Abstract
An olfactory biosensor based on a reduced graphene oxide (rGO) field‐effect transistor (FET), functionalized by the odorant‐binding protein 14 (OBP14) from the honey bee (Apis mellifera) has been designed for the in situ and real‐time monitoring of a broad spectrum of odorants in aqueous solutions known to be attractants for bees. The electrical measurements of the binding of all tested odorants are shown to follow the Langmuir model for ligand–receptor interactions. The results demonstrate that OBP14 is able to bind odorants even after immobilization on rGO and can discriminate between ligands binding within a range of dissociation constants from K d=4 μm to K d=3.3 mm. The strongest ligands, such as homovanillic acid, eugenol, and methyl vanillate all contain a hydroxy group which is apparently important for the strong interaction with the protein.
Keywords: biosensors, immobilization, odorant‐binding protein, olfaction, reduced graphene oxide
Odorant‐binding proteins (OBPs) are small acidic proteins (ca. 13–16 kDa) highly concentrated in the lymph of the chemosensillae of insects or in the nasal mucus of vertebrates.1 They act as carriers for volatile organic compounds, VOCs, (air‐borne odorants) shuttling them from the air–water interface to the membrane‐integral odorant receptor. Although their full function has not been completely clarified yet, OBPs certainly play a major role in detecting and recognizing olfactory stimuli.2
A large number of OBPs has been expressed in bacterial systems and their ligand‐binding properties have been investigated in solution by a fluorescent ligand displacement assay.3 Generally, dissociation constants, K d, are in the upper nanomolar or lower micromolar range for strong odorants.1c,d An important characteristic of OBPs for technical applications is their stability to extreme temperatures, solvents, and proteolysis, making such proteins ideal elements for biosensors to be used in medical application, for example, in breath analysis for cancer diagnostics, for food quality control, for crop‐disease detection, or in general environmental monitoring.4a–f
Various biosensor devices mimicking the olfactory system (artificial noses) have been developed but only few studies have used OBPs as functional elements for the design of a “bio‐electronic nose”.5 Herein we present the fabrication and functional characterization of a label‐free biosensor for odorant detection based on a reduced graphene oxide field‐effect transistor functionalized with the odorant‐binding protein 14 (OBP14) from the honey bee Apis mellifera.
Reduced graphene oxide field‐effect transistor (rGO‐FET) devices were fabricated according to established methods, schematically given in Figure 1 A.6 A scanning electron microscopic image of rGO flakes assembled onto the gate substrate before any further surface functionalization is shown in Figure 1 B. IR spectra of the linker monolayer, 1‐pyrenebutanoic acid succinimidyl ester (PBSE), that is typically used for protein immobilization on graphene substrates,7 with partial covalent immobilization of OBP taken at different times during the assembly from solution to the gate are given in Figure 1 C, while Figure 1 D summarizes the quantitative analysis of the functionalization process by monitoring the time‐dependent increase of the amide I and II bands of the protein and the corresponding decrease of the band at 1738 cm−1, assigned to the cleavage of the active ester group of the linker molecules. The entire fabrication process of the olfactory biosensor device, including the successful reduction of GO to rGO, the linker binding, and more details of the protein attachment are described in the Supporting Information. The cloning, expression, and purification of OBPs, the preparation of odorant solutions and the performance of the electrical measurements are also described in the Supporting Information.
Figure 1.
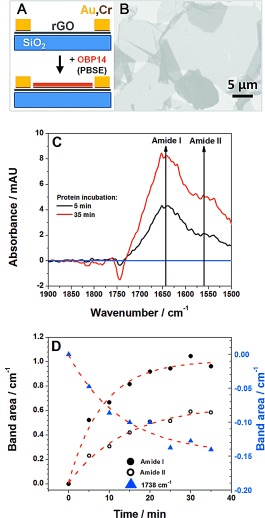
A) Schematic illustration of the individual fabrication steps of the graphene biosensor device. B) Scanning electron microscopic image of the rGO‐FET before the functionalization with 1‐pyrene‐butanoic acid succinimidyl ester (PBSE) linker. C) Infrared spectra of the PBSE linker attached to gate area of the rGO surface, and OBP14 immobilized for 5 and 35 min, respectively, as indicated (spectra measured in ATR configuration and water corrected). D) Time‐dependent increase of the amide I and II bands, upon binding of OBP14 to the linker molecules at the gate surface, and the corresponding decrease of the band at 1738 cm−1 upon cleavage of the active ester of the linker molecules during the protein immobilization (cf. also Figure S6). The dashed red curves are guides to the eye.
The mode of operation of the device as a field‐effect transistor is demonstrated in Figure S5. Recording the ISD versus VG scans under different bulk solution conditions, in particular, in aqueous solutions with different ligand concentrations results in a slightly modified slope of the cathodic branch. We attribute this change to a slight modification of the dipolar layer upon binding of the ligands to the free binding sites in the OBP which act as receptors. The OBP protein monolayer is immobilized on the graphene gate and ligand binding causes a the slight reorientation of its the alpha‐helical parts. At the selected gate voltage of V G=−0.6 V the concentration dependency of the slopes in the I SD versus V G curves could be used to measure the binding of odorants to OBP14 in real‐time, resulting in the quantitative determination of the kinetic rate constants for the association process, k on, and for the dissociation process, k off, as well as for the affinity constant K A and the dissociation constant K d. As an example, Figure 2 A shows a global analysis measurement, that is, the time dependence of the change of the source‐drain current, ΔI SD, of the FET as a function of time upon binding of methyl vanillate, a strong binder, from solution to the OBP14‐functionalized gate surface at increasing and decreasing odorant bulk concentrations. The error of the kinetic rate constants, k, obtained was estimated to be around 20 % (cf. also Figure S8 in the Supporting Information).
Figure 2.
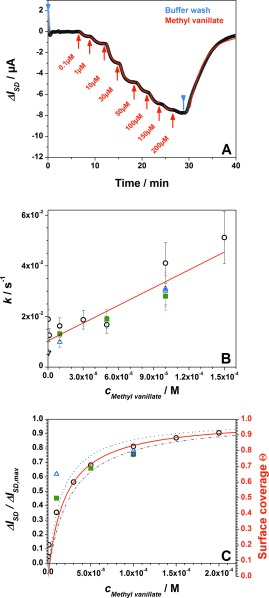
A) Real‐time biosensor measurement of the binding of methyl vanillate to OBP14: the current increases with the bulk concentration of methyl vanillate increasing (from 100 nm to 200 μm) and then saturates. Blue arrows indicate runs with pure buffer, red arrows indicate experiments with methyl vanillate solutions. Red curves are the fitting of the raw data by kinetic simulations of the association and dissociation processes based on the Langmuir model. (For the estimation of the error limits, see the Supporting Information). B) Analysis of the reaction rate constants, k, obtained from the fitted data in (A) as a function of methyl vanillate concentration; different symbols from three different devices; error bars are ±20 %. C) Langmuir adsorption isotherm, obtained for three different samples; the red fit curve gives K d=20 μm (plotted are also error limits for K d of 20 %).
Upon plotting the resulting reaction rate constants k, as they were derived from the fits to the kinetic traces as a function of the bulk ligand concentration, c 0=c Methyl vanillate, (Figure 2 B), gives a straight line, the slope of which, according to k=k on c 0+k off, with k on being the association and k off the dissociation rate constant, respectively, yields k on=235 m −1 s−1, and the intersection with the y‐axis gives k off=0.01 s−1. According to the Langmuir model the ratio k on/k off gives the affinity constant K A=2.3×104 m −1, which can be compared to the value derived from the equilibrium titration experiment given in Figure 2 C: according to θ=K A c 0/(1+K A c 0) the fit to the data (full red curve plus the error limits of ±20 %) gives the affinity constant K A=5×104 m −1 which compares quite well with the value obtained from the kinetic experiments thus confirming the Langmuir model for this binding process.
To exclude false signals arising from, in particular, non‐specific binding, several control experiments were performed. Firstly, a sensor was prepared which was covered only with linker molecules, without the coupling of the odorant‐binding protein. Even high concentrations of the strong binder homovanillic acid (cf. Figure S7A) or other ligands (Figure S7B) did not result in a significant signal. Coating the gate with OBP14 but exposing it to a totally uncorrelated small molecule, biotin, gave no signal (Figure S7C). And finally, immobilizing a protein (OBP9A from the red flour beetle, Tribolium castaneum) that is structurally similar but is not a receptor for these ligand odorants also gave a very weak signal (Figure S7D).
Further evidence for the specificity of the ligand receptor binding originates from a direct comparison of the binding of eugenol and methyl eugenol to the same chip, functionalized with OBP14. Figure 3 A shows a real‐time current trace recorded during the addition of methyl eugenol at 50 μm, then rinsing with pure buffer, and then a 5 μm solution of eugenol being rinsed through the flow cell. Despite the minor chemical variation between the two ligands a large difference in the affinity of their binding reaction to OBP14 confirms the specificity of the sensor. Figure 3 B summarizes the Langmuir isotherms for both ligands, demonstrating the significantly different dissociation constants: K d=40 μm for eugenol and K d=1400 μm for methyl eugenol.
Figure 3.
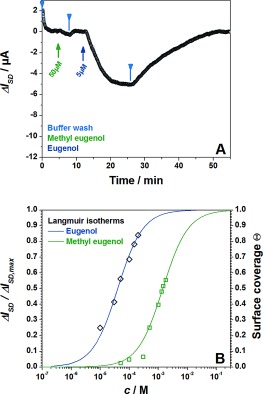
A) Real‐time sensor response to the injection of a 50 μm solution of methyl eugenol, subsequent buffer wash, and the injection of 5 μm eugenol solution. B) Langmuir isotherms of eugenol and methyl eugenol.
Table 1 summarizes the quantitative data measured for a series of ligands and gives the kinetic rate constants, k on and k off, obtained as well as the dissociation constant K d. It could be confirmed also for this label‐free sensing format that the odorant binding affinity to OBP14 gradually decreases from homovanillic acid to citral and to geraniol.8 Molecules which are structurally related to eugenol also showed strong affinities to OBP14.
Table 1.
Dissociation constants, K d, association rate constants, k on, and dissociation rate constants, k off, for a variety of odorants binding to OBP14 as obtained by the global analysis.
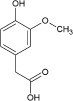
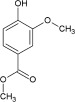
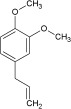
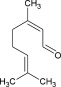
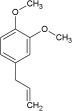
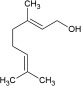
| Odorant | Homovanillic acid | Methyl vanillate | Eugenol | Citral | Methyl eugenol | Geraniol |
|---|---|---|---|---|---|---|
| structural formula |
|
|
|
|
|
|
| K d [×10−6 m] | 4 | 20 | 40 | 800 | 1400 | 3300 |
| k on [m −1 s−1] | 1130 | 235 | 170 | 9 | 6 | 3 |
| k off [s−1] | 0.008 | 0.01 | 0.006 | 0.003 | 0.006 | 0.008 |
Interestingly, also other odorant‐binding proteins bind their odorants with affinities in the same order of magnitude.9 It is remarkable to note that for all the ligands investigated the dissociation rate constants k off, differ by less than a factor of 2.5. The strongly differing affinity constants, varying by nearly three orders of magnitude, can be almost exclusively attributed to the differences in the association rate constants k on, (cf. Table 1).
The Spinelli group used X‐ray diffraction analysis to determine the structure of the eugenol–OBP14 complex. They found that the hydroxy group of eugenol interacts with the cavity wall of the OBP14 binding pocket by forming two hydrogen bonds.10
It has been speculated that the hydroxy group, together with the substituted aromatic backbone, plays a key role for the strong binding which is in agreement with the high affinities also of homovanillic acid and methyl vanillate (cf. Table 1).10 This hypothesis is further supported by the lower affinity measured for methyl eugenol, in which the hydroxy group is replaced by a methoxy group.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information
Acknowledgements
This research is supported by the European Science Foundation (ESF), Grant Number: 10‐EuroBioSAS‐FP‐005, by the FFG within the comet program, and from the governments of Lower and Upper Austria. We thank Prof. Lendl (Technical University of Vienna) and his group (Christoph Gasser, Georg Ramer, Karin Wieland) for technical support regarding Raman and FTIR measurements.
References
- 1.
- 1a. Vogt R. G. in Comprehensive Insect Science, Vol. 8 (Eds.: L. Gilbert, K. Iatrou, S. Gill), Elsevier, London, 2005, p. 753; [Google Scholar]
- 1b. Vogt R. G., Riddiford L. M., Nature 1981, 293, 161; [DOI] [PubMed] [Google Scholar]
- 1c. Leal W. S., Annu. Rev. Entomol. 2013, 58, 373; [DOI] [PubMed] [Google Scholar]
- 1d. Pelosi P., Zhou J. J., Ban L. P., Calvello M., Cell. Mol. Life Sci. 2006, 63, 1658; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1e. Fan J., Francis F., Liu Y., Chen J. L., Cheng D. F., GMR Genet. Mol. Res. 2011, 10, 3056. [DOI] [PubMed] [Google Scholar]
- 2.
- 2a. Xu P., Atkinson R., Jones D. N., Smith D. P., Neuron 2005, 45, 193; [DOI] [PubMed] [Google Scholar]
- 2b. Matsuo T., Sugaya S., Yasukawa J., Aigaki T., Fuyama Y., PLoS Biol. 2007, 5, e118; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2c. Swarup S., Williams T. I., Anholt R. R., Genes Brain Behav. 2011, 10, 648; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2d. Sun Y. F., De Biasio F., Qiao H. L., Iovinella I., Yang S. X., Ling Y., Riviello L., Battaglia D., Falabella P., Yang X. L., Pelosi P., PLOS ONE 2012, 7, e32759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campanacci V., Krieger J., Bette S., Sturgis J. N., Lartigue A., Cambillau C., Breer H., Tegoni M., J. Biol. Chem. 2001, 276, 20078. [DOI] [PubMed] [Google Scholar]
- 4a. Schwaighofer A., Kotlowski C., Araman C., Chu N., Mastrogiacomo R., Becker C., Pelosi P., Knoll W., Larisika M., Nowak C., Eur. Biophys. J. 2014, 43(2–3), 105–112; [DOI] [PubMed] [Google Scholar]
- 4b. Paolini S., Tanfani F., Fini C., Bertoli E., Pelosi P., Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1999, 1431, 179–188; [DOI] [PubMed] [Google Scholar]
- 4c. Matsumura K., Opiekun M., Oka H., Vachani A., Albelda S. M., Yamazaki K., Beauchamp G. K., PLOS ONE 2010, 5(1), e8819; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4d. Ghasemi‐Varnamkhasti M., Mohtasebi S. S., Siadat M., J. Food Eng. 2010, 100, 377–287; [Google Scholar]
- 4e. Schütz S., Weißbecker B., Koch U. T., Hummel H. E., Biosens. Bioelectron. 1999, 14, 221–228; [DOI] [PubMed] [Google Scholar]
- 4f. Bourgeois W., Romain A.‐C., Nicolas J., Stuetz R. M., J. Environ. Monit. 2003, 5, 852–860. [DOI] [PubMed] [Google Scholar]
- 5.
- 5a. Park S. J., Kwon O. S., Lee S. H., Song H. S., Park T. H., Jang J., Nano Lett. 2012, 12, 5082; [DOI] [PubMed] [Google Scholar]
- 5b. Jin H. J., Lee S. H., Kim T. H., Park J., Song H. S., Park T., Hong S., Biosens. Bioelectron. 2012, 35, 335; [DOI] [PubMed] [Google Scholar]
- 5c. Kim T. H., Lee S. H., Lee J., Song H. S., Oh E. H., Park T. H., Hong S., Adv. Mater. 2009, 21, 91; [Google Scholar]
- 5d. Yoon H., Lee S. H., Kwon O. S., Song H. S., Oh E. H., Park T. H., Jang J., Angew. Chem. Int. Ed. 2009, 48, 2755; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2009, 121, 2793; [Google Scholar]
- 5e. Di Pietrantonio F., Cannatà D., Benetti M., Verona E., Varriale A., Staiano M., D’Auria S., Biosens. Bioelectron. 2013, 41, 328; [DOI] [PubMed] [Google Scholar]
- 5f. Sankaran S., Panigrahi S., Mallik S., Biosens. Bioelectron. 2011, 26, 3103; [DOI] [PubMed] [Google Scholar]
- 5g.S. D’Auria, V. Scognamiglio, M. Rossi, M. Staiano, S. Campopiano, N. Cennamo, L. Zeni in Conference on Biomedical Vibrational Spectroscopy and Biohazard Detection Technologies, Vol. 5321, SPIE—The International Society for Optical Engineering, San Jose California (USA), 2004, p. 258.
- 6.
- 6a. Huang J. F., Larisika M., Fam W. H. D., He Q. Y., Nimmo M. A., Nowak C., Tok I. Y. A., Nanoscale 2013, 5, 2945; [DOI] [PubMed] [Google Scholar]
- 6b. Su C. Y., Xu Y., Zhang W., Zhao J., Tang X., Tsai C. H., Li L. J., Chem. Mater. 2009, 21, 5674–5680. [Google Scholar]
- 7. Kodali V. K., Scrimgeour J., Kim S., Hankinson J. H., Carroll K. M., de Heer W. A., Berger C., Curtis J. E., Langmuir 2011, 27, 863–865. [DOI] [PubMed] [Google Scholar]
- 8. Iovinella I., Dani F. R., Niccolini A., Sagona S., Michelucci E., Gazzano A., Turillazzi S., Felicioli A., Pelosi P., J. Proteome Res. 2011, 10, 3439. [DOI] [PubMed] [Google Scholar]
- 9.
- 9a. Golebiowski J., Topin J., Charlier L., Briand L., Flavour Fragrance J. 2012, 27, 445; [Google Scholar]
- 9b. Briand L., Eloit C., Nespoulous C., Bézirard V., Huet J.‐C., Henry C., Blon F., Trotier D., Pernollet J.‐C., Biochemistry 2002, 41, 7241; [DOI] [PubMed] [Google Scholar]
- 9c. Briand L., Nespoulous C., Perez V., Rémy J.‐J., Huet J.‐C., Pernollet J.‐C., Eur. J. Biochem. 2000, 267, 3079. [DOI] [PubMed] [Google Scholar]
- 10. Spinelli S., Lagarde A., Iovinella I., Legrand P., Tegoni M., Pelosi P., Cambillau C., Insect Biochem. Mol. Biol. 2012, 42, 41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information