathione levels in human platelets display a circadian rhythm in vitro. Thromb. Res. 1985;40:823–31. doi: 10.1016/0049-3848(85)90319-6. [DOI] [PubMed] [Google Scholar]
- 112.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–54. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 113.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–12. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–37. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim Y-I, Dong G, Carruthers CW, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. USA. 2008;105:12825–30. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331:220–23. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 2005;24:1202–10. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wood TL, Bridwell-Rabb J, Kim Y-I, Gao T, Chang Y-G, et al. The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc. Natl. Acad. Sci. USA. 2010;107:5804–9. doi: 10.1073/pnas.0910141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 2008;22:1513–21. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Qin X, Byrne M, Xu Y, Mori T, Johnson CH. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 2010;8:e1000394. doi: 10.1371/journal.pbio.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zwicker D, Lubensky DK, Wolde ten PR. Robust circadian clocks from coupled protein-modification and transcription-translation cycles. Proc. Natl. Acad. Sci. USA. 2010;107:22540–45. doi: 10.1073/pnas.1007613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Teng SW, Mukherji S, Moffitt JR, de Buyl S, O’Shea EK. Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. Science. 2013;340:737–40. doi: 10.1126/science.1230996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jolley CC, Ode KL, Ueda HR. A design principle for a posttranslational biochemical oscillator. Cell Rep. 2012;2:938–50. doi: 10.1016/j.celrep.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 124.Olmedo M, O’Neill JS, Edgar RS, Valekunja UK, Reddy AB, Merrow M. Circadian regulation of olfaction and an evolutionarily conserved, nontranscriptional marker in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2012;109:20479–84. doi: 10.1073/pnas.1211705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins: structures, mechanisms and functions. FEBS J. 2009;276:2469–77. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rhee SG, Jeong W, Chang T-S, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int. 2007;72:S3–S8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- 127.Poole LB. The catalytic mechanism of peroxiredoxins. Subcell. Biochem. 2007;44:61–81. doi: 10.1007/978-1-4020-6051-9_4. [DOI] [PubMed] [Google Scholar]
- 128.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–56. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 129.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–21. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 130.Buxton OM, Cain SW, O’Connor SP, Porter JH, Duffy JF, et al. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci. Transl. Med. 2012;4:129ra43. doi: 10.1126/scitranslmed.3003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A, McKnight SL. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–87. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 132.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, et al. REV-ERBα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–89. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 133.Gupta N, Ragsdale SW. Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor REV-ERBα. J. Biol. Chem. 2011;286:4392–403. doi: 10.1074/jbc.M110.193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang J, Kim KD, Lucas A, Drahos KE, Santos CS, et al. A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol. Cell. Biol. 2008;28:4697–711. doi: 10.1128/MCB.00236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 136.Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, et al. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 1999;274:18157–60. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 137.Cai L, Tu BP. On acetyl-CoA as a gauge of cellular metabolic state. Cold Spring Harb. Symp. Quant. Biol. 2011;76:195–202. doi: 10.1101/sqb.2011.76.010769. [DOI] [PubMed] [Google Scholar]
- 138.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 139.Zhao S, Xu W, Jiang W, Yu W, Lin Y, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011;80:825–58. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wagner E, Frosch S. Cycles in plants. J. Interdisipl. Cycle Res. 1974;5:231–39. [Google Scholar]
- 142.Isaacs J, Binkley F. Glutathione dependent control of protein disulfide-sulfhydryl content by subcellular fractions of hepatic tissue. Biochim. Biophys. Acta. 1977;497:192–204. doi: 10.1016/0304-4165(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 143.Isaacs JT, Binkley F. Cyclic AMP--dependent control of the rat hepatic glutathione disulfide-sulfhydryl ratio. Biochim. Biophys. Acta. 1977;498:29–38. doi: 10.1016/0304-4165(77)90084-8. [DOI] [PubMed] [Google Scholar]
- 144.Robinson JL, Foustock S, Chanez M, Bois-Joyeux B, Peret J. Circadian variation of liver metabolites and amino acids in rats adapted to a high protein, carbohydrate-free diet. J. Nutr. 1981;111:1711–20. doi: 10.1093/jn/111.10.1711. [DOI] [PubMed] [Google Scholar]
- 145.Kaminsky YG, Kosenko EA, Kondrashova MN. Analysis of the circadian rhythm in energy metabolism of rat liver. Int. J. Biochem. 1984;16:629–39. doi: 10.1016/0020-711x(84)90032-6. [DOI] [PubMed] [Google Scholar]
- 146.Bélanger PM, Desgagné M, Bruguerolle B. Temporal variations in microsomal lipid peroxidation and in glutathione concentration of rat liver. Drug Metab. Dispos. 1991;19:241–44. [PubMed] [Google Scholar]
- 147.Scheer FAJL, Michelson AD, Frelinger AL, Evoniuk H, Kelly EE, et al. The human endogenous circadian system causes greatest platelet activation during the biological morning independent of behaviors. PLoS ONE. 2011;6:e24549. doi: 10.1371/journal.pone.0024549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Roenneberg T, Merrow M. Circadian systems and metabolism. J. Biol. Rhythms. 1999;14:449–59. doi: 10.1177/074873099129001019. [DOI] [PubMed] [Google Scholar]
- 149.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu. Rev. Biochem. 2002;71:307–31. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 150.Hirayama J, Cho S, Sassone-Corsi P. Circadian control by the reduction/oxidation pathway: Catalase represses light-dependent clock gene expression in the zebrafish. Proc. Natl. Acad. Sci. USA. 2007;104:15747–52. doi: 10.1073/pnas.0705614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Krishnan N, Davis AJ, Giebultowicz JM. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2008;374:299–303. doi: 10.1016/j.bbrc.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Beaver LM, Klichko VI, Chow ES, Kotwica-Rolinska J, Williamson M, et al. Circadian regulation of glutathione levels and biosynthesis in Drosophila melanogaster. PLoS ONE. 2012;7:e50454. doi: 10.1371/journal.pone.0050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, et al. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337:839–42. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kil IS, Lee SK, Ryu KW, Woo HA, Hu M-C, et al. Feedback control of adrenal steroidogenesis via H2O2-dependent, reversible inactivation of peroxiredoxin III in mitochondria. Mol. Cell. 2012;46:584–94. doi: 10.1016/j.molcel.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 155.Yoshida Y, Iigusa H, Wang N, Hasunuma K. Cross-talk between the cellular redox state and the circadian system in Neurospora. PLoS ONE. 2011;6:e28227. doi: 10.1371/journal.pone.0028227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yoshida Y, Maeda T, Lee B, Hasunuma K. Conidiation rhythm and light entrainment in superoxide dismutase mutant in Neurospora crassa. Mol. Genet. Genomics. 2008;279:193–202. doi: 10.1007/s00438-007-0308-z. [DOI] [PubMed] [Google Scholar]
- 157.Gyöngyösi N, Nagy D, Makara K, Ella K, Káldi K. Reactive oxygen species can modulate circadian phase and period in Neurospora crassa. Free Radic. Biol. Med. 2013;58:134–43. doi: 10.1016/j.freeradbiomed.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 158.Chang T-S, Jeong W, Woo HA, Lee SM, Park S, Rhee SG. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem. 2004;279:50994–1001. doi: 10.1074/jbc.M409482200. [DOI] [PubMed] [Google Scholar]
- 159.Yang K-S, Kang SW, Woo HA, Hwang SC, Chae HZ, et al. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J. Biol. Chem. 2002;277:38029–36. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 160.Murayama Y, Mukaiyama A, Imai K, Onoue Y, Tsunoda A, et al. Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J. 2011;30:68–78. doi: 10.1038/emboj.2010.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Paulose JK, Rucker EB, Cassone VM. Toward the beginning of time: Circadian rhythms in metabolism precede rhythms in clock gene expression in mouse embryonic stem cells. PLoS ONE. 2012;7:e49555. doi: 10.1371/journal.pone.0049555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–56. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hasegawa K, Saigusa T, Tamai Y. Caenorhabditis elegans opens up new insights into circadian clock mechanisms. Chronobiol. Int. 2005;22:1–19. doi: 10.1081/cbi-200038149. [DOI] [PubMed] [Google Scholar]
- 164.Ghosh A, Chance B. Oscillations of glycolytic intermediates in yeast cells. Biochem. Biophys. Res. Commun. 1964;16:174–81. doi: 10.1016/0006-291x(64)90357-2. [DOI] [PubMed] [Google Scholar]
- 165.Chance B, Schoener B, Elsaesser S. Control of the waveform of oscillations of the reduced pyridine nucleotide level in a cell-free extract. Proc. Natl. Acad. Sci. USA. 1964;52:337–41. doi: 10.1073/pnas.52.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Goldbeter A. Biochemical Oscillations and Cellular Rhythms. Cambridge Univ. Press; Cambridge, UK: 1997. [Google Scholar]
- 167.Frenkel R. Control of reduced diphosphopyridine nucleotide oscillations in beef heart extracts. Arch. Biochem. Biophys. 1968;125:151–56. doi: 10.1016/0003-9861(68)90649-8. [DOI] [PubMed] [Google Scholar]
- 168.Chou HF, Berman N, Ipp E. Oscillations of lactate released from islets of Langerhans: evidence for oscillatory glycolysis in β-cells. Am. J. Physiol. Endocrinol. Metab. 1992;262:800–5. doi: 10.1152/ajpendo.1992.262.6.E800. [DOI] [PubMed] [Google Scholar]
- 169.O’Rourke B, Ramza BM, Marban E. Oscillations of membrane current and excitability driven by metabolic oscillations in heart cells. Science. 1994;265:962–66. doi: 10.1126/science.8052856. [DOI] [PubMed] [Google Scholar]
- 170.Goodwin BC. Temporal Organization in Cells: A Dynamic Theory of Cellular Control Process. Academic; New York: 1963. [Google Scholar]
- 171.Sel’kov EE. Stabilization of energy charge, generation of oscillations and multiple steady states in energy metabolism as a result of purely stoichiometric regulation. Eur. J. Biochem. 1975;59:151–57. doi: 10.1111/j.1432-1033.1975.tb02436.x. [DOI] [PubMed] [Google Scholar]
- 172.Winfree AT. Biological rhythms and the behavior of populations of coupled oscillators. J. Theor. Biol. 1967;16:15–42. doi: 10.1016/0022-5193(67)90051-3. [DOI] [PubMed] [Google Scholar]
- 173.Pavlidis T. Populations of interacting oscillators and circadian rhythms. J. Theor. Biol. 1969;22:418–36. doi: 10.1016/0022-5193(69)90014-9. [DOI] [PubMed] [Google Scholar]
- 174.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–58. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 175.Klevecz RR, Bolen J, Forrest G, Murray DB. A genome-wide oscillation in transcription gates DNA replication and cell cycle. Proc. Natl. Acad. Sci. USA. 2004;101:1200–5. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, et al. Cyclic changes in metabolic state during the life of a yeast cell. Proc. Natl. Acad. Sci. USA. 2007;104:16886–91. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Murray DB, Beckmann M, Kitano H. Regulation of yeast oscillatory dynamics. Proc. Natl. Acad. Sci. USA. 2007;104:2241–46. doi: 10.1073/pnas.0606677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Červený J, Sinetova MA, Valledor L, Sherman LA, Nedbal L. Ultradian metabolic rhythm in the diazotrophic cyanobacterium Cyanothece sp. ATCC 51142. Proc. Natl. Acad. Sci. USA. 2013;110:13210–15. doi: 10.1073/pnas.1301171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol. Rev. 2003;83:1153–81. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- 180.Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat. Rev. Mol. Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- 181.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 182.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc. Natl. Acad. Sci. USA. 1996;93:10183–88. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hut RA, Beersma DGM. Evolution of time-keeping mechanisms: early emergence and adaptation to photoperiod. Philos. Trans. R. Soc. B. 2011;366:2141–54. doi: 10.1098/rstb.2010.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Vitalini MW, de Paula RM, Park WD, Bell-Pedersen D. The rhythms of life: circadian output pathways in Neurospora. J. Biol. Rhythms. 2006;21:432–44. doi: 10.1177/0748730406294396. [DOI] [PubMed] [Google Scholar]
- 185.Peskin AV, Low FM, Paton LN, Maghzal GJ, Hampton MB, Winterbourn CC. The high reactivity of peroxiredoxin 2 with H2O2 is not reflected in its reaction with other oxidants and thiol reagents. J. Biol. Chem. 2007;282:11885–92. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 186.Go Y-M, Jones DP. Redox compartmentalization in eukaryotic cells. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]