Abstract
In this review, we discuss the potential pharmacological targeting of a set of powerful epigenetic mechanisms: DNA methylation control systems in the central nervous system (CNS). Specifically, we focus on the possible use of these targets for novel future treatments for learning and memory disorders. We first describe several unique pharmacological attributes of epigenetic mechanisms, especially DNA cytosine methylation, as potential drug targets. We then present an overview of the existing literature regarding DNA methylation control pathways and enzymes in the nervous system, particularly as related to synaptic function, plasticity, learning and memory. Lastly, we speculate upon potential categories of CNS cognitive disorders that might be amenable to methylomic targeting.
Keywords: epigenetics, neuropharmacology, DNMT, Tet oxidase, cytosine methylation, demethylation, learning, memory, cognitive disorders, cognitive enhancement
INTRODUCTION
Although there is considerable information available concerning how DNA methylation may be regulated in the central nervous system (CNS), there is little existing literature concerning its pharmacological targeting or drug development in this area. Moreover, most of the available pharmacology literature on epigenetics in the nervous system is related to histone modifications, whereas here, we focus on DNA modifications. Thus, in contrast to the typical comprehensive review of the literature presented in the Annual Review of Pharmacology and Toxicology, in this review we present potential future pharmacology rather than established pharmacology.
The Unique Pharmacology of Epigenetically Targeted Drugs
Conceptually speaking, epigenetically targeted drugs in general and DNA methylation–targeted drugs in particular may have unique pharmacological and toxicological attributes. In this section and by way of introduction to the contents of the review, we very briefly highlight possible unique characteristics of hypothetical DNA methylation–targeted drugs in terms of their potential pharmacokinetics, pharmacodynamics, therapeutics, and toxicology. We intend to pique the interest of the reader by defining some ways in which epigenetically targeted drugs might act orthogonally to existing pharmacology dogma. This is not to say that all epigenetically targeted drugs will possess such idiosyncratic attributes, but rather that some might.
Pharmacokinetics
Once triggered, changes in DNA methylation can be self-perpetuating within the cell. Thus, transiently altering DNA methylation can trigger permanent effects. In this vein, even a single transient application of a methylation-altering drug may trigger lifelong effects. In classical pharmacology, irreversible inhibitors such as covalent modifiers of enzyme active sites are not even in the same ballpark—their effects last at most for the lifetime of the target protein or molecule. DNA methylation manipulation potentially takes irreversibility to a new level pharmacologically.
Pharmacodynamics
Epigenetic changes can trigger a state change for the entire cell, for instance, in the case in which DNA methylation regulates gene transcription. Methylation can completely silence a gene, and demethylation can render it fully functional at the transcriptional level. Thus, the effects of manipulating the methylome can be all-or-none. In classical pharmacodynamics terms, these agents can exhibit extreme positive cooperation. Theoretically, a single cytosine methylation event in a cell can completely silence the associated gene, one single molecular modification triggering a state change for the cell’s transcriptional complement.
Moreover, there is an emerging literature suggesting possible transgenerational effects of altered DNA methylation—not just teratogenic effects of drug exposure in utero, but possible heritable and self-perpetuating germline modifications that can be perpetuated across generations. This represents an entirely different type of pharmacodynamics: a drug effect in the absence of the organism ever having directly experienced the drug.
Therapeutics
Epigenetic drugs may enable extremely precise gene- or gene domain–specific targeting, possibly even reaching an unprecedented level of target specificity. However, the issue of off-target effects may be completely different with methylome-targeted pharmacological agents. This is because, to a first approximation (with a few exceptions in the immune system and germline), every cell in a particular individual has the same genome. Different cell types have different epigenomes (including different cytosine methylation patterns), but the underlying genome in every cell is the same even if half or two-thirds of the genome is silenced in that cell type. Thus, even gene-specific targeting of methylation could affect every cell in the body, because this targeting acts upon the conserved organism-wide genome sequence. This contrasts to traditional drugs targeted to a specific cell surface receptor, for example, where the target itself intrinsically generates some cell selectivity—only a subset of cells has the particular cell surface receptor expressed from their genome. In those cells where the gene for the drug target is silenced during development, the drug cannot act on that target. Gene targeting therefore presents a unique opportunity and challenge with methylomically targeted drugs.
However, promiscuous epigenomically targeted drugs might present unique therapeutic opportunities. There may be instances where broadly regulating swaths of the genome (hundreds of genes at once) could be particularly beneficial. In terms of epigenetic drugs currently under development, histone deacetylase (HDAC) inhibitors may be an example of this therapeutic advantage; where diseased cells (e.g., cancer cells) have broadly disrupted patterns of gene expression, agents acting broadly across the genome may be uniquely beneficial. In terms of CNS applications, where a disease process (e.g., drug addiction, depression, schizophrenia, stress, cognitive dysfunction) has resulted in extensive reorganization of the epigenome, broadly affecting the epigenome might be desirable or even required for therapeutic efficacy.
Toxicology
Finally, all the attributes described in this section can be either beneficial or detrimental, depending on whether the effects triggered are therapeutic or toxic. With this in mind, it will be even more important to screen epigenetic therapeutics for potentially toxic side effects, as the changes generated could potentially be long-lasting.
EPIGENETICS AS A STABLE MECHANISM FOR BEHAVIORAL CHANGE
Mechanisms of DNA Methylation
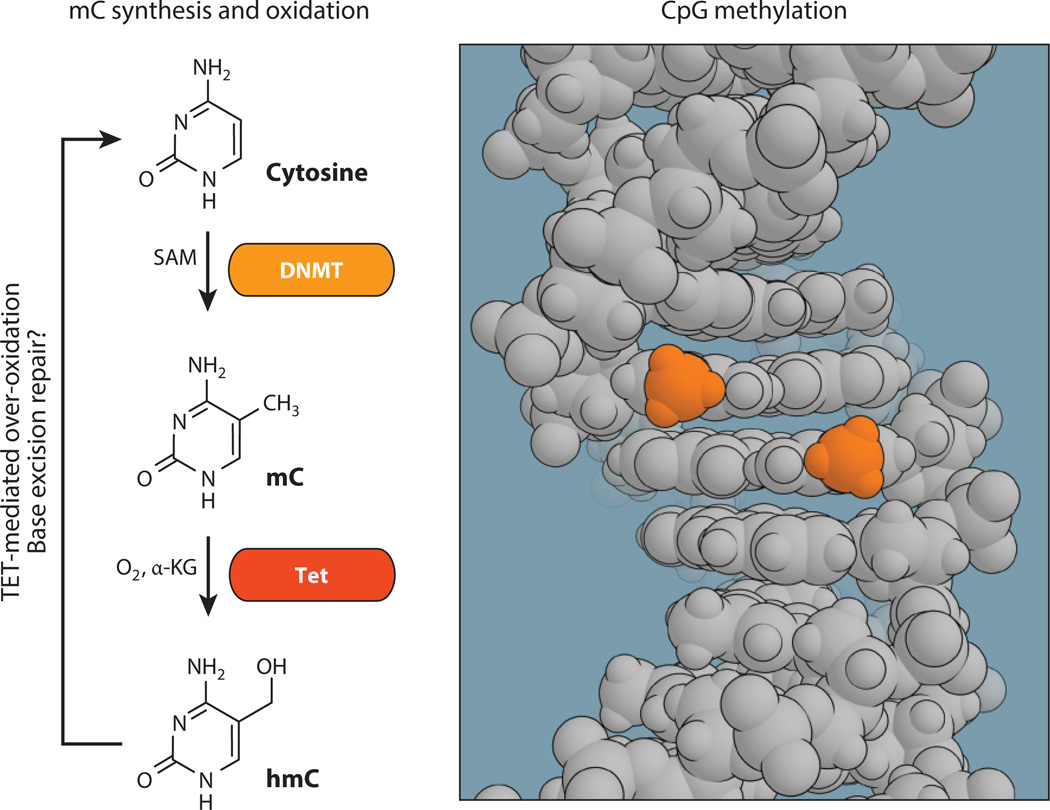
In the nucleus, DNA is wound tightly around histone proteins, forming a nucleosome, the smallest subunit of chromatin. A large variety of epigenetic modifications can occur on histones, including phosphorylation, acetylation, and alkylation, many of which have profound effects on DNA availability and the recruitment of transcriptional elements (1). Underneath the histone code, DNA methylation acts covalently on the gene to regulate its expression. DNA methylation is mediated by de novo DNA methyltransferases (DNMTs) to yield 5-methylcytosine (mC), primarily at cytosine-phosphate-guanine (CpG) dinucleoside sites (Figure 1) (2, 3). This epigenetic mark can then self-perpetuate through the activity of maintenance DNMTs, which recognize monomethylation and methylate the complementary strand of the CpG site, producing a dimethylated tag. Double-stranded methylation, in which the two methyl groups adopt a syn confirmation in the major groove, can act to regulate gene transcription by affecting chromatin structure. These regulatory effects appear to be mediated through the attraction of methyl-binding domain (MBD)-containing proteins that recruit histone modifiers (4–6), inhibit transcription by disrupting transcription factor binding (7, 8), or increase expression by blocking insulators or repressors (9–11).
Figure 1.
Cytosine is methylated in vivo by DNMTs, which use SAM as an electrophilic methyl source, to produce mC at CpG sites in double-stranded DNA. Maintenance DNMTs may then methylate the complimentary cytosine to produce double-stranded CpG methylation. An otherwise stable epigenetic mark, mC can be oxidized by the α-KG-dependent Tet family of dioxygenases to yield hmC, which is the first step in removing the methyl as an epigenetic mark. Abbreviations: α-KG, α-ketoglutarate; CpG, cytosine-phosphate-guanine; DNMT, DNA methyltransferase; hmC, 5-hydroxymethylcytosine; mC, 5-methylcytosine; SAM, S-adenosyl methionine; Tet, ten-eleven translocation.
Double-stranded methylation can be long-lived. In the event that one mC is damaged, metabolized, or excised to yield an unsubsidized cytosine, the memory of the mark is not necessarily lost and can be reestablished by maintenance DNMT activity. Indeed, it is by this mechanism that dividing cells pass down their methyl marks to their progeny. That is not to say demethylation cannot occur, and active CpG demethylation mechanisms have been an ongoing area of research (12). Ten-eleven translocation (Tet) mC dioxygenases oxidize the benzylic position of mC to produce 5-hydroxymethylcytosine (hmC) (13). The hydroxymethyl mark itself is stable and exists in relatively elevated levels in the brain; however, it is also the first step in active demethylation, and either over-oxidation by the Tet family of proteins to aldehyde or carboxylate products or deamination mechanisms followed by base excision repair then erases cytosine alkylation (14, 15).
DNA methyltransferases
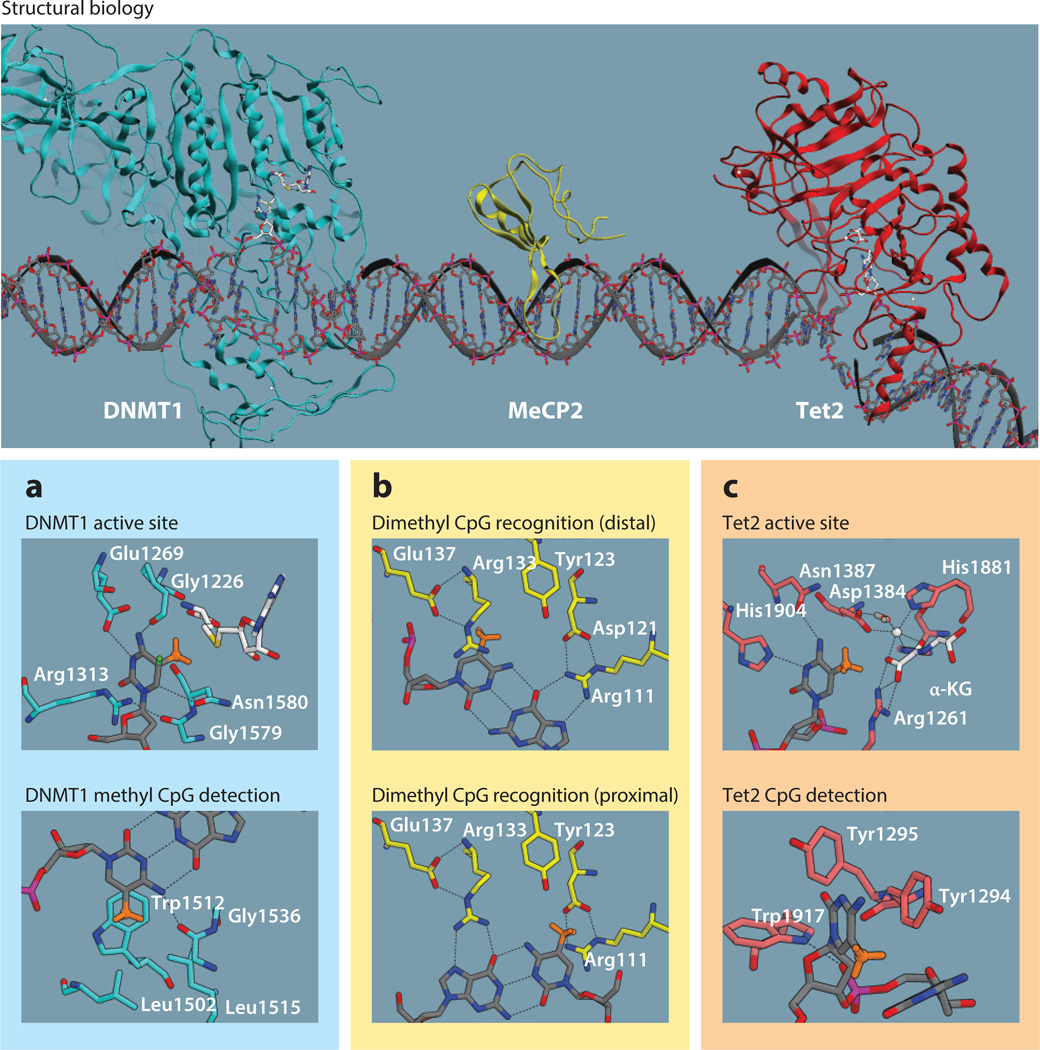
Three DNMTs (DNMT1, DNMT3a, and DNMT3b) function to mediate the methyl transfer from S-adenosyl methionine (SAM) to the 5 position of cytosine (9). A fourth (DNMT3l) is catalytically inactive yet is required for DNA methylation–dependent genomic imprinting through its recruitment of DNMT3a and b (16, 17). All three active DNMTs are capable of de novo methylation, but the structurally distinct DNMT1 is predominately responsible for maintenance methylation (18). The mechanism by which DNMT1 recognizes hemimethylation and facilitates the methyl transfer was recently solved through the clever application of a synthetic 5-fluorocytosine to trap the otherwise unstable tetrahedral intermediate (Figure 2) (19). DNMT1 flips out the target cytosine, activates the aromatic system of the base with electron-donating contacts, and delivers the electrophilic methyl group. The fidelity of DNMT1 for hemimethylated DNA is mediated by a hydrophobic pocket that detects the contiguous mCpG base pair through pi-stacking and hydrogen bond contacts as well as a pair of hydrophobic Leu residues. This small, hydrophobic pocket is ideal for accepting a methyl group and may explain why the presence of the polar hydroxymethyl reduces DNMT1 activity anywhere from 12- to 60-fold for hemi-hmC sites. These reaction rate differences could potentially then allow for active demethylation to occur once both CpG methyl groups have been oxidized by Tet proteins, in spite of local DNMT1 maintenance activity.
Figure 2.
DNA methyl (orange) addition, recognition, and oxidation. (top) Crystal structures of DNMT1, MeCP2, and TET2 bound to DNA. (a) Mouse DNMT1 (teal) trapped by double-stranded DNA containing 5-fluorocytosine to reveal active site catalysis and means of detecting hemimethylation (19). The coproduct SAH is shown in white. (b) Human MeCP2 (yellow) bound to methylated DNA (20). Hydrogen bond contacts showcase CpG recognition. (c) Human Tet2 (red) is bound to methylated DNA with a water-Fe complex rather than oxygen (26). α-KG is shown in white. Abbreviations: α-KG, α-ketoglutarate; CpG, cytosine-phosphate-guanine; DNMT, DNA methyltransferase; MeCP2, methyl CpG–binding protein 2; SAH, S-adenosyl-l-homocysteine; Tet, ten-eleven translocation.
Methyl-binding domain
Methylated CpG recognition is often mediated by MBD-containing proteins. MBD proteins can recruit HDACs and histone methyltransferases to sites of CpG methylation, leading to local chromatin rearrangement. These relatively small domains that consist of a twisted beta sheet and alpha helix scan DNA’s major grove and detect CpG sites through a pair of Arg residues that chelate the complimentary guanine rings (20). The crystal structure shown in Figure 2 is that of methyl CpG–binding protein 2 (MeCP2) that binds dimethyl CpG sites 50-fold more tightly than unmethylated sites and in which inactivating mutations cause the neurodevelopmental disorder Rett syndrome (21). The mC groups are detected via interactions with Tyr123 and Arg133, residues conserved in the MBD family of proteins with the exception of MBD3, which does not recognize CpG methylation and in which the equivalent Tyr residue is mutated. Perhaps not surprisingly, the stabilization of Arg133 by Glu137 is important for MeCP2’s function, and although the equivalent glutamic acid is present in MBD1, which also binds dimethyl CpG sites strongly, it is mutated in MBD2–4, which have weaker affinities. Mutations of Glu137 in MeCP2 cause mental retardation, X-linked, syndromic 13 rather than Rett syndrome (22). Interestingly, oxidation of one mC to hmC reduces the affinity of MeCP2, MBD1, and MBD2 for the CpG site, whereas dihydroxymethylation erases all selectivity over unmethylated CpGs.
Ten-eleven translocation
Methyl oxidation is the first step in the removal of DNA methylation; however, it is not rate limiting, and hmC is present at detectable levels in vivo. The prediction that the Tet family of proteins serves to oxidize DNA came from their sequence homology to parasitic proteins that produce hydroxymethyluracil (base J) (23, 24). There are three mammalian Tet proteins, each containing a core catalytic region and DNA-binding CXXC domain, with the exception of Tet2, in which the CXXC domain coding region is detached and inverted into the separate CXXC4 gene (25). These iron-mediated dioxygenases use α-ketoglutarate (α-KG) to oxidize mC to hmC by flipping the target mC out and into the active site (Figure 2) (26). The complimentary cytosine of the CpG site is then detected by three aryl residues [Tyr, Tyr(Phe), and Trp]. The exact mechanism by which hmC is then converted back into unalkylated cytosine remains somewhat enigmatic, but there is building evidence that Tets can overoxidize hmC to 5-formylcytosine and even 5-carboxylcytosine, which may then be excised by thymine-DNA glycosylase (15). In any event, hmC can be viewed as a meta-stable intermediate in the erasure of a methyl mark that itself (hmC) may perform some regulatory effect.
Activity- and Experience-Dependent Changes in DNA Methylation
Within the brain, DNA methylation regulates multiple aspects of neuronal development, function, and plasticity and has been shown to be an essential regulator of memory processes. Each of these topics has received extensive coverage in recent reviews (2, 3, 27–31), and we therefore limit our discussion of these topics here. However, we quickly note that neuronal activity induces alterations (both increases and decreases) in DNA methylation at specific sites within the genome (32), as does the formation of long-lasting memories and early-life experiences (2, 33–37). Likewise, DNA methylation is critically important for memory formation and maintenance, synaptic plasticity, behavioral phenotypes that result from early-life experiences, and even transgenerational inheritance of behavioral traits (36–43). Overall, these various results demonstrate that DNA methylation is dynamically regulated in the adult CNS in response to experience and that this cellular mechanism is a crucial step in memory formation.
THE UNIQUE EPIGENOMIC LANDSCAPE OF THE CENTRAL NERVOUS SYSTEM
Despite the potentially unique conceptual role of DNA methylation in activity- and experience-dependent transcriptional reorganization within nondividing neurons, it was once a basic assumption that the overall epigenetic complement of the brain (i.e., the types of modifications and their meaning with relation to gene expression) was not unique compared to other somatic organs. Indeed, the brain uses DNA methylation to silence genes in particular cells just as other organs do, and for the vast majority of the time that epigenetics has constituted its own subdiscipline, it was unclear precisely how DNA methylation could be reversed absent ongoing cell division (58–61). However, the past five years have seen remarkable advances in our understanding of the dynamic nature of DNA methylation processes. Overall, this has generated compelling reasons to believe that DNA methylation in neurons is mechanistically distinct from DNA methylation in almost all other cell types in the body.
The first of these advances was the finding that hmC was enriched in the brain compared to other somatic tissues (47) and the simultaneous discovery that this modification was catalyzed by a special group of mC hydroxylases from the Tet family of proteins (13). Intriguingly, this modification is critical for the removal of methylated cytosines from neurons (62, 63), and this has now been shown to be necessary for neurobiological processes including neurogenesis, activity-related gene transcription, and memory formation and extinction (40–42). Furthermore, it provides a mechanism to explain how established methylation marks in neurons can be removed despite the absence of cell division.
A second, more recent round of advances has demonstrated that, in addition to methylation in a CpG dinucleotide context, neurons also exhibit cytosine methylation in other dinucleotide contexts (CpH, where H represents A, C, or T) (48, 51, 64, 65). Strikingly, CpH methylation appears to be almost completely absent from almost all other somatic cells [excluding embryonic and pluripotent stem cells (52, 66)], suggesting clear neuronal relevance. In a landmark study, Lister and colleagues (51) revealed that CpH methylation is present at very low levels in fetal brains but accumulates with age and corresponds to transcriptional increases in DNMT3a levels across the lifespan. Furthermore, although almost absent from glia, CpH methylation is the predominant form of methylation in human neurons, and its emergence in the frontal cortex is correlated with synaptogenesis. Interestingly, this modification is also read by methyl-binding proteins such as MeCP2, is maintained in adulthood by DNMT3a, and is associated with gene repression (48).
Together, these critical findings reveal that a much larger fraction of the neuronal genome is subject to active DNA methylation and demethylation processes, and they have generated considerable interest in tools that could manipulate the neuronal methylome in the context of cognition and behavior.
THE DRUGGABLE EPIGENOME
Current Approaches
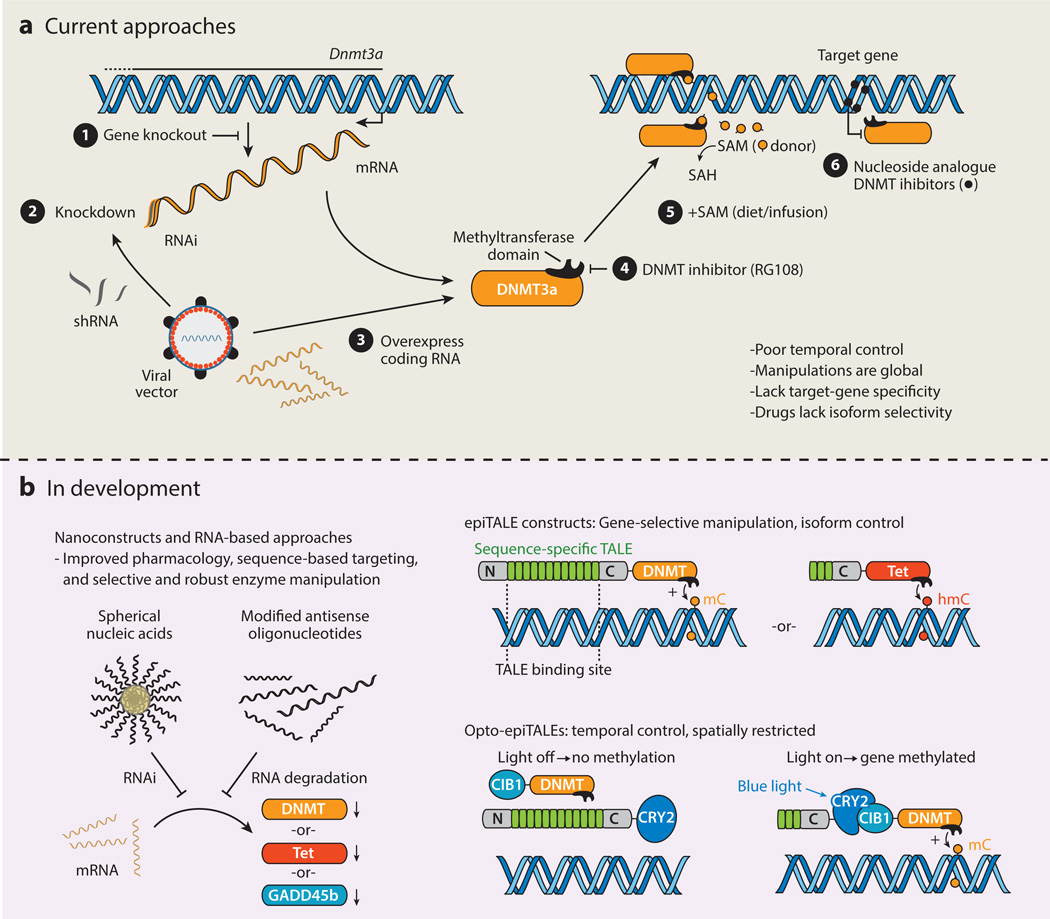
All current approaches to manipulating DNA methylation levels in some way target the endogenous enzymatic machinery that is responsible for the addition or removal of mCs from DNA. Constitutive and conditional gene knockout mouse models as well as viral-mediated RNA knockdown or overexpression techniques have elucidated much about the importance of active DNA methylation during neurodevelopment and in the functioning adult CNS (Figure 3). For example, researchers now understand that whereas DNMT3a/b are often responsible for de novo methylation and DNMT1 for its maintenance, these assignments are not exclusive, and the knockout of bothDNMT1 andDNMT3a in adult forebrain neurons is necessary to elicit dysfunction in long-term plasticity and deficits in learning and memory (67). Recently, a Tet1 knockout mouse model and RNA knockdown experiments demonstrated that mC oxidation by Tet1 is critical for memory and regulation of activity-related genes in the dorsal hippocampus, such as Fos and Arc (40, 41).
Figure 3.
Current and future pharmacological and genetic approaches to manipulating DNA methylation. (a) Current tools include ➊ traditional gene knockout transgenic mouse lines that lack a specific component of DNA methylation machinery; ❷ virally mediated knockdown or ❸ overexpression of DNA methylation components; ❹ small-molecule inhibitors such as RG108 that block the active site on DNMTs; ❺ direct infusion, injection, or dietary supplementation of the methyl group donor SAM; and ❻ nucleoside analogue DNMT inhibitors that incorporate into DNA and trap DNMT enzymes. Unfortunately, the approaches exhibit poor temporal control over DNA methylation and create changes that are genetically and cellularly global. (b) New approaches include spherical nucleic acids and antisense oligonucleotides, which can be used to selectively and robustly inhibit translation of DNA methylation components. Specific changes in DNA methylation at target genes can be accomplished using TALEs, which selectively bind a specific DNA sequence and serve as an anchor for DNA methylation enzymes such as DNMTs or methylcytosine hydroxylases. This system can also be adapted for use with optogenetic tools. For example, a sequence-specific TALE construct could be fused to CRY2, which changes conformation in the presence of blue light, causing it to recruit its binding partner CIB1. Thus, fusion of an effector enzyme such as a DNMT to CIB1 could generate DNA methylation changes with both genetic and temporal specificity. Abbreviations: C, C terminus; CIB1, cryptochrome-interacting basic-helix-loop-helix 1; CRY2, cryptochrome 2; DNMT, de novo DNA methyltransferase; epiTALE, epigenetically modified TALE; GADD45b, growth arrest and DNA damage-inducible protein 45; hmC, 5-hydroxymethylcytosine; mC, 5-methylcytosine; N, N terminus; opto-epiTALE, optogenetic epiTALE; RNAi, RNA interference; SAH, S-adenosyl-l-homocysteine; SAM, S-adenosyl methionine; shRNA, small hairpin RNA; TALE, transcriptional activator–like effector; Tet, ten-eleven translocation.
Small-molecule approaches, including DNMT inhibitors or manipulation of SAM concentrations by direct infusion, dietary supplementation, or restriction of methionine, have been invaluable in demonstrating the necessity of active DNA methylation in the hippocampus and prefrontal cortex during learning and consolidation (3, 34, 35). The nucleoside analogues Zebularine and 5-aza-2′-deoxycytidine, which are incorporated into double-stranded DNA and function by trapping the DNMT active site, and the SAM competitive inhibitor RG108 can be used to examine the effects of unselective inhibition of DNA methylation across the entire genome (Figure 3). The direct infusion of DNMT inhibitors into the hippocampus and ventral tegmental area have been shown to block contextual fear memory formation and stimulus-reward associations, respectively (33, 35). Unfortunately, no such class of potent and metabolically stable small-molecule inhibitors exists for the Tet family of proteins; however, similarly to the design and mechanism of DNMT inhibitors, Tet inhibitors will most likely be competitive mimics of cytosine and α-KG. Perhaps the most advanced area of the current druggable epigenome is the development of potent and selective HDAC inhibitors, which have been thoroughly reviewed elsewhere (68, 69). HDAC inhibitors in general work by decreasing nucleosome compaction and therefore the availability of nearby genes for transcription, but how HDAC inhibition may function to affect gene-specific DNA methylation patterns is not well understood.
These pharmacological and genetic tools to manipulate DNA methylation status have provided key insights into the role of DNA methylation in activity-related neuronal changes (32, 39), neurogenesis (70), learning and memory (33–35, 40, 41, 71, 72), and long-term behavioral plasticity (36, 37, 73, 74). However, each of these phenomena is comprised of temporally, cellularly, spatially, and genetically precise changes that arise from circuit interactions between and within defined neuronal structures (27, 75–77). Indeed, although recent tools such as fluorescence-activated cell sorting and next-generation sequencing have greatly enhanced our ability to measure epigenetic changes with cellular and genetic precision (see sidebar, Measuring the Epigenome), our approaches to manipulating DNA methylation are much less sophisticated. For example, pharmacological DNMT inhibitors promote global demethylation, presumably at all genes and in all cells affected. Thus, even where these drugs can be infused directly into a target brain region, they lack the specificity to demethylate specific genetic elements, such as the promoter region of a gene of interest. Furthermore, the nucleoside analogue DNMT inhibitors act to covalently trap DNMT enzymes, causing overt toxicity to mammalian cells (78). Likewise, these drugs lack specificity for particular DNMT isoforms (DNMT1, 3a, or 3b) and therefore do not allow selective assessment of how these different isoforms operate at DNA. In contrast, genetic approaches such as traditional gene knockout animal lines, small hairpin RNA knockdown, and virally mediated gene overexpression are capable of exhibiting complete isoform selectivity, as has been demonstrated for various DNMT proteins and Tet1 (40–42, 67, 71, 79, 80). These approaches still present problems in that they do not deliver precise temporal control over methylation status. Moreover, these approaches still lack target specificity, as they presumably affect methylation status in a genome-wide fashion.
Need for an Expanded Epigenetic Pharmacology
Together, these weaknesses emphasize the need for an expanded toolbox to control DNA methylation status in vivo and in vitro. Ideally, new tools for epigenetic control would have several properties that confer advantages over existing approaches. First, there is much need for a technique that can manipulate DNA methylation status with temporal precision. It is now well understood that gene expression programs, DNA methylation patterns, and the corresponding activity of enzymes related to DNA methylation can change on a rapid timescale (minutes to hours) within the brain (32, 33, 35, 41, 70, 81). In some cases, these changes can also be very transient, meaning that there is a defined temporal window wherein manipulation of DNA methylation could be critical to mechanistic interpretations. However, we presently lack the ability to modulate DNA methylation levels bidirectionally on the same timescale, which limits our ability to replicate or prevent these changes. Second, and perhaps most importantly, new tools for epigenetic control should be genetically selective. There is ample evidence for gene-specific and even nucleotide-specific DNA methylation profiles in the brain (32, 51). However, all the approaches mentioned above lack the ability to alter DNA methylation status at a specific target gene or at a specific location within that gene (e.g., proximal gene promoters, distal enhancer sites, intragenic locations). Finally, new methods for manipulation of DNA methylation should be spatially and perhaps even cellularly restricted. This is critical given that specific neuronal substructures contribute to unique behavioral and cognitive abilities. Furthermore, even within a defined brain region, it is often the case that specific cell types (e.g., neurons and glia) exhibit unique epigenetic patterns that contribute to phenotypic divergence (51). Therefore, in the future, the ability of researchers (and, eventually, clinicians) to target specific epigenetic responses in specific cell types will be essential to the success of any epigenetic pharmacology. Currently, a tool with all of these capabilities does not exist, which creates a significant barrier to the discovery of new epigenetic targets for manipulation of brain function.
Into the Future: Tools in Development for Manipulation of the Epigenome
The ability to selectively control DNA methylation levels at a single gene will be a truly transformative development for both neuroscience and epigenetics. For example, such a tool may enable derepression of imprinted gene loci, controlled expression of developmentally regulated genes, reversal of inherited epigenetic traits, selective restoration of genes critical to cognitive function in disease states, and perhaps even modulation of selected behavioral memories that are maintained by specific methylation profiles within defined brain circuits. With this in mind, this section examines some promising candidates for epigenetic control that provide some of the benefits that are currently absent in epigenetic pharmacology.
RNA-based approaches and nanoconstructs
One of the most promising avenues for rational pharmacology of any kind involves approaches that inhibit or degrade specific gene products, usually by manipulation of messenger RNAs. Recently, two such approaches have moved to the forefront for potential therapeutic applications. The first uses modified antisense oligonucleotides (ASOs) that can be customized to hybridize with and inhibit a specific RNA target, either through degradation by RNase H or steric hindrance of downstream RNA processing (82, 83). Although initial results with ASO technology were characterized by poor bioavailability and short half-life due to degradation, these challenges have been overcome by incorporation of chemically modified nucleic acid backbones that enhance stability in vivo (83). A recent report using this approach in the CNS demonstrated widespread brain activity of an ASO infused into cerebrospinal fluid and prolonged target gene knockdown with a single dose of ASO (84). A second approach targets specific RNA molecules using three-dimensional spherical nucleic acids (SNAs) (85). These constructs employ a gold nanoparticle core to serve as a docking site for covalent attachment of single- or double-stranded nucleic acids in a spherical array, which enhances the stability and cell penetrance of the molecule and therefore can achieve selective gene knockdown in vivo (86). This approach has recently been used to inhibit expression of an oncogene in a glioblastoma model using small interfering RNAs (87).
Theoretically, either SNAs or ASOs could be used to selectively target components of endogenous DNA methylation machinery in the CNS. Although this general category of tool has been available to researchers for many years in the form of virally mediated gene knockdown, these approaches confer a superior pharmacology: ease of entrance into cells without viral packaging or transfection reagents, stability of the ASO or SNA over time, and potentially prolonged gene knockdown with a single drug dose. Furthermore, the one-size-fits-all delivery method incumbent to both of these techniques is rapidly adaptable to new gene targets and could even support multiplexed targeting of several genes at the same time. Although the ability to target these constructs to specific brain locations within human populations using intrathecal or intracerebroventricular injections remains a challenge for pharmacotherapy, there are promising developments poised to solve this problem (88).
Transcriptional activator–like effectors and CRISPR/Cas systems
Accomplishing gene-specific manipulation of DNA methylation status requires the ability to target specific components of DNA methylation machinery to selected sequences of DNA. The ability to target DNA locations based on sequence alone has been an area of intense focus in genetic engineering and has recently been described with two unique sets of rapidly evolving tools. The first involves transcriptional activator–like effectors, or TALEs, which are short (33–34-amino-acid) bacterial proteins that, because of variable diresidues within the protein sequence, each bind to a specific nucleotide in DNA. Linking these short sequences together in a customized arrangement can therefore generate sequence-specific binding properties and serve as site-specific genomic anchor points (89–92). Using this approach, recent work by Zhang and colleagues (93) describes a robust, versatile tool that employs TALEs to target specific transcriptional machinery to selected genomic sites to modulate gene expression. For example, TALEs targeted to various gene loci have been shown to induce robust gene expression both in vitro and in vivo when fused to a generic transcriptional activator (93). Importantly, this activator can be replaced with epigenetic modifiers (so-called epi-TALEs) to cause histone deacetylation and gene repression (93). Thus, one possibility for the manipulation of DNA methylation at specific gene targets is to fuse DNA methyltransferases (e.g., DNMT3a) or mC hydroxylases (e.g., Tet1, Tet2, or Tet3) to customized TALEs, thereby targeting these proteins directly to selected DNA sites (Figure 3). The feasibility of this approach has already been demonstrated with TALE-Tet1 fusion proteins, which achieved nucleotide specific demethylation at selected gene targets (94).
Like TALEs, a second bacteria-derived system known as CRISPR (clustered regularly interspaced short palindromic repeats) also uses the sequence specificity of DNA to create customizable genomic anchor points. However, in contrast to TALEs, in which DNA sequences are targeted with repetitive protein sequences, CRISPR uses guide RNAs for this purpose (95). These guide RNAs can be similarly customized to recognize almost any DNA sequence (96) and also bind to a nuclease (Cas9). Thus, this system can be used to cut DNA directly, thereby enabling direct manipulation of genetic material (97). Additionally, by substituting a modified Cas9 protein without nuclease activity, this system to be used as an anchor point for epigenetic activators or repressors (98), as described above with DNMTs or Tet proteins.
One challenge to using these tools in vivo will be to restrict their function to certain genetically defined cell types, especially in a brain structure that contains a heterogeneous neuronal population. Typically, TALE- and CRISPR-based tools are expressed in mammalian cells using DNA constructs, which often involve the use of viral vectors for more complex systems. Although this approach generally targets cells nonselectively (unless a virus with a specific tropism is used), it can achieve robust expression in a relatively short time span and can be injected into specific brain regions for research applications. However, viral tools that incorporate loxP sites can be used in combination with Cre driver animal lines that express Cre recombinase only in genetically defined cell types, thereby limiting expression of specific constructs to those cells alone. This approach has recently been used with success to express discrete optogenetic actuators in distinct neuronal subtypes (99, 100) to enable precise interrogation of brain circuit function.
Optoepigenetics
As discussed above, the epigenetic toolbox would also ideally include a method that enables manipulations with some degree of temporal precision. Although this goal remains in an experimentally embryonic stage, it is worth noting that recent advances show much promise. Perhaps the greatest advance comes from the combination of optogenetic tools with the TALE systems described above (93). This approach uses cryptochrome 2 (CRY2), a light-sensitive protein from Arabidopsis thaliana, which exhibits a conformational change and binds to a partner protein [cryptochrome-interacting basic-helix-loop-helix 1 (CIB1)] in the presence of blue light. Thus, by fusing an effector protein such as a DNMT to CIB1 and CRY2 directly to a site-specific TALE domain, this system could be used as a light-guided switch to generate methylation of DNA with complete temporal control (Figure 3). This approach has already been employed to target generic transcriptional activators and repressive histone modifying enzymes to DNA (93) and is characterized by a very rapid response time (>1 h) that overlaps with the temporal dynamics of cognitive phenomena. This general technique could capitalize on growing optogenetic resources, allowing for a full set of optoepigenetic tools to manipulate DNA methylation in discrete brain regions using implanted fiber optic cables.
POTENTIAL APPLICATIONS FOR EPIGENETIC PHARMACOLOGY
Several neuropsychiatric areas can be identified at present that could represent where the future might lie in terms of mC-targeted therapeutics. We discuss these in more detail in the final section of this review. However, all these areas are highly speculative and not yet at the stage of actual drug development endeavors. As discussed above, the existing literature regarding how DNA methylation is altered in the CNS is much more broad and well developed than literature describing drugs designed to target these mechanisms. Indeed, literature describing the use of drugs that target DNA methylation in any human pathology is essentially absent outside of developing efforts using DNMT inhibitors to slow cancer progression (101–104).
A Conceptual Outlook
Thus, at this point there are few concrete examples of how mC might be targeted to achieve specific therapeutic effects. This is because the literature concerning specific roles for this form of epigenetic modification in neuroscience, neurology, and psychiatry is still in a very early stage. So, we start our consideration of the problem by considering first principles.
In an abstract sense, what are the theoretical ways in which one might target DNA methylation to achieve a therapeutic effect? We discuss four possibilities in a conceptual way to illustrate possible categories, and in each instance we give one example of a known disease gene target that might be amenable to such an approach.
Possibility 1: trigger demethylation to activate a suppressed gene
For this first example, we consider what may be the most straightforward potential therapeutic application of altering DNA methylation: activation of a suppressed imprinted gene through cytosine demethylation. For example, with a developmentally imprinted gene, one copy of the gene (the maternal or paternal allele) is silenced by methylation. So, in the case that the expressed copy is mutated, the imprinting of the remaining functional allele silences what would otherwise be a perfectly functional copy of the gene. De-imprinting and activating transcription of the imprinted allele could provide a strong beneficial effect. A good example of an application for this type of approach is Angelman syndrome (AS). AS is caused by an inactivating mutation of the maternal copy of the UBE3A ubiquitin ligase, and the paternal allele is silenced in many areas of the CNS. Thus, functionally, the AS brain manifests a homozygous UBE3A deficiency in critical learning and memory regions (105). Derepression of the paternal allele of UBE3A in the CNS, via demethylation of that allele, could restore a normal transcriptional level of an active UBE3A transcript.
Possibility 2: trigger methylation to suppress or silence a gene
Methylation-dependent gene silencing is the canonical role of cytosine methylation in development, and capitalizing on this mechanism for therapeutic benefit would be quite powerful. This might be especially applicable to diseases mediated by a gain-of-function mutation of a single allele of a gene. An example of a potential theoretical application of this type of approach is Huntington’s disease (HD). Mutant huntingtin (HTT) protein causes HD, which is likely mediated by a gain of function of the mutant gene. Silencing mutant HTT expression using cytosine methylation of the mutated allele could potentially lead to a treatment or even cure for HD. Yu et al. (106) used a conceptually similar but small interfering RNA–based approach to achieve allele-specific knockdown of mutant HTT in a mouse model, with therapeutic benefit demonstrated as a result.
Possibility 3: trigger increased methylation to activate a gene
It has become clear that, although cytosine methylation has historically been viewed as a gene suppression mechanism, methylation also has the capacity to increase transcription as well (10, 107). This may be most common for methylation events that occur within the gene body, i.e., within the coding region of the gene. Thus, as a third type of methylome-based therapeutic, one might also target the appropriate methylation sites within a gene to elicit gene-activating methylation. To illustrate this possibility, we can consider Pitt-Hopkins Syndrome (PTHS). PTHS is an autosomal dominant disease characterized by heterozygous inactivating mutation or deletion of the TCF4 transcription factor, which leads to pronounced intellectual disability, autistic behaviors, and loss of language acquisition (108, 109). Conceptually, one might trigger activating methylation of the remaining normal allele of TCF4 in PTHS patients to upregulate transcription of the remaining normal allele (i.e., move it above its normal transcriptional set point) to restore near-normal levels of active TCF4 in cells.
Possibility 4: trigger decreased methylation to attenuate expression of a gene
As mentioned above, methylation can be activating, and thus, demethylation could be repressing (in the case of activating intragenic methylation). By extension, then, one might trigger demethylation of a gene locus wherein intragenic methylation was operating normally to increase transcription at baseline and by that mechanism elicit decrease baseline transcription. Although purely speculative, let us suppose for illustrative purposes that intragenic activating methylation of one of the genes within the Down syndrome trisomic region controlled baseline transcription rates for that gene. By targeted loss of methylation of that baseline activating methylation site, one might diminish transcription across all three alleles, normalizing the cellular content of the gene product.
Potential Specific Applications of Methylcytosine-Targeted Pharmaceutics
In our opinion, in considering the available literature, there are three specific but broad areas that can be identified at this point in which mC-based pharmaceutics might first achieve therapeutic application. These are stress disorders, depression and schizophrenia, and cognitive function. In this final section, we briefly discuss each of these in turn.
Stress disorders
This is the area in which the most experimental work is available specifically indicating that cytosine methylation is correlated with and capable of triggering behavioral effects in vivo. Post-traumatic stress disorder (PTSD) is a debilitating anxiety disorder that develops in a subset of people after they experience psychological trauma. PTSD patients are commonly plagued by recurrent frightening thoughts and memories of the aversive experience and suffer from a host of persistent physiological and behavioral sequelae. These considerations make developing effective new therapies for PTSD an important priority for neurobehavioral biomedical researchers.
A contemporary hypothesis is that epigenetic mechanisms contribute to the formation and persistence of fear memories and that epigenetically based pharmacotherapy may provide a new avenue of drug treatment for PTSD-related cognitive symptoms (110). Potential domains in which to develop new epigenetic pharmacological agents that will allow novel treatments of PTSD-associated cognitive dysfunction include facilitated therapeutic relearning and enhanced extinction of conditioned and contextual fear. Thus, clinical stress disorders such as PTSD might be the first area wherein epigenetic pharmacology generally and DNA methylation specifically might be targeted for therapeutic effect. Indeed, numerous recent studies have indicated that HDAC inhibitors are beneficial for ameliorating specific conditioned fear responses in animal models.
Depression and schizophrenia
There are many interesting studies indicating persisting epigenetic marks can be acquired depending on early life environment and life experiences that can lead to subsequent altered adult behavior. For example, mother rats that exhibit strong nurturing behaviors toward their pups, manifest by frequently licking and grooming their offspring, produce lasting alterations in the patterns of DNA methylation in the CNS of their pups (36). These changes in methylation can persist throughout adulthood. In this vein, studies by Meaney and colleagues (37, 73) have presented evidence that these changes in DNA structure result in decreased anxiety and a strong maternal nurturing instinct in the adult offspring. These studies have recently been extended into studies of human experiential effects on DNA methylation and their potential role(s) in depression and suicide (111–113).
Schizophrenia as well has been associated with altered DNA methylation patterns, based on several studies available in the literature (114–117). Schizophrenia is a serious disorder of cognition, rendering sufferers unable to function normally in social situations and when performing everyday cognitive tasks. An emerging body of evidence suggests that alterations in DNA methylation contribute to transcriptional alterations in schizophrenia. Specifically, deficiencies in the extracellular matrix protein reelin have been associated with the etiology of schizophrenia—the promoter of the reelin gene contains several sites for DNA methylation, and inhibitors of HDAC and DNMT activity increase expression of reelin, indicating that epigenetic mechanisms govern the expression of reelin (115). This has led Grayson and colleagues (114) to specifically propose the hypothesis of an epigenetic disruption of reelin as contributing to schizophrenia.
Cognitive function
Whereas in an earlier section of this review we discussed animal model data implicating DNA methylation in memory, there is a considerable body of evidence implicating disruption of epigenetic mechanisms as a causal basis for human cognitive dysfunction. In this regard, several disorders of human cognition can be at least partly attributed to dysfunction in the mechanisms that underlie epigenetic marking of the genome. One example is Rett syndrome, an inherited, X-linked disease that is due, in the main, to inactivating mutations of MeCP2 (21). Fragile X syndrome, the most commonly inherited form of mental retardation, is brought about by an abnormal expansion of repeated trinucleotide sequences within one of two different fragile X genes: FMR1 and FMR2. Both FMR1 and FMR2 contain a polymorphic trinucleotide repeat—CGG and CCG, respectively—in their 5′ untranslated regions responsible for the loss of gene expression. Expansion of these repeats results in hypermethylation of these regions and flanking CpG islands, leading to transcriptional silencing of the FMR and surrounding genes (118, 119). The most widespread of senile dementias, Alzheimer’s disease, is associated with altered DNA methylation in both laboratory animal models and human postmortem brain tissue (120, 121).
All these observations indicate that dysfunction of the normal epigenetic status of the genome can have dramatic consequences on normal cognitive function. These studies also suggest that drugs that target the epigenome may represent viable therapies in treating various diseases affecting cognition, including those targeted to cytosine methylation.
SUMMARY AND CONCLUSIONS
In this brief overview we have tried to capture the excitement spawning from the rise of a new understanding of epigenetic molecular mechanisms and their relevance to CNS function and dysfunction. We consider this review to be a snapshot of where things stand at this point in time, with emerging new epigenetic mechanisms and targets surely to be over the horizon. The extent to which pharmacology will be able to capitalize on epigenetic processes and their regulation of the brain is certainly not yet clear. Nevertheless, the idiosyncratic power and properties of epigenetic mechanisms, and the unique pharmacology they may allow, hold great promise for future development.
MEASURING THE EPIGENOME.
Clearly, changes to the epigenome play a fundamental role in the development, baseline regulation, and experience-dependent alteration of the nervous system as a whole. However, this invites another question: How can we begin to understand which epigenetic modifications are important and which modifications we would seek to manipulate as part of a pharmacoepigenomic strategy? In essence, the answer to this question lies in our ability to link specific epigenetic modifications to discrete transcriptional outcomes, as defined in the section titled Druggable Epigenome.
Single-nucleotide resolution
The language of the DNA methylome is dominated by modifications that occur at single cytosine nucleotides. Therefore, it is necessary to measure epigenetic changes with techniques that capture this resolution. Scientists investigating the epigenome have long used bisulfite sequencing to investigate modifications at individual cytosine bases in DNA. This approach has proved to be useful for interrogating the methylation status of individual cytosine bases in DNA (32, 34, 36). Although bisulfite sequencing was once limited by the fact that both mC and hmC are preserved during bisulfite treatment (44), recent methodologies have built upon this approach to enable separate hydroxymethylation and methylation characterization (45, 46). A second, largely complementary method for estimation of global DNA methylation or hydroxymethylation involves high-performance liquid chromatography in tandem with mass spectrometry, which allows quantitative measurement of each modification in comparison to total cytosine (41, 47). However, this technique is limited by its inability to map cytosine modifications to any specific genomic locus.
Using next-generation sequencing to probe epigenetic landscapes
Although single-base changes in DNA methylation are clearly relevant to the transcriptional status of individual genes, the true power of this approach for pharmacotherapeutics comes from understanding DNA methylation principles across the entire genome. This ability not only enables the determination of potential off-target effects of a DNA methylation–related drug but also provides the opportunity to screen for and select new genetic or epigenetic targets for drug treatment. The advent of massively parallel next-generation sequencing platforms has made this idea a concrete reality, and it is now entirely feasible to sequence the entire methylome and transcriptome from neuronal populations to understand the precise functional relationship between DNA methylation and gene expression (48–52). As a practical matter, researchers should consider several issues, both in terms of which sequencing approach to use (53, 54) and how to interpret and analyze the resulting data (55). For example, sequencing approaches that rely on immunoprecipitation or capture of methylated DNA have proved to provide good genomic coverage and are relatively cheap, but they lack single-base resolution. Likewise, approaches that involve bisulfite conversion of DNA provide exquisitely detailed single nucleotide methylation maps but can be more expensive to generate and are bioinformatically more challenging because of the reduced complexity of DNA after conversion. Nevertheless, sequencing-based methylation assays have yielded enormous insights into the role of DNA methylation in the brain and will be required for therapeutic targeting of DNA methylation.
Genes in an epigenetic context
A final consideration is that DNA methylation does not occur in a vacuum but rather in the context of a wide variety of histone modifications and transcription factor binding sites that operate in concert to determine the ultimate transcriptional potential of a given gene. Therefore, going forward, it will be necessary to understand how these modifications influence each other. This will require the generation of composite data sets that include not only genome-wide methylation and RNA sequencing but also chromatin-immunoprecipitation sequencing to identify how these changes overlap with histone modifications and transcription factor binding (56) and Hi-C libraries to examine long-range DNA and chromatin interactions in a three-dimensional context (57).
Acknowledgments
We thank Cristin Gavin for help with the manuscript and members of the Sweatt lab for stimulating discussion. This work was supported by MH57014, MH091122, Civitan International, the Evelyn F. McKnight Brain Research Foundation, and the Pitt-Hopkins Syndrome (PTHS) Foundation (J.D.S.). J.J.D. is supported by the National Institute on Drug Abuse (DA034681). A.J.K. is supported by the PTHS Foundation.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Jeremy J. Day, Email: jjday@uab.edu.
Andrew J. Kennedy, Email: ajkennedy@uab.edu.
J. David Sweatt, Email: dsweatt@uab.edu.
LITERATURE CITED
- 1.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 2.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat. Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 5.Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 6.Fujita N, Watanabe S, Ichimura T, Ohkuma Y, Chiba T, et al. MCAF mediates MBD1-dependent transcriptional repression. Mol. Cell. Biol. 2003;23:2834–2843. doi: 10.1128/MCB.23.8.2834-2843.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J. Cell. Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 10.Wu H, Coskun V, Tao J, Xie W, Ge W, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 12.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito S, Shen L, Dai Q, Wu SC, Collins LB, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 17.Yokomine T, Hata K, Tsudzuki M, Sasaki H. Evolution of the vertebrate DNMT3 gene family: a possible link between existence of DNMT3L and genomic imprinting. Cytogenet. Genome Res. 2006;113:75–80. doi: 10.1159/000090817. [DOI] [PubMed] [Google Scholar]
- 18.Arand J, Spieler D, Karius T, Branco MR, Meilinger D, et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLOS Genet. 2012;8:e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335:709–712. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol. Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 21.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 22.Couvert PB, Aquaviva T, Poirier C, Moraine K, Gendrot C, et al. MECP2 is highly mutated in X-linked mental retardation. Hum. Mol. Genet. 2001;10:941–946. doi: 10.1093/hmg/10.9.941. [DOI] [PubMed] [Google Scholar]
- 23.Gommers-Ampt JH, Van Leeuwen F, de Beer ALJ, Vliegenthart JF, Dizdaroglu M, et al. β-d-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 24.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 25.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu L, Li Z, Cheng J, Rao Q, Gong W, et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 27.Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat. Neurosci. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatric. Res. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- 29.Meaney MJ, Ferguson-Smith AC. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat. Neurosci. 2010;13:1313–1318. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- 30.Akbarian S, Beeri MS, Haroutunian V. Epigenetic determinants of healthy and diseased brain aging and cognition. JAMA Neurol. 2013;70:711–718. doi: 10.1001/jamaneurol.2013.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zovkic IB, Guzman-Karlsson MC, Sweatt JD. Epigenetic regulation of memory formation and maintenance. Learn. Mem. 2013;20:61–74. doi: 10.1101/lm.026575.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo JU, Ma DK, Mo H, Ball MP, Jang MH, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, et al. DNA methylation regulates associative reward learning. Nat. Neurosci. 2013;16:1445–1452. doi: 10.1038/nn.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, et al. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 36.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol. Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 38.Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLOS ONE. 2011;6:e19958. doi: 10.1371/journal.pone.0019958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 40.Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, et al. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nat. Neurosci. 2013;16:42–47. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLOS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu M, Hon GC, Szulwach KE, Song CX, Jin P, et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat. Prot. 2012;7:2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo JU, Su Y, Shin JH, Shin J, Li H, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr. Opin. Plant Biol. 2009;12:107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:6146. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bock C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 56.Consortium EP. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 60.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- 62.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie W, Barr CL, Kim A, Yue F, Lee AY, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLOS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng J, Zhou Y, Campbell SL, Le T, Li E, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gräff J, Tsai L-H. Histone acetylation: molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 69.Gräff J, Tsai L-H. The potential of HDAC inhibitors as cognitive enhancers. Annu. Rev. Pharmacol. Toxicol. 2013;53:311–330. doi: 10.1146/annurev-pharmtox-011112-140216. [DOI] [PubMed] [Google Scholar]
- 70.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, et al. Neuronal activity–induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.LaPlant Q, Vialou V, Covington HE, III, Dumitriu D, Feng J, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maddox SA, Schafe GE. Epigenetic alterations in the lateral amygdala are required for reconsolidation of a Pavlovian fear memory. Learn. Mem. 2011;18:579–593. doi: 10.1101/lm.2243411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J. Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogerson T, Cai DJ, Frank A, Sano Y, Shobe J, et al. Synaptic tagging during memory allocation. Nat. Rev. Neurosci. 2014;15:157–169. doi: 10.1038/nrn3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frankland PW, Bontempi B. The organization of recent and remote memories. Nat. Rev. Neurosci. 2005;6:119–130. doi: 10.1038/nrn1607. [DOI] [PubMed] [Google Scholar]
- 77.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jüttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl. Acad. Sci. USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 80.Oliveira AMM, Hemstedt TJ, Bading H. Rescue of aging-associated decline inDnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 2012;15:1111–1113. doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- 81.Sultan FA, Sweatt JD. The role of the gadd45 family in the nervous system: a focus on neurodevelopment, neuronal injury, and cognitive neuroepigenetics. Adv. Exp. Med. Biol. 2013;793:81–119. doi: 10.1007/978-1-4614-8289-5_6. [DOI] [PubMed] [Google Scholar]
- 82.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 83.Southwell AL, Skotte NH, Bennett CF, Hayden MR. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol. Med. 2012;18:634–643. doi: 10.1016/j.molmed.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 84.Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 86.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 87.Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci. Transl. Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang K, Hao L, Hurst SJ, Mirkin CA. Antibody-linked spherical nucleic acids for cellular targeting. J. Am. Chem. Soc. 2012;134:16488–16491. doi: 10.1021/ja306854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hsu PD, Zhang F. Dissecting neural function using targeted genome engineering technologies. ACS Chem. Neurosci. 2012;3:603–610. doi: 10.1021/cn300089k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Briggs AW, Rios X, Chari R, Yang L, Zhang F, et al. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 2013;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mali P, Yang L, Esvelt KM, Aach J, Guell M, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jennings JH, Stuber GD. Tools for resolving functional activity and connectivity within intact neural circuits. Curr. Biol. 2014;24:R41–R50. doi: 10.1016/j.cub.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jennings JH, Rizzi G, Stamatakis AM, Ung RL, Stuber GD. The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science. 2013;341:1517–1521. doi: 10.1126/science.1241812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Graca I, Sousa E, Baptista T, Almeida M, Ramalho-Carvalho J, et al. Anti-tumoral effect of the non-nucleoside DNMT inhibitor RG108 in human prostate cancer cells. Curr. Pharma. Des. 2013;20:1803–1811. doi: 10.2174/13816128113199990516. [DOI] [PubMed] [Google Scholar]
- 102.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 103.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J. Natl. Cancer Inst. 2005;97:1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 104.Yang PM, Lin YT, Shun CT, Lin SH, Wei TT, et al. Zebularine inhibits tumorigenesis and stemness of colorectal cancer via p53-dependent endoplasmic reticulum stress. Sci. Rep. 2013;3:3219. doi: 10.1038/srep03219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, et al. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sweatt JD. Pitt-Hopkins Syndrome: intellectual disability due to loss of TCF4-regulated gene transcription. Exp. Mol. Med. 2013;45:e21. doi: 10.1038/emm.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blake DJ, Forrest M, Chapman RM, Tinsley CL, O’Donovan MC, Owen MJ. TCF4, schizophrenia, and Pitt-Hopkins Syndrome. Schizophr. Bull. 2010;36:443–447. doi: 10.1093/schbul/sbq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Graff J, Joseph NF, Horn ME, Samiei A, Meng J, et al. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 2014;156:261–276. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nature. Suicide watch. Nature. 2014;506:131. doi: 10.1038/506131a. [DOI] [PubMed] [Google Scholar]
- 112.Na KS, Chang HS, Won E, Han KM, Choi S, et al. Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLOS ONE. 2014;9:e85425. doi: 10.1371/journal.pone.0085425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kang HJ, Kim JM, Lee JY, Kim SY, Bae KY, et al. BDNF promoter methylation and suicidal behavior in depressive patients. J. Affect. Disord. 2013;151:679–685. doi: 10.1016/j.jad.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 114.Grayson DR, Chen Y, Costa E, Dong E, Guidotti A, et al. The human reelin gene: transcription factors (+), repressors (−) and themethylation switch (+/−) in schizophrenia. Pharmacol. Ther. 2006;111:272–286. doi: 10.1016/j.pharmthera.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 115.Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, et al. Reelin promoter hypermethylation in schizophrenia. Proc. Natl. Acad. Sci. USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLOS ONE. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kundakovic M, Chen Y, Guidotti A, Grayson DR. The reelin and GAD67 promoters are activated by epigenetic drugs that facilitate the disruption of local repressor complexes. Mol. Pharmacol. 2009;75:342–354. doi: 10.1124/mol.108.051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, et al. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 120.Coppieters N, Dieriks BV, Lill C, Faull RLM, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol. Aging. 2013;35:1334–1344. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- 121.Sanchez-Mut JV, Aso E, Panayotis N, Lott I, Dierssen M, et al. DNA methylation map of mouse and human brain identifies target genes in Alzheimer’s disease. Brain: J. Neurol. 2013;136:3018–3027. doi: 10.1093/brain/awt237. [DOI] [PMC free article] [PubMed] [Google Scholar]