Abstract
Background
Increased glutamine uptake is known to drive cancer cell proliferation, making tumor cells glutamine-dependent. Glutamine provides additional carbon and nitrogen sources for cell growth. The first step in glutamine utilization is its conversion to glutamate by glutaminase (GLS). Glutamate is a precursor for glutathione synthesis, and we investigated the hypothesis that glutamine drives glutathione synthesis and thereby contributes to cellular defense systems.
Methods
The importance of glutamine for glutathione synthesis was studied in H460 and A549 lung cancer cell lines using glutamine-free medium and Bis-2-(5-phenyl-acetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES) a GLS inhibitor. Metabolic activities were determined by targeted mass spectrometry.
Results
A significant correlation between glutamine consumption and glutathione excretion was demonstrated in H460 and A549 tumor cells. Culturing in the presence of [13C5]glutamine demonstrated that by 12 hrs >50% of excreted glutathione is derived from glutamine. Culturing in glutamine-free medium or treatment with BPTES, a glutaminase (GLS)-specific inhibitor, reduced cell proliferation and viability, and abolished glutathione excretion. Treatment with glutathione-ester prevented BPTES induced cytotoxicity. Inhibition of GLS markedly radiosensitized the lung tumor cell lines, suggesting an important role of glutamine-derived glutathione in determining radiation sensitivity.
Conclusions
We demonstrate here for the first time that a significant amount of extracellular glutathione is directly derived from glutamine. This finding adds yet another important function to the already known glutamine dependence of tumor cells and probably tumors as well.
General significance
Glutamine is essential for synthesis and excretion of glutathione to promote cell growth and viability.
Keywords: glutaminase, glutathione synthesis, glutamine utilization, H460, A549
Introduction
Lung cancer is the leading cause of cancer-related death in the US, with an expected 228,000 new cases and 160,000 deaths per year [34]. Despite immense research efforts, the overall 5-year survival rate of <17% remains poor compared to other cancers. In the past, treatment of advanced lung cancer followed a straightforward algorithm of platinum-based combination therapies or third-generation cytotoxic drugs, irrespective of histopathology subtypes [7,30,32]. More recently, treatment efficacies have improved due to pre-selection based on histopathology sub-types and identification of specific driver mutations [3]. Including a patient’s tumor biology in therapy selection (personalized medicine) is transforming the diagnosis and treatment of lung cancer [17,18]. We recently studied metabolite profiles in lymph node aspirates containing malignant lung tumor cell lines and found a strong correlation between glutamine consumption and glutathione excretion (Sappington et al. submitted Lung Cancer).
Mechanistic studies in tissue culture and animal models suggest glutamine utilization and glutathione synthesis are important in cancer promotion and progression [16,24,36,38,42]. Glutamine (Gln) is an essential nutrient and the most abundant free amino acid in human serum [4]. The first step in utilization of glutamine is its conversion to glutamate by glutaminase (GLS). Glutamate dehydrogenase converts glutamate to alpha-ketoglutarate an important tricarboxylic acid cycle (TCA) metabolite. The glutamine dependence of tumor cells has been known and GLS inhibitors are under development [37,38]. In parallel, tumor cells are known to increase glutathione synthesis, and intracellular glutathione concentrations in tumor cells are reported to be as high as 10 mM [1,10,22,28,40]. Glutathione, the most abundant thiol-compound, (i) functions as an antioxidant, (ii) is a precursor for conjugation reactions, (iii) participates in maintenance of the cysteine pool and (iv) contributes to the regulation of cellular processes, including apoptosis [20].
Glutamate is a precursor for glutathione synthesis, which is carried out by two conjugation reactions catalyzed by glutamate cysteine ligase and glutathione synthase [37]. Therefore, we hypothesize that glutamine-derived glutamate might also contribute to cellular defense by supplying necessary glutamate for glutathione synthesis. To test this hypothesis, glutamine consumption and glutathione synthesis and excretion patterns of H460 and A549 lung tumor cells were established. The importance of glutamine-derived glutamate for glutathione synthesis and cell proliferation is demonstrated by (i) culturing in glutamine-free media, (ii) utilization of stable isotope-labeled [13C5]-glutamine, (iii) inhibition of GLS by BPTES, a known GLS inhibitor [33] and (iv) the presence of glutathione-ester, the bioavailable form of glutathione, prevents BPTES-induced cytotoxicity. Lastly, sensitivity to radiation treatment demonstrated the role of glutamine-derived glutathione in radiotherapy resistance.
Materials and Methods
Cell culture
H460 and A549 lung tumor and MRC-5 alveolar fibroblast cell lines were purchased from ATCC and cultured in standard incubation conditions using 89% RPMI Medium 1640 + L-Glutamine (Sigma St. Louis, MO), 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin (complete medium) at a humidified 37°C with 5% CO2. Each cell line was propagated from an initial concentration of 100,000 cells per flask. Sub-cultures of each cell line were then seeded into 25-T flasks at 100,000 cells/flask. Cells were allowed to attach for 24 hours.
Glutamine-free medium experiments
Media were removed from all flasks (Time 0hrs) and 5 replicates from each line were given 5 mL complete medium and 5 replicates were given 5 mL glutamine-free complete medium (89% RPMI Medium 1640 without glutamine, 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin) (Sigma St. Louis, MO). Media aliquots of 100 μl were removed from each flask at 12 hour intervals to monitor the metabolomic footprint [12]. After 48 or 84 hours, media were removed, cells were washed with water (5 sec), and immediately flash frozen by the addition of 15 ml liquid nitrogen into the culture flasks and stored at −80°C as describe previously [19].
BPTES experiments
In the mechanistic experiments with BPTES, cells (7500) were seeded (Time -24hrs) into 96-well plates containing 100 μl complete medium, and allowed to attach for 24 hrs. Complete medium was added (Time 0hrs) containing various concentrations of BPTES to give the final concentrations of 1, 2, 5, 10, 20, 40, and 100 μM BPTES. Media aliquots of 25 μl were removed from each well at 24, 36, and 48 hrs to monitor the time course of metabolite consumption and production (metabolomic footprint). Cells were maintained at 37°C, 5% CO2 and 40% humidity until time of harvest.
[13C5]-glutamine experiments
To determine the fraction of glutathione that is derived directly from glutamine, A549 and H460 cell lines were cultured in 96-well plates in glutamine-free complete medium supplemented with 0.3 g/L [13C5]-glutamine. Cell media aliquots were harvested at 12, 24, 36, and 48 hrs. Light microscopy was used at 12 hours to visualize the cells at each dosage. After 48 hours the CellTiter 96, CellTiter-Glo® 2.0, and CellToxTM Green Cytotoxicity assays, (Promega, Madison, WI) were utilized to respectively assess cell proliferation, cell viability, and cell death of both the BPTES and [13C5]-glutamine experiments.
Glutathione-monoethyl ester (GSHE) experiments. Cells (7500) were seeded into 96 well plates containing 100 μl complete media. Cells were treated with 10 μM BPTES in DMSO and 0, 1, 5 and 10 mM GSHE. After 48 hrs cells were harvested and cell viability and toxicity were assessed as described above.
Radiation exposure of lung tumor cell lines
Radiation sensitivity was determined by methods described previously [39]. In brief, for colony formation A549 and H460 cells were seeded in 6-well plates containing 3 ml standard complete medium and allowed to attach for 24 h. Media were changed and cells were grown for another 24h in standard medium, glutamine-free medium, medium containing 2 μM BPTES (to deplete glutathione), and medium containing only the vehicle. Cells were subsequently radiated using a Faxitron X-ray Generating System (CP-160, Faxitron X-Ray Corp., Wheeling, IL). Single-doses of 4 or 8 Gy (Gray) were delivered at a dose rate of 1 Gy/min (150 kVp and 6.6 mA). Cells were then placed in standard complete RPMI medium and allowed to form colonies for 8 days. The surviving colonies (containing > ~50 cells) were stained and counted on a stereomicroscope. Plating efficiency (PE) of cells with each treatment were determined and normalized to that of control untreated cells and the surviving fractions were calculated by dividing the PE of the treated cells by the PE of the control untreated cells.
LC-MS quantitation by liquid chromatography tandem mass spectrometry (LC-MS/MS)
Metabolites were extracted from cell pellets by the addition of 3 ml of 50% methanol/0.2% formic acid to the frozen cells in the culture flasks. Cells were scraped from the culture flasks and transferred to 15 ml tubes. Proteins were precipitated from 750 μl cell pellet suspensions by addition of 1050 μl acetonitrile/0.2% formic acid. Metabolites were extracted from 25 or 100 μl culture media from 96 well plates or 25-T flasks, respectively, and with the respective addition of 275 μl or 1300 μl of 40% methanol/60% acetonitrile/0.2% formic acid. Cell pellets or media suspensions were incubated on ice for 30 min, followed by centrifugation for 10 min at 13000 g. Supernatants were transferred to new vials, solvents were removed in a speed vac and the concentrated metabolites stored at −80°C until analysis.
Samples were reconstituted in 0.2% formic acid and analyzed by LC-MS/MS (Agilent, 1290 Infinity LC coupled to an Agilent 6490 triple quadrupole mass analyzer). An Agilent Poroshell 2.7μm C18 (2.1mm × 100 mm) column was operated with a linear gradient from 2% Methanol/0.01% formic acid to 95% Methanol/0.01% formic acid in 10 min. Individual metabolites were monitored in multiple reactions monitoring mode, monitoring specific ion transitions shown in Supplemental Table 1 and Supplemental Figure 1 shows representative extracted ion chromatograms. The specific ion transitions and retention times for each metabolite were established experimentally using commercially available standard compounds. Quantitation of the individual metabolites were based on external calibration curves that were generated with each set of samples. In the stable isotope tracing experiment, 13C5-incorporation into subsequent metabolites was determined by monitoring the corresponding m/z ion transitions (Supplemental Table 1).
Statistical Analysis
Media and cell pellets were analyzed for differences between cell lines and glutamine treatment and the surviving fractions of cell colonies were compared among each cell line via Conover’s Kruskal-Wallis method [9]. The time-dependent relationships between glutamine consumption and glutathione production were analyzed by linear regression on each cell line. The resulting regression slopes (with their standard errors) were interpreted as “glutathione production ratios,” i.e., μM glutathione produced per mM glutamine consumed. Glutathione production ratios were compared for differences between cell lines via t-test using the combined standard errors of the ratios. All post-hoc comparisons employed a P<0.05 significance level despite the multiple comparisons, in order not to inflate Type II error in this study.
Results
The two lung carcinoma cell lines (H460 and A549) and one alveolar fibroblasts (MRC-5) were grown in complete or glutamine-free media (Figure 2). Their doubling times, calculated based on propagation over 48 hours in complete media, had means ± standard deviations (SDs) of 12.4 ± 0.02, 16.2 ± 0.08 and 29.0 ± 0.61 hours for H460, A549 and MRC-5, respectively. The doubling times for H460 and A549 are reported as 17.8 and 22.9 hours, suggesting that the cells grow slightly faster in the complete medium chosen for this study than in the recommended F12-K media [26]. In glutamine-free media, cell growth was reduced and the doubling times had means ± SDs of 15.1 ± 0.43, 24.3 ± 0.36 and 38.4 ± 0.84 hours (H460, A549 and MRC-5, respectively). Of these, A549 was the most glutamine dependent, with a 33% growth reduction in glutamine-free medium compared to 17% and 23% growth inhibition of H460 and MRC-5, respectively (Figure 2). The different cell lines showed distinct variability in size and morphology (data not shown).
Figure 2.
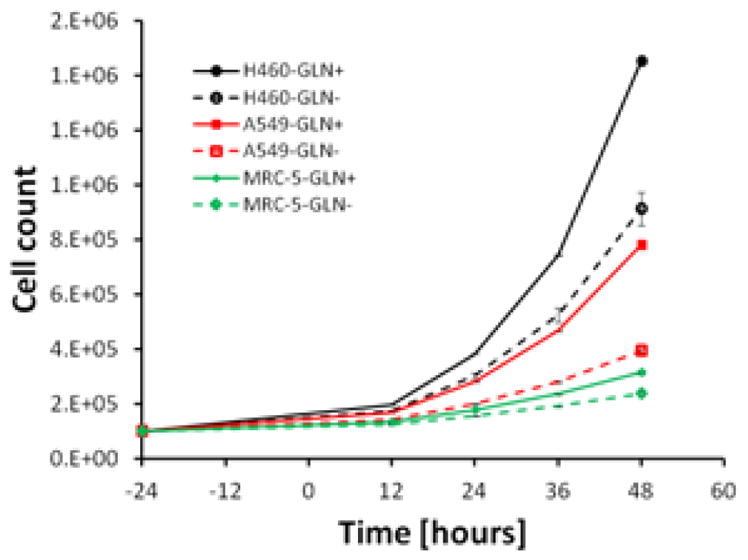
Growth curves of cell lines in complete and glutamine-free media. Cells were seeded in 96 well plates and incubate at standard condition see Material and Methods section. Some error bars are smaller than the symbol and therefore are not visible. Cells grown in glutamine-free media grew statically significant slower than cells grown in complete medium (P<0.05).
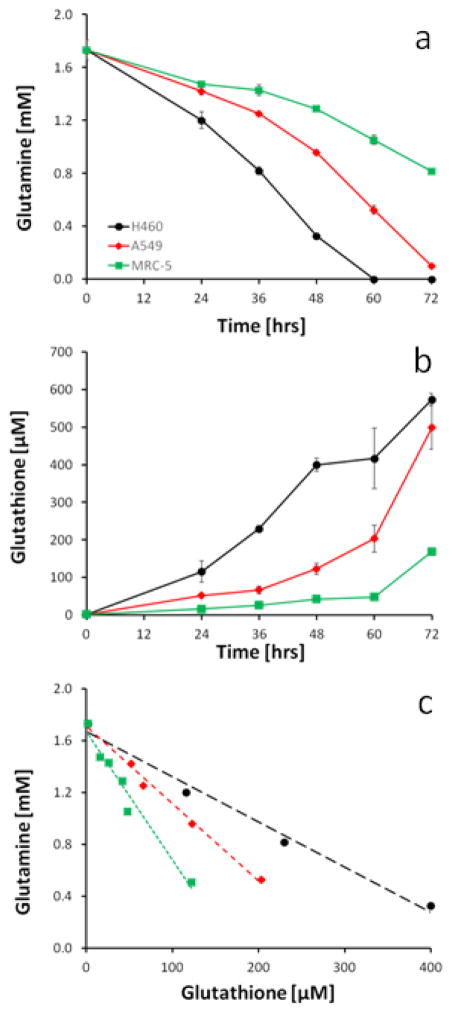
To study the potential mechanistic link between glutamine uptake and glutathione synthesis, the time courses for glutamine consumption and glutathione excretion were established in H460, A549 and MRC-5 cell lines (Figure 3a and 2b). The fastest growing H460 cell line was the most efficient in glutamine uptake, while the slower growing A549 and MRC-5 lines used much less glutamine during the same time period. For glutamine uptake all pairwise comparisons were statistically significant except between A549 and MRC-5 at 24 hrs. Likewise, the H460 cell line was the most efficient in glutathione synthesis, and MRC-5 the least efficient (Figure 3b). All pairwise comparisons at each time point for glutathione were statistically significant. There was a statistically significant correlation between glutamine consumption and glutathione excretion (Figure 3c). Regression analysis revealed glutathione production ratios ± standard errors (in μM/mM of glutamine) of 278 ± 19.9 for H460, 166 ± 12.0 for A549, and 95 ± 6.7 for MRC-5, all of which were statistically significantly different from each other.
Figure 3.
Time course of extracellular (a) glutamine and (b) glutathione in lung tumor cell lines. (c) Correlation of glutamine consumption to glutathione production in H460, A549 and MRC-5 cells, respectively. At the indicated time points, 100 μl of medium from cultured cells was analyzed. Some error bars are smaller than the symbol and therefore are not visible. Metabolite concentrations (a and b) were statistically different between cell lines at each time point except for glutamine between MRC-5 and A549 at 24 hrs (p<0.05). Regression analysis of glutamine consumption and glutathione synthesis (c) and excretion showed strong statistical correlation with R2 > 0.95 and paired-slope comparisons were significantly different with p <0.0001.
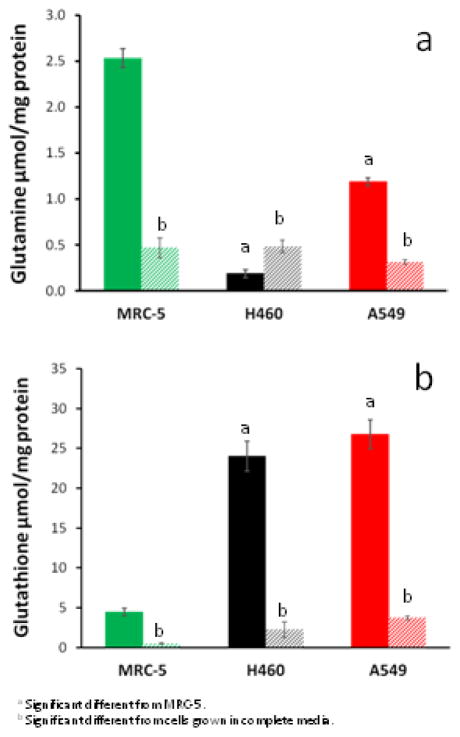
From the time course experiment, 48 hrs was chosen for subsequent studies, because glutamine is essentially depleted at 60 hrs in the fast growing H460 cell line. The excretion of glutamate increased in all cell lines over time (Supplemental Figure 2a). Interestingly, H460 cells start consuming glutamate after 48 hrs, coinciding with the depletion of glutamine. All cell lines receiving complete media showed significant differences in intracellular glutamine (Figure 4a), glutamate (Supplemental Figure 2b), γ-glutamylcysteine (Supplemental Figure 2c), and glutathione (Figure 4b) per mg protein as compared to glutamine-free complete media. Intracellular glutathione per mg protein were significantly higher (about 5-fold) in the carcinomas H460 and A549 cells than in MRC-5 fibroblasts, although no significant differences were observed between the tumor cell lines. This was true for the complete and glutamine-free media cultures. In presence of glutamine, the amounts of intracellular glutamate were not different between A549 and MRC-5 cells (Supplemental Figure 2b). Glutamate amounts were ~40% lower in H460 cells, a difference that was significant for cultures in complete or glutamine-free media.
Figure 4.
Intracellular concentrations of (a) glutamine, and (b) glutathione at 48 hrs. Solid columns indicate culturing in complete media and striped columns indicate culturing in glutamine-free media. Intracellular amounts were significantly different in cells grown in glutamine-free media compared to cells grown in complete media (p<0.001).
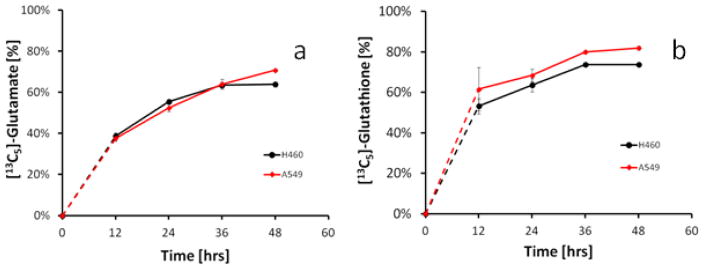
To test the hypothesis that glutathione is in part derived from glutamine, we incubated H460 and A549 cells with glutamine-free medium supplemented with [13C5]-glutamine and monitored 13C5-incorporation into glutamate and glutathione at various time points (Figure 5). After 12 hrs, excreted glutamate was >38% labeled in both cell lines. Excreted glutathione were 53% and 62% [13C5] labeled for H460 and A549 cells, respectively, indicating that it is derived from the [13C5]-glutamine. The cell-free medium control did not show any 13C-labeled glutamate or glutathione. [13C5]-glutamine-supplemented media did not affect cell growth compared to complete media (data not shown).
Figure 5.
Time course of (a) [13C5]-glutamate and (b) [13C5]-glutathione derived from [13C5]-glutamine in A549 and H460 cells. Cells were grown in glutamine-free medium supplemented with 2 mM [13C5]-glutamine. At the indicated time points, 25 μl of culture medium was analyzed for the presence of stable isotope-enriched metabolites.
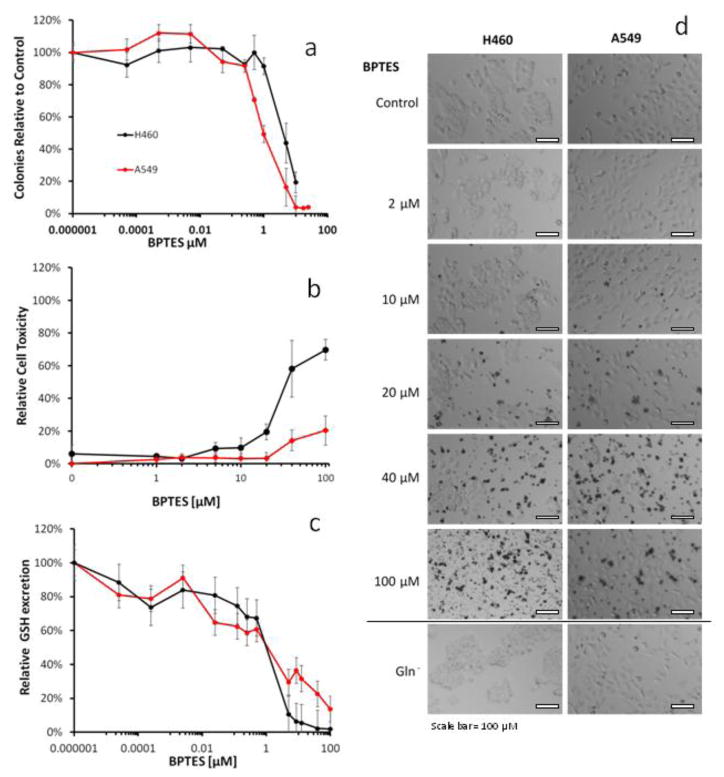
GLS catalyzes the conversion of glutamine to glutamate, a precursor of glutathione synthesis and an essential step in glutamine utilization. To determine the role of glutamine as a pre-cursor for glutathione synthesis, cells were treated with BPTES, a known GLS inhibitor [33]. BPTES treatment led to significant growth inhibition, toxicity, and abolished glutathione excretion (Figure 6a–c). The effects of GLS inhibition were similar to culturing cells in glutamine-free medium (data not shown). Cell responses to BPTES were assessed by four different assays (CellTiter, CellTiter-Glo, CellTox, colony formation), of which the colony formation showed a dose response with ED50 of 4.2 and 1.0 μM BPTES for H460 and A549, respectively (Figure 6a). The metabolic response to BPTES showed an ED50 of 7.2 and 4.0 μM BPTES for A549 and H460, respectively. At greater BPTES concentrations formation and excretion of glutathione and the intermediate metabolites glutamate and γ-glutamyl-cysteine were essentially abolished (data not shown). H460 and A549 responses to BPTES treatments demonstrate a visual increase in cell death at concentration of 10, 20, 40, and 100 μM, but no cell death was observed in cells cultured in glutamine-free medium. (Figure 6d).
Figure 6.
Effect of GLS inhibition by BPTES on cell (a) clonogenic viability (b) cell toxicity and (c) glutathione excretion in H460 and A549 cells. Shown are mean ± SD of three independent experiments. (d) Representative bright field images (40x magnification) of H460 and A549 cells after 48 hrs treatment with various concentrations of BPTES or culturing in glutamine-free (Gln−) media.
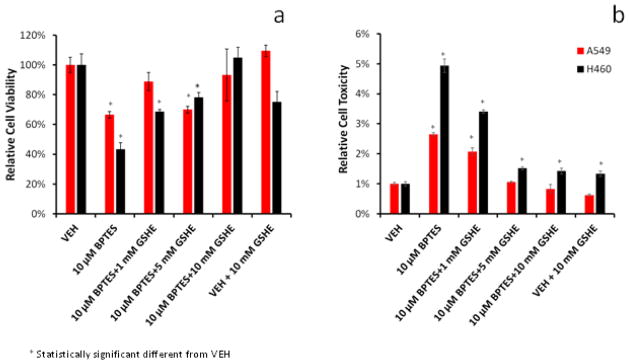
The importance of glutamine-derived GSH for cell viability and against cytotoxicity was investigated using media supplemented with GSHE, the bioavailable form of GSH [2,6,27,29]. Initial treatment with 10 μM BPTES reduced viability and induced cytotoxicity (Figure 7a and 7b) in both tumor cell lines. Addition of 1–10 mM GSHE showed a dose-dependent protection against BPTES-induced cytotoxicity in both cell lines. As in the previous experiment the BPTES effects were less pronounced in the A549 compared to the H460.
Figure 7.
Protective effect of GSHE on BPTES-treated cells. Shown are the (a) viability (b) cell toxicity responses. H460 and A549 cells were treated with 10 μM BPTES in presence of 1–10 mM GSHE. Shown are mean ± SD of three independent experiments. BPTES treatment significantly reduced cell viability and increased cell toxicity (p<0.01).
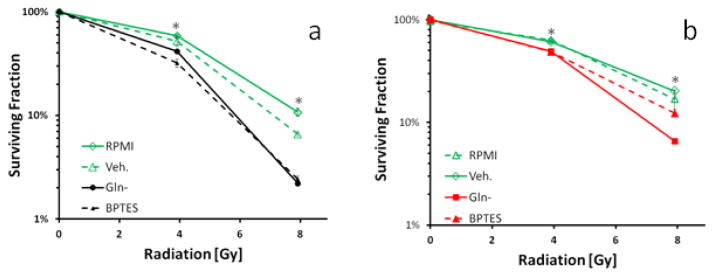
To demonstrate that the glutamine-derived glutathione is biologically, and likely clinically relevant, responses to ionizing radiation were determined in both tumor cell lines after culturing in glutamine-free media or after treatment with BPTES, a known GLS inhibitor. The results demonstrate the biological importance of glutamine-derived glutathione in defense against oxidative stress induced by ionizing radiation. First, lack of glutamine significantly increased radiation sensitivity for both A549 and H460 as compared to control at both the 4 Gy and 8 Gy levels (p<0.01 for all) suggesting an essential role for glutamine-derived glutathione in defense against radiation-induced injury (Figure 8a and 6b). Second, inhibition of GLS by BPTES produced a radiation-induced reduction in colony formation in H460 and A549 cells at 4 Gy and 8 Gy levels as compared to control cells (p<0.001). Although sensitization was slightly less pronounced in A549 cells compared to H460 (Figure 8a and b). These results suggest that GLS is intimately linked to GSH production and defense against the oxidative stress created by ionizing radiation exposure, and further implicates glutamine in the response to therapeutics.
Figure 8.
Effect of culturing in glutamine-free media or 2 μM BPTES treatment on radiation response in (A) H460 and (B) A549 tumor cells. Radiation sensitivity was determined utilizing the clonogenic assay and normalizing to the plating efficiency in untreated cells. Shown are mean ± SD of three independent experiments. Some error bars are smaller than the symbol and therefore are not visible. BPTES treatment or culturing in glutamine-free media significantly increased radiation sensitivity (*, p<0.05).
Discussion
Glutamine utilization has been shown to promote tumor cell proliferation and GLS activity has been identified as a viable target for cancer therapy [15]. The common belief is that glutamine provides additional building blocks for biosynthesis and energy production [37,38]. We herein provide strong evidence that glutamine is essential for glutathione production, and therefore contributes directly to cellular defense systems.
Studying the uptake and release of nutrients, we were not surprised to observe greater glutamine uptake and glutamate excretion in the faster growing carcinoma cell lines (H460 and A549) compared to the MRC-5 alveolar fibroblasts (Figure 3a and a). Under our culture conditions H460 cells consume glutamine and excrete glutamate over the first 48 hrs. After that, glutamine is depleted in H460 cultures, and H460 cells start to utilize glutamate as a carbon source. These results suggest that the initial abundance of glutamine leads to its rapid uptake and conversion to glutamate. However, the intracellular glutamate pool is limited and excessive glutamate is excreted into the medium (Supplemental Figure 2b). Adjusting for doubling time, each cell line utilizes about 4 μmoles of glutamine per doubling. For example, H460 reduced glutamine concentration in culture medium from 2 mM to 1.2 mM, which translates to 0.8 μmoles /ml. Therefore, H460 cells cultured in 5 ml media use 4 μmoles (5 ml x 0.8 μmoles/ml) in 24 hours corresponding to approximately 2 doubling times.
Surprisingly, both tumor cell lines excreted large amounts of glutathione (Figure 3b). The H460 and A549 cultures starting with 100,000 cells produced and excreted over the first 48 hrs a minimum of 2.0 ± 1.10 and 0.62 ± 0.09 μmoles of glutathione, respectively. In contrast, MRC-5 fibroblast excreted 0.12 ± 0.00 μmoles during same time period. The amounts of glutathione excreted by 48 hours corresponded to 28.3% and 15.6% of total glutamine utilized by H460 and A549, respectively. The isotope-labeled glutamine experiment indicates that at least 50% of the excreted glutathione is replaced within 12 hrs, demonstrating rapid glutathione turnover. These steady state amounts do not account for any glutathione consumed or utilized for cellular defense during that time period and therefore underestimate the actual amount synthesized and excreted. Together these results emphasize the fact that lung tumors spend huge efforts on glutathione synthesis and excretion. The high glutathione turnover is consistent with the short half-life (2–6 hrs) of glutathione reported in lung tumors, and suggests that glutathione synthesis and cycling in lung tumors and tumor cells is much higher than estimated from steady state measurements [21,23].
Correlation analysis clearly suggests a mechanistic link between the glutamine uptake and glutathione excretion (Figure 3c). Subsequent culturing in glutamine-free media abolished glutathione synthesis and excretion (data not shown) indicating that glutathione synthesis and excretion are tightly linked to, and even dependent on, available glutamine. Culturing in glutamine-free medium significantly reduces cell growth, suggesting that glutathione excretion is important for cell proliferation. Intracellular concentrations of glutathione per mg protein mirrored the observation in culture media, with glutathione being higher in tumor cells compared to MRC-5 fibroblasts, however effects were less pronounced (Figure 4). This was not unexpected since intracellular metabolite concentrations are highly controlled through synthesis and degradation processes, and therefore remain within biologically acceptable ranges. In contrast, the extracellular metabolites corresponding to cellular activity are less stringently regulated, and therefore their concentration ranges are much wider than could be tolerated within the cell.
The first step in glutamine utilization for energy production is its conversion to glutamate by GLS. Culturing in glutamine-free media drastically reduced cell proliferation and might have confounded the results. Therefore, to test whether glutamine as an energy and nutrient source was essential for cell proliferation and a precursor for glutathione synthesis, cells were treated with a specific GLS inhibitor [24,35]. The dose-response of BPTES treatment showed decreased cell viability based on the colony formation, CellTiter, CellTiter-Glo®, and CellToxTM assays, while glutathione excretion was essentially abolished at >5 μM BPTES (Figure 6c). Together these results provide evidence for a mechanistic link between GLS activity and glutathione excretion. While the effect of BPTES on cell proliferation has been reported previously [24,25] the fact that glutathione excretion was affected demonstrates for the first time the essential roles of glutamine and GLS-activity for glutathione synthesis and excretion. GLS knockdown in cervical cancer cells reduced intercellular glutathione in radiation resistant HeLa cells to relative levels as normal HeLa cells suggesting that GLS and glutathione synthesis are mechanistically linked in other tumors as well[41]. The presented results extend this observation to lung cancer cells and establish that glutamine-derived glutamate is in fact utilized for glutathione synthesis and excretion.
The ED50 for the metabolic effect (~>5.0 μM BPTES) was similar to the ED50 for biological responses, reduction in cell proliferation, suggesting a mechanistic link between glutathione excretion and cell viability. Glutathione depletion has been reported previously by inhibition of γ-glutamyl-cysteine ligase using 100 to 10,000 μM buthionine sulfoximine [8,13]. The presented results suggest that BPTES is at least 100-fold more effective in depletion of glutathione, making it a better candidate for combination therapies [8,13].
Glutamate is a common metabolite and can be derived from various sources such as glucose metabolism, protein degradation or amino acid metabolism [25]. Based on the media time courses of glutamate, H460 and A549 cells excrete significant amounts of glutamate, suggesting that these cell lines synthesize or liberate large amounts of glutamate (Supplemental Figure 1a). Glutamate is a known cofactor for membrane transports such as the Xc− antiport which imports cystine in exchange for one glutamate[5]. Therefore the increased amounts of extracellular glutamate may reflect increased cystine uptake. Surprisingly, the glutamate excretion and glutamate independence depends on the availability of glutamine as a carbon source.
To determine the source of glutamate that is used for glutathione synthesis, cells were cultured in glutamine-free media supplemented with [13C5]-glutamine. Glutathione derived from the [13C5]-glutamine was distinguished from glutathione that is derived from other sources using mass spectrometry. This experiment provided stunning proof that the majority of glutathione is, in fact, synthesized from glutamine-derived glutamate. After 12 hours, approximately 40% 13C-carbon incorporation was observed in the intermediate metabolite glutamate (Figure 5a). Excreted glutathione contained the >50% 13C-carbon backbone (Figure 5b), confirming the active utilization of the metabolic pathway proposed in Figure 1. The stable isotope tracing experiment clearly demonstrates that glutamine is utilized for glutathione synthesis and excretion. It is interesting to note that the lung tumor cells seem to use mainly glutamine–derived glutamate for glutathione synthesis suggesting that the glutamine-derived glutamate does not mix with the glutamate pool. The question of why tumor cells spend so much effort on glutathione synthesis and excretion needs to be further investigated.
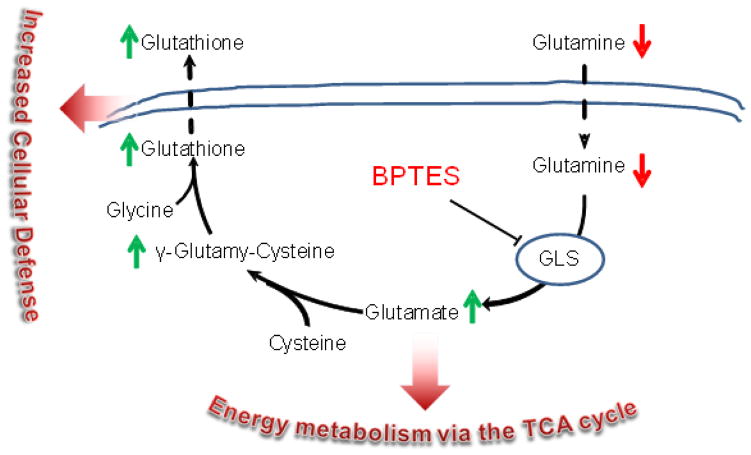
Figure 1.
Metabolic scheme of the glutamine uptake, metabolism to glutamate and utilization for GSH synthesis. Green upward arrows indicate metabolites increased in fast proliferating lung tumor cell lines. Red downward arrows indicate metabolites consumed in fast proliferating lung tumor cell lines. Also shown are the glutaminase (GLS) enzyme and its inhibitor BPTES.
These findings have wide implications in regard to the suitability of a GLS inhibitor in chemotherapy. It is expected that low dose treatments with BPTES, and probably other GLS inhibitors, may be sufficient to significantly reduce glutathione synthesis and thereby increase sensitivity to radiotherapy and reduce multi-drug resistance. If this is true, it will make GLS inhibitors a prime component of combination therapies [31]. To test the former, we determined the radiosensitivity of the lung tumor cell lines after culturing in glutamine-free media or after BPTES treatment. Both pretreatments caused glutathione depletion and increased radiosensitivity (Figure 8a and 8b). This provides additional evidence of the mechanistic link between glutamine consumption and glutathione synthesis and their importance for cell viability.
The study was not designed to distinguish which of the known glutathione functions will be most affected by GLS inhibition, but based on the μM amounts of excreted glutathione it is unlikely that the increased glutathione production in lung tumor cells functions as intracellular antioxidants. Endogenous concentration of reactive nitrogen or oxygen species have been estimate to reach a maximum of 100 nM [11], which is orders of magnitude lower than the >3 μM glutathione in the lung tumor cell lines. This strongly suggests that biochemical turnover of glutathione is more important than spontaneous biochemical detoxification of reactive oxygen species [14].
In conclusion, we demonstrate here for the first time that a significant amount of glutathione is directly derived from glutamine. This finding adds yet another important function to the already known glutamine dependence of lung tumor. Glutaminolysis has been identified as a suitable target for cancer therapies, and the findings presented herein suggest that a GLS inhibitor leads to glutathione depletion and marked changes in response to radiation treatment. Depleting glutathione in this manner is expected to reduce multi-drug resistance and increase sensitivity to oxidative stress caused by radiation therapy, leading to greater cytotoxicity and tumor response [8].
Supplementary Material
Highlights.
Glutathione synthesis is dependent of available glutamine and glutaminase activity.
Acknowledgments
Support has been provided in part by the National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program, grants UL1TR000039 and KL2TR000063 and the Arkansas Bioscience Institute, and the Envoys, an advocacy group of 4The Winthrop P. Rockefeller Cancer Institute Foundation.
Abbreviations
- BPTES
2-(5-phenyl-acetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide
- GLS
glutaminase
- LC-MS/MS
liquid chromatography tandem mass spectrometry
Footnotes
Authors’ Contributions
DRS contributed to the study design and carried out all cell culture experiments, metabolite extraction and mass spectrometry analyses. GH and AD performed the BPTES inhibition experiments. AJP conduced the radiation sensitization experiments colony formation assays. RJG supervised and designed the radiation treatments. ES and RBP participated in the design of the study and performed the statistical analysis. GB conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ames BN, Gold LS, Willett WC. The causes and prevention of cancer. Proc Natl Acad Sci U S A. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson ME, Powrie F, Puri RN, Meister A. Glutathione monoethyl ester: preparation, uptake by tissues, and conversion to glutathione. Arch Biochem Biophys. 1985;239:538–548. doi: 10.1016/0003-9861(85)90723-4. [DOI] [PubMed] [Google Scholar]
- 3.Ausborn NL, Le QT, Bradley JD, Choy H, Dicker AP, Saha D, Simko J, Story MD, Torossian A, Lu B. Molecular profiling to optimize treatment in non-small cell lung cancer: a review of potential molecular targets for radiation therapy by the translational research program of the radiation therapy oncology group. Int J Radiat Oncol Biol Phys. 2012;83:e453–e464. doi: 10.1016/j.ijrobp.2012.01.056. [DOI] [PubMed] [Google Scholar]
- 4.Baltar VT, Xun WW, Johansson M, Ferrari P, Chuang SC, Relton C, Ueland PM, Midttun O, Slimani N, Jenab M, Clavel-Chapelon F, Boutron-Ruault MC, Fagherazzi G, Kaaks R, Rohrmann S, Boeing H, Weikert C, Bueno-de-Mesquita B, Boshuizen H, van Gils CH, Onland-Moret NC, Agudo A, Barricarte A, Navarro C, Rodriguez L, Castano JM, Larranaga N, Khaw KT, Wareham N, Allen NE, Crowe F, Gallo V, Norat T, Krogh V, Masala G, Panico S, Sacerdote C, Tumino R, Trichopoulou A, Lagiou P, Trichopoulos D, Rasmuson T, Hallmans G, Roswall N, Tjonneland A, Riboli E, Brennan P, Vineis P. A structural equation modelling approach to explore the role of B vitamins and immune markers in lung cancer risk. Eur J Epidemiol. 2013;28:677–688. doi: 10.1007/s10654-013-9793-z. [DOI] [PubMed] [Google Scholar]
- 5.Bhutia YD, Babu E, Ramachandran S, Ganapathy V. Amino Acid transporters in cancer and their relevance to "glutamine addiction": novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75:1782–1788. doi: 10.1158/0008-5472.CAN-14-3745. [DOI] [PubMed] [Google Scholar]
- 6.Campbell KC, Larsen DL, Meech RP, Rybak LP, Hughes LF. Glutathione ester but not glutathione protects against cisplatin-induced ototoxicity in a rat model. J Am Acad Audiol. 2003;14:124–133. [PubMed] [Google Scholar]
- 7.Carney DN. Lung cancer--time to move on from chemotherapy. N Engl J Med. 2002;346:126–128. doi: 10.1056/NEJM200201103460211. [DOI] [PubMed] [Google Scholar]
- 8.Clark EP, Epp ER, Morse-Gaudio M, Biaglow JE. The role of glutathione in the aerobic radioresponse. I. Sensitization and recovery in the absence of intracellular glutathione. Radiat Res. 1986;108:238–250. [PubMed] [Google Scholar]
- 9.Convoer W. Chapter 5, Practical Nonparametric Statistics. John Wiley & Sons; New York: 1999. [Google Scholar]
- 10.Conway JG, Neptun DA, Garvey LK, Popp JA. Carcinogen treatment increases glutathione hydrolysis by gamma-glutamyl transpeptidase. Carcinogenesis. 1987;8:999–1004. doi: 10.1093/carcin/8.7.999. [DOI] [PubMed] [Google Scholar]
- 11.Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Arch Biochem Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fath MA, Ahmad IM, Smith CJ, Spence J, Spitz DR. Enhancement of carboplatin-mediated lung cancer cell killing by simultaneous disruption of glutathione and thioredoxin metabolism. Clin Cancer Res. 2011;17:6206–6217. doi: 10.1158/1078-0432.CCR-11-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flohe L. The fairytale of the GSSG/GSH redox potential. Biochim Biophys Acta. 2013;1830:3139–3142. doi: 10.1016/j.bbagen.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 16.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim ES, Pandya KJ. Advances in personalized therapy for lung cancer. Expert Opin Med Diagn. 2013;7:475–485. doi: 10.1517/17530059.2013.826645. [DOI] [PubMed] [Google Scholar]
- 18.Langer CJ, Besse B, Gualberto A, Brambilla E, Soria JC. The evolving role of histology in the management of advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:5311–5320. doi: 10.1200/JCO.2010.28.8126. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz MA, Burant CF, Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal Chem. 2011;83:3406–3414. doi: 10.1021/ac103313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lushchak VI. Glutathione homeostasis and functions: potential targets for medical interventions. J Amino Acids. 2012;2012:736837. doi: 10.1155/2012/736837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 22.Meister A, Tate SS. Glutathione and related gamma-glutamyl compounds: biosynthesis and utilization. Annu Rev Biochem. 1976;45:559–604. doi: 10.1146/annurev.bi.45.070176.003015. [DOI] [PubMed] [Google Scholar]
- 23.Meister A, Tate SS. Glutathione and related gamma-glutamyl compounds: biosynthesis and utilization. Annu Rev Biochem. 1976;45:559–604. doi: 10.1146/annurev.bi.45.070176.003015. [DOI] [PubMed] [Google Scholar]
- 24.Mohamed A, Deng X, Khuri FR, Owonikoko TK. Altered glutamine metabolism and therapeutic opportunities for lung cancer. Clin Lung Cancer. 2014;15:7–15. doi: 10.1016/j.cllc.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newsholme P, Procopio J, Lima MM, Pithon-Curi TC, Curi R. Glutamine and glutamate--their central role in cell metabolism and function. Cell Biochem Funct. 2003;21:1–9. doi: 10.1002/cbf.1003. [DOI] [PubMed] [Google Scholar]
- 26.NIH. Cell Lines. The In Vitro Screen. 2014 Http://Dtp.Nci.Nih.Gov/Docs/Misc/Common_Files/Cell_List.Html Discovery Services.
- 27.Nishida K, Ohta Y, Ito M, Nagamura Y, Kitahara S, Fujii K, Ishiguro I. Conversion of gamma-glutamylcysteinylethyl ester to glutathione in rat hepatocytes. Biochim Biophys Acta. 1996;1313:47–53. doi: 10.1016/0167-4889(96)00054-7. [DOI] [PubMed] [Google Scholar]
- 28.Ortega AL, Mena S, Estrela JM. Glutathione in cancer cell death. Cancers (Basel) 2011;3:1285–1310. doi: 10.3390/cancers3011285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajasekaran NS, Sathyanarayanan S, Devaraj NS, Devaraj H. Chronic depletion of glutathione (GSH) and minimal modification of LDL in vivo: its prevention by glutathione mono ester (GME) therapy. Biochim Biophys Acta. 2005;1741:103–112. doi: 10.1016/j.bbadis.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 30.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709–719. doi: 10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 31.Rocha CR, Garcia CC, Vieira DB, Quinet A, de Andrade-Lima LC, Munford V, Belizario JE, Menck CF. Glutathione depletion sensitizes cisplatin- and temozolomide-resistant glioma cells in vitro and in vivo. Cell Death Dis. 2014;5:e1505. doi: 10.1038/cddis.2014.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 33.Shukla K, Ferraris DV, Thomas AG, Stathis M, Duvall B, Delahanty G, Alt J, Rais R, Rojas C, Gao P, Xiang Y, Dang CV, Slusher BS, Tsukamoto T. Design, synthesis, and pharmacological evaluation of bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. J Med Chem. 2012;55:10551–10563. doi: 10.1021/jm301191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 35.van den Heuvel AP, Jing J, Wooster RF, Bachman KE. Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol Ther. 2012;13:1185–1194. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 37.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vander Heiden MG, Lunt SY, Dayton TL, Fiske BP, Israelsen WJ, Mattaini KR, Vokes NI, Stephanopoulos G, Cantley LC, Metallo CM, Locasale JW. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb Symp Quant Biol. 2011;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Yu M, Xiao L, Xu S, Yi Q, Jin W. Radiosensitizing effect of oleanolic acid on tumor cells through the inhibition of GSH synthesis in vitro. Oncol Rep. 2013;30:917–924. doi: 10.3892/or.2013.2510. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Ballatori N. Endogenous glutathione conjugates: occurrence and biological functions. Pharmacol Rev. 1998;50:335–356. [PubMed] [Google Scholar]
- 41.Xiang L, Xie G, Liu C, Zhou J, Chen J, Yu S, Li J, Pang X, Shi H, Liang H. Knock-down of glutaminase 2 expression decreases glutathione, NADH, and sensitizes cervical cancer to ionizing radiation. Biochim Biophys Acta. 2013;1833:2996–3005. doi: 10.1016/j.bbamcr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.